Abstract
The chromodomain helicase DNA-binding (CHD) family of enzymes is thought to regulate gene expression, but their role in the regulation of specific genes has been unclear. Here we show that CHD8 is expressed at a high level during early embryogenesis and prevents apoptosis mediated by the tumour suppressor protein p53. CHD8 was found to bind to p53 and to suppress its transactivation activity. CHD8 promoted the association of p53 and histone H1, forming a trimeric complex on chromatin that was required for inhibition of p53-dependent transactivation and apoptosis. Depletion of CHD8 or histone H1 resulted in p53 activation and apoptosis. Furthermore, Chd8−/− mice died early during embryogenesis, manifesting widespread apoptosis, whereas deletion of p53 ameliorated this developmental arrest. These observations reveal a mode of p53 regulation mediated by CHD8, which may set a threshold for induction of apoptosis during early embryogenesis by counteracting p53 function through recruitment of histone H1.
Although apoptosis has a key role in organization of the developing embryo, it is not fully understood how apoptosis is regulated during embryogenesis. The tumour suppressor protein p53 mediates the induction of apoptosis in response to DNA damage caused by genotoxic stress. It activates the transcription of numerous genes, and thereby triggers cell-cycle arrest, senescence or apoptosis to prevent tumorigenesis1–5.
Activation of transcription by p53 is regulated, at least in part, by the amount of p53 as well as by post-translational modifications of p53 (refs 6–8). In addition, certain chromatin-associated proteins that change chromatin configuration interact with p53 and thereby modulate its transactivation activity5,9–12. Although these findings suggest that chromatin configuration may affect the transactivation activity of p53, the mechanism by which the structure of chromatin changes, as well as the biological outcome of such regulation, have remained largely unknown.
Certain classes of molecules recognize modified histones and are thought to translate the modification code into specific functions. Such proteins include members of the chromodomain helicase DNA-binding (CHD) family of enzymes, which also belong to the SNF2 superfamily of ATP-dependent chromatin remodellers13–15. Chd1 of Saccharomyces cerevisiae is a component of the multi-subunit histone acetyltransferase complexes SAGA and SLIK16, and is required for methylation of histone H3 at Lys 4 (H3K4; ref. 17). Human CHD1 catalyses the ATP-dependent transfer of histones from the NAP-1 chaperone to DNA, resulting in the assembly of active chromatin18,19. Nine genes for CHD1-related proteins have been identified in mammalian species.
Among these proteins, CHD8 (Duplin) was originally isolated as a negative regulator of the Wnt–β-catenin signalling pathway20. The carboxy-terminal region of CHD8 interacts with the insulator-binding protein CTCF, and this interaction is important for insulator activity21. We previously generated Chd8−/− mice and showed that these animals die in utero between embryonic day (E) 5.5 and E7.5, manifesting widespread apoptosis22. However, Wnt activation was not seen in the Chd8−/− embryos before their death. Although these observations suggest that CHD8 may possess anti-apoptotic activity that is independent of Wnt signalling inhibition, it has been unclear how the loss of CHD8 induces apoptosis. We now show that CHD8 binds to both p53 and histone H1, and that these interactions facilitate recruitment of histone H1 to p53 target genes, resulting in suppression of their expression and of apoptosis induced by genotoxic insults. These results thus suggest that loss of CHD8 in mice induces apoptosis as a result of unrestrained p53 activity at the early stage of embryonic development. Consistent with this conclusion, deletion of p53 in Chd8−/− mice ameliorated the developmental arrest. The physiological role of CHD8 may thus be to prevent such unwanted apoptosis during early embryogenesis.
RESULTS
CHD8 suppresses p53-dependent apoptosis
Mouse Chd8 consists of 37 exons spanning about 40 kb. Alternative splicing of exon 9 generates two transcripts that encode a protein (CHD8L) with a relative molecular mass of 280,000 (Mr 280K), containing two chromodomains (a helicase/ATPase domain and a DNA binding domain) and a 110K protein (CHD8S, also known as Duplin), which contains only the amino-terminal chromodomain (Supplementary Information, Fig. S1a, b). These molecules are expressed in most cell lines and tissues (Supplementary Information, Fig. S1c, d), but the ratio of CHD8L to CHD8S varies.
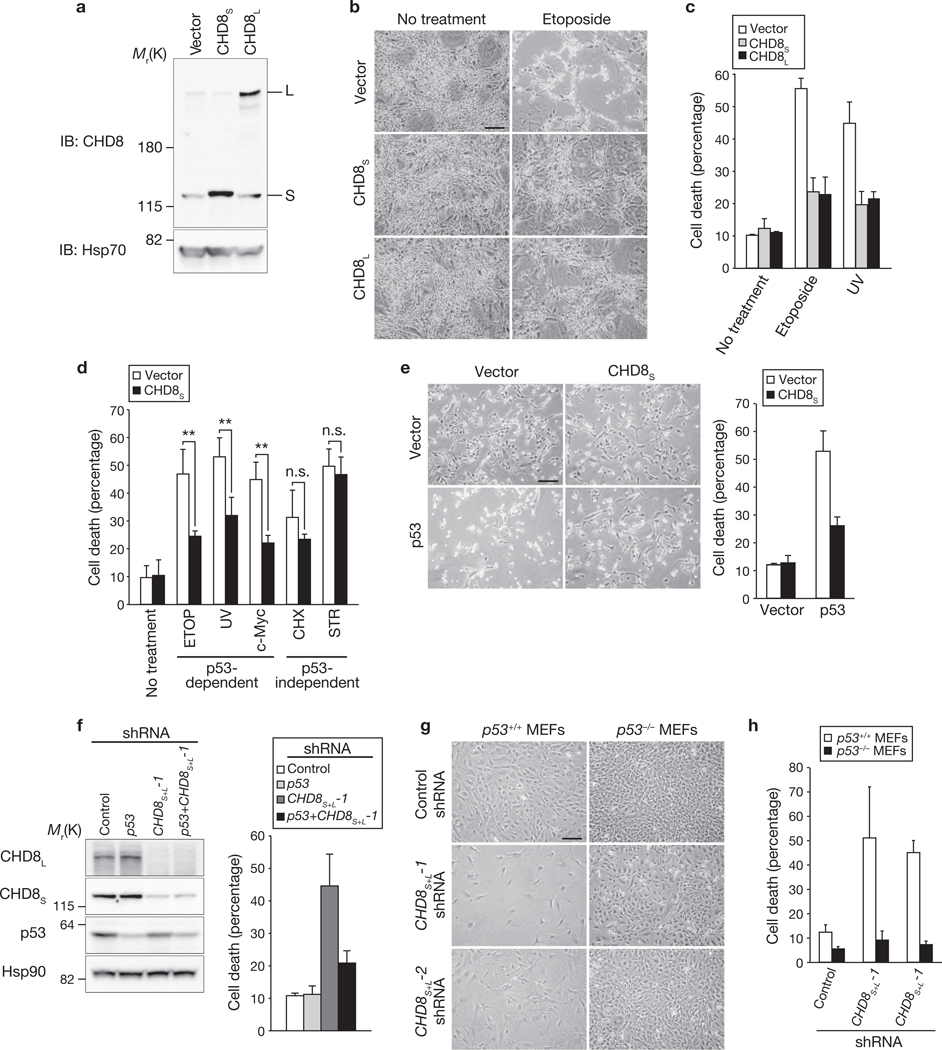
Given that lack of both CHD8S and CHD8L results in widespread apoptosis in mouse embryos22, we hypothesized that CHD8 may possess anti-apoptotic activity. Indeed, overexpression of either CHD8S or CHD8L inhibited the induction of apoptosis in mouse NIH 3T3 cells exposed to etoposide or ultraviolet (UV) radiation (Fig. 1a–c; Supplementary Information, Fig. S2a). The apoptosis-related cleavage of both caspase-3 and poly(ADP-ribose) polymerase (PARP) was also inhibited by overexpression of CHD8 (Supplementary Information, Fig. S2b). CHD8 markedly suppressed apoptosis induced by etoposide, UV radiation or c-Myc, all of which are dependent on p53, but did not affect apoptosis induced by cycloheximide or staurosporine, which are independent of p53 (Fig. 1d). Furthermore, apoptosis induced by overexpression of p53 was inhibited by CHD8S (Fig. 1e) or CHD8L (Supplementary Information, Fig. S3a).
