Abstract
Grape expressed sequence tags (ESTs) are a new resource for developing simple sequence repeat (SSR) functional markers for genotyping and genetic mapping. An integrated pipeline including several computational tools for SSR identification and functional annotation was developed to identify 6,447 EST-SSR sequences from a total collection of 215,609 grape ESTs retrieved from NCBI. The 6,447 EST-SSRs were further reduced to 1,701 non-redundant sequences via clustering analysis, and 1,037 of them were successfully designed with primer pairs flanking the SSR motifs. From them, 150 pairs of primers were randomly selected for PCR amplification, polymorphism and heterozygosity analysis in V. vinifera cvs. Riesling and Cabernet Sauvignon, and V. rotundifolia (muscadine grape) cvs. Summit and Noble, and 145 pairs of these primers yielded PCR products. Pairwise comparisons of loci between the parents Riesling and Cabernet Sauvignon showed that 72 were homozygous in both cultivars, while 70 loci were heterozygous in at least one cultivar of the two. Muscadine parents Noble and Summit had 90 homozygous SSR loci in both parents and contained 50 heterozygous loci in at least one of the two. These EST-SSR functional markers are a useful addition for grape genotyping and genome mapping.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-010-9477-2) contains supplementary material, which is available to authorized users.
Keywords: SSR, EST, Marker, Genotyping, Grape
Introduction
Microsatellites, or simple sequence repeats (SSRs), are short (1–6 bp) repeat DNA motifs that are usually single locus markers with characteristics of hypervariability, abundance and reproducibility. The variation of the SSR repeat units can be easily differentiated by PCR products amplified with primers flanking the SSR motif. SSRs have been widely used for bacteria screening (Lin et al. 2005), plant genotyping (Chen et al. 2006), linkage mapping (Zhang et al. 2002), gene tagging (Roy et al. 2002), and map-based gene cloning (Tekeoglu et al. 2002).
The availability of ESTs greatly accelerates the systematic identification of SSRs and corresponding marker development based on computer analytical approaches (Varshney et al. 2002; Gao et al. 2003; Thiel et al. 2003; Chen et al. 2006). EST-derived SSRs have been well documented in some plant species including Arabidopsis (Depeiges et al. 1995), sugarcane (Cordeiro et al. 2001), cereal species (Kantety et al. 2002), cacao (Lima et al. 2008), and rubber tree (Feng et al. 2009). Using homology searches, putative functions can be deduced for the SSRs and thereby provide a new resource that can further aid in genetic and evolutionary studies (Cho et al. 2000; De Keyser et al. 2009).
EST-SSR and genomic SSR markers should be considered as complementary to plant genome mapping, with EST-SSR being less polymorphic but concentrated in the gene-rich regions (Varshney et al. 2006). With hundreds of thousands of ESTs available in the public domain, the process of developing EST-SSR markers has been greatly accelerated by using optimized computational pipelines and high-throughput genotyping techniques.
SSR markers have been widely used in grape genotyping. The high polymorphism of Vitis-derived microsatellite loci has been reported extensively in the literature and used for fingerprinting (Thomas and Scott 1993; Bowers et al. 1996, 1999a, b; Sefc et al. 1998; Arroyo García et al. 2002; Di Gaspero et al. 2005, 2007; Merdinoglu et al. 2005; Lamoureux et al. 2006; Costantini et al. 2007; De Mattia et al. 2007; Cipriani et al. 2008; Bocharova et al. 2009; Riaz et al. 2009). Several publications have also demonstrated transferability of SSR markers across the Vitis genus (Lin and Walker 1998; Tessier et al. 1999; Di Gaspero et al. 2000; Fernández et al. 2008).
SSR markers have been used for construction of grape genetic maps (Dalbò et al. 2000; Doligez et al. 2002; Grando et al. 2003; Adam-Blondon et al. 2004; Doucleff et al. 2004; Riaz et al. 2004, 2006; Lowe and Walker 2006; Di Gaspero et al. 2007; Vezzulli et al. 2008). While the majority of the loci in grape linkage maps are microsatellite markers developed from genomic DNA libraries, the availability of EST-SSRs will serve as new genetic markers to be included into the linkage map (Decroocq et al. 2003; Akkak et al. 2006; Salmaso et al. 2008). EST-SSRs have been reported to be less polymorphic but to have higher transferability than genomic SSRs in grape and other plants because of greater DNA sequence conservation in transcribed regions (Scott et al. 2000; Cho et al. 2000; Chabane et al. 2005).
Traditionally, SSR PCR products are separated by polyacrylamide gels (Thiel et al. 2003) or Metaphor Agarose gels (Chani et al. 2002). The electrophoresis-based technology is low-throughput and comes with imprecise sizing at times. Automatic capillary sequencing using fluorescently-labeled primers (Eujayl et al. 2002) provides more accurate and high-throughput genotyping results but the cost of dye-labeling each forward primer is high. Using M13 universal labeled primers with automatic capillary sequencing can not only reduce the cost but also provide fast and precise genotyping results (Oetting et al. 1995, Chen et al. 2006).
Here we report the identification and characterization of 1,701 unique grape EST-SSRs derived from a total of 215,609 grape ESTs. A set of SSR markers was developed from this analysis and validated by using M13 universal primers and an automatic capillary sequencing system.
Materials and methods
Plant materials
For PCR amplification, genotyping and polymorphism analysis, we selected four genotypes which are parents of two mapping populations: V. vinifera cvs. Cabernet Sauvignon and Riesling, and V. rotundifolia cvs. Noble and Summit. Genomic DNA was extracted from young leaves/shoot tips of these grape cultivars using a modified CTAB protocol (Qu et al. 1996).
Grape EST and genomic sequences retrieval from NCBI
All grape EST sequences available in the NCBI database on 10 February 2006 were retrieved. Among the total of 215,609 ESTs, 194,200 were from V. vinifera, 10,704 were V. shuttleworthii, 2,177 were V. aestivalis, 1,995 were V. riparia, and 6,533 were V. hybrids (V. rupestris A. de Serres × V. spp. b42-26).
A total of 31,910 genomics sequences were also retrieved from NCBI on 12 June 06. Among them, 30,832 were from a BAC library of V. vinifera cv. Pinot Noir, and 1,078 from V. vinifera cvs. Syrah and Maxxa.
Computer programs for mining SSRs from ESTs
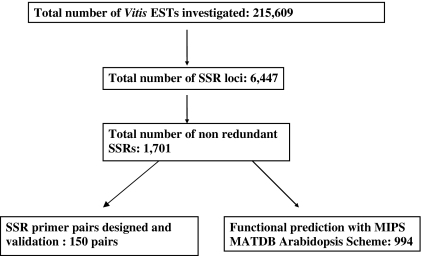
A Perl script program named Microsatellite (MISA) developed by Thiel et al. (2003, http://pgrc.ipk-gatersleben.de/misa) was used to identify EST-SSRs. The SSRs are between 2 and 6 nucleotides in size. The minimal length of SSR repeats was defined as 2 × 9 = 18 bp for dinucleotides, 3 × 6 = 18 bp for trinucleotides, 4 × 5 = 20 bp for tetranucleotides, 5 × 4 = 20 bp for pentanucleotides, and 6 × 4 = 24 bp for hexanucleotides. ESTs containing SSRs were assembled in Sequencher® version 4.2 (Genecodes, Ann Arbor, Michigan, USA) under criteria of 40% minimum overlap and 90% minimum match percentage. A flow chart for mining and developing the grape EST-SSR markers is provided in Fig. 1.
Fig. 1.
Flow chart of Vitis EST-SSR identification and validation
Functional annotation of EST-SSRs
HTGOFAT, a data mining and annotation tool kit developed in Microsoft NET 2003, was utilized to functionally annotate the assembled EST-SSRs sequences (Dowd and Zaragoza 2005). The putative functional genes were classified using the Munich Information Centre for Protein Sequences (MIPS) Arabidopsis thaliana functional catalogue (MATDB, http://mips.gsf.de).
PCR and fragment analysis
EST sequences flanking the microsatellite motifs were used to design PCR primers using the program Primer3®. A total of 150 primer pairs (Table 3) were screened for assessment of polymorphisms among the four parents using a CEQ Genetic Analyzer (Beckman Coulter, California, USA).
Table 3.
