SUMMARY
How polycomb group proteins repress gene expression in vivo is not known. While histone-modifying activities of the polycomb repressive complexes (PRCs) have been studied extensively, in vitro data have suggested a direct activity of the PRC1 complex in compacting chromatin. Here, we investigate higher-order chromatin compaction of polycomb targets in vivo. We show that PRCs are required to maintain a compact chromatin state at Hox loci in embryonic stem cells (ESCs). There is specific decompaction in the absence of PRC2 or PRC1. This is due to a PRC1-like complex, since decompaction occurs in Ring1B null cells that still have PRC2-mediated H3K27 methylation. Moreover, we show that the ability of Ring1B to restore a compact chromatin state and to repress Hox gene expression is not dependent on its histone ubiquitination activity. We suggest that Ring1B-mediated chromatin compaction acts to directly limit transcription in vivo.
INTRODUCTION
Transcriptionally inactive chromatin is generally considered to have a compact structure, while active chromatin is open and decondensed. The inference is that compact chromatin structure inhibits gene expression. However, while histone modifications and the proteins that deposit, remove, or bind them are increasingly well understood, mechanisms that control higher-order chromatin structure are poorly characterized.
Proteins implicated in driving higher-order chromatin compaction include variant and linker histones, HP1, and polycomb group (PcG) proteins. Linker histone H1 is thought to be important in forming 30 nm chromatin fibers (Allan et al., 1981; Bates et al., 1981), and its downregulation in Drosophila results in misaligned polytene chromatids and dispersed heterochromatin (Lu et al., 2009). In mammals, reducing the stoichiometry of linker histone to nucleosomes by a knockout of 3 of the 6 somatic H1 genes (herein called ΔH1) results in an altered nucleosome repeat length and a widespread decondensation of chromatin fibers, but misexpression of only a few genes (Fan et al., 2005). H1:nucleosome stoichiometry also varies between cell types, with the ratio in pluripotent cells, such as undifferentiated embryonic stem cells (ESCs), being lower than that in differentiated cells.
PcG proteins are key regulators of developmentally regulated loci in flies and mammals and are found in two broad classes of complex. The mammalian polycomb repressive complex 2 (PRC2) complex (Ezh/Suz12/Eed) trimethylates histone H3 at lysine 27 (H3K27me3) (Cao et al., 2002) through the activity of the histone methyltransferases (HMTases) Ezh2 and Ezh1 (Shen et al., 2008). The PRC1 complex can ubiquitinate H2AK119 through the E3 ligase activity of Ring1A/B (de Napoles et al., 2004; Wang et al., 2004a; Buchwald et al., 2006).
Even appreciating that PRCs have histone-modifying activities, it remains unclear how they actually repress gene expression (Simon and Kingston, 2009). In vitro, PRCs can decrease the accessibility of Hox genes to enzymes (Fitzgerald and Bender, 2001), inhibit chromatin remodeling (Francis et al., 2001), and block transcription (King et al., 2002). PRC components can also compact a nucleosomal array in vitro into a form that is refractory to chromatin remodeling (Francis et al., 2004; King et al., 2005; Lo et al., 2009; Margueron et al., 2008). This also occurs on nucleosome templates assembled from tail-less histones (Francis et al., 2004; Margueron et al., 2008) suggesting that this chromatin compaction is independent of histone tail modifications.
There has been little evidence to date to suggest that PRC components might function in vivo to change chromatin packaging in a way that is independent from their histone-modifying activities, and this remains a major gap in our understanding of polycomb function. PRC components are bound at, modify the chromatin of, and repress sets of developmentally regulated genes in human and mouse ESCs (Azuara et al., 2006; Boyer et al., 2006; Jørgensen et al., 2006; Lee et al., 2006; Stock et al., 2007; Endoh et al., 2008). Among these targets are the Hox loci, encoding key players in early developmental cell fate and cell identity decisions.
We have previously shown that Hox loci visibly decompact and undergo nuclear reorganization as their genes are activated, both during ESC differentiation (Chambeyron and Bickmore, 2004; Morey et al., 2007) and in the embryo at sites of Hox activation (Chambeyron et al., 2005; Morey et al., 2007), consistent with a function for PRCs in maintaining a compact chromatin state at silent loci. Here, we show that there is a specific visible decompaction of chromatin at the murine Hoxb and d loci in ESCs lacking functional PRC1 or 2 complexes. This contrasts with the apparently genome-wide chromatin decompaction in cells deficient in H1. We attribute chromatin compaction activity to PRC1 components, not PRC2, since decompaction is seen in Ring1B mutant cells in which H3K27me3 still blankets the Hox loci. We show that addition of Ring1B can restore in vivo chromatin compaction, and moreover, using a catalytically inactive Ring1B, we present evidence that this is independent of Ring1B’s ability to ubiquitinate histone H2A. Finally, we demonstrate that Ring1B-mediated chromatin compaction is functionally significant, as it represses the expression of Hox genes. We suggest that Ring1B, likely through a PRC1-like complex, functions in vivo to maintain gene repression through a change in higher-order chromatin structure, not just by histone modification.
RESULTS
Chromatin Decompaction and H3K27me3 Loss during Differentiation
Unlike most PRC targets, where PRC binding and H3K27me3 are restricted to within ~1 kb of the transcriptional start site (TSS), Hox loci are demarked by broad domains of this PRC2-mediated histone modification (Bernstein et al., 2006; Lee et al., 2006; Bracken et al., 2006; Soshnikova and Duboule, 2009). Such an extensive domain of modification would be compatible with a large-scale change in chromatin structure.
