Abstract
X-chromosome inactivation (XCI) results in the transcriptional silencing of one X-chromosome in females to attain gene dosage parity between XX female and XY male mammals. Mammals appear to have developed rather diverse strategies to initiate XCI in early development. In placental mammals XCI depends on the regulatory noncoding RNA X-inactive specific transcript (Xist), which is absent in marsupials and monotremes. Surprisingly, even placental mammals show differences in the initiation of XCI in terms of Xist regulation and the timing to acquire dosage compensation. Despite this, all placental mammals achieve chromosome-wide gene silencing at some point in development, and this is maintained by epigenetic marks such as chromatin modifications and DNA methylation. In this review, we will summarise recent findings concerning the events that occur downstream of Xist RNA coating of the inactive X-chromosome (Xi) to ensure its heterochromatinization and the maintenance of the inactive state in the mouse and highlight similarities and differences between mammals.
Introduction
Sex chromosomes differ significantly in their gene content (Ohno 1967), which has led to the evolution of mechanisms of dosage compensation (Lyon 1961, 1962). At least three different mechanisms have been described to adjust sex chromosome gene expression dosage between males and females. The single male X-chromosome is hypertranscribed in flies (Park and Kuroda 2001), both X-chromosomes are partially repressed in worms (Meyer 2000), while in mammals one of the two X-chromosomes is silenced in each cell (Lyon 1961). It was 50 years ago that cytological observations in mouse, rat, opossum and human (Lyon 1962; Ohno and Hauschka 1960) resulted in the Lyon hypothesis (1961), whereby dosage of X-chromosomal gene products is equalised between male and female mammals, by inactivating one of the X-chromosomes during early development in females (Lyon 1961, 1962). Fifty years on, we are still seeking the exact mechanisms that trigger the initial differential treatment of the two X-chromosomes, as well as the spread and maintenance of the inactive state. In this review, we will discuss recent studies that have contributed to our understanding of how an entire chromosome is transformed, in a step-wise fashion, from an active into an inactive and heterochromatic entity and how this state is then maintained through cell division and life span. While describing this process in the mouse, we will consider possible differences in the XCI process and the Xi chromosome itself in other mammals.
Regulation of X-chromosome inactivation during early development of eutherian mammals
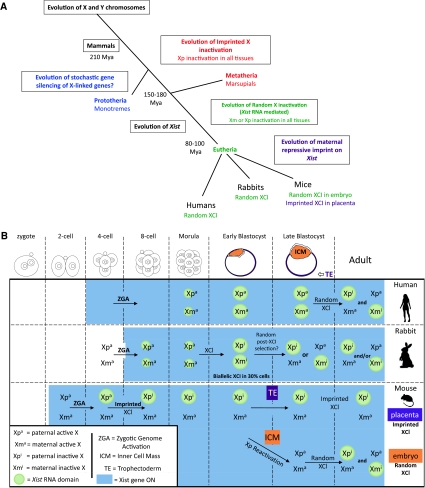
Over the years the mouse has been the privileged mammal for investigation of XCI given the ease of genetics and embryology in this species. It is now apparent, however, that the XCI strategy found in the mouse may represent just one of many modes of regulating X-chromosome gene dosage in mammals (Fig. 1). In marsupials, XCI is subject to imprinting, with the paternal X-chromosome (Xp) being chosen for inactivation. This form of imprinted X inactivation is very leaky being tissue- and locus-specific, the extent to which remains to be carefully assessed (Deakin et al. 2009). In the egg-laying monotremes such as the platypus, which have five X and Y-chromosomes, dosage compensation appears to be only partial and may be locus specific, similar to birds. Some genes on the platypus X-chromosome are not dosage compensated, while others show some form of compensation, possibly via stochastic inactivation (Deakin et al. 2008), suggesting that monotremes have retained an ancestral compensation system (Deakin et al. 2009).
Fig. 1.
Strategies and timing of XCI regulation in eutherians. a Hypothetical time line of events during evolution for the appearance of different XCI strategies in different mammals. b Developmental timing of Xist/XIST up-regulation and XCI in humans, rabbits and mice during pre-implantation embryogenesis. In humans, XIST is not imprinted and upregulation is observed on both X-chromosomes from the morula stage with partial coating of both chromosomes. Random XCI initiates at or after the blastocyst stage in both extra and embryonic lineages—the exact timing is not clear. In the rabbit, Xist is not imprinted and upregulation occurs on one or both of the X-chromosomes in some cells of female embryos, and eventually leads to XCI. By the late blastocyst only one of the X-chromosomes shows Xist coating and X-linked gene inactivation. How XCI goes from a biallelic to monoalleic situation is unclear. In the mouse, Xist is imprinted to stay silent on the maternal X-chromosome. Paternal XCI is initiated by triggering Xist RNA coating from 4 to 8 cell stage onwards exclusively on the paternal chromosome. This imprinted form of XCI is found in the extra-embryonic tissues such as placenta. However, in the ICM of the late blastocyst, the Xp is re-activated, so that one of the two X-chromosomes then has an equal chance of being inactivated in the lineages that will give rise to the adult mouse
In eutherians, XCI is generally thought to be random, with the paternal and maternal X-chromosomes having an equal chance of being silenced. For example, in humans, random XCI is thought to occur in both embryonic and extra-embryonic derivatives (Migeon 2002; Moreira de Mello et al. 2010). However, in the mouse, XCI is initially imprinted (Takagi and Sasaki 1975) with the silencing of the Xp from the 4–8 cell stage (Donohoe et al. 2009; Navarro et al. 2008, 2010; Okamoto et al. 2004). The Xp remains inactive in the trophectoderm (Takagi and Sasaki 1975) but is reactivated in the inner cell mass (ICM) of the blastocyst (Mak et al. 2004; Okamoto et al. 2004), followed by random XCI (McMahon and Monk 1983). This second, random wave of XCI is accompanied by a complex interplay of events including downregulation of pluripotency factors such as Nanog, Oct4/Pou5f1 and Sox2, activation of Xist by dosage-sensitive X-linked competence factors such as Rnf12 and random monoallelic regulation of Xist via its antisense transcription unit, Tsix, as will be described in the section on initiation of random X-chromosome inactivation in mice.
The status of the X-chromosome and the regulation of XCI during early female development in mammals other than mice have been unclear until recently. Although mice are clearly a powerful model for dissecting the actors involved in XCI, different strategies for regulating the onset of XCI exist in other mammals as seen in rabbits and humans (Okamoto et al. 2011) (Fig. 1). In both these species, XCI was found to begin later than in mouse, and furthermore Xist was found not to be subject to imprinting. This reinforces the previously proposed notion that imprinted XCI, via a Xist imprint may be a recent evolutionary event, occurring in mice due to their rather rapid pre-implantation development (Okamoto et al. 2005). Another major difference observed between mammals is that in rabbit and human pre-implantation embryos, Xist is up-regulated on both X-chromosomes in a high proportion of blastomeres, a situation rarely observed in the mouse, in vivo. This up-regulation has different consequences on gene expression. In rabbits, XCI appears to be triggered on both X-chromosomes transiently, in a subset of cells, but this situation rapidly resolved to monoallelic XCI. In humans, although XIST is expressed from both X-chromosomes in pre-implantation embryos, chromosome-wide XCI only begins later, after the blastocyst stage (Okamoto et al. 2011). A previous study on human pre-implantation embryos concluded that XCI in human starts by the blastocyst stage (van den Berg et al. 2009). In contrast to Okamoto et al. (2011), this study showed XIST monoallelic accumulation from the 8-cell-stage and only 5% of blastomeres accumulate XIST on both X-chromosomes by morula and blastocyst stages (van den Berg et al. 2009). The discrepancy between this and the Okamoto et al. (2011) study could be due to technical differences, such as RNA fluorescence in situ hybridization (FISH) efficiency, which can influence the detection of biallelic versus monoallelic expression—Okamoto et al. (2011) provided high-efficiency RNA FISH data showing biallelic expression of XIST and of four independent X-linked genes in female blastocysts. The discrepancy may also be due to differences in embryo in vitro culture conditions.
A further species difference concerns the X-chromosome reactivation process observed in the mouse ICM, which is presumed to be due to pluripotency factor-mediated repression of Xist in pre-epiblast cells. In rabbits, the X-chromosome actually initiates XCI in cells of the ICM, rather than being reactivated, while in humans, XIST is highly expressed but does not trigger XCI in the ICM, suggesting that in both species Xist/XIST is rather differently controlled in the ICM (Okamoto et al. 2011). Whether this is due to differences in Xist regulatory sequences and/or to differences in the exact combination of pluripotency factors and other Xist/XIST regulators in the ICM in other mammals remains to be found. In summary, there is substantial diversity in the timing and mode of regulation of XCI initiation between mammals, even though the outcome, dosage compensation, seems to be the same. The reasons for such diversity are unclear but the most likely explanation is that the XCI process has had to adapt to the considerable variety between mammals in developmental dynamics. For example, the remarkably early zygotic gene activation found in mice compared with other mammals may have led to pressure to evolve an imprint on Xist in mice, in order to prevent precocious initiation of XCI on the maternal and paternal X-chromosomes, at a stage of development when gene dosage might be less easy to evaluate.
In mice, the random form of XCI follows Xp reactivation in the ICM, at around the time of implantation (day 5). A major difficulty in the study of random XCI is the small size of the embryo and the limited amount of material available at this stage. For this reason, in vitro model systems, such as embryonic stem (ES) cells (derived from the ICM of blastocysts) provide a valuable alternative. In mouse ES cells, both X-chromosomes are active (Xa), while the cells are maintained in an undifferentiated state and the XCI process can be initiated by simply inducing ES cell differentiation (Martin et al. 1978). Such cell lines have greatly facilitated the investigation of the different steps in the process of random XCI, from the establishment through to the maintenance of the inactive state.