Figure 1.
Anti-apoptotic activity of CHD8. (a–c) NIH 3T3 cells overexpressing CHD8S or CHD8L were subjected to immunoblot (IB, a) analysis with anti-CHD8 and exposed to genotoxic stress. Cells were examined by phase-contrast microscopy (b) and the percentage of dead cells was determined by trypan blue staining (c). Data in c are mean ± s.d., n = 3. (d) NIH 3T3 cells overexpressing CHD8S were exposed to etoposide (ETOP, 50 µM), cycloheximide (CHX, 100 µg ml−1), staurosporine (STR, 1 µM), UV radiation or c-Myc overexpression. Data in d are mean ± s.d., n = 3 (**P < 0.01; n.s., not significant; P > 0.05; Student’s t-test). (e) U2OS cells were infected with retroviral vectors for CHD8S or p53 and were stained with trypan blue (left panel). Data shown in the right panel are mean ± s.d., n = 3. (f) U2OS cells were infected with retroviral vectors encoding shRNAs specific for p53, both CHD8S and CHD8L (CHD8S+L-1) or EGFP (control), subjected to immunoblotting (left panel), stained with trypan blue and the percentage of dead cells determined (right panel). Data shown in the right panel are mean ± s.d., n = 3. (g, h) p53+/+ or p53−/− MEFs infected with retroviral vectors for CHD8S+L-1 or CHD8S+L-2 shRNAs were examined by phasecontrast microscopy (g) and the percentage of dead cells determined by trypan blue staining (h). Data in h are mean ± s.d., n = 3. Scale bars are 100 µm (b, e, g).
Conversely, depletion of both CHD8S and CHD8L by RNA interference (RNAi) induced cell death in U2OS human osteosarcoma cells (Fig. 1f), which harbour wild-type p53 alleles, as well as in HeLa and HCT116 cells (Supplementary Information, Fig. S3b–d). Depletion of CHD8L alone had no such effect. We confirmed that cell death triggered by depletion of CHD8 is due to apoptosis (Supplementary Information, Fig. S2c, d). Together, these observations indicate that both CHD8S and CHD8L possess anti-apoptotic activity and that the presence of CHD8S alone in cells is sufficient to prevent apoptosis, suggesting that the anti-apoptotic activity of CHD8 is dependent on the common region of CHD8S and CHD8L. Apoptosis induced by depletion of CHD8 was blocked by the caspase inhibitor Z-VAD–fmk and was associated with retardation of cell growth (Supplementary Information, Fig. S2e, f).
To investigate whether the apoptosis induced by CHD8 depletion was dependent on p53, we depleted U2OS cells of both CHD8 and p53 and found that additional depletion of p53 in cells depleted of CHD8 restored cell survival (Fig. 1f). Depletion of CHD8 resulted in a marked increase in apoptosis in p53+/+ MEFs, but had virtually no effect in p53−/− MEFs (Fig. 1g, h), indicating that CHD8 depletion results in p53 activation. These data thus indicate that CHD8 is an anti-apoptotic molecule and a negative regulator of p53.