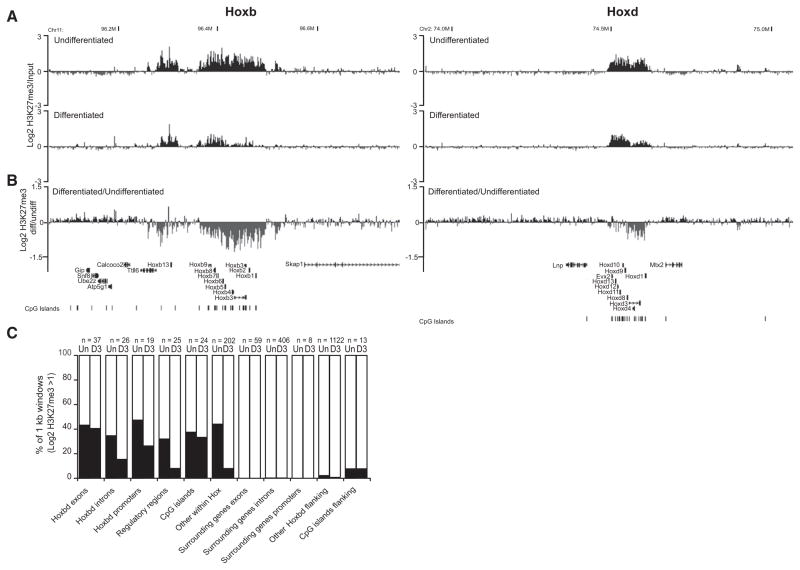
To confirm this, we hybridized micrococcal nuclease (MNase)-digested chromatin from undifferentiated ESCs (input) together with the H3K27me3-modified chromatin immunoprecipitated from this digested chromatin to a custom tiling array encompassing the murine Hoxb and d loci, flanking regions, and control loci. We indeed detect extensive domains of H3K27me3 over Hoxb and d (Figure 1A). At Hoxb, the major domain of modification is a 140 kb region encompassing Hoxb1-Hoxb9, with a separate 50 kb modified region over the most 5′ gene, Hoxb13. Across Hoxd, a 145 kb domain of H3K27me3 includes Evx2 5′ of Hoxd13 and appears to be bipartite, with subdomains that encompass Hoxd1-d4 and Hoxd8-Evx2.
Figure 1. Loss of H3K27me3 at Hox Loci during ESC Differentiation.
(A) Log2 H3K27me3/input at Hoxb (left) and Hoxd (right), by microarray hybridization of ChIPed material from undifferentiated (top) and differentiated (bottom) OS25 ESCs. Running mean of three biological and two technical replicates (n = 6) with 1 kb window size and a 1 kb increment is shown.
(B) Log2 differentiated/undifferentiated H3K27me3. Map position (Mb) and RefSeq gene annotations are from the August 2005 (mm7) Build 35 assembly of the mouse genome (http://genome.ucsc.edu).
(C) Percent of 1 kb running mean windows with log2 H2K27me3/input >1 in undifferentiated (Un) and differentiated (D3) ESCs for different categories of probes from the tiling array, either within Hox clusters (Hox exons, introns, promoters, CpG islands, known regulatory regions, and other) or in flanking regions (surrounding exons, introns, promoters, CpG islands, and other probes).
H3K27me3 stops abruptly outside of Hoxb and d. However, there is a small 16 kb area of H3K27me3 just 3′ of Hoxb1, and 5′ there are small blocks of modification within Tt116. There is a 10 kb block 5′ of Hoxd at a conserved region between Evx2 and Lnp, but we found no evidence for H3K27me3 modification downstream of Lnp over the conserved global control region (GCR), which is responsible for the regulation of 5′ Hoxd gene expression in digits and in the CNS (Spitz et al., 2003). The functional significance of small blocks of H3K27me3 ~300 kb 3′ of Hoxd and beyond Mtx2 (Figure 1A) is not known.
Within Hox loci, H3K27me3 is enriched at all sequence classes (Figure 1C), though it is the promoters and exons of Hox genes and, surprisingly, other oligonucleotide probes of no known function from within the clusters that have higher levels of modification. Consistent with previous findings, there is a peak of H3K27me3 <2 kb from the TSS of Hox genes (Figure S1) (Boyer et al., 2006; Pan et al., 2007).
Retinoic acid (RA)-directed ESC differentiation activates temporally regulated Hox expression (Papalopulu et al., 1991; Morey et al., 2007). After 3 days of differentiation, the most pronounced decrease in H3K27me3 was over the 3′ genes (Hoxb1-b5 and Hoxd1-d8) (Figures 1A and 1B), consistent with the polarized activation of more 3′ Hox genes at this time point (Morey et al., 2009). Proportionately, the biggest changes occur over the introns, known regulatory regions, and other sequences from within the Hox clusters. The smallest changes were over the exons of Hox genes and CpG islands (Figure 1C). Loss of hybridization signal could formally be due to loss either of the histone modification, or of the histone itself. However, a comparative hybridization of MNase-digested chromatin and MNase-digested naked DNA did not reveal any obvious large-scale loss of nucleosomes from the Hox loci during differentiation (data not shown). Therefore, we conclude that we are detecting the specific loss of H3K27me3 from nucleosomes at Hox loci.
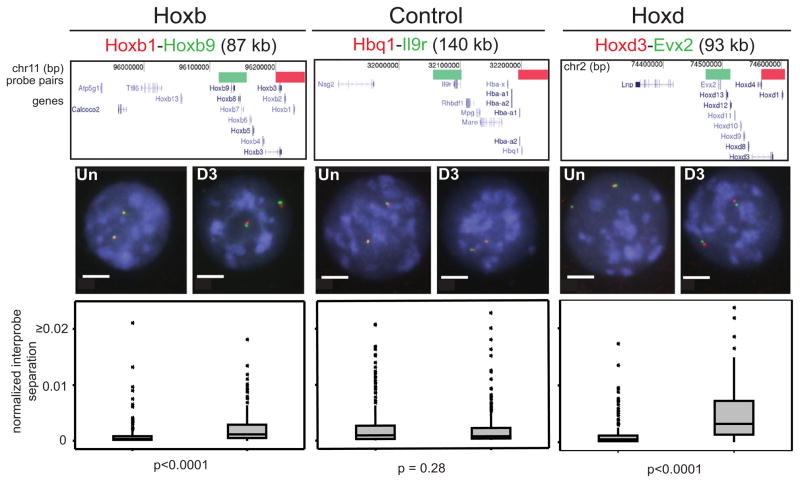
To assay chromatin compaction, we used fluorescence in situ hybridization (FISH) with closely apposed pairs of fosmid probes. Squared interprobe distances (d2) are generally linearly related to genomic separation (in kb) (van den Engh et al., 1992) and can be used to identify differences in chromatin compaction, either between different regions of the genome in a particular cell type (Yokota et al., 1997) or at specific loci between cells at different stages of differentiation (Chambeyron and Bickmore, 2004). Any change attributable solely to altered nuclear size is assessed by normalizing d2 by the nuclear radius (r2). Chromatin decompaction was seen across Hoxb and d loci upon differentiation, but not across the control α-globin locus, which is not a polycomb target but is on the same chromosome (MMU 11) as Hoxb (Garrick et al., 2008) (Figure 2). Chromatin decompaction during differentiation was contained within the domain of H3K27me3, unlike looping out of Hox loci from their chromosome territories, which extends beyond the H3K27me3 domain (Morey et al., 2009).
Figure 2. Chromatin Decompaction within Hox Loci during Differentiation.