Initiation of random X-chromosome inactivation in mice
Studies of XCI patterns in mouse embryos and cells carrying translocated or truncated X-chromosomes revealed the existence of an X-linked region, the X-inactivation centre (Xic), that is required to trigger this process (reviewed in Augui et al. 2007). The Xic locus is thought to ensure that only a single X-chromosome will remain active per diploid autosome set; it also underlies the “choice” of which X-chromosome will be inactivated (Brown et al. 1991b) and it produces the signal that triggers cis-inactivation of the X-chromosome, in the form of the non-coding Xist transcript (Brown et al. 1991a). Xist is responsible for initiating silencing of the chromosome from which it is expressed (Borsani et al. 1991; Marahrens et al. 1997; Penny et al. 1996) and also seems to underlie many of the chromatin changes that take place during XCI. Functional studies have shown that Xist is clearly the essential trigger in mice and given that it is conserved in eutherians, it is fair to imagine that is would play a similar role, although functional proof of its role in XCI in other mammals has not so far been demonstrated.
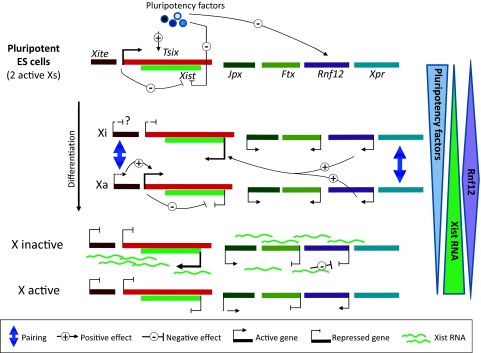
The regulatory network that controls Xist expression in mice has recently begun to be unravelled, revealing a complex interplay of cis-acting elements and trans-acting factors or RNAs, all of which are tightly integrated with the early steps of differentiation (Fig. 2). The downregulation of pluripotency and stem cell factors, such as Oct/Pou5f14, Nanog, Rex1 and others, during ES cell differentiation coincides with Xist up-regulation. Genetic and developmental studies suggest that such factors play a central role in this Xist regulatory network, at least in mice. Although some of these factors were originally proposed to repress Xist directly, through binding sites in its first intron (Navarro et al. 2008), more recent studies suggest that these sites alone are not essential for the control of Xist and that pluripotency factors control Xist’s regulators, such as the X-linked activator of Xist, Rnf12 (Barakat et al. 2011; Navarro et al. 2011), or the repressive antisense transcript to Xist, Tsix (Navarro et al. 2010). Although downregulation of pluripotency factors can partly explain Xist activation during differentiation, it does not explain why Xist is upregulated only in female, not male cells. Recently, several X-linked loci have been implicated in XX-specific activation (sensing, competence) of Xist. These include the Rnf12 gene, which encodes an ubiquitin ligase protein that can directly act on the Xist promoter (Barakat et al. 2011; Jonkers et al. 2009). In the context of increased dose of the RNF12 protein, Xist expression can be triggered even in male cells. However, RNF12 is probably not the only dosage-sensitive activator as its deletion on one allele does not abolish Xist activation (Jonkers et al. 2009). Other loci that have been proposed to be implicated in XX-specific Xist expression include the X-pairing region (Xpr) (Augui et al. 2007) and the Jpx non-coding RNA locus (Tian et al. 2010).
Fig. 2.
Initiation of random XCI and the monoallelic regulation of Xist in the mice. In pluripotent ES cells, Xist is expressed at low levels and this repression is partly due to pluripotency factors that repress Xist. Xist regulation via pluripotency factors could be direct through binding to the intron 1 of Xist gene and/or indirect, via activation of Xist’s repressive antisense transcript, Tsix or repression of Xist activators such as the X-linked gene Rnf12. During differentiation the levels of pluripotency factors decrease, while the levels of Xist activators such as Rnf12 increase (the only activator reported so far). Other long-range elements possibly involved in this process are the non-coding RNAs (Xite, Jpx, Ftx) or Xpr (X-pairing region). Xist activation on only one allele is believed to be partly regulated by monoallelic regulation of Tsix and its enhancer Xite. The Xpr and Xite/Tsix regions have been shown to undergo transient “pairing” events that could potentially help in establishment the monoallelic expression of Xist. Other mechanisms could underlie monoallelic Xist regulation such as stochastic events and negative feedback loops involving X-linked activators such as Rnf12. Xist upregulation results in cis-coating of the X-chromosome and results in silencing of genes, including the activator Rnf12
XX-specific activation of Xist via sensing or competence mechanisms does not explain how only one of the two X-chromosomes up-regulates Xist during random XCI, a process known as “choice”. As mentioned earlier, monoallelic regulation of Xist seems to be less well coordinated in rabbits and humans, compared with the mouse. How is such tight monoallelic regulation of Xist expression achieved in the mouse? The Xist antisense transcription unit, Tsix, seems to play a critical role in choice of Xist allele to be upregulated. Tsix transcription across the Xist promoter is accompanied by repressive chromatin modifications (Navarro et al. 2005; Sado et al. 2005; Sun et al. 2006), although it is not clear whether this is simply a result of antisense transcription across the Xist locus or due to a role for the Tsix RNA, for example in recruiting the chromatin modifying machinery to silence the Xist gene in cis. Deletion of Tsix in the mouse leads to preferential up-regulation of Xist on the deleted allele (Lee and Lu 1999). Furthermore, deletion of both Tsix alleles in female ES cells leads to chaotic Xist up-regulation (Lee 2005), implying a critical role for Tsix in the coordination of monoallelic Xist expression. However, it is important to note that Tsix is poorly conserved in other eutherians and antisense transcription across XIST has not been found in humans for example (Chang and Brown 2010; Chureau et al. 2002; Migeon 2002; Migeon et al. 2001). This apparent lack of Tsix in other eutherians might explain the species differences in Xist’s monoallelic regulation in rabbits and humans (Okamoto et al. 2011). In the mouse, the precise manner in which Tsix controls monoallelic expression of Xist is still unclear, but one hypothesis, recently supported by live-cell imaging data, is that transient pairing events at the level of Tsix result in asymmetric Tsix expression, and this in turn facilitates asymmetric Xist up-regulation (Bacher et al. 2006; Masui et al. 2011; Xu et al. 2006).
Xist RNA-mediated silencing and heterochromatinization of the inactive X-chromosome
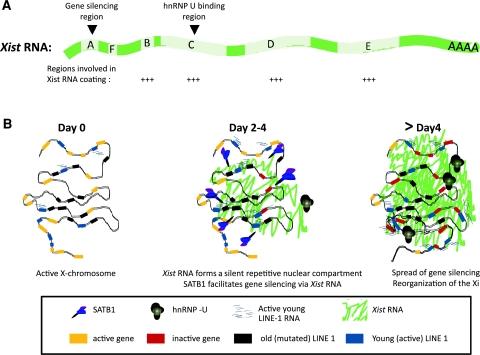
In the mouse, the upregulation of Xist is critical for downstream events in XCI leading to gene silencing and heterochromatinization. Deletions of Xist have demonstrated its essential role in inducing chromosome-wide inactivation in cis (Marahrens et al. 1997; Penny et al. 1996). This long, non-coding transcript has fascinated researchers since its discovery two decades ago (Brown et al. 1991a), yet very little is known about its unusual behaviour and mechanism of action. Xist/XIST is a polyadenylated, capped, spliced RNA, devoid of any significant open reading frame. It is retained in the nucleus as a 15- to 17-kb-long RNA and coats the chromosome from which it is expressed (Brockdorff et al. 1992; Brown et al. 1992; Clemson et al. 1996). To date, Xist is the only gene expressed exclusively from the Xi in somatic cells. Although the structure and organisation of the Xist gene does not differ greatly between mammals, its overall sequence conservation is poor (Brockdorff et al. 1992; Brown et al. 1992). The exception to this concerns a series of conserved repeats motifs in Xist, termed A to F (Fig. 3a), although their sequence and size can also be quite variable between species (Yen et al. 2007). The A repeat, lying at the 5′ end of Xist exon 1, is the most conserved and has been shown to be essential for the silencing function of Xist (Wutz et al. 2002) (discussed in “Xist RNA coating”).
Fig. 3.
Xist and the formation of the inactive compartment. a Structure of the Xist gene showing the conserved repeat regions labelled A to F. Specific functions and characteristics of the different regions of Xist are highlighted by arrowheads. b Model for the formation of the silent nuclear compartment by Xist RNA: Xist RNA coating occurs immediately upon differentiation and forms a nuclear silent compartment where X-linked genes are recruited as they become silent in a locus-specific manner. The formation of this silent and heterochromatic compartment is probably helped by the silent repetitive elements, especially LINE elements that are abundant on the X-chromosome; this occurs when the matrix attachment protein SATB1 is present and might facilitate gene relocation into the Xi. At this stage of differentiation (day 4), young LINE-1 elements are highly expressed and they might play a role in the spread of silencing into regions prone to escape, but which will eventually become silenced later (not shown). The hnRNP U matrix attachment protein becomes enriched on the Xi around days 4–5 of differentiation and participates in the maintenance of Xist RNA coating
Xist RNA coating
The precise manner in which Xist RNA coats the chromosome from which it is expressed in cis remains mysterious. This capacity does not appear to be restricted to the X-chromosome as Xist transgenes located on autosomes and X:autosome translocations display Xist RNA coating of autosomal regions in cis (Heard et al. 1999; Herzing et al. 1997; Lee and Jaenisch 1997; Lyon 1998a; Tang et al. 2010), although the degree of Xist coating and gene silencing can be less efficient on autosomes compared with the X-chromosome (Duthie et al. 1999; Popova et al. 2006). This implies that intrinsic characteristics of the sequence or chromatin environment of the X-chromosome might facilitate Xist RNA coating and/or spreading of the inactive state and possibly also its maintenance. Repeats such as LINEs are clearly enriched on the X-chromosome compared with autosomes (Bailey et al. 2000; Lyon 1998b). However, these elements do not seem to provide a scaffold for Xist RNA coating, but rather they help in the formation of heterochromatin, as will be discussed in the section on the formation of a silent nuclear compartment by Xist RNA (Chow et al. 2010). The precise nature of the DNA sequences to which Xist RNA might bind still remains a mystery. In fact, as DNAse treatment does not disrupt the Xist RNA nuclear signal in any way (Clemson et al. 2006), suggesting that Xist RNA may interact only indirectly with chromosomal components.