Genotyping and allelic details of the 145 EST-SSRs in four grape cultivars
| Marker ID | Accession no. | Repeat type | Forward primer | Reverse primer | Expected size (bp) | Riesling | Cabernet | Summit | Noble | R × C Allele | S × N Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FAM01 | CA817092 | (ga)9 | TTACCCGACACTGGACAC | ACTTACCACCGAGATGAGG | 310 | 967/983 | 967/983 | 989 | 989 | ab × ab | |
| FAM02 | CB973719 | (tc)10 | GCCTTGGACCGAACTATC | CTAAGAAACACCATTCATCAG | 199 | 218/236 | 208/216 | 218 | 218 | ab × cd | |
| FAM03 | CF371950 | (tca)6 | CACCGAAAGAGCACAAGA | CACCGAAAGAGCACAAGA | 232 | 239/266 | 243/251 | 262 | 252/280 | ab × cd | aa × bc |
| FAM04 | CF405747 | (ct)10 | GTGACTTACAATCCTTCCAAA | AGGGAGAGAGAGAGAGAGAGA | 180 | 198 | 196 | 189/191 | 189/191 | aa × bb | ab × ab |
| FAM05 | CB035928 | (tat)6 | ATTTCACCACCTGTCAATAAA | CCACTTCCATACACACATACA | 331 | 470 | 470/476 | 458 | 454 | aa × ab | aa × bb |
| FAM06 | CB915165 | (aag)11 | AGATAATGACCGCTATGTGAA | CAACAATCCCTACCCAAAC | 296 | 313 | 313 | 304/335 | 316/322 | ab × cd | |
| FAM07 | CV092730 | (ctt)6 | ACTTGTCTCCAAATCATCACA | CATCAGCAGGGTAGAAATAGA | 236 | 360/369 | 369 | 362 | 362/365 | ab × aa | aa × ab |
| FAM08 | CV093018 | (tcg)6 | TCATCATCCACCACAACAC | AGTCTCTTCGCATTAGGGA | 235 | 248 | 248 | 254 | 254 | ||
| FAM09 | BQ106736 | (ga)9 | TGAGGCCTACATCTTGTTCT | TCTGTGTGTCTCTCTGGTGA | 277 | 282/310 | 286/294 | 306 | 296/324 | ab × cd | aa × ab |
| FAM10 | CA814604 | (aat)13 | TGAAGCACTGATGCTTATTG | ACAATGTCACACACAAGGTG | 126 | 117 | 120/147 | 147 | 120/147 | aa × bc | aa × bc |
| FAM11 | CB348477 | (cgat)11 | TACTCTAGGTCCATTGTGGG | CGAATAACAATCTGGCTACC | 367 | 568 | 566 | 570 | 570 | aa × bb | |
| FAM12 | CB346585 | (cct)6 | GAGAGAAGAGTGGTGGTGAA | CTCCGTGTAGCACCTTAAAT | 352 | 379 | NA | 356/371 | 356/368 | ab × ac | |
| FAM13 | CB974681 | (cag)6 | CTCTTCAGGAAACACTGGAG | CCTGGAGTTCCTGGTAGATT | 195 | 214 | 214 | 185/211 | 187/211 | ab × ac | |
| FAM14 | CF208236 | (ctt)8 | AGACCACCATGGATCACTT | CTTGATATTCTTAATGGGCG | 196 | 212 | 212/215 | 198/201 | 198/201 | aa × ab | ab × ab |
| FAM15 | CF208572 | (tc)13 | TCATCCTTTCCATACAGACC | CTCCATTGGAAGACACTCAT | 113 | 123 | 125/131 | 133/139 | NA | aa × bc | |
| FAM16 | CF214062 | (tgc)6 | GTTATGAAGCTGGAGGTGAG | AAACTGGAGGACATTGCTAA | 325 | 333/342 | 342 | 330 | 330 | ab × aa | |
| FAM17 | CF214143 | (ttc)7 | TTTGCTTCTCCATTTGATCT | CTTCAATCCTTGACAGGAAG | 286 | NA | NA | 517 | 517 | ||
| FAM18 | CF211871 | (aga)7 | AGAGAGCAAAGGAACATGAA | ACAAACCCTAACCCTAGCTC | 204 | 220 | 220 | 220/225 | 229 | ab × cc | |
| FAM19 | CF211908 | (caa)6 | TTATCAGAGACGAGTCCACC | TAAGTTATGGACTTGGACGG | 302 | 314 | 314/316 | 314 | 314 | aa × ab | |
| FAM21 | CF403813 | (ag)9 | TTCCAGAGACCTGTTTGTTT | TGGAGGAGTAGGATGAGCTA | 309 | 332 | 326 | 316/320 | 320 | aa × bb | ab × aa |
| FAM22 | CF415755 | (ttc)11 | CTTGCTTCTCATACTCGTCC | GAATCACCCATGGTTTCTAA | 274 | 280/290 | 284/288 | 276 | 276 | ab × cd | |
| FAM23 | DT026156 | (aaaat)5 | AGATCCTCCGAAACAAACTT | CAAGATCAAGGAGAAACTGC | 371 | 388/392 | NA | 276 | 276 | ||
| FAM24 | CF512631 | (tatg)7 | TCCATCTTCTTCTCGTGTTT | TTTGAAGAAACAGGGACTTG | 271 | 281/288 | 281/288 | 302 | 298/300 | ab × ab | aa × bb |
| FAM25 | CV095500 | (acc)9 | CCACTATCACCACTACCACC | CTTGTTCTTGGTCTGAGAGG | 333 | 348 | 348 | 348/356 | 348 | ab × aa | |
| FAM26 | CF515612 | (gttt)5 | CTCTCCACATTACGTCTTCC | ATCAGGGCAAGTCTCTTGTA | 290 | 310/318 | 310/318 | 316 | 316 | ab × ab | |
| FAM28 | CF516198 | (tc)21 | TGGCCTTATATGCAGTTTCT | AGGCTCAATTCCAACTGTTA | 371 | 392 | 392 | 356 | 356/392 | aa × ab | |
| FAM29 | BM437681 | (att)6 | TATAGTGGTCAATGCAACCA | GGTGAGTCCACATGGTAAAT | 117 | 135 | 135 | 133 | 133 | ||
| FAM30 | CB003274 | (tc)9 | ACTCAGCCAAACCAAGTAAA | TTAGATCAAGCCCAGTCATT | 277 | 294/302 | 298 | 298 | 298 | ab × cc | |
| FAM31 | CD012233 | (ta)10 | CTTGGTGTCCTAAGGTTTCA | AGAATTGCTGTCAGCTTCAT | 231 | 240 | 248 | 241 | 241 | aa × bb | |
| FAM32 | BM437023 | (cac)7 | AAACTGGACTCCACTGTCTG | GTGGAGATGGCACTAATAGC | 211 | 227 | 221/227 | 213 | 213 | aa × ab | |
| FAM33 | CB912861 | (ct)14 | TACAGAAACCGAGTCACACA | AATTCAAACTTGCAATCCAT | 142 | 148/152 | 148/150 | 166/168 | NA | ab × ab | |
| FAM34 | CB914464 | (cca)7 | GTATGGGTTTGAGCAGAAAG | GTTGTGGTGGTCGTGAAG | 145 | 153 | 153 | 153 | 153/163 | aa × ab | |
| FAM35 | CB005343 | (cag)7 | CACTCTCCAACTCCAGATGT | ATGTTTCCCATATTCACAGC | 156 | 160/182 | 172/182 | 180 | 180 | ab × ac | |
| FAM36 | CB920562 | (tct)6 | TATCATTGTTTCCCTTCTGG | GAAGAATTCAAGAGTGTCCG | 359 | 380/386 | 396 | 381 | 381 | ab × aa | |
| FAM37 | CB915120 | (ct)16 | CTTTGATTTGGGATGTGTCT | TGGAAGCTCTTGATGAAGTT | 121 | 139 | 139 | 138 | 138 | ||
| FAM38 | CB005457 | (ct)10 | GCTTCCATACGAGAAACTCA | TAGGGTAATCCACAGTTTGC | 225 | 235/238 | 238/242 | 231/235 | 231/235 | ab × cd | ab × ab |
| FAM39 | CB915484 | (ag)53 | TACGTTCTGTTATCCAGGCT | CAATATTTCAGTAGGGCCAG | 388 | 388/414 | NA | 388 | 388 | ||
| FAM40 | CB920839 | (ctt)7 | AAGGTACTCAGCTTCCCTTC | CACCATCTCTTCTCCACAAT | 320 | 328/334 | 328/338 | 326 | 326 | ab × ac | |
| FAM41 | CB922482 | (agc)6 | CAGAAGTTGAGAAGTCAGGG | ACTTTGGCATTCCTAACTGA | 175 | 185/191 | 185/191 | 185 | 185/200 | ab × ab | aa × ab |
| FAM42 | CB916884 | (gat)7 | AAATTGATTTCATCAGTGCC | CTATCATTGTCGCTTTCCTC | 292 | 412 | 397/412 | 402 | 402 | aa × ab | |
| FAM43 | CB911681 | (cag)6 | AATAGGGAAAGAGAACAGGC | TCAATGTATGCACCCAAGTA | 340 | 489 | 489 | 491 | 491 | ||
| FAM44 | CB922984 | (aag)6 | GAGGAGGTGGAAGGAGAA | TTTGATAAGGTTGATGGTCC | 113 | 129/135 | 135 | 135/141 | 132/141 | ab × aa | ab × ac |
| FAM45 | CD801743 | (ga)10 | ATATAAGCCAAAGGTTCACA | CAAAGGATGGAAAGCATAAG | 379 | 395 | 395 | 381/398 | 383 | ab × cc | |
| FAM46 | CD801804 | (aaagg)4 | TAACCTCACATCACATCCCT | TATTAGGGTCTGCTGCAAAT | 144 | 154/160 | 154/160 | 172/182 | 172/182 | ab × ab | ab × ab |