FISH with probe pairs at Hoxb, Hoxd, and a control locus on MMU 11, in MAA-fixed nuclei of undifferentiated (Un) and differentiated (D3) OS25 ESCs counter-stained with DAPI (blue). Scale bar = 5 μm. Above the images, diagrams show the positions of probes in the UCSC browser (February 2006 Assembly, mm8 Build 36). Genome position is shown in bp. Below the images, box plots show the distribution of interprobe distances2 (d2) normalized for nuclear radius2 (r2) for Un and D3 cells. The shaded boxes show the median and interquartile range of the data; asterisks indicate outliers. n = 2 × biological replicates, each of 100 loci. The statistical significance of differences between Un and D3 were examined by Mann-Whitney U tests.
Specific Chromatin Decompaction at Hox Loci in the Absence of H3K27me3
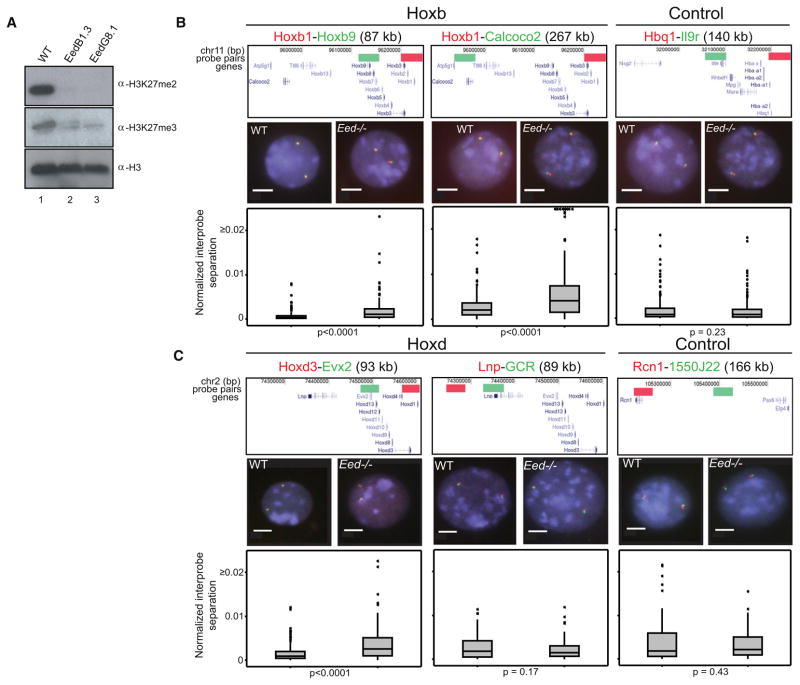
To determine whether decompaction is a direct consequence of the loss of H3K27me3/PRC2, rather than an indirect effect of differentiation, we analyzed Hox loci in undifferentiated wild-type (WT) ESCs and in the corresponding ESCs that are null for the Eed component of PRC2 (Eed−/−). Results from two independent Eed−/− cell lines (G8.1 and B1.3) (Azuara et al., 2006) were indistinguishable, and H3K27me3 is lacking from these cells (Montgomery et al., 2005; Shen et al., 2008) (Figure 3A).
Figure 3. Chromatin Decompaction at Hox Loci in the Absence of Eed.
(A) Western blot of H3K27me2 and me3 in Eed−/− (Lane 2 and 3) and matched wild-type (WT) ESCs (Lane 1). Levels of H3 are shown for comparison. Weak H3K27me3 signal in Eed−/− lanes is due to remnant feeder cells.
(B and C) 2D FISH with probe pairs at Hoxb and a control locus on MMU 11 n = 2 biological replicates, each of 100 loci (B), and probes across the Hoxd locus (n = 200 loci) or 5′ flanking the Hox cluster and a control locus on MMU 2 (n = 100 loci each) (C), in nuclei of WT (left) and Eed−/− (right) cells counterstained with DAPI (blue). Scale bar = 5 μm. Probe position and the distribution of normalized interprobe distances for WT and Eed−/− cells are indicated as in Figure 2A.
We found significant chromatin decompaction (d2/r2) within (Hoxb1-b9) and across (Hoxb1-Calcoco2) Hoxb in Eed−/− ESCs compared to WT (p < 0.001 in Mann-Whitney U analysis) in three independent experiments scored by two observers (R.E. and S.B.) (Figure 3B). There was no corresponding decompaction at the α-globin control locus. In Eed−/− cells, there was also decompaction within Hoxd (Hoxd3-Evx2) (Figure 3C), but not at the 5′ GCR, consistent with the absence of H3K27me3 from this long-range regulatory element (Figure 1A). There were no significant changes in chromatin compaction across a control region of the chromosome (MMU 2) in which Hoxd resides. The same conclusions were reached considering simple interprobe separations, i.e., without normalization to nuclear size (Table S5).
No Specific Chromatin Decompaction in Cells with Reduced H1
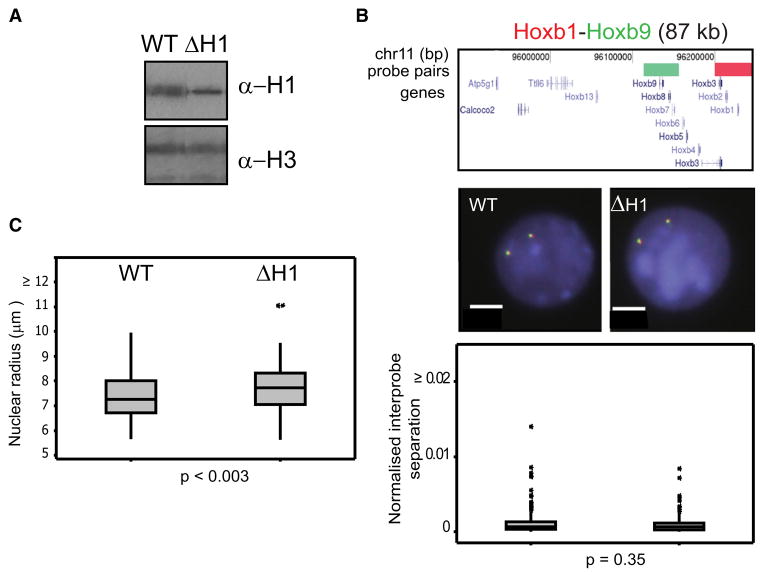
Although the specific decompaction of Hox loci in Eed−/− cells is compatible with a direct effect of PRC2 activity, it is possible that Hox loci are especially susceptible to any general change in higher-order chromatin structure. To assess this, we investigated chromatin compaction in ESCs deleted for the linker histones H1c, d, and e (ΔH1) (Fan et al., 2003, 2005) (Figure 4A).
