Abstract
Genome modifications that occur at the initial interspecific hybridization event are dynamic and can be consolidated during the process of stabilization in successive generations of allopolyploids. This study identifies the number and chromosomal location of ribosomal DNA (rDNA) sites between Secale cereale, Dasypyrum villosum, and their allotetraploid S. cereale × D. villosum hybrids. For the first time, we show the advantages of FISH to reveal chromosome rearrangements in the tetraploid Secale × Dasypyrum hybrids. Based on the specific hybridization patterns of ribosomal 5S, 35S DNA and rye species-specific pSc200 DNA probes, a set of genotypes with numerous Secale/Dasypyrum translocations of 1R/1V chromosomes were identified in successive generations of allotetraploid S. cereale × D. villosum hybrids. In addition we analyse rye chromosome pairs using FISH with chromosome-specific DNA sequences on S. cereale × D. villosum hybrids.
Keywords: C-banding, FISH, pSc200, Secale × Dasypyrum hybrids, 5S rDNA, 35S rDNA
Introduction
In qualitative plant breeding programmes, agronomic traits are improved through transfer of alien genes responsible for disease resistance, mainly from related species. Wild species from the Triticeae tribe have played an important role in increasing the genetic variation of bread wheat (Triticum aestivum L.) and rye (Secale cereale L.) (see Grądzielewska 2006 for review). The genus Dasypyrum comprises many agronomically important traits including disease resistance, salinity, drought and freezing tolerance, high protein content and quality, and therefore is a valuable resource for crop improvement (De Pace et al. 2001; Mariani et al. 2003). Dasypyrum villosum (L.) P. Candargy (syn. Haynaldia villosa (L.) Schur) is an annual, open pollinated, wild diploid grass (2n = 2x = 14), with its genome designated as VV (Sears 1953). The plants of this species were successfully hybridized with different forms of Triticum spp. (Sears 1953; von Bothmer and Claesson 1998; Minelli et al. 2005) producing amphiploids and wheat addition, substitution and recombination lines. Direct hybrids of D. villosum with S. cereale tend not to yield hybrid plants (Lucas and Jahier 1988), suggesting a low degree of homoeologous chromosome pairing. Tetraploid S. cereale × D. villosum hybrids have been obtained (Łapiński M and Gruszecka 1997), but these hybrids are of limited value due to the partial infertility of spikes; however, their contribution in rye improvement cannot be definitely excluded. Diploid Secale × Dasypyrum hybrids, which might be directly used in rye breeding, have been successfully obtained by Gruszecka (1997) and investigated morphologically (Gruszecka 1997), cytologically (Apolinarska and Gruszecka 2001), biochemically (Makarska et al. 2004) and molecularly (Grądzielewska 2009). Plant breeding strictly requires the characterisation of interspecific hybrids. Fluorescence and genomic in situ hybridization (FISH and GISH) techniques have been widely used for this purpose (Zhou et al. 2001; Molnár et al. 2009). The first cytogenetic study of allotetraploid Secale × Dasypyrum hybrids is presented in this paper that demonstrates the advantages of FISH in revealing chromosome rearrangements that could not be detected with molecular markers or by conventional means.
The present study aimed to characterize the chromosome constitution of the tetraploid S. cereale × D. villosum hybrids, maintained for long periods in successive generations and (i) to assign parental chromosome origins to the hybrid chromosomes and (ii) to identify the translocations, by use of repetitive and tandemly organized ribosomal DNA (rDNA) sequences as well as the rye-genomic-specific sequence (pSc200). These goals were achieved using Ag-NOR, C-banding and fluorescence and genomic in situ hybridization techniques. We provide initial information about the physical location of rDNA sites in allotetraploid Secale × Dasypyrum amphidiploids, which, together with pSc200, may be used as chromosome landmarks to describe the translocations more precisely. This first molecular cytogenetic analysis of allotetraploid S. cereale × D. villosum hybrids may contribute to the broader characterisation of intergenomic translocations between parental genomes.