CHD8 interacts with and inhibits p53
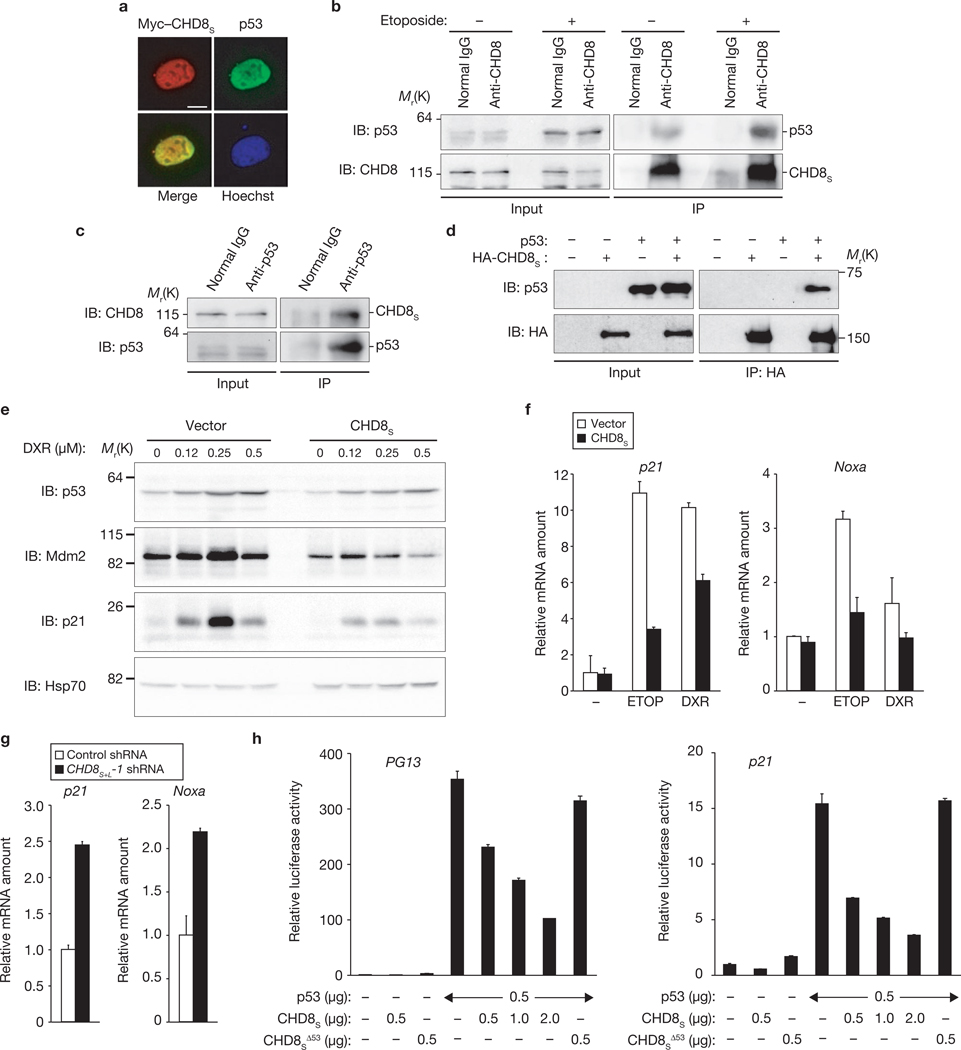
The subcellular localization of CHD8S was found to be almost identical to that of p53, with both proteins being largely restricted to the nucleus (Fig. 2a). Reciprocal co-immunoprecipitation analysis revealed that endogenous CHD8 specifically interacted with endogenous p53 in U2OS cells (Fig. 2b, c; Supplementary Information, Fig. S3e). Similar experiments with a series of deletion mutants of CHD8S revealed that the N-terminal region of CHD8 is responsible for binding to p53 (Supplementary Information, Fig. S4a). Reciprocal analysis showed that CHD8 binds to the central core domain of p53 (Supplementary Information, Fig. S4b). Furthermore, pulldown assays revealed that recombinant CHD8S and recombinant p53 bound to each other in vitro, suggesting that the interaction is direct (Fig. 2d).
Figure 2.
CHD8 interacts with and inhibits transactivation by p53. (a) HeLa cells expressing Myc–CHD8S were immunostained with anti-Myc or p53. Scale bar, 5 µm. (b) U2OS cells were incubated with etoposide and then subjected to immunoprecipitation (IP) with an anti-CHD8 antibody, and immunoblot analysis with an anti-p53 antibody. (c) Immunoprecipitation of U2OS lysates with anti-p53 and immunoblot analysis with an anti-CHD8 antibodies. (d) In vitro binding assay for recombinant CHD8S and p53. (e) U2OS cells overexpressing CHD8S were incubated with doxorubicin (DXR) and then subjected to immunoblot analysis with the indicated antibodies. (f) U2OS cells overexpressing CHD8S were treated with the genotoxic agents DXR (0.5 µM) and etoposide (ETOP, 20 µM) and then subjected to qRT–PCR. (g) U2OS cells infected with a retroviral vector encoding CHD8S+L-1 shRNA were subjected to qRT–PCR. (h) Luciferase assay using either wild-type or mutant CHD8SΔ53. Data are mean ± s.d., n = 3 (f–h).
We next examined whether CHD8 affects transcriptional activation by p53 in U2OS cells. Overexpression of CHD8S or CHD8L markedly inhibited the etoposide- or doxorubicin-induced upregulation of Mdm2, p21 and Noxa, all of which are encoded by p53 target genes, in U2OS cells (Fig. 2e, f; Supplementary Information, Figs S3f, S5a–c). In contrast, depletion of CHD8 resulted in an increase in the expression of p53 target genes, including those for p21 and Noxa (Fig. 2g), suggesting that CHD8 antagonizes p53 function. Furthermore, overexpression of CHD8S inhibited in a concentration-dependent manner the p53- induced increase in luciferase activity in human osteosarcoma SaOS2 cells (p53-null) harbouring a luciferase gene fused to the promoter of the p53 target genes for p21 or PG13 (Fig. 2h). CHD8S overexpression did not affect the activity of a p21 promoter in which the two p53 binding sites are mutated (Supplementary Information, Fig. S5d). The inhibitory effect of CHD8S was not mimicked by the CHD8SΔ53 mutant (Fig. 2h; Supplementary Information, Fig. S5d), which lacks the p53-binding domain (Supplementary Information, Fig. S5e), suggesting that binding of CHD8 to p53 is required for suppression of p53 function. CHD8S did not affect the activity of the transcription factor NF-κB induced by TRAF2 (Supplementary Information, Fig. S5f), suggesting that it is not a general inhibitor of transcription but rather, a specific inhibitor of p53-dependent transactivation. Together, these data indicate that CHD8 interacts with p53 and thereby negatively regulates its function.