In order to define the regions of the Xist transcript that are involved in chromosome coating, gene silencing and chromatin modifications, various deletion derivatives of an inducible Xist cDNA were stably integrated into the X-linked locus (Hprt) in male ES cells. In this way, it was shown that several distinct regions of Xist can act cooperatively and redundantly to ensure the coating of the chromosome (Wutz et al. 2002) (Fig. 3a). These regions are also important for inducing some of the chromatin modifications associated with the Xi. Failure of Xist RNA to coat the Xi results in failure to inactivate the chromosome (Wutz et al. 2002). Importantly, Xist RNA lacking the highly conserved A-repeat region was capable of coating the X-chromosome but did not lead to chromosome-wide gene silencing (Chow et al. 2010; Wutz et al. 2002). The Xist A-repeat mutant could also induce histone modifications and other chromatin changes on the X-chromosome. This uncouples the properties of coating and silencing by Xist RNA and also demonstrates that Xist RNA is likely to have multiple, possibly independent, roles in gene silencing and heterochromatin formation of the Xi. The above studies were performed in differentiating ES cells, using a Tet-inducible Xist transgene. Another report used locked nucleic acid (LNA) interference probes to interfere with different parts of the endogenous Xist RNA in differentiated female ES cells and transformed mouse embryonic fibroblasts (MEFs). This revealed that targeting of the Xist repeat C region causes Xist RNA to be displaced from the Xi (Sarma et al. 2010). Interestingly, this region has been shown to interact with RGG domain of the hnRNP U/SP120/SAF-A, a matrix attachment protein whose localisation to the Xi depends on this RGG domain (Hasegawa et al. 2010; Helbig and Fackelmayer 2003). Depletion of the hnRNP U results in the dissociation of Xist RNA from the Xi and its dispersion into the nucleoplasm in differentiated female cells (Hasegawa et al. 2010). Furthermore, differentiating ES cells depleted for hnRNP U initiate XCI in a smaller percentage of cells due to failure of Xist to coat the X-chromosome properly (Hasegawa et al. 2010). This is the only factor so far shown to be crucial for Xist localization to the Xi. It remains to be clarified whether hnRNP U is actually required to initiate the Xist RNA coating process or instead to maintain it. The latter currently would be the likely scenario given that hnRNP U is only recruited to the Xi several cell cycles after Xist coating during ES cell differentiation (Pullirsch et al. 2010) (Figs. 3b, 4b).
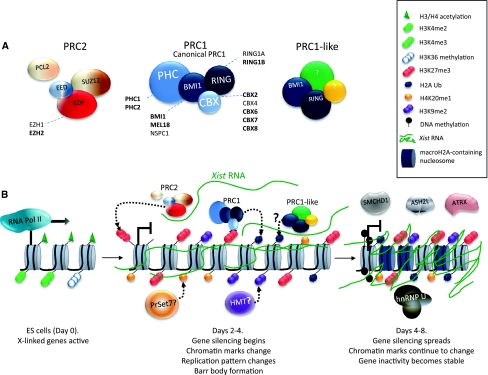
Fig. 4.
Chromatin dynamics at the initiation of the XCI. a Composition of the polycomb group 2 and the polycomb group 1 complexes. PRC2 is composed of three core elements SUZ12, EED and EZH and several co-factors including PCL2. The PRC1 complex has four core components: CBX, BMI1, RING and PHC. RING and the BMI1 component can also be found in other kind of complexes that we mention as PRC1-like complexes. All the alternative components are shown and in bold highlights the ones that have been documented as enriched in the Xi. b Schematic representation of the epigenetics events during XCI. In undifferentiated ES cells, the chromatin around the promoter of an X-linked gene is shown transcribed, with the presence of euchromatic marks such as H3K4me2/3, H3K36 methylation and H3 and H4 acetylation. Days 2–4: when differentiation is induced, Xist is upregulated on one X-chromosome and coats the X-chromosome from which it is expressed in cis. Gene silencing is initiated and Xist-mediated chromatin modifying complexes such as PRC1 and PRC2 and others are recruited to the Xi, resulting in the accumulation of PTMs typical of facultative heterochromatin such as H3K27me3, H2AK199ub, H3K9me2 and H4K20me1. The replication pattern of the Xi changes and the condensation of the Xi become apparent. Days 4–8: during this period, gene silencing is spread and maintained thereafter. Further epigenetic events take place such as the acquisition of DNA methylation at the CpGs of the 5′ end of genes and the incorporation of macroH2A histone variant; chromatin modifications acquired previously are maintained; there is also the recruitment of different proteins such as SMCHD1, ASH2L, ATRX and hnRNP U that will lock in the inactive state
Developmental time window for Xist RNA induction of XCI
Xist RNA upregulation during early development leads to XCI. On the other hand, aberrant Xist expression from the active X-chromosome in somatic cells following treatment with DNA demethylating agents does not cause gene silencing (Tinker and Brown 1998). Such studies suggested that Xist might only be capable of triggering XCI during an early developmental time window. The use of inducible Xist cDNA transgenes, whereby Xist can be expressed or repressed at different time points during ES cell differentiation, demonstrated that Xist is only competent to initiate XCI when expressed in undifferentiated and early differentiating ES cells (first 72 h of differentiation). Induction of Xist after this time, or in fully differentiated MEFs, no longer resulted in XCI (Wutz and Jaenisch 2000). Recently, it was found that ectopic Xist induction might also be able to induce XCI in certain types of adult cells, such as pro-B cells (Savarese et al. 2006), or in some lymphomas (Agrelo et al. 2009). The latter study in fact enabled the identification of a factor, SATB1, that might participate in Xist’s silencing capacity (Agrelo et al. 2009). SATB1 is a nuclear protein that is highly expressed in thymocytes and has previously been implicated in regulating chromatin structure (Cai et al. 2003). It has also been shown to interact physically with chromatin remodelling factors (Yasui et al. 2002). Although previously considered to be rather tissue specific, SATB1 and the closely related SATB2 protein are in fact expressed transiently during an early window of ES cell differentiation that exactly matches the period during which Xist is competent to trigger XCI. Indeed, knock-down of Satb1 and/or Satb2 in mouse ES cells prevented Xist-mediated silencing and ectopic expression of these factors in MEFs, where Xist RNA is artificially induced, enables Xist to repress transcription to some extent (Agrelo et al. 2009). The exact role of Satb1/2 for XCI in vivo remains to be defined, however. Furthermore, the development time window and factors involved in Xist RNA functions in other mammals have not yet been evaluated. However, findings in pre-implantation human embryos, where XIST RNA is expressed and yet chromosome-wide XCI is not triggered (Okamoto et al. 2011), suggest that the XIST RNA competence time window can be dissociated from the onset of XIST up-regulation found in mice.
Cell cycle time window for Xist RNA coating
In addition to the developmental time window during which Xist RNA can induce silencing, an important question concerns whether Xist RNA can coat the chromosome throughout the cell cycle or whether events downstream of Xist coating are sufficient to maintain the inheritance of the inactive state during early development. Following Xist RNA coating in cis, downstream events are triggered in a step-wise manner over a period of several cell cycles during differentiation and many of these seem to be implicated in helping to lock in the inactive state and provide cellular memory (Kohlmaier et al. 2004). Once established, the Xi no longer needs Xist RNA to maintain global inactivity (Csankovszki et al. 1999). However, during early development, gene silencing is initially Xist dependent and is totally reversible if Xist RNA coating is removed. Studies investigating the cell cycle kinetics of Xist RNA association with the Xi have reported that it remains associated throughout interphase but is displaced during mitosis in human and mouse cells (Clemson et al. 1996; Duthie et al. 1999; Hall and Lawrence 2003; Smith et al. 2004). Initial reports suggested that whereas human XIST RNA is rapidly lost from the chromosome by metaphase, murine Xist RNA remains associated up to telophase, but then dissociates. However, recent re-evaluation of Xist RNA coating during the cell cycle in differentiating female mouse ES cells (Jonkers et al. 2008) revealed that a significant number of female differentiated ES cells and MEFs exhibit Xist RNA coating even at telophase. Furthermore, experiments using MS2-tagged Xist RNA suggest that it is retained on the Xi throughout the cell cycle and does not diffuse into the nucleoplasm (Jonkers et al. 2008). Thus, Xist RNA could indeed provide a “memory” of the inactive state at least during an early developmental time window, which is when Xist expression is essential for XCI.
Formation of a silent nuclear compartment by Xist RNA
Soon after Xist is upregulated and accumulates on the X-chromosome in cis, the formation of a transcriptionally inert compartment within the Xist RNA domain can be observed within a relatively short period of time. Notably, the Xist RNA-coated chromosome shows rapid loss of the basic transcriptional machinery such as RNA POL II, TAF10 and TBP proteins, as well as a depletion of Cot1 repetitive RNA and a loss of euchromatic histone modifications (Chaumeil et al. 2002, 2006; Okamoto et al. 2004). Importantly, the X-chromosome sequences lying within the newly formed Xist domain are believed to consist mainly of silent repeats (Chaumeil et al. 2006; Chow et al. 2010). X-linked genes on the other hand are initially located outside or on the edge of this Xist RNA compartment but are moved into it as they become inactivated (Chaumeil et al. 2006; Chow et al. 2010) (Fig. 3b). Whether this relocalisation of genes into the Xist RNA compartment is a cause or a consequence of Xist RNA-mediated silencing, is still an open question. Interestingly, the appearance of this compartment does not depend on the silencing activity of Xist, since it is still formed in A-repeats Xist mutant. However, relocation of the genes into the compartment as they get silenced is dependent on the A-repeats (Chaumeil et al. 2006). Once XCI has been accomplished, the Xi is organised as a repetitive core surrounded by a rim of silent genes both in humans and mice (Chaumeil et al. 2006; Clemson et al. 2006). Only genes that escape XCI (including Xist itself) are located at the edge or outside of the Xist RNA domain in somatic cells (Chaumeil et al. 2006). Indeed, escape genes may be flanked by sequences permissive for their externalisation or looping out from the Xist compartment to be in contact with the transcription factor in the nucleoplasm (reviewed in Heard and Bickmore 2007).