Materials and methods
Seeds of diploid forms of S. cereale L. (2n = 2x = 14; R-genome) ‘Smolickie’, D. villosum (L.) P. Candargy (syn. Haynaldia villosa (L.) Schur) (2n = 2x = 14; V-genome), and S. cereale × D. villosum amphidiploid (RRVV) were provided by the Institute of Genetics Polish Academy of Sciences (Poznań, Poland) and the West Pomeranian University of Technology (Szczecin, Poland), where the plant genetic stocks are deposited and subjected to availability. Intergeneric hybrids of S. cereale × D. villosum (2n = 2x = 14; RV) were generated in 1966 by intercrossing diploid forms of both species. The term ‘Senaldia’ (Se cale + Hay naldia ; M. Łapiński, unpubl. data) is applied to describe the corresponding intergeneric S. cereale × D. villosum hybrid plants (2n = 4x = 28; RRVV) for the remainder of this paper. S. cereale ‘Smolickie’ was used as the female parent and D. villosum as the pollinator. Only one hybrid seed producing a single plant was achieved as a result of 518 crosses. This plant was cloned and 6 of F1 plants were doubled using colchicine as described by Łapiński and Gruszecka (1997). One recovered plant that was open-pollinated produced 15 C1 progeny plants. All C1 allopolyploid plants were male and female fertile and were sibling-pollinated under controlled conditions to produce the successive generations. Over 20 generations of Senaldia plants were performed in isolation and without selection (M. Łapiński, unpubl. data). The plants derived from available successive generations of intergeneric S. cereale × D. villosum hybrids (>C20) were used in our studies. Seeds of amphidiploids and each of the parents were germinated. Silver staining (Ag-NOR) was performed according to Hizume et al. (1980), and the C-banding procedure was carried out as described by Lukaszewski and Gustafson (1983). The Ag positive bands and the C-banding patterns were analyzed on 3–5 well-spread metaphases. For in situ hybridization, root tips of hybrids and parental forms were collected in ice water, refrigerated for 24–28 h, fixed in ethanol with glacial acetic acid (3:1, v/v), and then stored at −20°C until use. Chromosome preparations were made from an individual root per plant of 48 randomly chosen hybrids and of 10 randomly chosen plants of the parental forms. Chromosome analyses were conducted on 5–10 well-spread metaphases. FISH was carried out according to Książczyk et al. (2010), using the ribosomal DNA probes 5S rDNA (pTa794) (Gerlach and Dyer 1980) isolated from wheat, 25S rDNA (Unfried and Gruendler 1990) isolated from Arabidopsis thaliana, and the repetitive DNA probe pSc200 that contains highly repetitive DNA sequences specific to S. cereale (Bedbrook et al. 1980). The GISH procedure was adapted from Kosmala et al. (2006) with minor modifications (Książczyk et al. 2010), using total genomic DNA, extracted according to Lombard and Delourme (2001) from S. cereale and D. villosum, as a probe and/or a block. The 5S and 25S rDNA probes were isolated from plasmids (the latter being used for detection of 35S rDNA loci). The 5S rDNA was labelled with tetramethyl-rhodamine-5-dUTP (Roche) by PCR, and 25S rDNA was labelled with tetramethyl-rhodamine-5-dUTP and digoxygenin-11-dUTP (Roche; ratio 1:1) or only digoxygenin-11-dUTP by nick translation. The pSc200 probe was amplified using universal M13 sequencing primers and labelled also by PCR with digoxygenin-11-dUTP (Roche). The genomic DNA from D. villosum was labelled by nick translation with digoxygenin-11-dUTP (Roche). Post-hybridization washes (an equivalent of 51%-81% stringency) were followed by immunodetection of the digoxigenated probes by FITC-conjugated anti-digoxigenin antibodies (Roche). For FISH, the preparations were mounted and counterstained in Vectashield (Vector Laboratories) containing 2.5 μg/ml of DAPI (Sigma) or 1.5 μg/ml of propidium iodide (Vector Laboratories); for GISH - the preparations were mounted and counterstained in Vectashield containing 1.5 μg/ml of propidium iodide. C-banding patterns were analyzed using a Nikon HFX-DX microscope, photographed on Fuji 800 film and the photographic images were then scanned into a computer. Ag-NOR and FISH/GISH images were acquired using an Olympus XM10 CCD camera attached to an Olympus BX 61 automatic epifluorescence microscope. Image processing and superimpositions were done using Olympus Cell-F imaging software and Micrographx Picture Publisher software. The recombinant rye chromosomes and the rDNA-bearing-rye and D. villosum chromosomes were compared to previous karyotypes and/or ideograms, reported by Lukaszewski and Gustafson (1983), Galasso et al. (1997), Alkhimova et al. (1999) and Minelli et al. (2005).