| FAM47 | CD798867 | (aata)5 | GGGATGATGCTACCAGTCT | CAAGTATAACAGGGTCCCAA | 399 | 417 | 417 | 417 | 417 | ||
| FAM48 | CD798949 | (cac)6 | CTCAACAACGAATACCCACT | AAGCATCGTTTCAAGTGTTT | 241 | 519 | 519 | 523 | 523 | ||
| FAM49 | CD799025 | (aaaac)5 | AGCCTGAACACAATTTCTTT | CAGCAAGAACTGAAGTGTGA | 217 | 204/230 | 204/230 | 196 | 196 | ab × ab | |
| FAM50 | BQ794329 | (ag)14 | CACAAAGCATGTCCATAAAC | GGCTTATGCATTACTGGACT | 178 | 185 | 185 | 197 | 185 | aa × bb | |
| FAM51 | BQ798187 | (ct)9 | CTTGAGTCCTCACTCCAAAG | TGTGACCATGGTGGACTT | 101 | 129/130 | 129/130 | 126 | 126 | ab × ab | |
| FAM52 | BQ800590 | (ta)21 | AAATCACACCCTACCATATACT | AGGTGCACTAGCTTGAGTTC | 142 | 133 | NA | 134 | NA | ||
| FAM53 | CF603660 | (aacaag)4 | AACCTCTGCCACCACAAC | CCACACCTCATCGAATATCT | 385 | 394/400 | 394/400 | 390 | 390 | ab × ab | |
| FAM54 | CF605507 | (cca)7 | ATCTCAAGCCTCTTCTTTCC | ATCAAGAATATCATCCACGC | 319 | 326/335 | 326/335 | 326 | 326 | ab × ab | |
| FAM55 | CF605791 | (acc)6 | GCACCCACTCACAATGTT | AGGGAAGGAGTAGTAGGTGG | 271 | 284/290 | 290 | 293 | 287/293 | ab × aa | aa × ab |
| FAM56 | DT019642 | (ttc)6 | GCAGAACCCAAGTCTCATAC | CAGGTATGAGAGGACTGAGC | 356 | 368 | 368 | 368 | 368 | ||
| FAM57 | CB968692 | (ct)16 | CCATCTACCATCACCTTTGT | GGAGAAGTGGTATTTGGTGA | 152 | 155 | 173 | 158/163 | 171 | aa × bb | ab × cc |
| FAM58 | CF403802 | (at)18 | TAGACGTTTGCCCTATTTGT | CCTCTAACATGTCCCATTTC | 158 | 154 | 154 | NA | 154 | ||
| FAM59 | CB917857 | (gca)7 | GATGGTATACGACGGAGAAA | AGAGTACGACCCTTCGATCT | 175 | 186/192 | 192 | 180 | 180 | ab × aa | |
| FAM60 | CB916170 | (caa)6 | CCTCATCTGGCTTTCATAAC | CTGGACAGAACTTGGATCAT | 143 | 152/158 | 132/152 | 155 | 155 | ab × cd | |
| FAM61 | CF608950 | (ct)9 | GCTACTTCTGGGAATGTTCA | AGTCCTCATAAATTCTCAAACA | 375 | 395 | 395 | 406 | 406 | ||
| FAM62 | CB342303 | (gca)6 | CTAATCTCCAGCGAAACAAC | GTCAGCAATGTTGTCATTTG | 274 | 257/289 | NA | 253/286 | 286 | ab × aa | |
| FAM63 | CB346557 | (tca)6 | AAATGCACTCGTCTCTTCAT | CGCTTGACCTTACATACTCC | 334 | 350 | 350 | 348/360 | 348/360 | ab × ab | |
| FAM64 | CN007369 | (aag)8 | TATACTTCACCGCAATTCCT | TGATCAGCTCCTCGATATTT | 261 | 277 | 274/286 | 291 | NA | aa × bc | |
| FAM65 | DT019756 | (gca)6 | CCCTATCCAGCAGACACTAC | TTCATCTGGCTATACATCCC | 129 | 145/148 | 145 | 142 | 142 | ab × aa | |
| FAM66 | BQ794995 | (at)10 | TTTATCTCAAACCTTCACATCT | GTTGTTAGGAGTGACTTCCG | 200 | 210 | 210 | 210 | 207/212 | aa × bc | |
| FAM67 | CB977433 | (atttt)4 | GGACTTCATCCTGGAGTACA | CTTGCAGGACACCTAAATTC | 376 | 395 | 395 | 387 | 387 | ||
| FAM68 | CF607255 | (aga)6 | AAACCCTACCGAAGTCTCTC | CTTCTTCTTCGCCTCTGTTA | 307 | 321 | 321 | 321 | 321 | ||
| FAM69 | CF608166 | (gaa)6 | GATACATAAGATGCCAAGGG | CATCCTCGTCTTCAATCTTC | 335 | 352 | 352 | 352 | 352 | ||
| FAM70 | CN545596 | (ga)10 | TGGCAATAGAAGAGGAGTGT | AACAACACTTCCAGTATGGG | 281 | 389/392 | NA | 380/397 | 397/403 | ab × ac | |
| FAM71 | CN548152 | (at)9 | AGTCTCTTCAAGTGCCTCAG | CTGCATAGACTGACGAAACA | 180 | 202 | 195 | 195 | 197 | aa × bb | aa × bb |
| FAM72 | CN549034 | (ct)14 | TCAGTCCAGATTTACCTTGC | TCATGTGGTTCTGCAATAGA | 171 | 172/188 | 172/188 | 180 | 170/180 | ab × ab | aa × ab |
| FAM73 | CO818811 | (tttc)5 | GGCATATGGAAAGGGATAA | ATTTGGTCAGATGGATCAAG | 359 | 390/399 | 377/399 | 369 | 369 | ab × ac | |
| FAM74 | CO819364 | (ag)8 | GGCCTCCAGATCAACTAGTAA | GCGCCTCTGTCATAGAATAC | 289 | 307 | 307 | 234/318 | 309 | ab × cc | |
| FAM75 | DT004860 | (ctt)11 | CCTGTAAACGCTTCAAATCT | ATGGCTGAGTCATAGAGAGG | 169 | 184/187 | 187 | 172/175 | 172/175 | ab × aa | ab × ab |
| FAM76 | DT009858 | (cag)6 | ATTAACGAGGATGTGTTTGG | AAGGATCCATTTCACATACG | 394 | 409 | 409 | 427 | 433 | aa × bb | |
| FAM78 | DT010742 | (tgaag)5 | TTCATGACAATTGTGTTTGG | CGGACTCATCAGAGAAGAAG | 303 | 322 | 322 | 339 | 339 | ||
| FAM79 | DT011109 | (ag)12 | GCAGAAGCAAGAAGTGAAGT | AGATTCAAAGCCACTGAAGA | 147 | 162/172 | 162/172 | 153/157 | 153/157 | ab × ab | ab × ab |
| FAM80 | DT011686 | (ct)9 | AACTCATTCAGACAGACCCA | ATGATTTCCTCAGCTTTCAA | 316 | 424 | 424 | 428 | 428 | ||
| FAM81 | DT011972 | (gcc)8 | TTCTCTCAACATACATGGCA | GCACTGAATACACTTGGGTT | 203 | 218 | 218 | 226 | 226 | ||
| FAM82 | DT012098 | (tc)8 | AGAAGCACTCCATCTGAGAA | GAAGGCATAATCATCCTGAA | 344 | 357/361 | 361 | 348 | 348 | ab × aa | |
| FAM83 | DT012268 | (gagagg)4 | CTCCGTGAGAGAAGGTTATG | CATTCCTGACAACCATGC | 249 | 251/275 | 255 | 244/255 | 244/255 | ab × cc | ab × ab |
| FAM84 | DT014885 | (ga)18 | CCCATATTCTCAACCAAG | TCCCAATATGTAGAACCTGG | 177 | 195 | 195/210 | 184 | 184 | aa × ab | |
| FAM85 | DT015345 | (ta)10 | GAATTCAAGGAGAGGACACA | TATATATTGCGAGGCAACAA | 283 | 300 | 300/304 | 308 | 308 | aa × ab | |
| FAM86 | DT016122 | (at)11 | AGAAACCAGCTGCCAATA | GGAGGAGACCATAGACATGA | 267 | 276 | 284 | 277 | NA | aa × bb | |
| FAM87 | DT026426 | (gat)6 | TCCGAAGAAGAAGAAGAAGA | CTGGCCATACTGTTTAAAGG | 375 | 388/400 | 388 | 421/436 | 421/433 | ab × aa | ab × ac |
| FAM88 | DV220116 | (at)14 | AATGTCAAAGATTCACCAGG | CAGTTGCAGCTCATAGAACA | 237 | 230/242 | 254/256 | 223 | 226 | ab × cd | aa × bb |
| FAM89 | DV220950 | (ag)9 | CCTTGTTTGGACTTTGGAG | CTAATGGCTTCTGATATGGC | 193 | 211 | 211 | 211 | 211 | ||
| FAM90 | DV221456 | (ct)10 | GATCAAAGATTATTGCGAGG | AACAAGCAAACAGAGGGTTA | 368 | 358/387 | 387 | 392/400 | 400 | ab × cd | |
| FAM91 | DV222762 | (tct)8 | ATTATCGCAACCAAGATGTC | ATCAGCCTCTGTAACTGGAA | 159 | 176 | 173/176 | 161 | 161 | aa × bc | |
| FAM92 | DV939679 | (ta)9 | TTGTACTTTGGTGCACCTTT | ACCTTTGATAACCATTGGG | 388 | 405 | 405 | 417 | 417 | ||
| FAM93 | DV940288 | (ag)12 | ATTACATCTCATCCCGGTAA | ATGCTCTCAGAGGAGTCTCA | 242 | 258 | 244/258 | 238/243 | 238/243 | aa × ab | ab × ab |
| FAM94 | CF206303 | (ttc)6 | GGCAATGCAAGGCTATTT | ATCTTCATATGCAGCACCTT | 190 | 277 | 277 | 277 | 277 | ||
| FAM95 | CF205720 | (ga)10 | ATCATCTTCTGCCTCGAATA | TGAAACTGTGCATTCATCAT | 176 | 191 | 191 | 191 | 191 | ||