Figure 4. Chromatin Compaction in H1-Depleted Cells.
(A) Western blot showing reduced levels of H1 in ΔH1 compared to WT ESCs.
(B) 2D FISH with probes for Hoxb1 (red) and Hoxd9 (green) in nuclei of WT and ΔH1 cells counterstained with DAPI (blue). Scale bar = 5 μm. Probe position and the distribution of normalized interprobe distances between WT and ΔH1 cells are as in Figure 2A. n = 2 biological replicates, each of 100 loci.
(C) Box plot showing the distribution of nuclear sizes (plotted as radius in μm of a circle of equal area to that of the nucleus) for WT and ΔH1 cells. n = 100 cells.
ΔH1 ESCs have a larger mean nuclear size (radius r = 7.74 ± 0.07 μm) compared to WT (7.47 ± 0.08 μm) (p < 0.0003) (Figure 4C), and this is not just a consequence of a differential response to the FISH procedure, as it is also seen in cells that have not been subject to FISH (data not shown). However, we found no significant change in the normalized interprobe distances at Hox loci between WT and ΔH1 cells (Figure 4B).
Specific Chromatin Decompaction at Hox Loci in the Absence of Ring1B
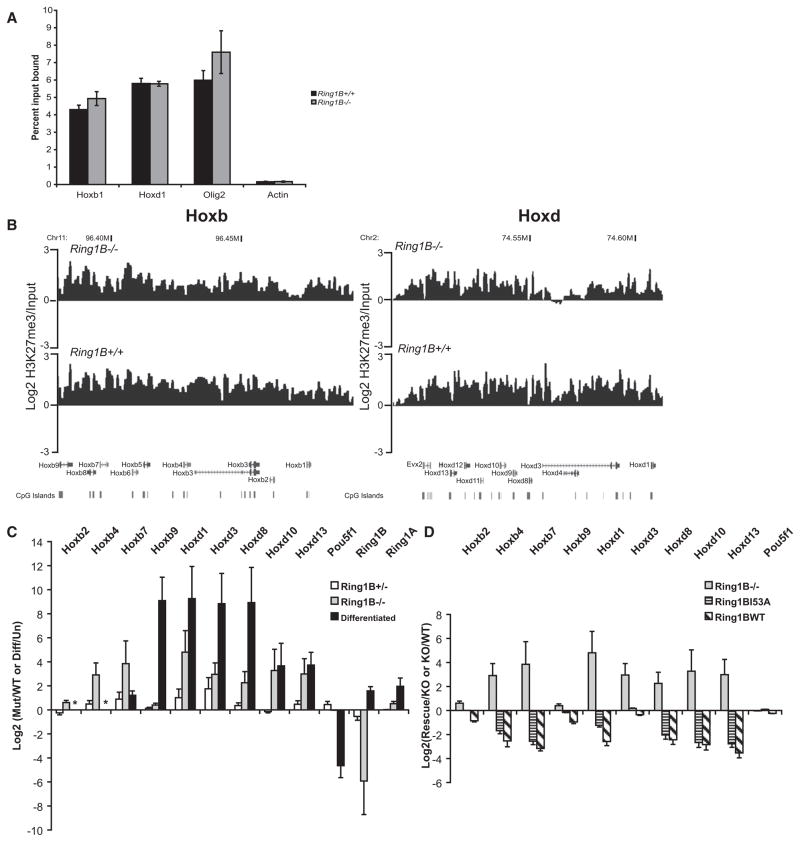
PRC2 H3K27 HMTase activity is considered necessary to recruit PRC1 to many target loci (Ku et al., 2008). Indeed, as previously described, Ring1B binding at Hox loci is significantly reduced in Eed−/− ESCs (Figure S2) (Boyer et al., 2006). Hence, chromatin decompaction in the absence of Eed could be a direct consequence of the loss of PRC2/H3K27me3 on chromatin structure, or it could be due to the inability to recruit PRC1 in the absence of H3K27me3. To distinguish between these possibilities, we analyzed ESCs mutant for the PRC1 component Ring1B. In the absence of Ring1B (Ring1B−/−), other PRC1 components are also depleted, and levels of H2AK119 ubiquitination (H2AK119ub) decrease (Leeb and Wutz, 2007; van der Stoop et al., 2008). Residual levels of H2AK119ub are likely due to Ring1A or remnant feeder cells. By chromatin immunoprecipitation (ChIP) and quantitative RT-PCR (qRT-PCR), we confirmed that H3K27me3 is not decreased at the promoters of Hox genes in Ring1B−/− ESCs (Figure 5A), and hybridization to tiling arrays confirmed that H3K27me3 still blankets Hox loci in mutant cells (Figure 5B).
Figure 5. H3K27me3 and Gene Expression at Hox Loci in Ring1B Mutant Cells.
(A) ChIP for H3K27me3 at the promoters of Hoxb1, Hoxd1, Olig2, and β-actin, assayed by qRT-PCR, in WT (+/+) (black) or Ring1B−/− cells (gray). Enrichment is shown as mean percent input bound ± SEM over three biological replicates.
(B) Mean log2 H3K27me3/input over Hoxb (left) and Hoxd (right), established by microarray hybridization of ChIPed material from Ring1B3−/− (top) or WT (+/+) (bottom) ESCs. Running mean of three biological and technical replicates (n = 5), with 1 kb window size and a 1 kb increment, is shown. Map position and RefSeq gene annotations are as in Figure 1B.
(C) Log2 ratios ± SEM for Hox expression, assayed by qRT-PCR, in Ring1B mutant cells (+/−, white bars; −/−, gray bars) relative to WT. This is compared to the log2 changes in gene expression during the differentiation (diff/un) of OS25 ESCs (black bars). Asterisks indicate where levels of Hoxb2 and Hoxb4 induction during differentiation could not be analyzed by qRT-PCR because the transcripts in undifferentiated cells were only detectable after 40 cycles of PCR.
(D) Log2 ratios ± SEM for Hox gene expression, from three biological replicates assayed by qRT-PCR, in Ring1B3−/− cells relative to WT (KO/WT, gray bars) (the same as in C) or in cells rescued with WT or I53A mutant relative to the Ring1B−/− cells (Rescue/KO, hatched and striped bars).