The silent Xist RNA compartment initially encompasses X-chromosome repeats. Repetitive elements have long been implicated in silencing and heterochromatin formation (Eymery et al. 2009). In the context of XCI, LINEs and possibly other repeats within Xist RNA repressive compartment might have an important role by helping to nucleate heterochromatin of the chromosome. Consistent with this, genes in LINE-rich regions show much more efficient silencing and relocalization into the inactive Xist RNA compartment than genes in LINE-poor regions based on studies on ectopic Xist expression on autosomes (Chow et al. 2010; Tang et al. 2010). Genes escaping silencing in LINE-poor regions remain at distances far from the repressive compartment (Chow et al. 2010). Therefore, the high density of repeats, in particular, LINEs on the X-chromosome might provide a helpful environment for heterochromatin formation and the spread of inactivation. Interestingly, in regions of the X-chromosome that are prone to escape silencing (for example, around the Huwe1/Jarid1c locus), expression of young, active LINE-1 elements has been observed on the X-chromosome undergoing inactivation during ES cell differentiation (Chow et al. 2010). It has been proposed that these transcribed LINEs might facilitate the local propagation of XCI, especially into regions of the X-chromosome that are otherwise prone to escape.
Chromatin changes during X-chromosome inactivation
The chromatin of the Xi is dramatically altered following Xist RNA coating. The alterations appear to occur in a step-wise fashion and two major waves of events have been described which may or may not be inter-dependent. The initial wave of chromatin changes is tightly coupled to Xist coating, occurring within the first 2–3 days of ES cell differentiation. The second wave (4–8 day of differentiation) corresponds to the shift from reversible to irreversible XCI and may depend on the changes induced during the first wave (Wutz and Jaenisch 2000) and is accompanied by the acquisition of a chromosomal memory of unknown nature (Kohlmaier et al. 2004). Extinction of Xist expression after this time window (first 3 days of differentiation) does not cause major reactivation of genes, even if some of the characteristics of the Xi chromatin are disrupted (Csankovszki et al. 2001; Kohlmaier et al. 2004; Pullirsch et al. 2010; Wutz and Jaenisch 2000; Zhang et al. 2007).
It remains largely unknown how Xist mediates these chromatin events and whether they are direct or indirect consequences of Xist RNA coating. Nevertheless, it is important to consider the types of processes that could be implicated in the generation of facultative heterochromatin as a consequence of Xist coating. One mechanism is the recruitment of histone modifying complexes that would lay down post-translational modifications (PTM) on histones, mainly on their N-terminal tails that protrude out from the nucleosome, the fundamental unit of chromatin. Several PTMs have been described such as methylation, acetylation and ubiquitylation among others that have a direct impact on transcription, repair, replication or condensation (reviewed in Kouzarides 2007). PTMs could either disrupt the contacts between adjacent nucleosomes or recruit nonhistone proteins, their “readers” (Bannister and Kouzarides 2011). Indeed, the notion of a histone code (Strahl and Allis 2000; Turner 2000) whereby combinations of histone modifications could be targets for specific proteins is attractive, particularly in the case of the Xi to provide a specific signature of facultative heterochromatin that would distinguish it from constitutive heterochromatin and from euchromatin. A second mechanism that could be implicated in the heterochromatinization of the Xi is the replacement of histones by new synthesised histones including new histone variants. Histone variants have sequence variations providing a means to generate diversity in the chromatin structure (Banaszynski et al. 2010). A third mechanism could be the recruitment of chromatin remodelling machinery that changes accessibility to the DNA sequence by remodelling nucleosomes in an ATP-dependent manner. Finally, the recruitment of structural chromosome proteins, such as condensins and cohesions, able to affect higher order chromatin conformation and the compaction state of the chromosomes (Hirano 2006), as has been found for X-chromosome dosage compensation in C. elegans (Meyer 2010) might confer a fourth mechanism through which Xist coating might cause the heterochromatinization of the Xi.
Early chromatin changes during X-chromosome inactivation
Soon after Xist RNA accumulates a depletion in the histone modifications generally associated with gene activity, such as H3K4me2/3, H3K36 methylation and H3 and H4 acetylation is observed for the part of the X-chromosome within the Xist RNA domain, based on immunofluorescence (IF) detection (Chaumeil et al. 2002, 2006; Heard et al. 2001; Keohane et al. 1996; O’Neill et al. 2008). Shortly after this loss of active marks, an enrichment of repressive marks is observed—including H3K27me3, H3K9me2, H4K20me1, H2AK119ub and others (Fig. 4b). This first wave of alterations in the chromatin landscape of the X-chromosome chosen for inactivation happens within a relatively small time window lasting no more than 1–2 cell cycles. Indeed, whether cell division is actually required for some of these chromatin changes to take place remains an open question. It is also during this time window that gene silencing initiates on the X-chromosome, with genes shifting from outside to inside the Xist RNA compartment, which is depleted for actively marked chromatin and enriched for inactive chromatin, as they become silenced. The dynamics of gene silencing (and relocation) differs greatly among X-linked genes during ES cell differentiation (Chow et al. 2010; Lin et al. 2007; Patrat et al. 2009). The Xi also shifts its replication timing (Casas-Delucchi et al. 2011) and assumes a more compact conformation during this time window. Thus, the early wave of chromatin changes affecting the Xist RNA-coated X-chromosome in the mouse is preceded by a change in nuclear organisation of the X-chromosome and followed by a temporal change in replication. The interdependence between these events still remains to be defined (Chadwick and Willard 2002; Okamoto and Heard 2009; Okamoto et al. 2004) (Figs. 3b, 4b).
Potential mechanisms involved in early chromatin changes on the inactive X-chromosome
The role of chromatin modifications in XCI and the mechanisms by which these changes are induced are still largely mysterious. First we consider the mechanisms by which euchromatic marks are lost from the Xi. H3K4 methylation is a mark normally associated with active transcription, with H3K4me3 and H3K4me2 being strongly enriched at active promoters, whereas H3K4me1 has been more associated with enhancers (Heintzman et al. 2007). Rapid global loss of H3K4 methylation marks, based on immunufluorescence detection, is one of the earliest chromatin events observed during XCI (Chaumeil et al. 2002; Heard et al. 2001; O’Neill et al. 2008). Curiously, the three different forms behave differently during XCI, with the H3K4me3 mark being lost before H3K4me2, while H3K4me1 is in fact depleted on both the Xa and Xi compared with autosomes, during ES cell differentiation (O’Neill et al. 2008; da Rocha unpublished observations). Loss of these marks could either be passive through DNA replication and the exclusion in some way of the H3K4 methyltransferase complexes following Xist coating. The rapidity of this H3K4 methylation loss—maybe even within a single cell cycle—might imply active mechanisms, however. For example, histone exchange might affect the Xist RNA-coated X-chromosome; an alternative, not mutually exclusive mechanism, could involve histone demethylases (Christensen et al. 2007; Shi et al. 2004). The latter would be consistent with the more rapid loss of H3K4me3 compared with H3K4me2. Chromatin-modifying factors recruited to the X-chromosome in a Xist-dependent manner could play a part in such active mechanisms of histone methylation loss.
The loss of histone acetylation on the Xi is another characteristic change during early XCI. Acetylation of histone lysine residues results in the neutralisation of the basic charge of this amino acid and is believed to open up chromatin present at promoters of active genes (reviewed in Kouzarides 2007). Histone hypoacetylation on the Xi is first observed for histone H3K9 and subsequently for histone H4 (H4K5ac, H3K8ac, H3K12ac) (Chaumeil et al. 2002) (Fig. 4b). This difference in timing could imply different mechanisms that may be involved. Studies using general deacetylase inhibitors such as Trichostatin A (TSA) suggest the implication of histone deacetylases of the HDAC family in XCI (Casas-Delucchi et al. 2011; Csankovszki et al. 2001; Do et al. 2008; O’Neill et al. 1999) in establishing the depletion, or in maintaining the deacetylated state, a hallmark of the Xi. To date, however, no specific implication of any of the 11 HDACs has been found during XCI, and indeed, some degree of redundancy might be anticipated among these enzymes.
Several histone PTMs become enriched on the Xist RNA-coated X-chromosome during XCI. The mechanisms underlying the recruitment of heterochromatic marks on the Xi are better understood. The enrichment of H3K9me2, H3K27me3, H4K20me1 and H2AK119ub are the best documented Xi marks in the mouse (de Napoles et al. 2004; Fang et al. 2004; Heard et al. 2001; Kohlmaier et al. 2004; Peters et al. 2002; Plath et al. 2003). Certain candidate modifying complexes responsible for these modifications are also observed to be recruited transiently at early stages of XCI (Kohlmaier et al. 2004; Schoeftner et al. 2006): in particular, the Polycomb repressive complex 2 (PRC2) that catalyses the tri-methylation of H3K27 (Plath et al. 2003; Silva et al. 2003) and the PRC1 complex that mono-ubiquitinates the K119 at H2A (de Napoles et al. 2004; Plath et al. 2004). These complexes and their recruitment to the Xi will be discussed below.