Results and discussion
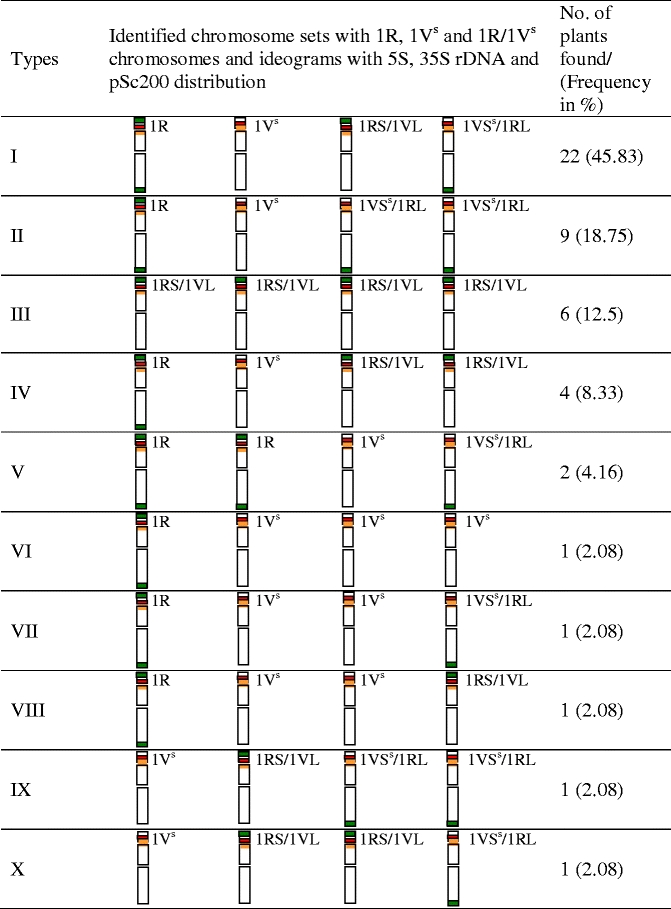
The FISH experiments with 5S and 25S rDNA probes provided chromosomal landmarks in the diploid genomes of S. cereale (Fig. 1a) and D. villosum (Fig. 1b), showing a number of different 5S rDNA sites between these two species. The 5S rDNA sites were found in the short arms of two pairs of chromosome 1R and 5R in S. cereale, and in the short arms of a pair of chromosome 5V in D. villosum. The 35S rDNA sites were found in the short arms of a pair of chromosome 1R in S. cereale and in the short arms of a pair of chromosome 1V in D. villosum. rDNA-FISH in the tetraploid genome of Senaldia (Fig. 1c) revealed four instead of the expected three pairs of chromosomes carrying 5S rDNA sites in these hybrids. Therein, two pairs of rDNA-bearing chromosomes with colocalisation of 5S and 35S rDNA sites were found (Fig. 1c). This suggests the presence of four homologues of chromosome 1R, based on the distribution patterns of rDNA loci in S. cereale chromosomes, and a lack of 1V chromosome pair. The possibility of 1R/1V translocated chromosomes in Senaldia genome cannot be excluded, but rDNA-FISH is not able to detect them. We report the use of GISH to cytogenetically investigate Senaldia hybrids (Fig. 1e) was unsuccessful. The high degree of S. cereale and D. villosum genome homoeology makes the genomes indistinguishable. However, the presence of green distinct signals at the termini of some chromosomes in the hybrid genome may indicate the concentration of constitutive heterochromatin in the terminal C-bands of rye and possible intergenomic chromosome rearrangements between parental genomes, based on the distribution patterns of rye chromosome-specific subtelomeric sites in S. cereale chromosomes. A detailed amphiploid genome analysis was carried out using a silver staining method (Ag-NOR). This offers an opportunity to study the number of active and inactive ribosomal 35S DNA loci and determine the genomic origin of possible dominant and suppressed 35S rRNA genes. No nucleolar dominance in Senaldia genomes was revealed (Fig. 1c and d). It is worth mentioning that with four homologues of chromosome 1R present in Senaldia plants, the lack of nucleolar dominance appears to be as expected. Highly reliable diagnostic banding patterns allowing rye chromosome identification has been achieved by Lukaszewski and Gustafson (1983), using the C-banding technique. The application of this technique enabled the identification of some rye chromosomes in Senaldia that do not show the typical bands reported for S. cereale by Lukaszewski and Gustafson (1983). This observation suggests the presence of S. cereale/D. villosum translocations (Fig. 1f and g). The 1R chromosome pair and 1R/1V translocated chromosomes were important because of their ease of identification and traceability by FISH with rDNAs and rye species-specific sequence pSc200 as probes. Triple-FISH with both rDNAs and pSc200 to chromosomes of root-tip-cells of diploids revealed the presence of rDNA-bearing chromosomes in D. villosum (Fig. 1h) without rye chromosome-specific subtelomeric (pSc200) sites, while in S. cereale (Fig. 1i), apart from known