| FAM96 | CF205720 | (ga)10 | ATCATCTTCTGCCTCGAATA | TGGTGAAGGTTAGTGTGATCT | 302 | 300 | 300 | 304 | 304 | ||
| FAM97 | CF205251 | (aat)6 | AGGTCTCCAGCTTGTACTCA | ATATGTAGCCAGACGTGTCC | 301 | 326 | 318/326 | 320 | 320/324 | aa × ab | aa × ab |
| FAM98 | CF205081 | (acc)6 | AAAGGGTCTTCTGAACTTCC | TCCTAACTGAAACGAAAGGA | 382 | 395 | 395 | 395 | 395 | ||
| FAM99 | CF204388 | (aaaat)6 | ATTCCAAACAAAGCAGGTAA | GGATTGTGAATAAGCCCATA | 247 | 261/266 | 261 | NA | NA | ab × aa | |
| FAM100 | CF203674 | (tc)13 | CATTTCACGAGCTCTAAACC | GGATGAGACCAAATTCAAGA | 185 | 186 | 186 | 182/191 | 182 | ab × aa | |
| FAM101 | CF202410 | (tatg)5 | TGTGACTATGTTTCTTTGTATGT | CAAATCTGATTGTTCCAGGT | 251 | 283/286 | 283 | 292/294 | 292/298 | ab × aa | ab × ac |
| FAM102 | CF201608 | (tatg)5 | ACCCATGTTCTCTTCAACAC | CGAGAGATTGGAGAGTATCG | 147 | 145 | 145 | 125 | 125 | ||
| FAM103 | CV092439 | (gga)7 | GGAGCTTCTTGACATCATTC | CGAATTTCATCATTCTCACA | 245 | 347 | 347 | 350 | 344/350 | aa × ab | |
| FAM104 | CV092870 | (agc)7 | TGGATCCTATTCTCTCCTCA | AAATATTTCCTAATACCCGACT | 113 | 132 | 132 | 132 | 132 | ||
| FAM105 | CV092969 | (atc)8 | CCCTCTCACTCTTTGAAATC | ATGATTGGATGGTCATGTCT | 279 | 291 | 291 | 285 | 285 | ||
| FAM106 | CV093018 | (tcg)6 | TCATCAACATCATCATCCAC | GCACTCTTCTCACCTTTGTT | 202 | 216 | 216 | 222 | 219/225 | aa × bc | |
| FAM107 | CV094376 | (cag)11 | ATCAGGTCGAAATAATGGTG | GACCATTGTTAACCGTAGGA | 302 | 301 | 301 | 304 | 304 | ||
| FAM108 | CV094448 | (ctt)8 | CTCTTCTCAAACTCCAATGC | AGGAGTCACCAATGATGAAG | 151 | 156 | 156 | 156 | 156 | ||
| FAM109 | CV094765 | (at)9 | GTTAACATCTAGGCGGTTTG | GCTTGCACATGTTAACAGAA | 321 | 333 | 333 | 333 | 333 | ||
| FAM110 | CV095258 | (gct)6 | GGCTATTGATTCAGCTCCTA | TACAAGCCGTTCTATCCATT | 192 | 287/303 | 303 | 290 | 290 | ab × aa | |
| FAM112 | CV097295 | (gaa)8 | AGTTTCGTATTCGAAGTCCC | TCTTGAAATCGACTGAGGTT | 205 | 206 | 206 | 199 | 199 | ||
| FAM113 | CV098232 | (gga)6 | ACTTCCATCTACCGTCCTCT | GACTTCCTTCCAGTCCTTCT | 246 | 275 | 275/291 | 260 | 260 | aa × ab | |
| FAM115 | CV098402 | (agg)6 | AACTAACTCAGCCAAGGACA | CACAGCCTTGTACATTATGC | 318 | 345 | 345 | 333 | 330 | ||
| FAM116 | CV099053 | (ag)16 | CTTCTATTTCTGGCACCCTT | CTTCTGTGGAGGAAGAGTTG | 330 | 333 | 333/341 | 333 | 333 | aa × ab | |
| FAM117 | CV099069 | (tga)7 | TAGTGGAATACCAGAGTGGG | AGTCGTTCAGATTGATCACC | 225 | 229 | 229 | 229 | 229 | ||
| FAM118 | CV100438 | (aag)11 | AAAGCTTAAGCAACACCTTG | AACAAATCACACGTTATCCC | 301 | 300 | 305/325 | 312 | NA | aa × bc | |
| FAM119 | CA810326 | (cca)7 | GCAAATGAGTTACCAGAAAG | GTAGAAAGGAGGAAGGACCA | 311 | 428/444 | 424/444 | 420 | 420 | ab × ac | |
| FAM120 | CA810919 | (agg)7 | CGCATCAGAAGTCATCAAC | ACCCTCACTCTCACACTCAC | 324 | 430 | 426/430 | 432 | 432 | aa × ab | |
| FAM121 | CA816978 | (gat)6 | CCCTTCCATACTCCAACATAC | CCTCAATCTTAGTCGCTCC | 348 | 348 | 348 | 348/351 | 348/351 | ab × ab | |
| FAM122 | CB343602 | (ct)9 | AGAGGAAGAAGCACAAATCTC | AAAGAGTGGAGGAATCGG | 354 | 370 | 370 | 385 | 385/391 | aa × ab | |
| FAM123 | CB343426 | (ga)9 | GTAGCCAACAGAACCAGAGA | CAAACACATCCTCACCCTT | 516 | 534 | 530/534 | 492 | NA | aa × ab | |
| FAM124 | CB349648 | (tga)6 | TAAGGAAGCATTAGAAACAAG | AACCAAGAAGGAAGAAGAGAA | 312 | 308/315 | 315 | 313 | 313 | ab × aa | |
| FAM125 | CB969938 | (cag)6 | TCTTGTCATCTACCTCATCTTG | CACAGTCCCTCCTCCTCT | 215 | 238 | 238 | 238 | 238 | ||
| FAM126 | CB973643 | (ag)10 | CGACCTAAGAAACACCATTC | CCTTGGACCGAACTATCTG | 202 | 220/236 | 208/218 | 218 | 218 | ab × cd | |
| FAM127 | CB982007 | (gga)6 | ACGGAAGAAGAGAAGAAAGAG | ATCCACCGAAACAAACTTAC | 197 | 216/222 | 216 | NA | NA | ab × aa | |
| FAM128 | CF214574 | (act)6 | TACAAGAGCCAAGAGGGATT | GGATAACGAAGGAGACAGAGT | 315 | 784 | 786 | 782 | 782 | aa × bb | |
| FAM129 | CF212154 | (aaagg)4 | ACATCCCTTTGTTGTCTTCTT | ATTTGTGCTGTTGTCTGTTGT | 119 | 130 | 130 | 141 | 141 | ||
| FAM130 | CF405979 | (ctc)6 | ACAAAGCAGGTAAGTAGCAAA | AAGACGGAAGAAGAGAAGAAA | 272 | 286 | 286 | 378 | 378 | ||
| FAM131 | CF514744 | (atg)6 | TGACTGGCATACTGATTTACC | CCCAATGAACTACCTTTACCT | 259 | 272/275 | 275 | 272/275 | 272/275 | ab × aa | ab × ab |
| FAM132 | CF518394 | (tca)6 | ACCCAATGAACTACCTTTACC | AGGAACAAGACAAACAATACACT | 133 | 149 | 149 | 146/149 | 146/149 | ab × ab | |
| FAM133 | CA812979 | (ga)13 | GGGAGATTGAAAGGAAGTG | GGAGACCGACGAGGATAA | 373 | 385 | 385 | 388 | 388 | ||
| FAM134 | CA813367 | (cct)7 | GTAGCCAACAGAACCAGAGA | AAACACATCCTCACCTTCC | 514 | 533 | 529/533 | 493 | 493 | aa × ab | |
| FAM135 | CB004296 | (cat)6 | AGGGTTGTGTCTCTTCTCAA | GATACTTCATCTGTTGCTTCTG | 400 | 413 | 417/419 | 413 | 413 | aa × bc | |
| FAM136 | CB919516 | (ctt)14 | AGGGAGATGACAAAGATGAAG | CCAAACACCGTAGGAGAGA | 245 | 249/264 | NA | 261/264 | 234/261 | ab × ac | |
| FAm137 | CB920177 | (tc)9 | CAAACTGTCCAATCCTCATAGT | AGTAGAGCCAAGTGTCAAACC | 146 | 157 | 155 | 157 | 157 | ||
| FAM138 | CB005751 | (gca)9 | CGAGTGGTAGAGAGGAGAGAG | GTTGAGGGTGATGGTAAGG | 208 | 223 | 223/237 | 237 | 237 | aa × ab | |
| FAM139 | CD013208 | (atttt)4 | GGCAGAAAGGCATAAATAGTC | TGGGCACTCTCCAACCTG | 377 | 395 | 395 | 395 | 395 | ||
| FAM140 | CB916384 | (tttc)6 | AAGGGAAGAGAGGTATCGG | CCATAAACGAGAAGAAACAAA | 253 | 274 | 265/279 | 269 | 269 | aa × bc | |
| FAM141 | CD711290 | (ttc)6 | GAACCATAGACAAGACAAACAA | AAGAGAGAAGCAACGAAGAAC | 182 | 192 | 186/198 | 192 | 192 | aa × bc | |
| FAM142 | CN603827 | (aga)5 | AAGACCGAAGAAGAGAAGAAA | TAATACCGTGGAAATCACAAA | 246 | 271 | 267 | 263 | 263/267 | aa × bb | aa × ab |
| FAM143 | CV099313 | (ctc)6 | CTCTTTGACCGTTTCCAG | CCCACTACCTCTTACCTTCTT | 199 | 217 | 217 | 217 | 217 | ||
| FAM144 | CV092545 | (acc)9 | CACCACTATCACCACTACCAC | AGGAGGCGAATGAAGGTC | 193 | 211 | NA | 211/220 | 211/220 | ab × ab | |
| FAM145 | CV093192 | (caa)7 | TCCAACAACAACAACTACTAC | AGGAATCTCGTGTCGCTC | 215 | 238/247 | 238/241 | 230/236 | 230/236 | ab × ac | ab × ab |