The H3K27me3 mark is strongly enriched on the Xi both at interphase and during mitosis (Plath et al. 2003; Silva et al. 2003). Recent ChIP-seq data in differentiating mouse female ES cells has shown that this mark is enriched fairly uniformly across the Xi, but preferentially in gene-rich regions. H3K27me3 is present at promoters, but not restricted to them and does not seem to spread from the Xist locus (Marks et al. 2009). Far less is known about the mechanisms of Xi enrichment of H3K9me2 and H4K20me1. Several H3K9 MTases are known to exist in the mouse and one or more of these might be implicated. H4K20me1 seems to be cell-cycle regulated, only appearing on the Xi during late S phase and mitotis (Oda et al. 2009). This closely matches the expression pattern of the only H4K20 monomethyltransferase known, Pr-SET7 (Oda et al. 2010), suggesting that this could indeed be responsible for this mark. The precise role of these different histone modifiers and modifications to Xi heterochromatin formation remains largely to be determined. Functional studies involving knock outs are confounded by redundancy on the one hand and lethality due to pleiotropic effects on the other (Oda et al. 2009).
Considerable progress has been made in recent years on our understanding of the Polycomb complexes in XCI. Genetic studies point to a critical role for PRC2 in the maintenance of the inactive state (Wang et al. 2001) at least in extra-embryonic tissues of the mouse. The PRC2 complex is composed of three major components, the histone methyltransferase EZH2 (or EZH1), the zinc-finger-containing SUZ12 and the WD40 repeat protein EED that are essential both for the stabilisation and for the methyltransferase activity of EZHs proteins (reviewed in Margueron and Reinberg 2011). Several other PRC2 co-factors have recently been reported in ES cells and other proliferative cells such as the Polycomblike 2 protein—PCL2/MTF2 (Fig. 4a). PRC2 components are very quickly recruited to the X-chromosome, after Xist RNA coating and at the same time as H3K27me3 enrichment is observed (Fig. 4) (Plath et al. 2003; Silva et al. 2003). Interestingly, the enrichment of PRC2 complex is transient—only being observed in the first 2–5 days of differentiation, even though H3K27me3 persists thereafter (Plath et al. 2003; Silva et al. 2003). The Polycomb complex 1 is recruited at very similar stages to PRC2 (de Napoles et al. 2004; Fang et al. 2004; Plath et al. 2004). PRC1 is composed of four subunits, containing an E3 ubiquitin ligase (either RING1A/RING1 or RING1B/RNF2) responsible for the ubiquitylation of K119 at histone H2A. In contrast to PRC2, there are several PRC1 complexes due to the great number of paralogs of its subunits (Fig. 4a) (reviewed in Simon and Kingston 2009). Given the numerous putative combinations of PRC1 complexes, it is not surprising that the individual proteins may have different kinetics, or degree of recruitment to the Xi. For example, among the five CBX subunits, CBX7 seems to be preferentially associated with the Xi (Bernstein et al. 2006). Unlike PRC2 complex, PRC1 proteins do not seem to be enriched on all cells with a Xi (de Napoles et al. 2004; Plath et al. 2004; Schoeftner et al. 2006). Intriguingly, the ubiquitin ligase subunit RING1B has been found enriched in 50% of Xist RNA domains during interphase and on mitotic chromosomes (Fang et al. 2004; Schoeftner et al. 2006), but the mark that this complex lays down, H2AK119ub, is present on the Xi in the majority of Xist RNA domains throughout the cell cycle (de Napoles et al. 2004; Plath et al. 2004; Schoeftner et al. 2006).
PRC1 recruitment to chromatin has been classically defined to rely on PRC2-dependent deposition of H3K27me3. The tri-methylation of H3K27 is then thought to be recognised by one of the Polycomb homolog, CBX proteins, via its chromodomain, which will bring with it the PRC1 complex, leading to both RING1B/RING1A-mediated H2AK119ub, the establishment of chromatin condensation and the maintenance of stable gene silencing (Fig. 4b) (reviewed in Simon and Kingston 2009). This series of events may hold true for the Xi in extra-embryonic tissues, as embryos mutated for the PRC2 component EED fail to recruit PRC1 and H2AK119ub to their Xi (Kalantry et al. 2006). Interestingly, the same does not hold true in the ES cell system. Loss of EED in a male cell line, in which ectopic Xist expression can be induced from an autosome, results in failure to recruit some PRC1 subunits, but does not affect RING1B recruitment or enrichment of H2AK119ub (Schoeftner et al. 2006). Thus, RING1B can be recruited to the Xi in the absence of H3K27me3—perhaps in a Xist RNA-mediated manner by some other protein complex. In this context it is noteworthy that RING1B is part of other PRC1-like complexes (Gearhart et al. 2006; Sanchez et al. 2007) (Fig. 4a). It remains to be seen if PRC2-independent recruitment of PRC1 components also occurs in a normal female context where Xist is not ectopically induced at high levels.
The possible mechanism of PRC2 recruitment to the Xi has been subject of high interest. EZH2 has been extensively studied as the putative binder to long non-coding RNAs that might be important for PRC2 recruitment to target genomic sites (Kaneko et al. 2011; Zhao et al. 2010) and in the case of XCI, both EZH2 and SUZ12 have been shown to bind to the A-repeat region of the Xist transcript in undifferentiated and differentiating ES cells (Kaneko et al. 2011; Kanhere et al. 2010; Maenner et al. 2010; Zhao et al. 2008) (Fig. 4). The A-repeat element is unlikely to be the only means by which PRC2 is recruited to the Xi, however, since an inducible Xist gene deleted for the A-repeat region still results in global H3K27me3 accumulation, in early differentiating ESCs, although with a slower kinetics (Kohlmaier et al. 2004; Plath et al. 2003). A recent study has shown that the Polycomb-like 2 protein (PCL2/MTF2), a co-factor of PRC2 in ES cells and proliferative cells (Li et al. 2010; Walker et al. 2010) affects, to some extent, PRC2 recruitment or maintenance and the deposition of H3K27me3 to the Xi (Casanova et al. 2011) (Fig. 4). The relative contributions of the Xist A-repeat PRC2 association, versus a PCL2-mediated mechanism for PRC2 accumulation on the Xi remain unclear. One hypothesis could be that PRC2 is recruited directly by Xist RNA via its A-repeat region (and perhaps helped by other parts of the RNA) and the association with the Xi is then stabilised by the recognition of specific histone modifications, via the two PHD domains or the TUDOR domain of PCL2 (Fig. 4). However, it is still not clear whether EZH2 binding to the Xist RNA is a direct association, nor whether the role of PCL2, or other PRC2 co-factors, is to recruit or to maintain PRC2 deposition on the Xi.
Chromatin landscape of the inactive X-chromosome in other mammals
Although most studies on chromatin changes during the onset of XCI have been conducted in the mouse, there are several reports on the characteristics of the Xi in somatic cells of other mammals, even though this was not during the onset of XCI (Table 1). In general, depletion of euchromatic marks is consistently observed on the Xi in all mammals (Chadwick and Willard 2003; Chaumeil et al. 2002; Koina et al. 2009; Rens et al. 2010). On the other hand, some of the repressive marks associated with the Xi in humans and marsupials are different (Table 1). For example, in humans it has been suggested that the Xi heterochromatin can be subdivided into two sub-types. One of these closely resembles the mouse Xi, characterised by Xist RNA coating, H3K27me3 and macroH2A; the second resembles constitutive heterochromatin characterised by H3K9me3, H4K20me3 and binding of heterochromatin protein 1 (HP1) (Chadwick and Willard 2004), which are not observed to be particularly enriched on the mouse Xi. In the case of the marsupial Xi, H3K9me2 and H3K27me3 are present but its enrichment seems to be less striking compared with mouse and human, being restricted to specific windows of the cell cycle. Constitutive heterochromatin marks such as H3K9me3, H4K20me3 and HP1 are also present on the marsupial Xi (Chaumeil et al. 2011; Koina et al. 2009; Mahadevaiah et al. 2009; Rens et al. 2010; Zakharova et al. 2011). All in all, these differences between mammals suggest that the Xi can be constituted of several types of heterochromatin. The exact constitution of these chromatin signatures may be dependent on differences in X-chromosome sequence content between species on the one hand and on differences in the nature and recruitment of chromatin-modifying enzymes on the other.
Table 1.
Comparison of the epigenetic marks that characterised the facultative heterochromatin of the Xi in the mouse, humans and marsupials
| Epigenetic marks/factors | Mouse | Human | Marsupials |
|---|---|---|---|
| Xist coating |
+ (Interphase and mitosis) |
+ (Interphase) |
− |
| H3K4 hypomethyaltion |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) |
| H3/H4 hypoacetylation |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) |
| H3K36 hypomethylation | + | + | nd |
| H3R17 hypomethylation | + |
+ (Interphase only) |
+ |
| H3K9me2 |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) |
+ (Late S phase and G2) Enrichment less striking compared to mouse and humans |
| H3K9me3 | − |
+ (Interphase and mitosis) Banded pattern |
+ (Interphase and mitosis) |
| H3K27me3 |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) Banded pattern |
+ (Late S phase, G2 and mitosis) Enrichment less striking compared to mouse and humans |
| H4K20me1 |
+ (Interphase and mitosis) |
nd | − |
| H4K20me3 | − |
+ (Interphase and mitosis) Banded pattern |
+ (Interphase and mitosis) |
| H2AK119ub |
+ (Interphase and mitosis) |
nd | nd |
| macroH2A |
+ (Interphase and mitosis) |
+ (Interphase and mitosis) Banded pattern |
nd |
| HP1 | − |
+ (Interphase) |
+ (Interphase) |
| DNA methylation at 5′ end of genes | + |
+ (Less in the placenta) |
− |
+: presence; −: absence; nd: not determined; in brackets is the presence or absence of the mark during cell cycle when determined
Replication timing of the inactive X-chromosome
One of the most conserved and well-documented features of the Xi concerns its replication timing. The Xi has been shown to replicate asynchronously with respect to its active homologue in the human, mouse, rabbit and marsupial (Gartler and Riggs 1983; Kinsey 1967; Robinson et al. 1994; Sharman 1971) Asynchronous replication of the Xi compared with its active counterpart could provide temporal segregation of the two X-chromosomes and minimise the exposure of the Xi to transcription factors, thus facilitating the maintenance of transcriptional silencing. Indeed, asynchronous replication is the most evolutionarily conserved feature of the Xi (reviewed in Heard and Bickmore 2007). In mouse, in adult somatic cells and in most tissues of the embryo, the Xi is thought to replicate during late S phase, except in extra-embryonic tissues, where early replication has been found (Takagi and Sasaki 1975). In a recent publication, evidence was found that the Xi can replicate during early to mid S phase in mouse myoblast C2C12 cells and MEFs (Casas-Delucchi et al. 2011). The general conclusion seems to be that the bulk of the Xi tends to replicate as a single block, which is often, but not always, asynchronous in its replication timing when compared with the Xa and the rest of the genome.