CHD8 recruits histone H1 to inhibit p53
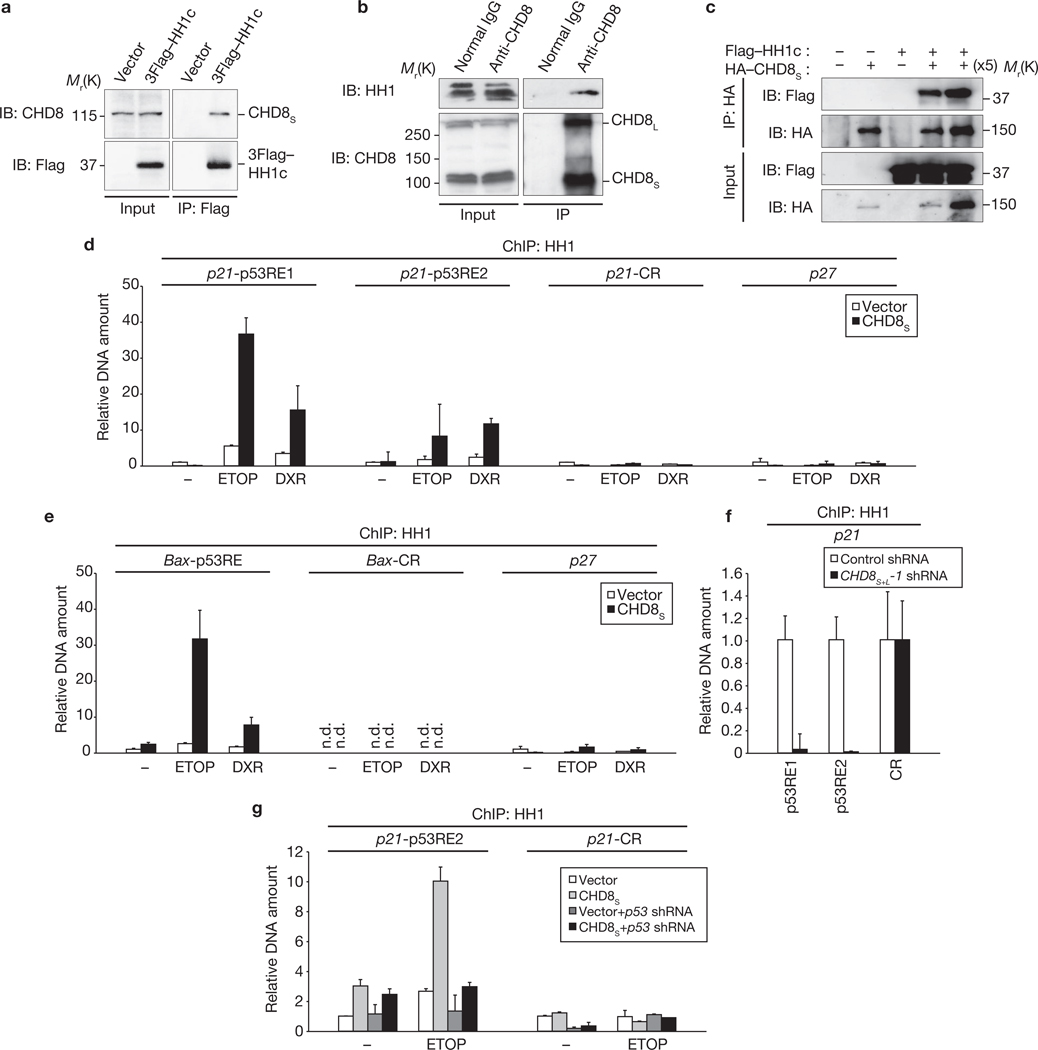
We next investigated the possible association of CHD8 and p53 with the promoter regions of p53 target genes using chromatin immunoprecipitation (ChIP; Supplementary Information, Fig. S6). ChIP using anti-CHD8 antibodies revealed that CHD8 specifically associated with p53-responsive elements 1 and 2 of the p21 promoter in U2OS cells treated with etoposide or doxorubicin but not in untreated cells (Fig. 3a). No substantial association of CHD8 with a control region of the p21 promoter that does not bind p53 or with the p27 promoter was detected. Moreover, depletion of p53 by RNAi reduced the extent of CHD8 association with the p53-responsive elements of the p21 promoter, suggesting that p53 mediates the association of CHD8 with chromatin. ChIP using anti-p53 antibodies revealed that CHD8S overexpression did not affect the association of p53 with chromatin in response to genotoxic stress (Fig. 3b), suggesting that p53 binds to the p21 promoter in a CHD8-independent manner. A sequential ChIP experiment revealed that p53 and CHD8 occupy the p21 promoter region simultaneously (Fig. 3c).
Figure 3.
CHD8 binds to the promoters of p53 target genes. (a) U2OS cells infected with a retroviral vector encoding p53 shRNA were incubated with the genotoxic agents etoposide (ETOP, 20 µM) and doxorubicin (DXR, 0.5 µM). The cells were then subjected to ChIP with an anti-CHD8 antibody and the precipitated DNA was quantified by real-time PCR with primers specific for p53-responsive elements (p53REs) 1 or 2 or a control region (CR) of the p21 promoter or for the p27 promoter. (b) U2OS cells overexpressing CHD8S were treated with ETOP (20 µM) and DXR (0.5 µM) and subjected to ChIP with an anti-p53 antibody. Data are mean ± s.d., n = 3 (a, b). (c) U2OS cells overexpressing CHD8S were incubated with ETOP and then subjected to ChIP with an anti-CHD8 antibody. The immunoprecipitates were subjected to ChIP with an anti-p53 antibody.
To elucidate the mechanism by which CHD8 inhibits p53 function, we attempted to identify molecules that associate with Flag–CHD8S in HEK293T cells using a ‘shotgun’ proteomics approach. The results of several independent experiments revealed that histone H1 was consistently the most prominent of the proteins associated with immunoprecipitated Flag–CHD8S (Supplementary Information, Table S1). Reciprocal co-immunoprecipitation analysis showed that Flag-tagged histone H1c interacted with endogenous CHD8 (Fig. 4a). We also confirmed that endogenous CHD8 associated with endogenous histone H1 (Fig. 4b). Furthermore, pulldown assays revealed that recombinant CHD8S and recombinant histone H1c bound to each other in vitro, suggesting that the interaction is direct (Fig. 4c). We examined which histone H1 subtypes interact with CHD8 in a co-immunoprecipitation assay. This analysis revealed that all five major H1 subtypes (H1a, H1b, H1c, H1d, H1e) interacted with CHD8S (Supplementary Information, Fig. S4c). In vitro binding assays with a series of deletion mutants showed that residues 500–600 is the region of CHD8S required for binding to histone H1 (Supplementary Information, Fig. S4d). The histone H1-binding domain thus includes the nuclear localization signal of CHD8S and is distinct from the chromodomain at the C terminus.
Figure 4.
CHD8 recruits histone H1 to the promoters of p53 target genes. (a) HEK293T cells expressing 3 × Flag–histone H1c were subjected to immunoprecipitation with anti-Flag and immunoblot analysis with anti-CHD8 antibodies. (b) Immunoprecipitation of HEK293T lysates with anti-CHD8 and immunoblot analysis with anti-histone H1 antibodies. (c) In vitro binding assay for recombinant CHD8S and histone H1c. (d) U2OS cells overexpressing CHD8S were treated with the genotoxic agents etoposide (ETOP, 20 µM) and doxorubicin (DXR, 0.5 µM) and subjected to ChIP with an anti-histone H1 antibody. (e) ChIP was performed for the BAX promoter (n.d., not detected). (f) U2OS cells infected with a retroviral vector for CHD8S+L-1 shRNA were subjected to ChIP. (g) U2OS cells overexpressing CHD8S were infected with a retroviral vector for p53 shRNA, incubated with ETOP and then subjected to ChIP. Data are mean ± s.d., n = 3 (d–g).