chromosomes carrying 5S and 35S rDNA, the distribution of pSc200 sites in most rye chromosome arms was similar to those previously observed by Alkhimova et al. (1999) and Hasterok et al. (2002). A lack of visible pSc200 patterns in the chromosomes of D. villosum was surprising when compared with the observations reported by Galasso et al. (1997) and Alkhimova et al. (1999), suggesting intravarietal variation of pSc200 homologous loci leading to the loss of DAPI positive bands and thus low level of resolution of FISH with pSc200 signal in D. villosum chromosomes. Somewhat fortuitously the lack of pSc200 signals in D. villosum allowed the differentiation between rye-genome-like and D. villosum-genome-like chromosomes in allotetraploid hybrids (Fig. 1j-m). When used as part of triple-FISH this allowed the re-evaluation of four chromosome 1R homologues of Senaldia hybrids by rDNA-FISH for 1R/1V translocations. The root meristems of 48 Senaldia hybrids were then screened by FISH with rDNAs and pSc200 for 1R/1V translocations. Using the third pSc200 probe, the images of triple-FISH on Senaldia metaphase chromosomes revealed that the variability of chromosomes carrying rDNA loci may involve the 1V chromosome. A new chromosome type (1Vs) carrying 5S and 35S rDNA loci was identified by triple-FISH and may be considered as a variant of the Dasypyrum-genome-like 1V chromosome. This observation indicates that chromosomal variation involving rDNA-containing regions had occurred and confirmed reports of inter- and/or intrachromosomal changes and numerous rDNA rearrangements in newly formed allopolyploids (Volkov et al. 1999; Pontes et al. 2004; Kotseruba et al. 2010; Książczyk et al. 2010). Subsequent observations also suggest that some rDNA loci can be unstable in recently formed S. cereale × D. villosum hybrids and can participate in a variety of alternative rearrangements. All hybrid plants studied had detectable rearrangements in 1R and 1Vs chromosomes, which were classified into distinct types (Table 1), suggesting an extensive chromosome rearrangements resulted from genome imbalance during polyploid formation (Volkov et al. 1999; Pontes et al. 2004; Malinska et al. 2010). The assumption is that translocations in these cases are reciprocal because the shape of the chromosomes 1R/1Vs appears unchanged. Ten different Senaldia chromosome complements having intact chromosomes 1R, 1Vs and 1R/1Vs translocations have been observed and the most prevalent types of 1R/1Vs translocations detected in Senaldia hybrids are presented in Fig. 1j (Table 1, type I), k (Table 1, type III) and l-m (Table 1, type IV). Other Senaldia chromosome complements with numerous structural variants of 1R/1Vs chromosomes (Table 1, types VI-X) were relatively uncommon and constituted 10.4% of those observed. The chromosomes 1R and 1V showed modifications in all plants studied (Table 1, Fig. 1c, f, g and j-m) and thus appeared to be unstable; the recombinant chromosomes with 1V chromosome arms were reported by Minelli et al. (2005) in Triticum × Dasypyrum hybrids, while 1R chromosome arms were identified as translocated chromosome in wheat/rye translocations (Lukaszewski and Gustafson 1983).