The time of onset of asynchronous replication during the initiation of XCI corresponds to the first 2–3 days of differentiation which is the time window in which Xist-mediated gene silencing and chromatin changes occur (Chaumeil et al. 2002; Keohane et al. 1996) (Fig. 4). A recent study suggested that histone hypoacetylation could be causal in the change in replication timing of the Xi although this remains to be further investigated as the study was based on TSA treatment with arguably pleiotropic effects (Casas-Delucchi et al. 2011). What exactly the replication profile of the Xi represents in terms of origin firing and usage and whether it plays a role in the initiation or maintenance of XCI still remain open questions. At the single gene level in humans, late replication timing is highly correlated with the transcriptional activity of specific genes, rather than being a global chromosome-wide characteristic of the Xi, as late replicating X-linked genes were found to be transcriptionally inactive, whereas genes that escape XCI are synchronously replicated on the Xa and the Xi (Boggs and Chinault 1994).
Nuclear compartmentalization of the inactive X-chromosome
In addition to chromatin marks and asynchronous replication, the spatial segregation of the two X-chromosomes into different nuclear compartments might also help in the differential treatment required to adopt and maintain opposite epigenetic and transcriptional states (Cremer et al. 2006). Indeed, as discussed above, the Xi in both mouse and human is organised as a repetitive inner core surrounded by genes and Xist RNA localised often in contact with the nuclear periphery or with the nucleolus (Chaumeil et al. 2006; Clemson et al. 2006; Rego et al. 2008; Zhang et al. 2007). It has also been proposed that the Xi migrates to a perinucleolar compartment that is specialised in replicating condensed chromatin (SNF2H-enriched) during mid-to-late S phase, in order to faithfully duplicate its epigenetic marks and stably repress resident genes (Zhang et al. 2007). Whether this location is a cause or consequence of the inactive state is still not clear. Nevertheless, we can conclude that the Xi is not only epigenetically marked at the level of its chromatin, but also in terms of its temporal and spatial segregation.
Inactive X-chromosome compaction and higher order chromatin conformation
The Xi is detected cytologically as a condensed DNA mass using common nucleic acid dyes. Barr and Bertram (1949) first observed that this sub-nuclear structure is kept in many female cells in different organisms, and hence it is commonly called the Barr body. Analysis at the ultra-structural level by electron microscopy has revealed that the Xi does not appear as a solid mass similar to pericentric heterochromatin, but rather it appears as a series of separate, but tightly packed fibres (Rego et al. 2008). This difference in structure may reflect the different chromatin composition of facultative heterochromatin of the Xi when compared with constitutive heterochromatin. In terms of volume, the Xi may be slightly smaller than the Xa (Naughton et al. 2010), consistent with Xi being more condensed, although other studies have reported that the volume is similar but the overall texture is different (Chaumeil et al. 2006; Clemson et al. 2006). Compaction of the Xi is an early event as the DAPI-dense Barr body is detected early in differentiation following Xist coating (Chaumeil et al. 2006). It may undergo a further degree of compaction and/or higher order organisation at later stages based on FISH studies (Chow et al. 2010). Biochemical approaches, involving chromatin fractionation after micrococcal nuclease digestion, have shown that both the Xa and Xi have similar bulk 30 nm chromatin fibre structures in human somatic cells, its differences only being detected locally at the promoters of genes that are expressed on the Xa and silent on the Xi. Analysis of large-scale chromatin compaction using 3D RNA/DNA FISH with probes 2 Mb apart revealed that only in gene-rich regions, the Xi indeed shows more compaction than the Xa (Naughton et al. 2010).
The molecular basis of the more compact state of the Xi remains to be elucidated but is likely to depend on multiple aspects of its chromatin content and associated proteins and nucleic acids. One candidate could be the PRC1 component, RING1B, which has recently been shown to compact chromatin in vivo in the Hox cluster independently of its ubyquitilation activity (Eskeland et al. 2010). So far no such role has been found on the Xi (de Napoles et al. 2004), but this could also be due to redundant roles of different chromatin factors.
Late epigenetic changes and chromosome structure of the Xi
Although major alterations in gene activity, chromatin status, nuclear compartmentalisation and replication timing of the Xi are observed in the first few days of differentiation, XCI is only fully accomplished several days later (Chow et al. 2010).
In this second wave (day 4 of differentiation onwards), many of the changes initiated during the first wave are maintained or extended to other regions (Marks et al. 2009). Interestingly, marks such as H3K27me3 and H2AK119ub are maintained even though the enrichment of PRC2 and PRC1 complexes are no longer detectable by IF (de Napoles et al. 2004; Silva et al. 2003). Instead, new players such as the Trithorax protein ASH2L and the chromatin remodelling factor ATRX (Baumann and De La Fuente 2009; Pullirsch et al. 2010) appear on the Xi (Fig. 4b). The roles of these proteins in XCI remain to be assessed. At first sight, the recruitment of ASH2L is counter-intuitive since it belongs to a family of H3K4 methyltransferases—which lead to histone modifications that are actually depleted on the Xi. However, it cannot be excluded that transient H3K4 methylation may be followed by rapid demethylation on the Xi during the cell cycle. Alternatively, ASH2L on the Xi could reflect another function of this protein. Another protein that is enriched during this period is the matrix attachment protein hnRNP U, important for RNA coating (Pullirsch et al. 2010). During this later phase, DNA methylation at promoters of X-linked genes, a step that seems to be very important for the maintenance of the inactive state, is also believed to occur. DNA methylation is somehow dependent on the recently discovered structural chromosome protein SMCHD1, which results in loss of DNA methylation and some degree of X-linked gene reactivation in mice mutated for this gene (Blewitt et al. 2008). Another chromatin change during this second phase is the recruitment of the histone variant macroH2A (Costanzi and Pehrson 1998). Finally, the organisation of the Xi also seems to become altered between days 4 and 8 of differentiation (Chow et al. 2010) (Figs. 3b, 4b). The changes during this second phase of the XCI process are likely to be involved in maintenance and stable locking in of the Xi.
DNA methylation and XCI
DNA methylation has long been considered to be an important feature of the Xi and the maintenance of the inactive state (Riggs and Xiong 2004). Methylation patterns can be inherited from cell to cell, thanks to maintenance DNA methyltransferase activity that recognises and methylates hemimethylated CpG sites during DNA replication (reviewed in Altun et al. 2010). Although promoters and CpG islands of X-linked genes tend to be methylated on the Xi (Norris et al. 1991; Tribioli et al. 1992), the overall methylation levels have been shown to be less than those on the Xa by cytogenetic studies in both humans and marsupials (Loebel and Johnston 1993; Viegas-Pequignot et al. 1988). This has been confirmed by recent studies using global microarray analyses involving methylated DNA immunoprecipitation (MeDIP) to assess the DNA methylation status of the Xi relative to the Xa in human primary cells (Weber et al. 2007). Intriguingly, the Xa shows increased methylation levels in gene bodies compared with the Xi, although the significance of such gene-body methylation still remains to be elucidated (Hellman and Chess 2007). The precise kinetics of DNA hypermethylation at promoters during XCI has never been systematically investigated, but should now be feasible thanks to next generation sequencing technologies that enable allele-specific assessment of epigenomic states.
Although the mechanism by which DNA methylation becomes recruited to different genes on the Xi is still not clear, this mark is an important player in the maintenance of the inactive state in somatic cells of eutherian mammals, as shown by studies involving 5-aza-C treatment or deletion of the maintenance DNA methyltransferase, Dnmt1 (Csankovszki et al. 2001; Graves and Young 1982; Mohandas et al. 1981). Furthermore, instability of silencing of the Xi has been reported in patients suffering immunodeficiency centromeric instability facial anomalies (ICF), a syndrome caused by a defect in the de novo DNA methyltransferase DNMT3B (Hansen et al. 2000). In both cases, the effects observed in derepression were minimal and gene-specific, arguing that other repressive mechanisms are sufficient to maintain silencing of most loci. Indeed, in extra-embryonic tissues the promoters of X-linked genes do not appear to be methylated on the Xi even though it is globally inactive. This may account for the greater dependence on Polycomb-mediated maintenance of Xi inactivity in these tissues (Wang et al. 2001). Furthermore, although DNA methylation on the Xi is found in all eutherians examined to date, it is not a feature of promoters of X-linked genes on the Xi in marsupials (Hornecker et al. 2007; Kaslow and Migeon 1987; Loebel and Johnston 1996). However, the precise extent to which this mark is present on the Xi in other mammals needs to be systematically elucidated.
XCI and the histone variant macroH2A
The only histone variant to date that has been found to be enriched on the Xi is macroH2A. This is an unusual histone variant consisting of a domain similar to that of the conventional H2A (H2A-like domain) fused to a large nonhistone region (Pehrson and Fried 1992). The enrichment of macroH2A during XCI suggests that it might play a role in this process. The presence of this histone variant in chromatin is correlated with transcriptional repression. The chromatin-remodelling complex SWI/SNF is unable to remodel in the presence of macroH2A. MacroH2A also represses RNA polymerase II transcription (Doyen et al. 2006). In agreement with this, macroH2A is depleted from active genes (Changolkar and Pehrson 2006). Three macroH2A variants were described (Changolkar and Pehrson 2006); two of them have been shown to be enriched on the Xi (Chadwick et al. 2001; Costanzi and Pehrson 1998) from around day 4–5.