Given that CHD8 was found to interact with histone H1, we also examined whether histone H1 is recruited to the p53-responsive elements of the p21 promoter. ChIP using an anti–histone H1 antibody showed that histone H1 was indeed recruited to the p21 promoter on treatment of U2OS cells with etoposide or doxorubicin, and this effect was markedly enhanced by overexpression of CHD8S (Fig. 4d). Similar results were obtained with the promoter of the p53 target gene BAX (Fig. 4e). Conversely, depletion of either CHD8 or p53 markedly reduced the amount of histone H1 associated with the p21 promoter (Fig. 4f, g). This series of ChIP experiments thus suggests that a p53–CHD8–histone H1 complex forms at the promoters of p53-inducible genes in response to genotoxic stress.
p53–CHD8–histone H1 complex is necessary for suppression of p53 function
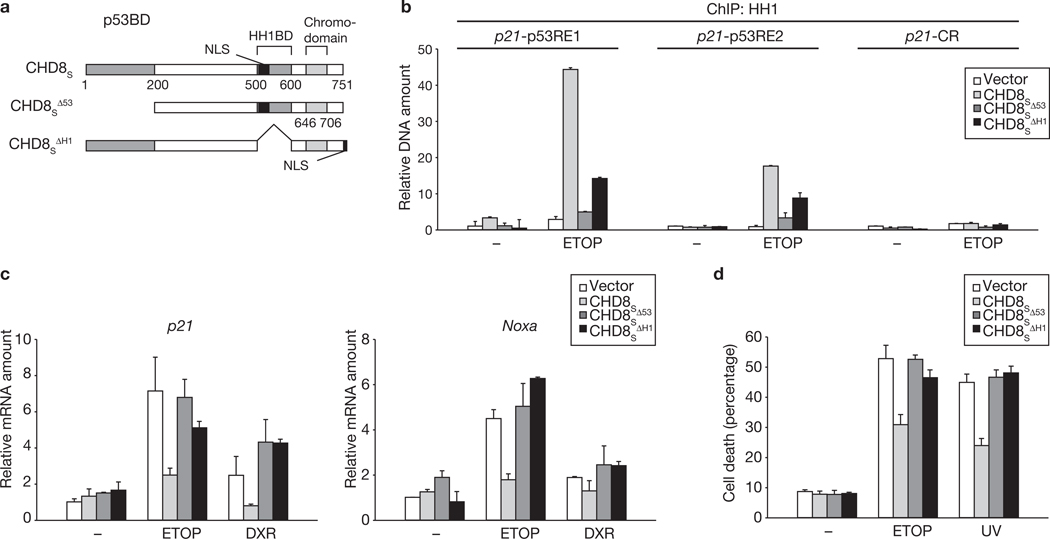
We next examined the ability of CHD8 mutants that lack either the p53 binding domain (CHD8SΔ53) or the histone H1 binding domain (CHD8SΔH1) to recruit histone H1 to the promoters of p53 target genes (Fig. 5a). Given that the CHD8SΔH1 mutant lacks residues 500–600, which include the nuclear localization signal, we added the nuclear localization signal of the large T antigen of simian virus 40 to the C terminus of the mutant protein. Deletion of either of the two binding domains markedly impaired the ability of CHD8S to recruit histone H1 to the p53-responsive elements of the p21 promoter (Fig. 5b). Furthermore, neither CHD8SΔ53 nor CHD8SΔH1 had an inhibitory effect on p53-dependent transactivation (Fig. 5c) or apoptosis (Fig. 5d). Together, these data suggest that CHD8 recruits histone H1 to p53-responsive elements, and that such recruitment of histone H1 results in suppression of transcriptional activation by p53.
Figure 5.
Interaction of CHD8 with p53 and histone H1 is necessary for recruitment of histone H1 to the promoters of p53 target genes. (a) Schematic representation of CHD8S derivatives. (b–d) U2OS cells overexpressing CHD8S derivatives were exposed to genotoxic stress (etoposide, ETOP, 20 µM and doxorubicin, DXR, 0.5 µM) and then subjected to ChIP (b), qRT–PCR (c) and trypan blue staining (d). Data are mean ± s.d., n = 3.
Histone H1 is essential for repression of p53-dependent transcription
Our observation that the CHD8SΔH1 mutant is unable to repress p53 function led us to investigate the requirement for histone H1 in such repression. To this end, we adopted three independent approaches. First, CHD8-dependent suppression of p53 function was examined in HH1c−/−HH1d−/−HH1e−/− triple-knockout (TKO) embryonic stem (ES) cells23. Expression of p21 and Noxa genes (Fig. 6a), as well as the level of apoptosis (Fig. 6b) induced by genotoxic stress, were markedly increased in TKO cells, compared with those in control cells. Second, U2OS cells were subjected to RNAi with a short hairpin RNA (shRNA) designed to deplete all five major subtypes of histone H1 (Fig. 6c). Such depletion of histone H1 resulted in a substantial increase in etoposide- or doxorubicin-induced upregulation of p21 or Noxa mRNA levels (Fig. 6d). Third, expression of a dominant-negative mutant of histone H1 (N-fusion) reduced the amount of endogenous histone H1, as described previously24, and also increased that of p21, resulting in suppression of cell proliferation (Supplementary Information, Fig. S7). Expression of a histone H1 mutant that does not have dominant-negative activity (C-fusion) did not affect the abundance of p21 or cell proliferation. Together, these three lines of evidence support our conclusion that CHD8 negatively regulates p53 function by recruiting histone H1 to the promoters of p53 target genes, resulting in suppression of apoptosis triggered by p53.
Figure 6.
Requirement for histone H1 in repression of p53-mediated transcription. (a, b) Wild-type (WT) or HH1c−/−HH1d−/−HH1e−/− triple-knockout (TKO) ES cells were incubated with etoposide (ETOP, µ2 M) and doxorubicin (DXR, 0.05 µM) and then subjected to qRT–PCR (a) and trypan blue staining (b). (c) U2OS cells were infected with a retroviral vector for histone H1 (HH1) shRNA and then subjected to immunoblot analysis. (d) The cells were incubated with ETOP (2 µM) and DXR (0.05 µM) and then subjected to qRT–PCR. Data are mean ± s.d., n = 3 (a, b, d).