Fig. 1.
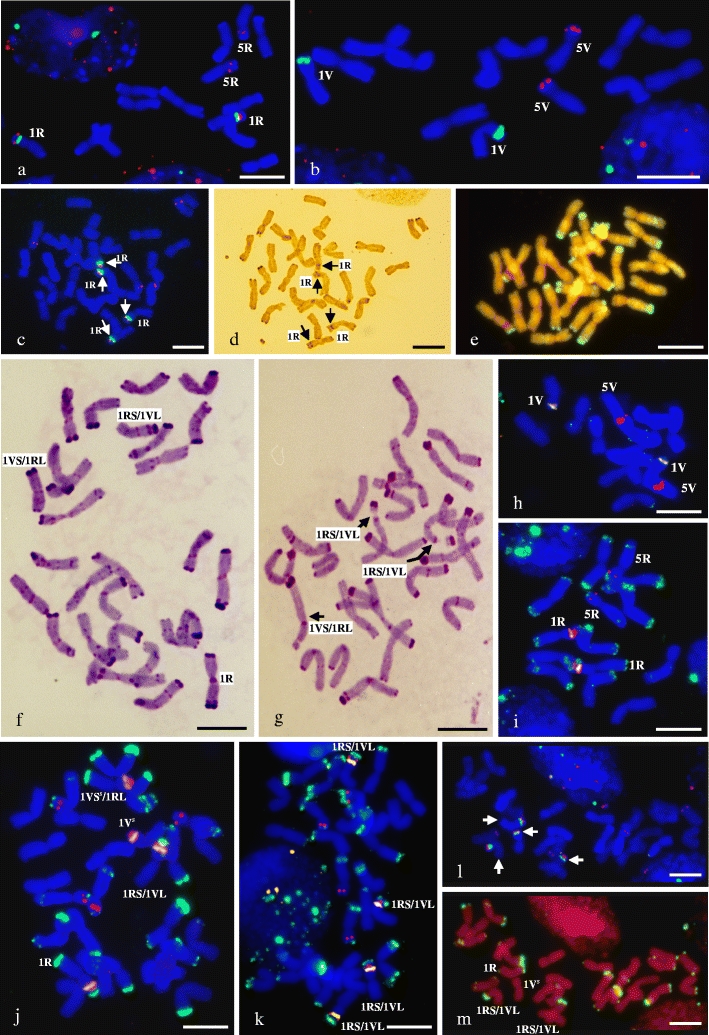
Root-tip metaphase chromosomes from diploid S. cereale (a, i), D. villosum (b, h) and allotetraploid Senaldia (c-g, j-m), after FISH (a, b, c, h-m), GISH (e), Ag-NOR staining (d) and C-banding (f, g). FISH with 5S rDNA labelled with rhodamine (red) and 25S rDNA labelled with digoxigenin and detected by anti-digoxigenin conjugated with FITC (green) to chromosomes of S. cereale (a), D. villosum (b) and Senaldia hybrid with 2 pairs of 5S and 35S rDNA-bearing chromosomes supplemented by the white arrows and 2 pairs of 5S rDNA-bearing chromosomes (c) and active rDNA loci in the same metaphase chromosome spread detected by silver staining; the black arrows indicate Ag-NORs (d). The chromosomes were counterstained with DAPI (blue) (a-c). GISH with a total genomic DNA from D. villosum as a probe labelled with digoxigenin and detected by anti-digoxigenin conjugated with fluorescein (green/yellow), with blocking genomic DNA of S. cereale (orange/red); chromosomes were counterstained with propidium iodide (e). C-banding patterns of two Senaldia hybrids with examples of Secale/Dasypyrum translocations – 1RS/1VL and 1VS/1RL (f) and 2x 1RS/1VL and 1VS/1RL (g). FISH with 5S rDNA labelled with rhodamine (red), 25S rDNA labelled with digoxigenin (green) and rhodamine (red) (1:1; orange/white) and repetitive DNA probe pSc200 labelled with digoxigenin (green) and detected by anti-digoxigenin conjugated with FITC (green) to chromosomes of D. villosum (h), S. cereale (i) and three Senaldia hybrids with the most prevalent types of 1R/1Vs translocations (j, k and m). FISH with 5S rDNA labelled with rhodamine (red) and 25S rDNA labelled with digoxigenin (green) and rhodamine (red) (1:1; orange/white) and detected by anti-digoxigenin conjugated with FITC (green) to chromosomes of Senaldia hybrid with 2 pairs of 5S and 35S rDNA-bearing chromosomes supplemented by the white arrows and 2 pairs of 5S rDNA-bearing chromosomes (l). The identified Senaldia chromosome sets with type I of 1R/1Vs translocations in (j), type III in (k) and type IV in (l-m); the white arrows indicate 2 pairs of 5S and 35S rDNA-bearing chromosomes (l), reprobing