| FAM146 | CB920177 | (tc)9 | CAAACTGTCCAATCCTCATAGT | AGTAGAGCCAAGTGTCAAACC | 146 | 157/160 | 155 | 157 | 157 | ab × aa | |
| FAM147 | CV092326 | (gat)6 | TACAACCACATAGAGGCACTT | TCTTCTTCAGTTTCTTCACCA | 185 | 606/612 | 606 | 602 | 606 | ab × aa | aa × bb |
| FAM148 | CV099379 | (caa)7 | TCCTCCTTGTTATCCTCTTCT | TAGTAGTTGTTTCGGTTGGAC | 400 | 413 | 413 | 410 | 410 | ||
| FAM149 | CV094327 | (ctt)9 | TAGACCTCCACCACTCTCTC | GTCATCAGCGAAAGCATC | 343 | 362/365 | 359/365 | 335/353 | 332/335 | ab × ac | ab × ac |
| FAM150 | CF519163 | (gct)6 | ATCTGACAAAGGAAAGGAGAA | GTAACATACCGAGGAAGGCA | 235 | 244 | NA | 237 | 237/244 | aa × ab |
NA not available
To save cost, a 20-bp long universal M13 forward primer sequence GTT GTAAAA CGA CGG CCA GT (Oetting et al. 1995) was added as a common tail to the 5′ end of all 180 SSR forward primers. All SSR primers, including regular and M13-tailed forward primers, were synthesized by Operon Technologies (Huntsville, Alabama, USA). The universal M13 primers were labeled by Sigma-Genosys (USA) and used for CEQ Genetic Analyzer Fragment Analysis.
PCR reactions were performed in a 20-μl reaction mix including 30 ng of genomic DNA, 10 × PCR buffer (Promega), 2 μl of 2 mM dNTP (Promega), 1.0 U Taq DNA polymerase, 2.8 μl of 25 mM MgCl2 (Promega), and 0.3 μM primers. The PCR reactions were carried out in a PTC-200 thermal cycler (MJ Research) with the following thermal profile: 3 min at 94°C followed by 30 cycles of 1 min denaturation at 94°C, 1 min annealing at 48 to 58°C (based on the T m of the different primer sets), and 2 min extension at 72°C, followed by a final step of 6 min extension at 72°C. The same conditions were also used for labeling the primers.
For fragment analysis using the CEQ Genetic Analyzer, 0.25 μl of each M13 labeled PCR product was mixed with 40 μl Sample Loading Solution (Beckman Coulter 608087) with 0.2 μl 400-bp DNA size standard (Beckman Coulter 608098) and overlaid with one drop of light mineral oil, then loaded into the 96-well sample microtiter plates (Beckman Coulter 609801). CEQ Sequencing Separation Buffer (Beckman Coulter 608012) were also loaded into the 96-well separation plate (Beckman Coulter 609844). Dye-labeled amplicons were automatically sized by running on “Frag-3” separation and the GenomeLab software (Beckman Coulter) and then visually examined.
Results and discussion
Identification and characterization of grape EST-SSRs
A total of 6,447 out of 215,609 (3%) grape ESTs retrieved from NCBI on 1 February 2006 contained SSRs (Table 1). With some of them having multiple SSR sites, a total of 6,815 SSR motifs were identified among these 6,447 EST sequences. The percentage of EST-SSRs varied slightly among different Vitis species, ranging from 2.98% for V. vinifera (5,782 of 194,200), 3.50% for V. aestivalis (74 of 2,116), 3.55% for V. shuttleworthii (389 of 10,933), to 5.43% for V. riparia (59 out of 1,087). The EST-SSRs accounted for 2.71% for a Vitis hybrid of (V. rupestris A. de Serres × V. spp. b42-26) (177 of 6,533; Electronic Supplementary Material 1).
Table 1.
Characterization of grape redundant and non-redundant EST and genomic SSRs
| Redundant SSR-ESTs | Non-redundant SSR-ESTs | Genomic SSR sequences | |
|---|---|---|---|
| Total % | 6,447 | 1,701 | 1,346 |
| Di- | 1,835(28.5%) | 664(39.0%) | 699(51.9%) |
| Tri- | 3,235(50.2%) | 710(41.7%) | 341(25.3%) |
| Tetra- | 391(6.1%) | 125(7.3%) | 154(25.2%) |
| Penta- | 618(9.6%) | 134(7.9%) | 99 (7.4%) |
| Hexa- | 736(11.4%) | 179(10.5%) | 53 (3.9%) |
| Abundant type | |||
| Di- | AG/CT(18.9%) | AG/CT(26.9%) | AT/AT (33.0%) |
| Tri- | AGG/CCT(14.8%) | AAG/CTT (11.3%) | AAT/ATT (18.6%) |
| Tetra- | AAGG/CCTT(1,7%) | AAAT/ATTT(2.4%) | AAAT/ATTT(7.0%) |
| Penta- | AAAAT/ATTTT(2.8%) | AAAAT/ATTTT(2.1%) | AAAAT/ATTTT(3.8%) |
| Hexa- | AGGGTC/AGTCCC(2.6%) | ACCCTG/ACTGGG(1.1%) | AAAAAT/TAAAAA(1.7%) |
Among the redundant EST-derived SSR repeats, tri-nucleotide, which accounted for 50.2% of total SSRs, was the most abundant repeat unit followed by di (28.5%), hexa (11.4%), penta (9.6%), and tetranucleotide (6.1%; Table 1). These findings are in agreement with previous observations on abundance of SSR repeat units in barley, maize, rice, sorghum, and wheat (Kantety et al. 2002). The dominance of trinucleotide SSRs was viewed as the result of a frame shift in size of one amino acid read, or three nucleotides, a selection against possible frame shift mutations (Metzgar et al. 2000; Toth et al. 2000; Wren et al. 2000; Cordeiro et al. 2001). For the same reason, a higher percentage was also observed in hexanucleotide SSRs than tetra- and penta-repeats. In both non-redundant and redundant EST-SSRs (Table 1), di- and tri-repeats were accounted for about 80% of the total EST-SSRs for each group (redundant: di-28.5%, tri-50.2%; non-redundant: di-39%, tri-41.7%). Interestingly, the proportion of tri repeats dropped from 50.2% in redundant to 41.7% in non-redundant ESTs while di repeats increased from 28.5 to 39.0% after eliminating the redundancy by contig assembling (Table 1). The result was interpreted to suggest that tri-repeat SSRs were mainly found in coding regions (Yu et al. 2004) and many of these redundant EST-SSRs were eliminated because these sequences contain tri-repeats representing putative amino acid runs (Li et al. 2004) as overexpressed ESTs representing the same set of genes. Another explanation is the effect of gene duplication and paralogy. Depending on the parameters used for clustering, untranslated regions of paralogous genes, which are more divergent and contain all types of SSR, might have remained separated, while ESTs covering exons of paralogous genes, which are more conserved and highly enriched in tri-nucleotide SSR, might have collapsed more frequently into a “single” redundant EST.