Survival of Chd8−/−p53−/− embryos
The abundance of CHD8 mRNA or protein in mouse embryos was greater during early embryogenesis than at later embryonic stages or in newborns (Fig. 7a, b). In addition to mouse embryos, all human and mouse cancer cell lines tested in this study were found to express CHD8 at relatively high levels (Supplementary Information, Fig. S1c, e, f). Given that CHD8 is a negative regulator of p53, we reasoned that the early embryonic death of Chd8−/− mice may be attributable to unscheduled activation of p53. To test this hypothesis, we examined whether deletion of p53 ameliorated the abnormalities of Chd8−/− embryos. Most of the Chd8−/− mice died in utero between E5.5 and E7.5 and none survived beyond E8.5, whereas Chd8−/−p53−/− embryos survived until E10.5 (Supplementary Information, Table S2). Histopathological examination revealed that the growth retardation seen in Chd8−/− embryos was markedly less pronounced in Chd8−/−p53−/− and Chd8−/−p53+/− embryos at E7.5 and E8.5 (Fig. 7c). Cells with condensed nuclei were not seen in Chd8−/−p53−/− embryos at E8.5. The extent of recovery was greater in Chd8−/−p53−/− embryos than in Chd8−/−p53+/− embryos, suggesting that it was related to the reduction in the amount of p53. Mesoderm formation, a crucial event in early embryonic development that does not occur in Chd8−/− mice, was observed in Chd8−/−p53−/− mice (Fig. 7d). Chd8−/−p53−/− embryos died in utero at E10.5 and had severe haemorrhage, indicative of a defect in the cardiovascular system (M.N and K.I.N., unpublished data), suggesting that CHD8 also has another function (or functions) that is required for mid-stage embryonic development. We also observed growth of E3.5 embryos (blastocysts) in culture. Chd8−/− blastocysts degenerated between E5.5 and E7.5, whereas Chd8−/−p53−/− blastocysts, like wild-type controls, were alive at E9.5 (Fig. 7e). It is thus likely that the embryonic death of Chd8−/− mice is attributable to the unscheduled activation of p53-dependent apoptosis.
Figure 7.
Deletion of p53 rescues the phenotype of CHD8-deficient mice. (a) qRT–PCR for CHD8S and CHD8L mRNAs in mouse embryos at the indicated stages. Data are mean ± s.d., n = 3. (b) Immunoblot analysis of CHD8S in mouse embryos as well as in U2OS cells and those overexpressing (OE) CHD8S. (c) Histopathological examination of Chd8+/+p53+/+, Chd8−/−p53+/+, Chd8−/−p53+/− and Chd8−/−p53−/− embryos stained with haematoxylin and eosin. (d) Higher-magnification views of the boxed regions in c. Arrowheads indicate a layer of mesoderm. (e) In vitro culture of blastocysts of the indicated genotypes. Scale bars are 100 µm (c–e).
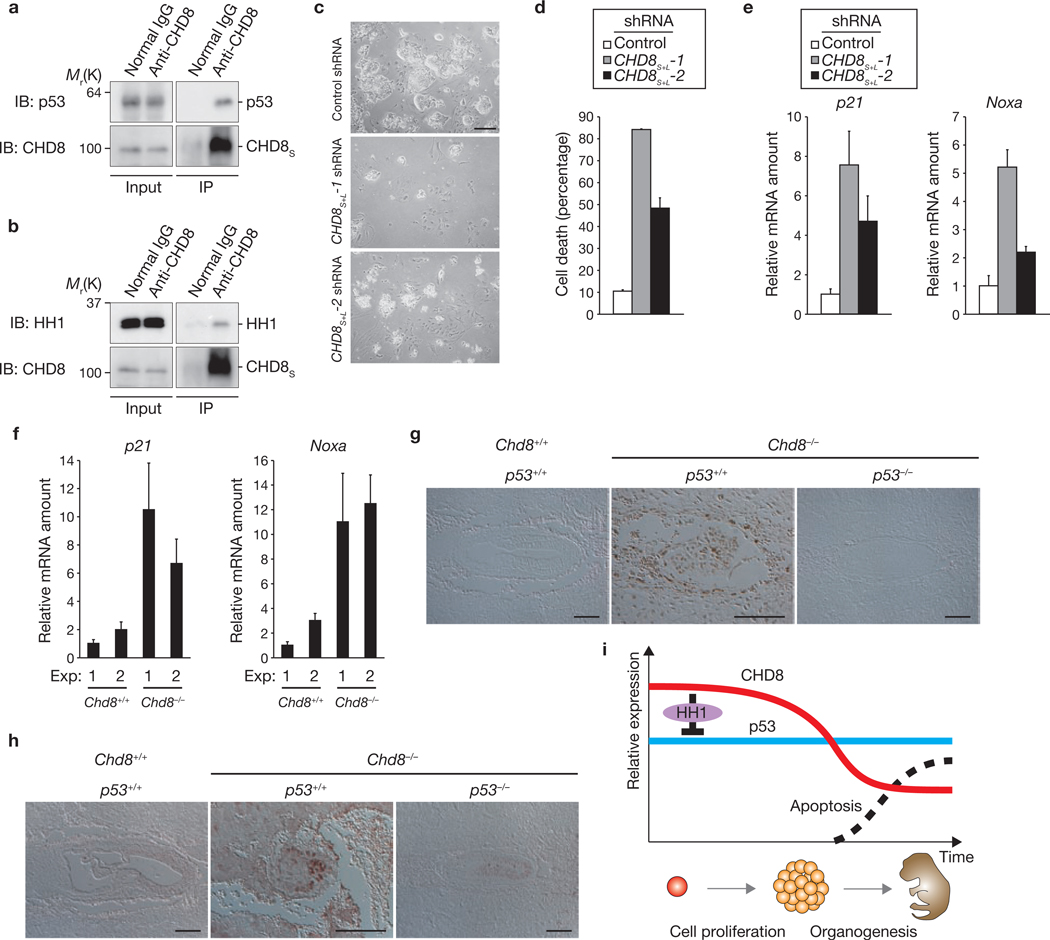
To investigate the physiological role of CHD8 during early embryogenesis, we examined embryos at various stages. In ES cells (equivalent to E3.5 embryos), endogenous CHD8 was shown to interact with both endogenous p53 (Fig. 8a) and endogenous histone H1 (Fig. 8b). Furthermore, RNAi-mediated depletion of CHD8 promoted both apoptosis (Fig. 8c, d) and expression of the p53 target genes p21 and Noxa (Fig. 8e). Cultured Chd8−/− blastocysts (equivalent to E5.5 embryos) also showed higher levels of p21 and Noxa mRNAs than did wild-type blastocysts (Fig. 8f). In E7.5 embryos, TUNEL assay showed substantial apoptosis in Chd8−/− embryos, whereas no apoptosis was observed in wild-type or Chd8−/−p53−/− embryos (Fig. 8g). Furthermore, Mdm2 expression was upregulated in Chd8−/− embryos in a p53-dependent manner (Fig. 8h). This genetic evidence supports the notion that CHD8 is a physiological antagonist of p53 in vivo, and that loss of CHD8 allows unrestrained p53 activity, which induces apoptosis at the early stage of embryonic development. We therefore propose that the biological role of CHD8 is to suppress unwanted apoptosis during early embryogenesis (Fig. 8i).
Figure 8.