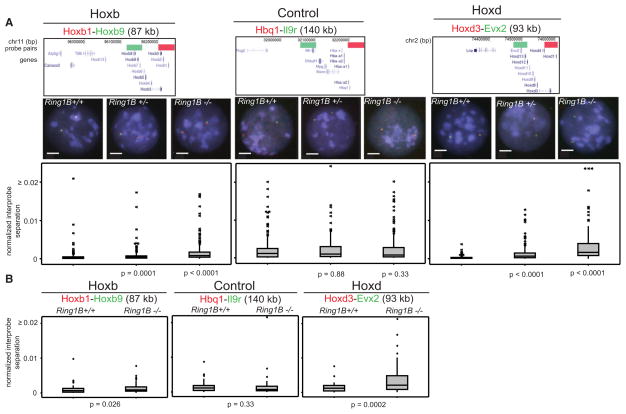
2D FISH revealed significant chromatin decompaction at Hox in Ring1B−/− compared with WT (Ring1B+/+) ESCs (Figure 6A and Table S5). This was confirmed by 3D FISH (Figure 6B). 3D interprobe separations at Hoxb and d, but not at a control locus, were significantly increased in the absence of Ring1B, although as we have previously reported (Morey et al., 2007), absolute distances scored in 2D preparations are generally larger than those seen in 3D.
Figure 6. Chromatin Decompaction in the Absence of Ring1B.
(A) 2D FISH with probe pairs at Hoxb and Hoxd and a control locus in nuclei of WT (+/+), Ring1B+/−, and Ring1B−/− cells, counterstained with DAPI (blue). Scale bar = 5 μm. Probe position and the distribution of normalized interprobe distances between WT and mutant cells are as in Figure 2A. n = 200 loci.
(B) Box plots showing the distribution of normalized interprobe distances (d2/r2) measured by 3D FISH. Shaded boxes show the median and interquartile range of the data. Asterisks indicate outliers in the data set. n > 60 loci each.
Since Hox loci decondense during differentiation, one possibility was that decompaction seen in the mutant cells is due to precocious differentiation, rather than loss of PRC1 per se. The colony morphology and expression of the pluripotency marker alkaline phosphatase in mutant cells suggest that this is not the case (Figure S3A). We also directly assessed the proportion of Oct4+ve (undifferentiated) cells in mutant and WT cultures by flow cytometry. In all cases (Figure S3B) of cells harvested as for assay by FISH >98% were Oct4+ve.
Chromatin Decompaction Is Sensitive to Ring1B Dosage and Is Not Just a Consequence of Transcriptional Activation
Loss of PRC2 activity in Eed−/− ESCs is known to result in some (2- to 50-fold) increased expression of Hox genes, especially those located toward the 5′ ends of the clusters (Boyer et al., 2006; Chamberlain et al., 2008). To determine whether loss of PRC1 has a similar effect, we assessed gene expression by qRT-PCR in WT, Ring1B+/−, and Ring1B−/− ESCs (Figure 5C). Expression of Hox genes was elevated ~10-fold in Ring1B−/− compared to WT cells (up to a maximum of 30-fold for Hoxd1). This is at least an order of magnitude less than upregulation of expression during the differentiation of WT ESCs (Figure 5C). There is no effect of Ring1B loss on Oct4 (Pou5f1) expression, consistent with the Oct4+ve state of the mutant cell population (Figure S3B).
In heterozygous (Ring1B+/−) mutant cells, Hox expression was not significantly increased (Figure 5C), and there were substantial levels of H2AK119ub in these cells (Figure 7A). However, relative to WT, there was still significant decompaction of chromatin at Hox loci in these cells, albeit less extensive than that seen in homozygous mutant cells (Figure 6A). We therefore conclude that chromatin decompaction is not just simply a consequence of transcriptional activation or the complete loss of H2AK119ub and suggest that it is directly due to the reduced Ring1B levels.
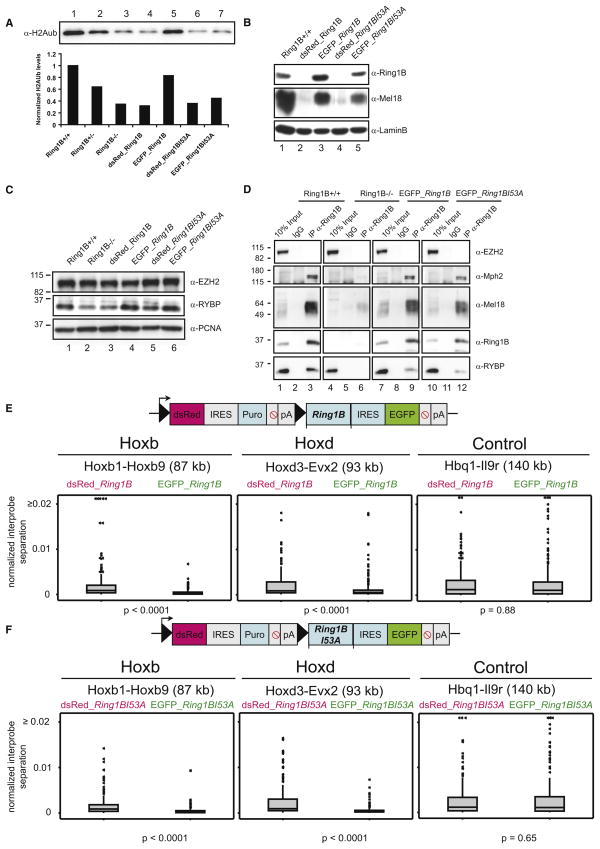
Figure 7. Rescue of Chromatin Compaction, but Not Histone Ubiquitination, by Ring1B I53A.
(A) Western blot of H2AK119ub levels in WT (Ring1B+/+) (Lane 1), Ring1B+/−(Lane 2), Ring1B−/− (Lane 3), dsRed-expressing cells with WT (Lane 4), or Ring1B I53A constructs (Lane 6) and EGFP cells expressing Ring1B WT (Lane 5) or Ring1B I53A (Lane 7). The graph displays H2Aub levels normalized to H3 and is set to 1 in Ring1B+/+ cells.
(B) Western blot analysis of LaminB, Mel18, and Ring1B in nuclear extracts of WT (Ring1B+/+) (Lane 1), control dsRed cells (Lanes 2 and 4), and rescued cells expressing EGFP and either WT (Lane 3) or Ring1B I53A (Lane 5).
(C) Nuclear extracts for WT (Ring1B+/+) (Lane 1), Ring1B−/− (Lane 2), control dsRed cells (Lanes 3 and 5), and rescued cells expressing EGFP and either WT (Lane 4) or Ring1B I53A (Lane 6) immunostained with EZH2, RYBP, and PCNA.
(D) Immunoprecipitation with anti-IgG or anti-Ring1B from WT (Ring1B+/+) (Lanes 1–3), Ring1B−/−(Lanes 4–6), EGFP WT (Lanes 7–9), or Ring1B I53A (Lanes 10–12) nuclear extracts. 10% of total protein extracts were loaded as inputs. Immunostaining was performed using antibodies against EZH2, Mph2, Mel18, Ring1B, and RYBP.