with the pSc200 sequence, after labeling with digoxigenin (green) and detected by anti-digoxigenin conjugated with FITC; chromosome were counterstained with propidium iodide (m). The chromosome nomenclature of S. cereale rDNA-bearing chromosomes (Arabic numerals) is acc. to Vershinin et al. (1995) (a, i). The chromosome nomenclature of D. villosum rDNA-bearing chromosomes (Arabic numerals) is acc. to Cremonini et al. (1994) (b, h). Upper case letters signify the genomic origin of tagged chromosomes. All scale bars represent 10 μm
Table 1.
Types of identified chromosome sets with Secale- and Dasypyrum-genome-like 1R and 1Vs chromosomes and ideograms depicting the location of hybridization sites of three repeated sequence probes on the 1R, 1Vs and 1R/1Vs chromosomes of Senaldia hybrids 5S rDNA (red), 35S rDNA (orange) and pSc200 (green)
The intergeneric hybrids between rye and wild diploid D. villosum are important for introgression-breeding programmes, and can be used to transfer abiotic and biotic stress resistance traits from Dasypyrum species into Secale species. Our results show the Senaldia hybrids have a stable number of rDNA-bearing chromosomes with colocalisation of 5S and 35S rDNA sites. On the other hand, these hybrids are highly unstable when considering Secale/Dasypyrum translocations. The 1R/1Vs translocation can be tracked by rDNA-FISH together with rye species-specific pSc200 sequence. It may be revealing to look more extensively at meiotic rye chromosome pairing using chromosome-specific FISH probes. This may allow the verification of their involvement in any rearrangements and thus genome reconstruction in Senaldia allopolyploids.
Acknowledgements
We are grateful to Dr. Ela Wolny (University of Silesia, Poland) for providing the pSc200 clone, to Dr. Magdalena Majewska (Institute of Plant Genetics PAS, Poznań) for technical assistance by GISH technique. We also thank Dr. Carol Ryder (Warwick University, UK) for comments and linguistic revision of the manuscript and the two anonymous referees for providing us with constructive comments and suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Mirosław Łapiński is the deceased author.