CHD8 sets a threshold for induction of apoptosis during early embryogenesis by counteracting p53 function. (a, b) Immunoprecipitation of ES cell lysates with an anti-CHD8 antibody, and immunoblot analysis with either anti-p53 (a) or anti-histone H1 (b) antibodies. (c–e) ES cells infected with retroviral vectors for CHD8S+L-1 or CHD8S+L-2 shRNAs were subjected to trypan blue staining (c, quantification shown in d) and qRT–PCR (e). (f) Chd8+/+ or Chd8−/− blastocysts were subjected to qRT–PCR for p21 and Noxa. Data are mean ± s.d., n = 3 (d–f). (g, h) Sections of Chd8+/+p53+/+, Chd8−/−p53+/+ and Chd8−/−p53−/− embryos were subjected to TUNEL assay (g) or to immunohistochemistry (h) with anti-Mdm2 antibody. Scale bars are 100 µm (c, g, h). (i) Model for the biological role of CHD8. When cells proliferate extensively during early embryogenesis, CHD8 is expressed at high levels and suppresses p53 function through histone H1 recruitment to prevent unwanted apoptosis. Once the level of CHD8 expression decreases, during mid to late embryogenesis, some cells undergo apoptosis for organogenesis.
DISCUSSION
We have shown that CHD8 negatively regulates p53 function by recruiting histone H1 to the promoters of p53 target genes. Formation of the p53–CHD8–histone H1 complex requires expression of CHD8 and stabilization of p53 by genotoxic stress. CHD8 is preferentially expressed in embryonic tissues and in cancer cell lines that are thought to reflect the embryonic state. Loss of CHD8 induced hyperactivation of p53, resulting in apoptosis, which was prevented by depletion of p53. The biochemical and genetic evidence provided by our study may explain how p53 function is regulated by histone H1.
Histone modification is a fundamental mechanism for epigenetic control of gene expression. The role of core histones in gene regulation has been studied extensively, but that of the linker histone H1 has remained unclear. In vitro studies suggest that linker histone molecules influence chromatin structure, nucleosome mobility and gene regulation25– 32. Histone H1 is not essential for growth or cell division in several unicellular eukaryotes33–36, but ablation of three of eight genes that encode isoforms of histone H1 in mouse ES cells, resulting in a 50% reduction in histone H1 content, led to embryonic death of the mutant mice23,37. Such downregulation of histone H1 elicited marked changes in chromatin structure, including a global reduction in nucleosome spacing, local reduction in chromatin compaction and changes in the modification of core histones. Despite these changes, microarray analysis of the mutant ES cells revealed that expression of only a few genes was altered23. These observations suggest that histone H1 participates in the regulation of specific genes that are essential for survival.
In response to genotoxic stress, mammalian cells activate a complex network of proteins, a key element of which is p53. Post-translational modification of p53 results in transcriptional activation of its target genes1,6,7. Regulation of p53 by checkpoint signalling was originally thought to be mediated almost exclusively at the post-translational level1,6,8. More recently, p53 has also been found to be regulated at the transcriptional38 and translational39 levels. We have now uncovered another mode of p53 regulation mediated by CHD8-dependent recruitment of histone H1 to the promoters of its target genes.
Cells proliferate extensively with short G1 and G2 phases during early embryogenesis. The DNA replication checkpoint may therefore be readily activated at this time, with the consequent risk of inducing p53-dependent apoptosis. We propose that the biological role of CHD8 is to suppress unwanted apoptosis during early embryogenesis (Fig. 8i). Consistent with this notion, we found that the amounts of CHD8 mRNA and protein are higher during the early and mid phases of mouse embryonic development than in newborns. Genetic ablation of CHD8 results in extensive p53-dependent apoptosis in mouse embryos at a stage when Chd8 is expressed at high levels in wild-type mice. On the other hand, apoptosis is necessary for organogenesis, which occurs mainly during mid to late embryogenesis, when the level of Chd8 expression decreases. CHD8 may therefore regulate a threshold for apoptosis induction in a developmental-stage-specific manner: The threshold for apoptosis triggered by p53 is high during early embryogenesis, whereas it is lower after the mid-stage. In cancer cell lines, the level of CHD8 is relatively high, suggesting that cancer cells generally show an undifferentiated phenotype and reflect embryonic stages of development, or that cells expressing CHD8 may have a selective advantage in terms of cell growth or acquisition of immortality as a result of the suppressing p53 function. Thus we suggest that p53 function is, at least in part, suppressed by CHD8 in such cancer cell lines. Furthermore, haploinsufficiency at the CHD8 locus was recently implicated in the pathogenesis of a human developmental anomaly40, providing further genetic evidence in support of a crucial role for CHD8 in development.
METHODS
Antibodies
Anti-p53 (FL393, Pab240), anti-Mdm2 (SMP14) and anti-His6 (H15) antibodies were obtained from Santa Cruz Biotechnology; anti-caspase-3 cleaved at Asp-175 and anti-PARP antibodies were from Cell Signalling; anti-histone H1 antibodies were from Abcam; anti-p21 antibodies from BD Pharmingen; anti-Hsp70 and anti-Hsp90 antibodies from Transduction Laboratories; anti-HA (HA11) antibodies from Babco; anti-Myc (9E10) antibodies from Roche; anti-Flag (M2) from Sigma; anti-α-tubulin (TU01) antibodies from Zymed; anti- glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1D4) from Stressgen; and anti-CHD8 antibodies were generated by A. Kikuchi (Hiroshima University). Alexa 488- or Alexa 546-conjugated goat antibodies to mouse or rabbit IgG were obtained from Molecular Probes. Antibodies were used at a dilution of 1:2,000.
Plasmids
Complementary DNAs encoding wild-type or mutant forms of mouse CHD8S or human CHD8L, each tagged at its N terminus with Flag, Myc or HA epitopes, were subcloned into pcDNA3 (Invitrogen). Complementary DNAs encoding wild-type or mutant forms of human p53, each tagged at its N terminus with the HA epitope, were subcloned into pCGN. Complementary DNAs encoding mouse histone H1 variants, each tagged at its N terminus with three copies of the Flag epitope, were subcloned into pcDNA3. His6-tagged proteins were expressed in Escherichia coli strain BL21(DE3)pLys(S) (Novagen).
Cell culture, transfection and infection
HEK293T, HeLa, HCT116, U2OS and SaOS2 cells were cultured in an atmosphere of 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). NIH 3T3 cells were cultured under the same conditions in DMEM supplemented with 10% bovine serum (Invitrogen). MEFs and ES cells were cultured as described previously41,42. HEK293T, HeLa and SaOS2 cells were transfected with vectors using FuGENE6 (Roche). For retroviral infection, cDNAs were subcloned into pMX-puro.