(E) Schematic representation of pTLC Ring1B expression vector (upper panel). Filled triangles = loxP sites. Lower panel: distribution from 2D FISH of normalized interprobe distances at Hoxb, Hoxd, and a control locus in nuclei of control Ring1B−/− ESCs expressing pTLC prior to Cre-mediated loxP excision (dsRed) versus the excised cells that express EGFP and WT Ring1B. 2× biological replicates, n > 95 nuclei.
(F) As in (E), but for the I53A mutant Ring1B.
Ring1B-Mediated Chromatin Compaction and Gene Repression Are Not Dependent on H2A Ubiquitination
The fact that we detect a heterozygous effect on chromatin compaction in Ring1B mutant cells would be consistent with a nonenzymatic function for the protein. Ring1B, in complex with Bmi1, is known to have E3-ligase activity for monoubiquitination of H2AK119. This activity resides in the RING domain located toward the N terminus (residues 50–103) of Ring1B (Buchwald et al., 2006), and a point mutant there (I53A) is known to abolish histone ubiquitination catalytic activity in vitro and is predicted to disrupt the interaction with the E2 UbcH5c (Buchwald et al., 2006; Elderkin et al., 2007). Therefore, we wished to determine whether this version of Ring1B had also lost the ability to ubiquitinate H2A and to compact chromatin structure in vivo. To this end, we introduced full-length WT Ring1B or the I53A mutant version into Ring1B−/− ESCs. The cells were transfected with a loxP-containing construct, which allows for fluorescence-activated cell sorting (FACS) of stable transfectants, without expression of the downstream cDNA (Figures 7E and 7F). Excision of the dsRed-puromycin cassette by Cre recombinase then brings both Ring1B and EGFP expression under the control of the CAG promoter. Levels of rescued Ring1B protein expression seem close to endogenous levels in Ring1B+/+ cells (Figure 7B). FACS was used to select for EGFP-expressing rescued cells, and analysis of Oct4 expression, colony morphology, and alkaline phosphatase staining (Figure S3) were used to confirm their undifferentiated state.
As expected, expression of WT Ring1B rescued both the levels of H2A ubiquitination (Figure 7A) and specific chromatin compaction at Hox loci (Figure 7E and Table S6). Consistent with its lack of E3 ligase activity in vitro (Buchwald et al., 2006; Elderkin et al., 2007), expression of Ring1B I53A did not rescue levels of H2A ubiquitination, but it did rescue Hox chromatin compaction (Figures 7A and 7F and Table S6). Expression of either form of Ring1B did not compact a control locus. We conclude that chromatin compaction in vivo is mediated by a direct effect of Ring1B that is independent of its histone ubiquitination activity.
It is known that in the absence of Ring1B, protein levels of several other PRC1 components are reduced (Leeb and Wutz, 2007). By western blot, we found that rescue of Ring1B null cells with either WT or mutant Ring1B also restores protein levels of the PRC1 components Mel18 (Figure 7B) and RYBP (Figure 7C). Moreover, a PRC-like complex is reassembled by the rescued Ring1Bs. Mph2, Mel18, and RYBP are all pulled down by immunoprecipitation for either the WT or the mutant Ring1B proteins (Figure 7D). As expected, the PRC2 component EZH2 is neither destabilized in the absence of Ring1B (Figure 7C) nor immunoprecipitated with the rescued Ring1B proteins (Figure 7D).
To determine whether the chromatin compaction restored by Ring1B is of functional significance, we assessed levels of Hox gene expression in Ring1B null (−/−) cells and in the rescued cells (Figure 5D). Expression of either WT Ring1B or Ring1B I53A mutant downregulates the expression of Hox genes that had been activated in Ring1B null cells. Therefore, Ring1B is able to both control the gene expression and compact the chromatin state at the Hox loci in ESCs independent of its H2A ubiquitin ligase activity.
DISCUSSION
Dynamic Changes in H3K27me3 at Mammalian Hox Loci during Differentiation
Recent studies have suggested that CpG islands are recruitment sites for PRC2 in pluripotent cells (Garrick et al., 2008; Ku et al., 2008) and that extensive PRC2-marked domains occur at CpG island regions that have unusually high levels of sequence conservation and are not necessarily associated with the 5′ ends of genes, such as at Hox loci (Tanay et al., 2007). We found that although all classes of sequence within murine Hoxb and d loci are enriched in H3K27me3 in undifferentiated ESCs, it is the promoters and exons of the Hox genes themselves and other as yet uncharacterized sequences within Hox loci that have the highest enrichment (Figure 1). In addition, a separate 16 kb block of H3K27me3 modification just 3′ of Hoxb1 and one ~300 kb 3′ of Hoxd do not contain any annotated CpG islands. Therefore, it is unlikely that CpG islands are the only motifs capable of recruiting PRC2.
PcG proteins regulate the ancestral function of Hox clusters in anterior-posterior patterning of the main body axis. Hoxd, but not Hoxb, was co-opted later during evolution to pattern a new axis: the limbs and digits. The elements regulating this facet of Hoxd function lie outside the cluster itself. We found no H3K27me3 enrichment over the GCR, which contains clusters of enhancers driving 5′ Hoxd expression in digits and the CNS (Spitz et al., 2003), suggesting that the GCR is not recognized as part of the regulatory landscape of Hoxd in ESCs.
H3K27me3 is lost from Hox loci during ESC differentiation, especially at regulatory sequences and other uncharacterized sequences within the clusters, rather than at CpG islands and the Hox genes themselves (Figure 1). Progressive changes in chromatin structure were proposed to underlie the temporal colinearity of Hox expression during early development (Kmita and Duboule, 2003). Paralleling the preferential activation of more 3′ Hox genes early in ESC differentiation (Morey et al., 2009), we also detect a greater loss of H3K27me3 from the 3′ side of the clusters than at the 5′ ends (Figure 1). The region of preferential H3K27me3 loss at Hoxb (Hoxb1-b5) corresponds with the domain of preferential RA sensitivity mapped at chicken Hoxb genes during neural tube differentiation (Bel-Vialar et al., 2002), while that at Hoxd corresponds with the region (Hoxd1-Hoxd4) where H3K27me3 is lost in the murine tail bud at early stages of embryogenesis (E8.5) before proceeding to more 5′ genes (Soshnikova and Duboule, 2009).