References
- Alkhimova AG, Heslop-Harrison JS, Shchapova AI, Vershinin AV. Rye chromosome variability in wheat-rye addition and substitution lines. Chrom Res. 1999;7:205–212. doi: 10.1023/A:1009299300018. [DOI] [PubMed] [Google Scholar]
- Apolinarska B, Gruszecka D. Transfer genów z Dasypyrum villosum (Haynaldia villosa L.) do Secale cereale L. Biotechnologia. 2001;2(53):63–65. [Google Scholar]
- Bedbrook JB, Jones J, O’Dell M, Thompson RD, Flavell RB. A molecular description of telomeric heterochromatin in Secale species. Cell. 1980;19:545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Cremonini R, Colonna N, Stefani A, Galasso I, Pignone D. Nuclear DNA content, chromatin organization and chromosome banding in brown and yellow seeds of Dasypyrum villosum (L.) P. Candargy. Heredity. 1994;72:365–373. doi: 10.1038/hdy.1994.53. [DOI] [Google Scholar]
- De Pace C, Snidaro D, Ciaffi M, Vittori D, Ciofo A, Cenzi A, Tanzarella OA, Qualset CO, Scarascia Mugnozza GT. Introgression of Dasypyrum villosum chromatin into common wheat improves grain protein quality. Euphytica. 2001;117:67–75. doi: 10.1023/A:1004095705460. [DOI] [Google Scholar]
- Galasso I, Blanco A, Katsiotis A, Pignone D, Heslop-Harrison JS. Genome organization and phylogenetic relationships in the genus Dasypyrum analyzed by Southern and in situ hybridization of total genomic and cloned DNA probes. Chromosoma. 1997;106:53–61. doi: 10.1007/s004120050224. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Dyer TA. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980;11:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grądzielewska A. The genus Dasypyrum – part 1. The taxonomy and relationships within Dasypyrum and with Triticeae species. Euphytica. 2006;152:429–440. doi: 10.1007/s10681-006-9232-2. [DOI] [Google Scholar]
- Grądzielewska A (2009) Identification of Dasypyrum villosum (L.) P. Candargy genetic material introgression to rye using RAPD and STS molecular markers. Zesz Probl Post Nauk Roln LXIV (3):29–40 (in Polish)
- Gruszecka D (1997) Otrzymanie i charakterystyka mieszańców Secale cereale L. z Dasypyrum villosum (Haynaldia villosa L.). Hodowla Roślin. I Krajowa Konferencja, Poznań 19–20 listopada, pp 135–139 (in Polish)
- Hasterok R, Jenkins G, Langdon T, Jones RN. The nature and destiny of translocated B-chromosome-specific satellite DNA of rye. Chrom Res. 2002;10:83–86. doi: 10.1023/A:1014278429178. [DOI] [PubMed] [Google Scholar]
- Hizume M, Sato S, Tanaka A. A highly reproducible method of nucleolus organizing regions staining in plants. Stain Technol. 1980;55:87–90. [PubMed] [Google Scholar]
- Kosmala A, Zwierzykowski Z, Gąsior D, Rapacz M, Zwierzykowska E, Humphreys MW. GISH/FISH mapping of genes for freezing tolerance transferred from Festuca pratensis to Lolium multiflorum. Heredity. 2006;96:243–251. doi: 10.1038/sj.hdy.6800787. [DOI] [PubMed] [Google Scholar]
- Kotseruba V, Pistrick K, Blattner FR, Kumke K, Weiss O, Rutten T, Fuchs J, Endo T, Nasuda S, Ghukasyan A, Houben A. The evolution of the hexaploid grass Zingeriakochii (Mez) Tzvel. (2n = 12) was accompanied by complex hybridization and uniparental loss of ribosomal DNA. Mol Phylogenet Evol. 2010;56:146–155. doi: 10.1016/j.ympev.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Książczyk T, Taciak M, Zwierzykowski Z. Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. J Appl Genet. 2010;51(4):449–460. doi: 10.1007/BF03208874. [DOI] [PubMed] [Google Scholar]