Comparison between genomic and EST derived SSRs
Unlike the tri-nucleotide repeats as the dominant type in SSR-ESTs, the number of genome sequence-derived SSRs were dominated by di-nucleotide repeats that accounted for 51.9% of total genomic SSRs, followed by tri- (25.3%), tetra- (25.2%), penta- (7.4%), and hexa-SSRs (3.9%; Table 1). Similar patterns for EST-SSRs having a higher proportion in tri-repeats than genomic SSRs were reported in the literacture (Cardle et al. 2000). Among the top 20 SSRs in ranking, the most abundant di-nucleotide repeat in non-redundant ESTs was AG/CT which accounted for 17.9% of total EST-SSRs, followed by AT/AT (8.4%; Table 2), while the most abundant di-nucleotide repeat in genomic sequences was AT/AT which accounted for 33.0%, followed by AG/CT (15.5%) and AC/GT (3.5%). The most common EST-derived tri-nucleotide repeat was AAG/CTT (14.0%), while AAT/ATT (18.6%) was the most abundant tri-nucleotide SSRs derived from genomic sequences. Among grape genomic sequences, around 67.1% of the SSRs belonged to three types of repeats: AT/AT (33.0%), AAT/ATT (18.6%), and AG/CT (15.5%; Tables 2 and 3).
Table 2.
Top 20 SSR motifs in grape ESTs and genomic sequences
| EST-SSR repeats | Total | % | Genomic SSR repeats | Total | % |
|---|---|---|---|---|---|
| AG/CT | 1,217 | 17.9 | AT/AT | 444 | 33.0 |
| AGG/CCT | 957 | 14.0 | AAT/ATT | 251 | 18.6 |
| AT/AT | 575 | 8.4 | AG/CT | 208 | 15.5 |
| AAG/CTT | 547 | 8.0 | AAAT/ATTT | 94 | 7.0 |
| AAT/ATT | 403 | 5.9 | AAAAT/ATTTT | 51 | 3.8 |
| ACC/GGT | 326 | 4.8 | AAG/CTT | 48 | 3.6 |
| AGC/CGT | 237 | 3.5 | AC/GT | 47 | 3.5 |
| ACG/CTG | 201 | 2.9 | AAAAAT/ATTTTT | 21 | 1.6 |
| AGT/ATC | 201 | 2.9 | AAAAG/CTTTT | 17 | 1.3 |
| AAAAT/ATTTT | 195 | 2.9 | AAAAAG/CTTTTT | 15 | 1.1 |
| ACT/ATG | 168 | 2.5 | AATT/AATT | 14 | 1.0 |
| AAC/GTT | 163 | 2.4 | AAAG/CTTT | 13 | 1.0 |
| AGGGTC/AGTCCC | 144 | 2.1 | AGT/ATC | 12 | 0.9 |
| AAAAG/CTTTT | 98 | 1.4 | ACAT/ATGT | 10 | 0.7 |
| AAAAC/GTTTT | 97 | 1.4 | ACT/ATG | 9 | 0.7 |
| AAAT/ATTT | 89 | 1.3 | AGAT/ATCT | 8 | 0.6 |
| AAGG/CCTT | 88 | 1.3 | AAC/GTT | 7 | 0.5 |
| AAAGG/CCTTT | 88 | 1.3 | AAAAC/GTTTT | 6 | 0.4 |
| ACCCTG/ACTGGG | 81 | 1.2 | AAATT/AATTT | 6 | 0.4 |
| AAAAAG/CTTTTT | 78 | 1.1 | AGG/CCT | 5 | 0.4 |
| Other motifs | 862 | 12.6 | Other motifs | 60 | 4.5 |
| Total | 6,815 | 100.0 | 1,346 | 100.0 |
Functional analysis of EST-SSR sequences
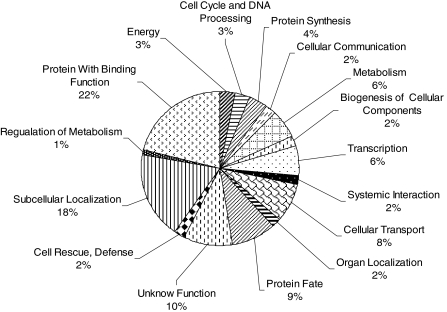
The 1,701 assembled non-redundant EST-SSRs were functionally annotated using the HTGOFAT program (Dowd and Zaragoza 2005). Fifty-eight percent (994 out of 1,701) of the EST-SSRs were annotated and grouped by the Biological Process Classification using the MIPS MATDB Arabidopsis Scheme. The most abundant EST-SSRs belonged to the categories of protein-binding (22%) and subcellular localization (18%
;
Fig. 2), which demonstrated a similar pattern to wheat, rice, maize and barley (Tang et al. 2006). The 150 validated markers were further functionally annotated (Electronic Supplementary Material 2) and estimation of their genomic/chromosome locations by comparison to grapevine genome assembly (Jaillon et al. 2007; Velasco et al. 2007) is given in Electronic Supplementary Material 3.
Fig. 2.
Functional prediction of 994 grape SSR-EST based on the MIP MATDB classification scheme
SSR marker development and validation
The Beckman CEQ8800 Genetic Analyzer was used for the SSR validation and analysis. This system can detect DNA fragment length polymorphism in a “single base pair”. A set of 150 primer pairs was initially screened for SSR marker development and validation. Parents of two mapping populations, V. vinifera Riesling × Cabernet Sauvignon (Riaz et al. 2004) and V. rotundifolia Summit × Noble (Ren et al. 2000) were used for the screening. Results showed that 145 out of 150 primers had well-amplified fragments among the four cultivars (Table 3). Some of the fragment sizes exceeded expected sizes possibly due to their having introns within the flanking regions or the length of the repeat being shorter than the source species, and less prone to polymerase slippage. Polymorphisms were found in 66 primer pairs between Riesling and Cabernet Sauvignon, and 40 between Summit and Noble. Only 16 of the polymorphic primers shared the same polymorphic lengths between these two parent pairs, reflecting the fact that the alleles between V. vinifera and V. rotundifolia grape are distinct (Riaz et al. 2008).
The homo and heterozygosity of these 145 loci were screened in the four testing cultivars; 92 of 144 were identified as homozygous and 52 were heterozygous loci in Riesling, while 86 of 136 were homozygous and 49 were heterozygous loci in Cabernet Sauvignon. Among the 92 Riesling and 86 Cabernet Sauvignon homozygous loci, 68 are common in both parents (Table 4). As for the muscadine grapes Noble and Summit, Noble showed 97 homozygous and 39 heterozygous loci and the respective number for Summit was 108 and 34 (Table 4). Some of those microsatellite loci were selected for having long stretches in V. vinifera grapes, and thus may show more polymorphisms than in muscadines. From this screening of 145 loci, muscadine grapes demonstrated a higher homozygosity compared to V. vinifera grapes.
Table 4.
Level of heterozygosity in two Vitis vinifera and two Muscadinia rotundifolia genotypes for a set of 145 SSR markers
| Genotype | Riesling | Cabernet Sauvignon | Summit | Noble | Riesling vs Cabernet Sauvignon | Summit vs Noble |
|---|---|---|---|---|---|---|
| Homozygous | 92 | 86 | 108 | 96 | 66(9)a | 86(6)b |
| Heterozygous | 52 | 49 | 34 | 40 | ||
| Failed | 1 | 10 | 3 | 9 |
a9 primers are homozygous in both Riesling and Cabernet Sauvignon, but are polymorphic between two cultivars
b6 primers are homozygous in both Submit and Noble, but are polymorphic between two cultivars
Pairwise comparison between Riesling and Cabernet Sauvignon showed that 72 loci were monomorphic with either one allele (60) or two (12). Seventy loci were heterozygous in at least one cultivar with either two (53), three (10), or four alleles (7; Table 5). The muscadine Noble and Summit showed 50 heterozygous loci in at least one parent with either two (31), three (17), or four alleles (2). Ninety homozygous loci were found in both cultivars with either one (84) or two alleles (6; Table 5). According to the results from these 145 EST-SSR loci, the percentage of polymorphisms is about 49% between Riesling and Cabernet Sauvignon, and 29% between Summit and Noble. However, those polymorphic SSRs that are homozygous (e.g. aa × bb) in both parents cannot be mapped in F1 populations although they are useful for mapping in F2 or backcross populations (Chen et al. 2006). The heterozygous monomorphic SSRs (e.g. ab × ab) can be used for mapping in F1 populations (Table 5). As a result, the estimated number of SSRs that can be mapped in the F1 populations between Riesling and Cabernet Sauvignon is about 46%, which means that out of the total 1,037 SSRs with successful primers designed, around 477 EST-SSR putative markers can be mapped in the F1 population, and about 33% of the total SSRs (342 EST-SSR loci) can be mapped in the F1 of Summit × Noble.