IF studies in human and mouse indicate that macroH2A is enriched on the Xi (Chadwick et al. 2001; Costanzi et al. 2000). MacroH2A was found to be uniformly distributed across the entire Xi chromosome by ChiP-on-chip analysis in human liver cells. This result suggests a potential role for macroH2A in large-scale chromosome structure, rather than directly inhibiting transcription and in genome stability (Mietton et al. 2009). Cell cycle analysis in human and mouse show high local concentration of macroH2A during S phase. This may be one of many redundant mechanisms to ensure the replication of the inactive state (Chadwick and Willard 2002). Interestingly, macroH2A like PTMs such as H2AK199ub and H3K27me3, remains associated with both the human and mouse Xi during mitosis. MacroH2A is detected as distinct bands that appear to mimic the bands seen with Xist RNA on the human Xi during mitosis (Chadwick and Willard 2004).
Deletion of Xist RNA leads to loss of macroH2A on the Xi (Csankovszki et al. 1999). Xist-dependent recruitment is supported by a Xist-inducible systems in XY cells (Wutz et al. 2002) in which cell differentiation and Xist induction were sufficient to recruit macroH2A (Pullirsch et al. 2010). However, macroH2A recruitment to the Xi does not depend on Xist transcript’s silencing function, since is still recruited in the A-repeats Xist mutant (Pullirsch et al. 2010). The rather late recruitment of macroH2A to the Xi (>day4) suggests that it has a role in the maintenance of the inactive state. However, just as other chromatin marks of the Xi, it is not essential since its deletion does not result in detectable re-activation of the genes on the Xi or impact in any measurable way on the XCI process (Changolkar et al. 2007). Nevertheless, conditional mutations of this variant might reveal a role at different stages of development.
Perspectives on epigenetic mechanisms and x-linked gene silencing during X-chromosome inactivation
Changes in chromatin composition and DNA methylation accompany the progressive gene silencing observed in the Xi, resulting in a remarkable locked state very refractory to reactivation and inherited through cell division. As we have seen, although some of these chromatin changes may be involved in the gene-silencing events during XCI, most of the evidence points to such changes representing epigenetic marks, that provide the cellular memory of the inactive state, at least, in certain developmental contexts.
The loss of euchromatic marks, such as H3K4 methylation and H3 acetylation are the first described chromatin modifications in the Xi. Therefore, we could speculate that they might be the initial triggering of silencing, resulting in loss of the transcriptional machinery that is then maintained by the recruitment of heterochromatic marks. This remains to be assessed. However, it could well be that loss of euchromatic marks is just a consequence of the removal of the transcriptional machinery, as genes become recruited into the silent Xist RNA compartment. On the other hand, removal of euchromatic marks on the Xi could be a crucial step for the subsequent recruitment of certain chromatin-modifying complexes. The coordinated regulation of gene repression by the H3K4 demethylase JARID1A together with PRC2 has been suggested to occur for a fraction of PRC2 target genes during ES cell differentiation (Pasini et al. 2008). It will be interesting to know whether, for example, the loss of H3K4 methylation has a similar impact on the efficiency of PRC2 recruitment in the context of XCI.
H3K27me3 is the histone mark that has been mostly extensively studied in the context of XCI. However, this modification clearly does not provide the initial trigger for gene silencing along the Xi. In the Eed−/− mouse, devoid of H3K27me3, the Xi remains silenced at the blastocyst stage in both ICM and trophoectoderm (Kalantry and Magnuson 2006; Kalantry et al. 2006; Wang et al. 2001). Xist-mediated silencing is also not significantly perturbed in ES cells lacking Eed (Schoeftner et al. 2006) or knockdown for the three core PRC2 proteins (Zhao et al. 2008). Furthermore, H3K27me3 is well recruited to the Xi by the Xist A-repeat mutant described as being incompetent for gene silencing (Kohlmaier et al. 2004). Therefore, global H3K27me3 enrichment is not sufficient to cause gene silencing. Importantly, however, a role for this mark in maintenance of X-linked gene silencing of the imprinted XCI in the murine extra-embryonic differentiated trophoblast has been established. Mice deleted for Eed can initiate imprinted XCI, but reactivation of X-linked genes and a GFP reporter transgene are observed in the extra-embryonic differentiated trophoblast (Kalantry et al. 2006; Wang et al. 2001). Curiously, general X-linked inactivation is still observed in undifferentiated trophoblast in these mutants, a stage when EED levels are normally highly enriched in the Xi. At this stage, Eed mutants begin to lose Xist RNA coating and heterochromatic marks (H3K27me3, H4K20me1, H2Aub and macroH2A), but reactivation of X-linked genes occurs only in differentiated trophoblast cells, coinciding with the time when H3K4me2 and H3 and H4 acetylation is regained in the Xi (Kalantry et al. 2006). The mechanism by which H3K27me3 loss affects maintenance of XCI in such cells remains unclear. Interestingly, reactivation of X-linked expression is not observed in other extra-embryonic lineages such as visceral endoderm and extra-embryonic ectoderm (Kalantry et al. 2006) and random XCI is not affected in the embryonic tissues (Kalantry and Magnuson 2006).
As discussed above, DNA methylation of X-linked promoters is considered a late epigenetic event in the process of XCI, acquired after initial transcriptional repression. Failure to maintain silencing through DNA methylation loss during development has been described in mice mutated for the Smchd1 gene, resulting in female-specific death at around E11.5 (Blewitt et al. 2008). The SMCHD1 protein has a SMC hinge domain, also present in proteins of the cohesion/condensin family. Female mutants for this protein display normal Xist RNA coating and H3K27me3 enrichment on the Xi, but show little sign of DNA methylation at 5′ end of X-linked genes when analysed at around E9.5–E10.5 (Blewitt et al. 2008). It remains to be understood whether the effect in DNA methylation is direct or indirect and whether this is happening at the level of recruitment of the DNA methyltransferase machinery or at the maintenance of the methylated state once established. Furthermore, we cannot rule out that SMCHD1 has effects other than affecting DNA methylation that could also lead to X-linked gene reactivation. Whatever is the reason for failure to keep XCI, SMCHD1 remains the only chromatin/chromosome structure factor so far described to cause failure of maintenance of the inactive state of Xi in the embryo proper.
Maintenance of the inactive state of the Xi in females clearly involves different layers of epigenetic regulation, including delayed replication kinetics, nuclear organisation and chromatin changes such as histone hypoacetylation, as well as heterochromatic marks, macroH2A and DNA methylation. With the exception of Smchd1, loss of any one of these marks is not sufficient to unlock the inactive state of the Xi in somatic, embryonic cells (Csankovszki et al. 2001; Hernandez-Munoz et al. 2005; Nusinow et al. 2007). Indeed, synergy between epigenetic mechanisms appears to preserve and reinforce the inactive state. Dissecting out the exact nature of these synergies and the molecular links between epigenetic marks remains an exciting, though challenging prospect as it will require sophisticated molecular genetics tools (conditional knock outs and/or transgenes) combined with biochemistry and single cell analyses. Such studies are clearly relevant from a medical point of view, as the X-chromosome harbours many genes that affect human health, particularly mental retardation disorders. For example, in the case of X-linked disorders such as Rett syndrome, which causes severe neurological effects in girls who are heterozygote for mutations in the X-linked MECP2 gene, understanding how the wild type, but silent allele of this gene on the Xi might be specifically induced to reactivate in affected cells, could provide a major step forward in terms of therapeutic strategies.
In conclusion, in the 50 years since Mary Lyon first proposed the process of XCI, we have a much deeper understanding of this phenomenon, but there is still a long road to its full understanding ahead of us. In particular, the major differences that are emerging in the X-inactivation strategies in different mammals suggest that this process will need to be explored not just in the mouse, but also in other species.
Acknowledgments
We would like to apologise to all those authors whose findings we could not cite due to space limitations. We wish to thank members of the Mammalian Development Epigenetics team for discussions and input and Rache Duffié for critical reading of the manuscript. ME is supported by the Fondation pour la Recherche Medical and SR is supported by a Marie Curie Fellowship (European Commission). Work in the Heard lab is funded by the Agence Nationale de la Recherche, an ERC Advanced Investigator Award, the INCA and the Fondation de France.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