- Lombard V, Delourme R. A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet. 2001;103:491–507. doi: 10.1007/s001220100560. [DOI] [Google Scholar]
- Lucas H, Jahier J. Phylogenetic relationships in some diploid species of Triticineae: cytogenetic analysis of interspecific hybrids. Theor Appl Genet. 1988;75:498–502. doi: 10.1007/BF00276756. [DOI] [Google Scholar]
- Lukaszewski AJ, Gustafson JP. Translocations and modifications of chromosomes in Triticale × Wheat hybrids. Theor Appl Genet. 1983;64:239–248. doi: 10.1007/BF00303771. [DOI] [PubMed] [Google Scholar]
- Łapiński M, Gruszecka D (1997) Charakterystyka płodnego mieszańca Secale cereale × Haynaldia villosa. Krajowa Konferencja „Mieszańce oddalone roślin zbożowych”, Poznań, 24 kwietnia, pp 4–5 (in Polish)
- Makarska E, Gruszecka D, Miąc A. Zmiany w składzie mineralnym ziarniaków nowych nowych translokacyjnych rodów żyta odmiany Amilo z Dasypyrum villosum L. Candargy. J Elementol. 2004;9(3):393–398. [Google Scholar]
- Malinska H, Tate JM, Matyasek R, Leitch AR, Soltis DE, Soltis PS, Kovarik A. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evol Biol. 2010;10(1):291. doi: 10.1186/1471-2148-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani M, Minelli S, Ceccarelli M, Cionini PG, Qualset CO, De Pace C (2003) Dasypyrum villosum chromosome segments introgressed in hexaploid wheat provide opportunities for prebreeding and preparing primary mapping populations for analyzing complex genetic traits. In: Proc. 10th Int. Wheat Genet. Symp., Instituto Sperimentale per la Cerealicoltura, Rome, Italy, pp 613–615
- Minelli S, Ceccarelli M, Mariani M, De Pace C, Cionini PG. Cytogenetics of Triticum × Dasypyrum hybrids and derived lines. Cytogenet Genome Res. 2005;109:385–392. doi: 10.1159/000082424. [DOI] [PubMed] [Google Scholar]
- Molnár I, Benavente E, Molnár-Láng M. Detection of intergenomic chromosome rearrangements in irradiated Triticum aestivum – Aegilops biuncialis amphiploids by multicolor genomic in situ hybridization. Genome. 2009;52:156–165. doi: 10.1139/G08-114. [DOI] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci USA. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. Addition of the genome of H. villosa to T. aestivum. Am J Bot. 1953;40:168–174. doi: 10.2307/2438774. [DOI] [Google Scholar]
- Unfried I, Gruendler P. Nucleotide sequence of the 5.8S and 25S rRNA genes and the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990;18:4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinin A, Schwarzacher T, Heslop-Harrison JS. The large-scale genomic organisation of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell. 1995;7:1823–1833. doi: 10.1105/tpc.7.11.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol. 1999;16:311–320. doi: 10.1093/oxfordjournals.molbev.a026112. [DOI] [PubMed] [Google Scholar]
- von Bothmer R, Claesson L. The hybrid Triticum turgidum ssp. dicoccum × Dasypyrum villosum and the cross and backcrosses to breadwheat, T. aestivum. Hereditas. 1998;128:47–52. doi: 10.1111/j.1601-5223.1998.00047.x. [DOI] [Google Scholar]
- Zhou A, Xia G, Zhang X, Chen H, Hu H. Analysis of chromosomal and organellar DNA of somatic hybrids between Triticum aestivum and Haynaldia villosa Schur. Mol Genet Genomics. 2001;265:387–393. doi: 10.1007/s004380000430. [DOI] [PubMed] [Google Scholar]