H3K27me3 Is Not Sufficient to Maintain Chromatin Compaction
The loss of H3K27me3 from Hox loci during differentiation parallels the visible decompaction of their chromatin structure (Figure 2). Using mutant ESCs, we show directly that the activity of the PRC2 complex is indeed required to maintain a compact chromatin state in vivo at Hox loci. In its absence, there is a visible increase in the interprobe separation between probes spanning either Hoxb or d (Figure 3 and Table S5). This is not seen at control loci, and moreover, it is restricted to the parts of Hoxd that are modified by H3K27me3 (Figures 1 and 3). We noted that the effects of H3K27me3 loss on chromatin compaction and indeed even the effects of Ring1B mutation (Figure 6) were larger at Hoxd than at Hoxb. We do not know the reason for this, but note that the genomic contexts of the two loci are very different from each other and so may impose different constraints on the structures that they can adopt once polycomb-mediated regulation is removed. While Hoxb is embedded within a region of genes that are active in ESCs, Hoxd is embedded in a large gene desert, and we have previously seen differences between the two loci in other aspects of their nuclear organization (Morey et al., 2009). Alternatively, this difference could be due to another PRC component.
PRC1 can be recruited to some regions destined for repression independent of PRC2 (Schoeftner et al., 2006; Terranova et al., 2008). However, at most PRC targets, including Hox, it is thought that H3K27me3 laid down by PRC2 provides the binding site for PRC1 recruitment (Wang et al., 2004b; Schwartz and Pirrotta, 2007; Ku et al., 2008). Hence, in Eed−/− ESCs, PRC1 binding is lost from Hox genes (Figure S2) (Boyer et al., 2006). The fact that we see decompaction of Hox loci in Ring1B−/− ESCs (Figure 6), even though H3K27me3 is still blanketing these domains (Figure 5), shows that this histone modification is not sufficient of itself to maintain a compacted chromatin state. Rather, chromatin decompaction in Eed−/− cells is due to a failure to recruit a PRC1-like complex (Figure S2) (Boyer et al., 2006). Neither is decompaction simply a downstream consequence of ectopic differentiation triggered by loss of polycomb, as both Eed and Ring1B mutant cells remain Oct4+ve (Figure S3).
This conclusion contrasts with the recent report that both PRC2 and PRC1 have independent roles in maintaining a contracted chromatin domain at the paternal Kcnq1 imprinted cluster in trophoblast stem cells and trophectoderm (Terranova et al., 2008). However, this imprinted cluster is controlled by a complex interplay of multiple epigenetic regulators that include both polycomb and H3K9 methylation mediated by the G9a HMTase, both recruited via a long noncoding RNA (Pandey et al., 2008; Redrup et al., 2009). There is no genetic evidence for a contribution of G9a to gene regulation at Hox loci, nor is there substantial H3K9 methylation at Hox loci in ESCs (Mikkelsen et al., 2007).
Ring1B Compacts Chromatin and Represses Gene Expression In Vivo, Independent of Histone Ubiquitination
Since we found some decompaction of Hox loci in Ring1B heterozygous mutant cells (Figure 6), which still have abundant levels of H2A ubiquitination (Figure 7A), we considered whether there might be a direct structural function of this PRC1 component in chromatin compaction in vivo, rather than compaction being just a downstream consequence of Ring1B’s catalytic function as a ubiquitin E3 ligase. Consistent with this hypothesis is the finding that PRC1-mediated compaction in vitro can occur on nucleosome templates assembled from tail-less histones (Francis et al., 2004; Margueron et al., 2008).
We have shown that the reintroduction of WT Ring1B into Ring1B−/− ESC lines is sufficient to restore targeted chromatin compaction at Hox loci (Figure 7). Levels of H2AK119ub are also restored in these cells. Reintroducing a mutant Ring1B that in vitro lacks E3 ligase activity (Buchwald et al., 2006) does not restore H2AK119ub but is nonetheless able to restore chromatin compaction at Hox loci. Hence, we conclude that Ring1B acts to compact higher-order chromatin structure independent of its known histone-modifying activity. We think it likely that this function of Ring1B is operating via its participation in a PRC1-like complex, as we do see restoration of protein levels for other PRC1 components—Mel18 and RYBP—in the rescued cells (Figures 7B and 7C), and the rescued Ring1B proteins immunoprecipitate together with Mph2, Mel18, and RYBP (Figure 7D). However, we cannot exclude that Ring1B may be operating via another complex, such as BCOR (Simon and Kingston, 2009).
Chromatin Conformation within PRC Domains
In Drosophila BX-C, there is evidence for long-range interactions between polycomb response elements and promoters that are abrogated by knockdown of polycomb (Cléard et al., 2006; Lanzuolo et al., 2007). Similarly, long-range contacts, detected by 3C methodology at the human GATA-4 locus, are somewhat abrogated by knockdown of Ezh2 (Tiwari et al., 2008). If a PRC-mediated, topologically complex, looped structure were resolved into a more linear one upon differentiation or in mutant cells, we would expect to see an altered distribution of interprobe distances, as well as of the mean distance. However, the distribution of interprobe distances across Hoxb and d (standard deviation d/mean d = ~0.5 and median d/mean d = ~0.9) that we have measured is similar both between WT and PRC mutant ESCs and to our previous analyses (Chambeyron and Bickmore, 2004; Morey et al., 2007). This is compatible with the chromatin of the Hox loci describing a random-walk path in both compact and decompact states (Sachs et al., 1995). We suggest that our data are more consistent with a PRC1-like complex functioning to further compact a randomly folded chromatin fiber, for example, by changing its persistence length, rather than it fundamentally changing the topology of target loci.
Despite the visible decompaction of chromatin at Hox loci that we report, a calculation of compaction ratios reveals that even in PRC mutant cells, there is still higher-order chromatin structure at target loci. This is consistent with the observation in living cells that reporter gene activation, though visibly accompanied by chromatin decompaction, nevertheless takes place in the context of chromatin that is still tightly packaged above the compaction ratio of the 30 nm fiber (Hu et al., 2009).
PRC-Mediated Chromatin Compaction and the Restriction of Gene Expression
Chromatin decompaction that we had previously reported in transgenic embryos carrying a rearranged Hoxd locus (Morey et al., 2008) had led us to conclude that chromatin decompaction occurs upstream of gene activation, rather than being just a passive consequence of transcription through the region. Our finding here of detectable Hox decompaction in Ring1B+/− cells (Figure 6) in the absence of transcriptional activation (Figure 5) corroborates this.