Table 5.
Distribution of the segregation types expected for the two mapping populations
| Alleles | R × Ca | Number | S × Nb | Number | Mappable in F1 |
|---|---|---|---|---|---|
| 1 | aa × aa | 57 | aa × aa | 80 | No |
| 2 | aa × bb | 9 | aa × bb | 6 | No |
| 2 | aa × ab | 15 | aa × ab | 13 | Yes |
| 2 | ab × aa | 18 | ab × aa | 4 | Yes |
| 2 | ab × ab | 12 | ab × ab | 13 | Yes |
| 3 | aa × bc | 8 | aa × bc | 4 | Yes |
| 3 | ab × cc | 2 | ab × cc | 4 | Yes |
| 3 | ab × ac | 6 | ab × ac | 8 | Yes |
| 4 | ab × cd | 9 | ab × cd | 1 | Yes |
| Total mappable | 70 | 47 |
aR × C: Riesling × Cabernet Sauvignon
bS × N: Summit × Noble
EST-SSR marker transferability was evaluated and the current research showed a high transferability across species. All but two of the 145 EST-SSR markers in Vitis vinifera appeared in the muscadine as well. This result indicated that development of EST-SSR markers is a cost-effective method for obtaining additional markers for grape genome typing and gene mapping.
EST-SSRs provided sources of additional markers for marker development. Compared to genomic-derived markers, EST-SSRs are highly transferable for detecting the gene-rich areas within the genome. We can utilize these markers to evaluate marker transferability across taxa, and conduct analysis in comparative mapping and gene functional diversity analysis, in addition to genotyping. The functional EST-SSR markers should be even more useful for developing a linkage map or tagging a viticulturally important trait. In addition, the polymorphic EST-SSR markers are much needed for genotyping, cultivar identification and development of a linkage map in muscadine grapes since they are genetically much less diversified than Vitis species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by USDA Capacity Building Grant (#0205031) and FAMU-ARS Science Center for Excellence. The authors thank two anonymous reviewers for invaluable comments on the manuscript. Special thanks go to Ms. Elisa Scott for helping us to prepare the DNA and PCR samples.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Hong Huang, Email: huanghon2003@gmail.com.
Jiang Lu, Email: jiang.lu@famu.edu.
Zhongbo Ren, Email: zhongbo.ren@famu.edu.
Wayne Hunter, Email: whunter@ushrl.ars.usda.gov.
Scot E. Dowd, Email: sdowd@pathogenresearch.org
Phat Dang, Email: pdang@nprl.usda.gov.
References
- Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P. Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet. 2004;109(5):1017–1027. doi: 10.1007/s00122-004-1704-y. [DOI] [PubMed] [Google Scholar]
- Akkak A, Lembo A, Torello Marinoni D, Gribaudo I, Botta R. Analysis and transferability of EST-SSR markers in grape. Acta Hort (ISHS) 2006;827:59–62. [Google Scholar]
- Arroyo García R, Lefort F, de Andrés M, Ibañez J, Borrego J, Jouve N, Cabello F, Martínez Zapater J. Chloroplast microsatellite polymorphisms in Vitis species. Genome. 2002;45(6):1142–1149. doi: 10.1139/g02-087. [DOI] [PubMed] [Google Scholar]
- Bocharova B, Kovaliova I, Mazurenko L. Identification of grapevine clone genotypes by use of microsatellite markers. Cytol Genet. 2009;43(6):371–378. doi: 10.3103/S0095452709060024. [DOI] [PubMed] [Google Scholar]
- Bowers J, Dangl G, Vignani R, Meredith C (1996) Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 39(4):628–633 [DOI] [PubMed]
- Bowers J, Boursiquot J, This P, Chu K, Johansson H, Meredith C. Historical genetics: the parentage of Chardonnay, Gamay and other wine grapes of northeastern France. Science. 1999;285(3433):1562–1565. doi: 10.1126/science.285.5433.1562. [DOI] [PubMed] [Google Scholar]
- Bowers J, Dangl G, Meredith C. Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic. 1999;50(3):243–246. [Google Scholar]
- Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabane K, Ablett G, Cordeiro G, Valkoun J, Henry R. EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genet Res Crop Evol. 2005;52:903–909. doi: 10.1007/s10722-003-6112-7. [DOI] [Google Scholar]
- Chani E, Ashkeni V, Hillel J, Veilleux R. Microsatellite marker analysis of an anther-derived potato family: skewed segregation and gene-centromere mapping. Genome. 2002;45(2):236–242. doi: 10.1139/g01-140. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhou P, Choi Y, Huang S, Gmitter F. Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet. 2006;112(7):1248–1257. doi: 10.1007/s00122-006-0226-1. [DOI] [PubMed] [Google Scholar]
- Cho Y, Ishii T, Temnykh S, Chen X, Lipovich L, Park W, Ayres N, Cartinhour S, McCouch S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) Theor Appl Genet 100(5):713–722
- Cipriani G, Marrazzo M, Di Gaspero G, Pfeiffer A, Morgante M, Testolin R (2008) A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol 8:127 [DOI] [PMC free article] [PubMed]
- Cordeiro M, Casu R, McIntyre C, Manners J, Henry R. Microsatellite markers from sugarcane (Saccharum spp) ESTs across transferable to erianthus and sorghum. Plant Sci. 2001;160(6):1115–1123. doi: 10.1016/S0168-9452(01)00365-X. [DOI] [PubMed] [Google Scholar]
- Costantini L, Grando M, Feingold S, Ulanovsky S, Mejía N, Hinrichsen P, Doligez A, This P, Cabezas J, Martínez-Zapater J. Generation of a common set of mapping markers to assist table grape breeding. Am J Enol Vitic. 2007;58:102–111. [Google Scholar]
- Dalbò M, Ye G, Weeden N, Steinkellner H, Sefc K, Reisch B. Gene controlling sex in grapevines placed on a molecular marker-based genetic map. Genome. 2000;43:333–340. doi: 10.1139/g99-136. [DOI] [PubMed] [Google Scholar]
- De Keyser E, De Riek J, Van Bockstaele E. Discovery of species-wide EST-derived markers in Rhododendron by intron-flanking primer design. Mol Breed. 2009;23(1):171–178. doi: 10.1007/s11032-008-9212-4. [DOI] [Google Scholar]
- De Mattia F, Imazio S, Grassi F, Lovicu G, Tardaguila J, Maitti C, Scienza A, Labra M. Genetic characterization of Sardinia grapevine cultivars by SSR markers analysis. Int J Vine Wine Sci. 2007;41:175–184. [Google Scholar]
- Decroocq V, Favé M, Hagen L, Bordenave L, Decroocq S. Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet. 2003;106:912–922. doi: 10.1007/s00122-002-1158-z. [DOI] [PubMed] [Google Scholar]
- Depeiges A, Goubely C, Lenoir A, Cocherel S, Picard G, Raynal M, Grellet F, Delseny M. Identification of the most represented repeated motifs in Arabidopsis thaliana microsatellite loci. Theor Appl Genet. 1995;91(1):160–168. doi: 10.1007/BF00220873. [DOI] [PubMed] [Google Scholar]
- Di Gaspero G, Peterlunger E, Testolin R, Edwards K, Cipriani G. Conservation of microsatellite loci within the genus Vitis. Theor Appl Genet. 2000;101(1):301–308. doi: 10.1007/s001220051483. [DOI] [Google Scholar]
- Di Gaspero G, Cipriani G, Marrazzo M, Andreetta D, Prado Castro M, Peterlunger E, Testolin R (2005) Isolation of (AC)n-microsatellites in Vitis vinifera L. and analysis of genetic background in grapevines under marker assisted selection. Mol Breed 15(1):11–20
- Di Gaspero G, Cipriani G, Adam-Blondon A-F, Testolin R. Linkage maps of grapevine displaying the chromosomal locations of 420 microsatellite markers and 82 markers for R-gene candidates. Theor Appl Genet. 2007;114(7):1249–1263. doi: 10.1007/s00122-007-0516-2. [DOI] [PubMed] [Google Scholar]
- Doligez A, Bouquet A, Danglot Y, Lahogue F, Riaz S, Meredith C, Edwards K, This P. Genetic mapping of grapevine (Vitisvinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor Appl Genet. 2002;105:780–795. doi: 10.1007/s00122-002-0951-z. [DOI] [PubMed] [Google Scholar]
- Doucleff M, Jin Y, Gao F, Riaz S, Krivanek A. A genetic linkage map of grape, utilizing Vitis rupestris and Vitis arizonica. Theor Appl Genet. 2004;109:1178–1187. doi: 10.1007/s00122-004-1728-3. [DOI] [PubMed] [Google Scholar]
- Dowd S, Zaragoza J. High throughput gene ontology functional annotation toolkit (Hit-Go-Fat) utilized for animal and plant. San Diego, CA: Plant and Animal Genome XIII Conference; 2005. [Google Scholar]
- Eujayl I, Sorrells M, Baum M, Wolters P, Powell W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet. 2002;104(2):399–407. doi: 10.1007/s001220100738. [DOI] [PubMed] [Google Scholar]