M. Escamilla-Del-Arenal, S. T. da Rocha have contributed equally.
References
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun G, Loring JF, Laurent LC. DNA methylation in embryonic stem cells. J Cell Biochem. 2010;109:1–6. doi: 10.1002/jcb.22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19:662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Gunhanlar N, Pardo CG, Achame EM, Ghazvini M, Boers R, Kenter A, Rentmeester E, Grootegoed JA, Gribnau J. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 2011;7:e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Baumann C, De La Fuente R. ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma. 2009;118:209–222. doi: 10.1007/s00412-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Chinault AC. Analysis of replication timing properties of human X-chromosomal loci by fluorescence in situ hybridization. Proc Natl Acad Sci USA. 1994;91:6083–6087. doi: 10.1073/pnas.91.13.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard HF, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, Ledbetter DH, Levy E, Craig IW, Willard HF. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Casanova M, Preissner T, Cerase A, Poot R, Yamada D, Li X, Appanah R, Bezstarosti K, Demmers J, Koseki H, Brockdorff N. Polycomblike 2 facilitates the recruitment of PRC2 polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development. 2011;138:1471–1482. doi: 10.1242/dev.053652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Delucchi CS, Brero A, Rahn HP, Solovei I, Wutz A, Cremer T, Leonhardt H, Cardoso MC. Histone acetylation controls the inactive X chromosome replication dynamics. Nat Commun. 2011;2:222. doi: 10.1038/ncomms1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Valley CM, Willard HF. Histone variant macroH2A contains two distinct macrochromatin domains capable of directing macroH2A to the inactive X chromosome. Nucleic Acids Res. 2001;29:2699–2705. doi: 10.1093/nar/29.13.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Brown CJ (2010) Identification of regulatory elements flanking human XIST reveals species differences. BMC Mol Biol 11:20 [DOI] [PMC free article] [PubMed]
- Changolkar LN, Pehrson JR. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol Cell Biol. 2006;26:4410–4420. doi: 10.1128/MCB.02258-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR. Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol. 2007;27:2758–2764. doi: 10.1128/MCB.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Waters PD, Koina E, Gilbert C, Robinson TJ, Marshall Graves JA. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals. PLoS One. 2011;6:e19040. doi: 10.1371/journal.pone.0019040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L (2002) Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res 12:894–908 [DOI] [PMC free article] [PubMed]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Stein P, Worrad DM, Schultz RM, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Marshall Graves JA. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 2008;4:e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Chaumeil J, Hore TA, Marshall Graves JA. Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Res. 2009;17:671–685. doi: 10.1007/s10577-009-9058-6. [DOI] [PubMed] [Google Scholar]
- Do JT, Han DW, Gentile L, Sobek-Klocke I, Stehling M, Scholer HR. Enhanced reprogramming of Xist by induced upregulation of Tsix and Dnmt3a. Stem Cells. 2008;26:2821–2831. doi: 10.1634/stemcells.2008-0482. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, Turner BM, Zakian SM, Brockdorff N. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol. 2009;53:259–268. doi: 10.1387/ijdb.082673ae. [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA, Young GJ. X-chromosome activity in heterokaryons and hybrids between mouse fibroblasts and teratocarcinoma stem cells. Exp Cell Res. 1982;141:87–97. doi: 10.1016/0014-4827(82)90071-4. [DOI] [PubMed] [Google Scholar]
- Hall LL, Lawrence JB. The cell biology of a novel chromosomal RNA: chromosome painting by XIST/Xist RNA initiates a remodeling cascade. Semin Cell Dev Biol. 2003;14:369–378. doi: 10.1016/j.semcdb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Stoger R, Wijmenga C, Stanek AM, Canfield TK, Luo P, Matarazzo MR, D’Esposito M, Feil R, Gimelli G, Weemaes CM, Laird CD, Gartler SM. Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum Mol Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Helbig R, Fackelmayer FO. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma. 2003;112:173–182. doi: 10.1007/s00412-003-0258-0. [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LB, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hornecker JL, Samollow PB, Robinson ES, Vandeberg JL, McCarrey JR (2007) Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 45:696–708 [DOI] [PubMed]
- Jonkers I, Monkhorst K, Rentmeester E, Grootegoed JA, Grosveld F, Gribnau J. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28:5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. The polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2011;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow DC, Migeon BR (1987) DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: evidence for multistep maintenance of mammalian X dosage compensation. Proc Natl Acad Sci USA 84:6210–6214 [DOI] [PMC free article] [PubMed]
- Keohane AM, O’Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kinsey JD. X-chromosome replication in early rabbit embryos. Genetics. 1967;55:337–343. doi: 10.1093/genetics/55.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koina E, Chaumeil J, Greaves IK, Tremethick DJ, Graves JA. Specific patterns of histone marks accompany X chromosome inactivation in a marsupial. Chromosome Res. 2009;17:115–126. doi: 10.1007/s10577-009-9020-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O’Neill LP, Turner BM. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel DA, Johnston PG. Analysis of DNase 1 sensitivity and methylation of active and inactive X chromosomes of kangaroos (Macropus robustus) by in situ nick translation. Chromosoma. 1993;102:81–87. doi: 10.1007/BF00356024. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Johnston PG (1996) Methylation analysis of a marsupial X-linked CpG island by bisulfite genomic sequencing. Genome Res 6:114–123 [DOI] [PubMed]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation spreads itself: effects in autosomes. Am J Hum Genet. 1998;63:17–19. doi: 10.1086/301940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, Visvikis A, Branlant C. 2D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol. 2009;19:1478–1484. doi: 10.1016/j.cub.2009.07.041. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR, Epstein CJ, Travis B, Tucker G, Yatziv S, Martin DW, Jr, Clift S, Cohen S. X-chromosome inactivation during differentiation of female teratocarcinoma stem cells in vitro. Nature. 1978;271:329–333. doi: 10.1038/271329a0. [DOI] [PubMed] [Google Scholar]
- Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupe P, Barillot E, Belmont AS, Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during es cell differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A, Monk M. X-chromosome activity in female mouse embryos heterozygous for Pgk-1 and Searle’s translocation, T(X; 16) 16H. Genet Res. 1983;41:69–83. doi: 10.1017/s0016672300021078. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Sex in the wormcounting and compensating X-chromosome dose. Trends Genet. 2000;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 2009;29:150–156. doi: 10.1128/MCB.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR. X chromosome inactivation: theme and variations. Cytogenet Genome Res. 2002;99:8–16. doi: 10.1159/000071568. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Chowdhury AK, Dunston JA, McIntosh I (2001) Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: implications for X inactivation. Am J Hum Genet 69:951–960 [DOI] [PMC free article] [PubMed]
- Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Moreira de Mello JC, de Araujo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Sproul D, Hamilton C, Gilbert N. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol Cell. 2010;40:397–409. doi: 10.1016/j.molcel.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- Navarro P, Oldfield A, Legoupi J, Festuccia N, Dubois A, Attia M, Schoorlemmer J, Rougeulle C, Chambers I, Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- Navarro P, Moffat M, Mullin NP, Chambers I (2011) The X-inactivation trans-activator Rnf12 is negatively regulated by pluripotency factors in embryonic stem cells. Hum Genet [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Norris DP, Brockdorff N, Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- O’Neill LP, Keohane AM, Lavender JS, McCabe V, Heard E, Avner P, Brockdorff N, Turner BM. A developmental switch in H4 acetylation upstream of Xist plays a role in X chromosome inactivation. EMBO J. 1999;18:2897–2907. doi: 10.1093/emboj/18.10.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LP, Spotswood HT, Fernando M, Turner BM. Differential loss of histone H3 isoforms mono-, di- and tri-methylated at lysine 4 during X-inactivation in female embryonic stem cells. Biol Chem. 2008;389:365–370. doi: 10.1515/BC.2008.046. [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno (1967) In: Sex chromosomes and sex-linked genes. Springer, Berlin
- Ohno S, Hauschka TS. Allocycly of the X-chromosome in tumors and normal tissues. Cancer Res. 1960;20:541–545. [PubMed] [Google Scholar]
- Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–669. doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM, Avner P, Heard E. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thépot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf J-P, Renard J-P, Duranthon Vr, Heard E. Eutherian mammals use different strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Park Y, Kuroda MI. Epigenetic aspects of X-chromosome dosage compensation. Science. 2001;293:1083–1085. doi: 10.1126/science.1063073. [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. Developmentally regulated alterations in polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova BC, Tada T, Takagi N, Brockdorff N, Nesterova TB. Attenuated spread of X-inactivation in an X; autosome translocation. Proc Natl Acad Sci USA. 2006;103:7706–7711. doi: 10.1073/pnas.0602021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullirsch D, Hartel R, Kishimoto H, Leeb M, Steiner G, Wutz A. The trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development. 2010;137:935–943. doi: 10.1242/dev.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci. 2008;121:1119–1127. doi: 10.1242/jcs.026104. [DOI] [PubMed] [Google Scholar]
- Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci USA. 2010;107:17657–17662. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD, Xiong Z. Methylation and epigenetic fidelity. Proc Natl Acad Sci USA. 2004;101:4–5. doi: 10.1073/pnas.0307781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson ES, Samollow PB, VandeBerg JL, Johnston PG. X-chromosome replication patterns in adult, newborn and prenatal opossums. Reprod Fertil Dev. 1994;6:533–540. doi: 10.1071/rd9940533. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman GB. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature. 1971;230:231–232. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Smith KP, Byron M, Clemson CM, Lawrence JB. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands. Chromosoma. 2004;113:324–335. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Tang YA, Huntley D, Montana G, Cerase A, Nesterova TB, Brockdorff N. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin. 2010;3:10. doi: 10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker AV, Brown CJ. Induction of XIST expression from the human active X chromosome in mouse/human somatic cell hybrids by DNA demethylation. Nucleic Acids Res. 1998;26:2935–2940. doi: 10.1093/nar/26.12.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribioli C, Tamanini F, Patrosso C, Milanesi L, Villa A, Pergolizzi R, Maestrini E, Rivella S, Bione S, Mancini M, et al. Methylation and sequence analysis around EagI sites: identification of 28 new CpG islands in XQ24-XQ28. Nucleic Acids Res. 1992;20:727–733. doi: 10.1093/nar/20.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- van den Berg IM, Laven JS, Stevens M, Jonkers I, Galjaard RJ, Gribnau J, van Doorninck JH. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009;84:771–779. doi: 10.1016/j.ajhg.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas-Pequignot E, Dutrillaux B, Thomas G. Inactive X chromosome has the highest concentration of unmethylated Hha I sites. Proc Natl Acad Sci USA. 1988;85:7657–7660. doi: 10.1073/pnas.85.20.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- Yen ZC, Meyer IM, Karalic S, Brown CJ. A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics. 2007;90:453–463. doi: 10.1016/j.ygeno.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Zakharova IS, Shevchenko AI, Shilov AG, Nesterova TB, Vandeberg JL, Zakian SM. Histone H3 trimethylation at lysine 9 marks the inactive metaphase X chromosome in the marsupial Monodelphis domestica. Chromosoma. 2011;120:177–183. doi: 10.1007/s00412-010-0300-y. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]