However, the complete absence of Ring1B does lead to the upregulation of Hox gene expression (Figure 5C), albeit to levels far lower than those seen during normal ESC differentiation. Expression of both WT and catalytically inactive Ring1B suppresses Hox gene expression. Therefore, Ring1B-mediated chromatin compaction, without accompanying H2AK119ub, has a direct functional consequence on gene expression.
A form of RNA polymerase II, detected by antibodies that recognize the Ser5 phosphorylated form (RNAPII Ser5P), is found at Hox genes in ESCs, and Ring1B is necessary to prevent this RNAPII proceeding to productive transcriptional elongation (Stock et al., 2007). Together with the reported presence in Drosophila of transcription initiation factors and RNAPII at the promoters of inactive Hox genes (Breiling et al., 2001) and at a polycomb repressed reporter (Dellino et al., 2004), these data suggest that PRC1 inhibits transcription downstream of RNAPII binding itself. This has led to the suggestion that H2A ubiquitination is directly involved in RNAPII pausing/stalling (Zhou et al., 2008) and that this is a key mechanism in polycomb-mediated silencing (Simon and Kingston, 2009). Our findings of H2Aub-independent Hox gene repression by Ring1B suggest that this mechanism demands further investigation and caution against the assumption that just because a protein has histone-modifying activity, this is the be-all and end-all of its functions.
EXPERIMENTAL PROCEDURES
ESC Culture and Differentiation
Tissue culture conditions, generation of rescue cell lines, alkaline phosphatase staining, and Oct4 flow cytometric analysis are described in the Supplemental Information.
Acid Extraction of Histones, Immunoprecipitation, and Western Blotting
Methods for preparation of ESC histones and nuclear extracts, immunoprecipitation, antibody dilutions for western blot analysis, and quantitative western analysis are found in the Supplemental Information.
FISH
For 2D FISH, nuclei were isolated in hypotonic buffer (0.25% KCl, 0.5% trisodium citrate) and fixed with 3:1 v/v methanol/acetic acid. For 3D FISH, cells were grown on slides, washed with PBS, and fixed in 4% paraformaldehyde (pFA) for 10 min. For details, see Supplemental Information. For coordinates and names of fosmid probes, see Table S1.
Image Capture and Analysis
Image capture and analysis were as previously described (Morey et al., 2009). For normalization and statistical data analysis, see Supplemental Information.
ChIP
A more detailed description of ChIP and relative quantification by qRT-PCR can be found in the Supplemental Information. Primer pairs are given in Table S2.
Microarray Design, Hybridization, and Analysis
Information on microarray design and hybridization can be found in the Supplemental Information, with oligos spanning genes described in Table S3.
Real-Time PCR and Analysis of Transcript Levels
Relative fold changes in transcription levels were measured in three biological replicates using primers (Table S4) as described in the Supplemental Information.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council, United Kingdom. C.K. was supported by the EU FP6 NoE Epigenome (LSHG-CT-2004-503433) and Y.F. and A.I.S. by NIH grant CA79057. We thank H. Jørgensen (MRC CSC, London) and R. John (Cardiff University) for Eed−/− cells, N. Brockdorff for Eed+/+ clone 2.21, M. Hughes (Liverpool Microarray Facility) for microarray printing, E. Brookes (MRC CSC) for advice on Ring1B ChIP, T. Sixma (NCI, Amsterdam) for Ring1B constructs, J. Brickman (ISCR, University of Edinburgh) for pTLC, H. Koseki (RIKEN Research Center for Allergy and Immunology) for antibodies, and our HGU colleagues I. Adams, H. Sutherland, E. Freyer, and P. Perry for advice, mouse embryos, FACS analysis, and help with image analysis.
Footnotes
ACCESSION NUMBERS
Data are accessible from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) through GEO Series accession numbers GSE20201 and GSE20213.
Supplemental Information includes Supplemental Experimental Procedures, Supplemental References, six tables, and three figures and can be found with this article online at doi:10.1016/j.molcel.2010.02.032.
References
- Allan J, Cowling GJ, Harborne N, Cattini P, Craigie R, Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981;90:279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bates DL, Butler PJ, Pearson EC, Thomas JO. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981;119:469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear reorganisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Cléard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V. Polycomb silencing blocks transcription initiation. Mol Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Elderkin S, Maertens GN, Endoh M, Mallery DL, Morrice N, Koseki H, Peters G, Brockdorff N, Hiom K. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol Cell. 2007;28:107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, Koseki H. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DP, Bender W. Polycomb group repression reduces DNA accessibility. Mol Cell Biol. 2001;21:6585–6597. doi: 10.1128/MCB.21.19.6585-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Garrick D, De Gobbi M, Samara V, Rugless M, Holland M, Ayyub H, Lower K, Sloane-Stanley J, Gray N, Koch C, et al. The role of the polycomb complex in silencing alpha-globin gene expression in nonerythroid cells. Blood. 2008;112:3889–3899. doi: 10.1182/blood-2008-06-161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H, Brockdorff N, Fisher AG. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle. 2006;5:1411–1414. doi: 10.4161/cc.5.13.2927. [DOI] [PubMed] [Google Scholar]
- King IF, Francis NJ, Kingston RE. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol. 2002;22:7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Emmons RB, Francis NJ, Wild B, Müller J, Kingston RE, Wu CT. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol Cell Biol. 2005;25:6578–6591. doi: 10.1128/MCB.25.15.6578-6591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SM, Ahuja NK, Francis NJ. Polycomb group protein Suppressor 2 of zeste is a functional homolog of Posterior Sex Combs. Mol Cell Biol. 2009;29:515–525. doi: 10.1128/MCB.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Kmita M, Duboule D, Bickmore WA. Ectopic nuclear reorganisation driven by a Hoxb1 transgene transposed into Hoxd. J Cell Sci. 2008;121:571–577. doi: 10.1242/jcs.023234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 anti-sense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Lovell-Badge R, Krumlauf R. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucleic Acids Res. 1991;19:5497–5506. doi: 10.1093/nar/19.20.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs RK, van den Engh G, Trask B, Yokota H, Hearst JE. A random-walk/giant-loop model for interphase chromosomes. Proc Natl Acad Sci USA. 1995;92:2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Tanay A, O’Donnell AH, Damelin M, Bestor TH. Hypercon-served CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci USA. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schübeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Engh G, Sachs R, Trask BJ. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science. 1992;257:1410–1412. doi: 10.1126/science.1388286. [DOI] [PubMed] [Google Scholar]
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS ONE. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004a;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004b;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Yokota H, Singer MJ, van den Engh GJ, Trask BJ. Regional differences in the compaction of chromatin in human G0/G1 interphase nuclei. Chromosome Res. 1997;5:157–166. doi: 10.1023/a:1018438729203. [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.