- Feng S, Li W, Huang H, Wang J, Wu Y. Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis) Mol Breed. 2009;23(1):85–97. doi: 10.1007/s11032-008-9216-0. [DOI] [Google Scholar]
- Fernández M, Núñez Y, Ponz P, Hernáiz S, Gallego F, Ibáñez J. Characterization of sequence polymorphisms from microsatellite flanking regions in Vitis spp. Mol Breed. 2008;22(3):455–465. doi: 10.1007/s11032-008-9189-z. [DOI] [Google Scholar]
- Gao L, Tang J, Li H, Jia J. Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed. 2003;113(3):163–185. [Google Scholar]
- Grando MS, Bellin D, Edwards KJ, Pozzi C, Stefanini M, Velasco R. Molecular linkage maps of Vitis vinifera L. and Vitis riparia Mchx. Theor Appl Genet. 2003;106:1213–1224. doi: 10.1007/s00122-002-1170-3. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon A-F, Weissenbach J, Quétier F, Wincker P. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Kantety R, La Rota M, Matthews D, Sorrells M. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol. 2002;48(5):501–510. doi: 10.1023/A:1014875206165. [DOI] [PubMed] [Google Scholar]
- Lamoureux D, Bernole A, Le Clainche I, Tual S, Thareau V, Paillard S, Legeai F, Dossat C, Wincker P, Oswald M, Merdinoglu D, Vignault C, Delrot S, Caboche M, Chalhoub B, Adam-Blondon AF. Anchoring of a large set of markers onto a BAC library for the development of a draft physical map of the grapevine genome. Theor Appl Genet. 2006;113:344–356. doi: 10.1007/s00122-006-0301-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Korol A, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Bio Evol. 2004;21(6):991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Lima L, Gramacho K, Gesteira A, Lopes U, Gaiotto F, Zaidan H, Pires J, Cascardo J, Micheli F (2008) Characterization of microsatellites from cacao—Moniliophthora perniciosa interaction expressed sequence tags. Mol Breed 22(2):315–318
- Lin H, Walker A. Identifying grape rootstocks with simple sequence repeat (SSR) DNA markers. Am J Enol Vitic. 1998;49(4):403–407. [Google Scholar]
- Lin H, Civerolo E, Hu R, Barros S, Francis M, Walker M. Multilocus simple sequence repeat markers for differentiating strains and evaluating genetic diversity of Xylella fastidiosa. Appl Environ Microbiol. 2005;71(8):4888–4892. doi: 10.1128/AEM.71.8.4888-4892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe KM, Walker MA. Genetic linkage map of the interspecic grape rootstock cross Ramsey (Vitis champinii) × Riparia Gloire (Vitis riparia) Theor Appl Genet. 2006;112:1582–1592. doi: 10.1007/s00122-006-0264-8. [DOI] [PubMed] [Google Scholar]
- Merdinoglu D, Butterlin G, Bevilacqua L, Chiquet V, Adam-Blondon AF, Decroocq S (2005) Development and characterization of a large set of microsatellite markers in grapevine (Vitisvinifera L.) suitable for multiplex. Mol Breed 15(4):349–366
- Metzgar D, Bytof J, Wills C. Selection against frame shift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10(1):72–80. [PMC free article] [PubMed] [Google Scholar]
- Oetting W, Lee H, Flanders D, Wiesner G, Sellers T, King R. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics. 1995;30(3):450–458. doi: 10.1006/geno.1995.1264. [DOI] [PubMed] [Google Scholar]
- Qu X, Lu J, Lamikanra O. Genetic diversity in Muscadine and American bunch grapes based on randomly amplified polymorphic DNA (RAPD) analysis. J Am Soc Hort Sci. 1996;121(6):1020–1023. [Google Scholar]
- Ren Z, Lamikanra O, Lu J. Identification of a RAPD marker closely linked to the fruit color in muscadine grape (Vitis rotundifolial) Acta Hort. 2000;528:263–266. [Google Scholar]
- Riaz S, Dangl G, Edwards K, Meredith C. A microsatellite marker based framework linkage map of Vitis vinifera L. Theor Appl Genet. 2004;108(5):864–872. doi: 10.1007/s00122-003-1488-5. [DOI] [PubMed] [Google Scholar]
- Riaz S, Krivanek AF, Xu K, Walker MA. ReWned mapping of the Pierce’s disease resistance locus, PdR1, and sex on an extended genetic linkage map of Vitis rupestris x V. arizonica. Theor Appl Genet. 2006;113:1317–1329. doi: 10.1007/s00122-006-0385-0. [DOI] [PubMed] [Google Scholar]
- Riaz S, Tenscher A, Smith B, Ng D, Walker M (2008) Use of SSR markers to assess identity, pedigree, and diversity of cultivated Muscadinia rotundifolia. J Am Soc Hort Sci 133:559–568
- Riaz S, Tenscher A, Graziani R, Krivanek A, Ramming D, Walker A. Using marker-assisted selection to breed Pierce’s disease-resistant grapes. Am J Enol Vitic. 2009;60:199–207. [Google Scholar]
- Roy J, Balyan H, Prasad M, Gupta P. Use of SAMPL for a study of DNA polymorphism, genetic diversity and possible gene tagging in bread wheat. Theor Appl Genet. 2002;104(2):465–472. doi: 10.1007/s001220100711. [DOI] [PubMed] [Google Scholar]
- Salmaso M, Malacarne G, Troggio M, Faes G, Stefanini M, Grando M, Velasco R. A grapevine (Vitis vinifera L.) genetic map integrating the position of 139 expressed genes. Theor Appl Genet. 2008;116:1129–1143. doi: 10.1007/s00122-008-0741-3. [DOI] [PubMed] [Google Scholar]
- Scott K, Eggler P, Seaton G, Rossetto M, Ablet E, Lee L, Henry R. Analysis of SSRs derived from grape ESTs. Theor Appl Genet. 2000;100(5):723–726. doi: 10.1007/s001220051344. [DOI] [Google Scholar]
- Sefc K, Steinkellner H, Glössl J, Kampfer S, Regner F. Reconstruction of a grapevine pedigree by microsatellite analysis. Theor Appl Genet. 1998;97(2):227–231. doi: 10.1007/s001220050889. [DOI] [Google Scholar]
- Tang J, Gao L, Cao Y, Jia J (2006) Homologous analysis of SSR-ESTs and transferability of wheat SSR-EST markers across barley, rice and maize. Euphytica 151:1–87
- Tekeoglu M, Rajesh N, Muehlbauer J. Integration of sequence tagged microsatellite sites to the chickpea genetic map. Theor Appl Genet. 2002;105(6):847–854. doi: 10.1007/s00122-002-0993-2. [DOI] [PubMed] [Google Scholar]
- Tessier C, David J, This P, Boursiquot J, Charrier A. Optimization of the choice of mocular markers for varietal identification in Vitis vinifera L. Theor Appl Genet. 1999;98(1):171–177. doi: 10.1007/s001220051054. [DOI] [Google Scholar]
- Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106(3):411–422 [DOI] [PubMed]
- Thomas M, Scott N. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs) Theor Appl Genet. 1993;86(8):985–990. doi: 10.1007/BF00211051. [DOI] [PubMed] [Google Scholar]
- Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10(7):967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7(2A):537–546 [PubMed]
- Varshney R, Granner A, Sorrells M. Genic microsatellite markers in plants: features and applications. Trends Biotech. 2006;23(1):48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright D, Cestaro A, Pruss D, Pindo M, FitzGerald L, Vezzulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma T, Facci M, Mitchell J, Perazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Demattè L, Mraz A, Battilana J, Stormo K, Costa F, Tao Q, Si-Ammour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett J, Sterck L, Vandepoele K, Grando M, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaramella V, Bhatnagar S, Fontana P, Gutin A, Van de Peer Y, Salamini F, Viola R. High quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli S, Troggio M, Coppola G, Jermakow A, Cartwright D, Zharkikh A, Stefanini M, Grando M, Viola R, Adam-Blondon AF, Thomas M, This P, Velasco R. A reference integrated map for cultivated grapevine (Vitis vinifera L.) from three crosses, based on 283 SSR and 501 SNP-based markers. Theor Appl Genet. 2008;117:499–511. doi: 10.1007/s00122-008-0794-3. [DOI] [PubMed] [Google Scholar]
- Wren J, Forgacs E, Fondon J, Pertsemlidis A, Cheng S, Gallardo T, Williams R, Shohet R, Minna J, Garner H. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am J Hum Genet. 2000;67(2):345–356. doi: 10.1086/303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Dake T, Singh S, Benscher D, Li W, Gill B, Sorrells M. Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome. 2004;47(5):805–818. doi: 10.1139/g04-057. [DOI] [PubMed] [Google Scholar]
- Zhang J, Guo W, Zhang T (2002) Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor Appl Genet 105(8):1166–1174 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.