Immunoprecipitation and immunoblot analysis
Cell lysis, immunoprecipitation and immunoblot analysis were performed as described previously43. For solubilization of chromatin-bound histone H1, cells were suspended in a solution containing 50 mM Tris-HCl (pH 7.5), 0.5% Triton X-100, 300 mM NaCl, 60 mM MgCl2, 10 mM CaCl2 and DNase I (167 U ml−1, Roche) and then incubated at 30 °C for 50 min.
Induction of apoptosis
Cells were incubated with etoposide (50 µM; Sigma) for 24 h, with cycloheximide (100 µg ml−1; Wako) for 24 h, with staurosporine (1 µM; Sigma) for 5 h or with doxorubicin (0.5 µM or the indicated concentrations; Sigma) for 24 h. Cells were exposed to UV radiation (50 J/m2) with an ultraviolet crosslinker UVC500 (Hoefer) and examined after 12 h.
RNAi
The retroviral vector for expression of shRNAs was described previously43. The hairpin sequences specific for human or mouse CHD8 (CHD8S+L-1, CHD8S+L-2, CHD8L-1, CHD8L-2), for human p53, for human histone H1A (also effective for H1B, H1C, H1D and H1E, given the high similarity of the target region) and for enhanced green fluorescent protein (EGFP, Clontech) mRNAs corresponded to nucleotides 138–158 (CHD8S+L-1), 814–834 (CHD8S+L-2), 3808–3828 (CHD8L-1), 4413–4433 (CHD8L-2), 775–793 (p53), 262–282 (histone H1A) and 126 –146 (EGFP) of the respective coding regions. The resulting vectors were used to transfect Plat E cells and thereby to generate recombinant retroviruses.
Protein identification by LC–MS/MS analysis
CHD8S-associated proteins were digested with Achromobacter protease I and the resulting peptides were analysed with a nanoscale liquid chromatography–tandem mass spectrometry (LC–MS/MS) system as described previously44.
Reverse transcription–polymerase chain reaction (RT–PCR) and luciferase assays
RT–PCR analysis and luciferase assays were performed as described previously45. Purification of mRNA from cultured blastocysts was performed with a TurboCapture mRNA kit (Qiagen). The primer sequences for RT–PCR are listed in Supplementary Information, Table S3. For luciferase assays, SaOS2 cells were transfected using FuGENE6 with expression vectors encoding human p53 or Flag-tagged wild-type or mutant versions of mouse CHD8S together with pRLTk (Promega) as an internal control and luciferase reporter plasmids containing promoter sequences of PG13 or human p21 (wild-type or mutant) genes. The cells were collected 24 h after transfection, lysed and assayed for luciferase activity with a dual-luciferase reporter assay system (Promega).
ChIP
ChIP assays were performed with a ChIP assay kit (Upstate Biotechnology) and 106 cells for each reaction. Precipitated DNA was quantified by real-time PCR as described previously45. The primer sequences are listed in Supplementary Information, Table S3.
Generation of Chd8−/−p53−/− mice, histopathology and culture of pre-implantation embryos
Chd8+/− mice generated in our laboratory22 were crossed with p53−/− mice (Taconic Biotechnology). All mice used in this study were backcrossed to the C57BL/6 background for more than six generations. Histopathological analysis, TUNEL assay, immunohistochemistry and culture of pre-implantation embryos were performed as described previously22. The primer sequences for genotyping of embryos by nested PCR are listed in Supplementary Information, Table S3. All animal experiments were performed in accordance with institutional guidelines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Kitamura for pMX-puro; J. M. Cunningham and K. Hanada for the mCAT-1 plasmid; S. Miyake for the PG13-, p21-, and p21 mutant–Luc plasmids; T. Takemori for the NF-κB Luc plasmid; F. Ishikawa and R. Funayama for the N-fusion and C-fusion plasmids; M. Kitagawa for HCT116 and SaOS2 cells; M. Sato, Y. Yamada, T. Moroishi, Y. Katayama, N. Nishimura and K. Oyamada for technical assistance; M. Kimura and A. Ohta for help with preparation of the manuscript; and T. Ushijima, K. Hayashi and members of the authors’ laboratories for discussion. K.I.N. was supported by Takeda Science Foundation. Y.F. and A.I.S. were supported by NIH grant CA79057. Y.F. is a GCC Cancer Scholar supported by Georgia Cancer Coalition.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
M.N. performed and planned all experiments, except some of ChIP and co-immunoprecipitation experiments, which were performed by K.O., Y.T. and T. Nakagawa; K.I.N. coordinated the study, oversaw the results and wrote the manuscript; S.I. and T. Natsume contributed to proteomic analysis; Y.F. and A.I.S. provided histone H1 triple-knockout cells and many suggestions; A.K. provided antibodies to CHD8. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nature Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 5.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 7.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Das S, et al. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee D, et al. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 12.Kim K, et al. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J. Biol. Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker PB. Nucleosome remodelers on track. Nature Struct. Mol. Biol. 2005;12:732–733. doi: 10.1038/nsmb0905-732. [DOI] [PubMed] [Google Scholar]
- 14.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat. Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 16.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Schroeder S, Fong N, Bentley DL. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 2005;24:2379–2390. doi: 10.1038/sj.emboj.7600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 19.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nature Struct. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto I, et al. A novel β-catenin-binding protein inhibits β-catenin-dependent Tcf activation and axis formation. J. Biol. Chem. 2000;275:32871–32878. doi: 10.1074/jbc.M004089200. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Nishiyama M, et al. Early embryonic death in mice lacking the β-catenin-binding protein Duplin. Mol. Cell. Biol. 2004;24:8386–8394. doi: 10.1128/MCB.24.19.8386-8394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Funayama R, Saito M, Tanobe H, Ishikawa F. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006;175:869–880. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vignali M, Workman JL. Location and function of linker histones. Nature Struct. Biol. 1998;5:1025–1028. doi: 10.1038/4133. [DOI] [PubMed] [Google Scholar]
- 26.Thomas JO. Histone H1: location and role. Curr. Opin. Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 27.Lusser A, Kadonaga JT. Strategies for the reconstitution of chromatin. Nature Methods. 2004;1:19–26. doi: 10.1038/nmeth709. [DOI] [PubMed] [Google Scholar]
- 28.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pennings S, Meersseman G, Bradbury EM. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl Acad. Sci. USA. 1994;91:10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimamura A, Sapp M, Rodriguez-Campos A, Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol. Cell. Biol. 1989;9:5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 33.Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 34.Ushinsky SC, et al. Histone H1 in Saccharomyces cerevisiae. Yeast. 1997;13:151–161. doi: 10.1002/(SICI)1097-0061(199702)13:2<151::AID-YEA94>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- 36.Ramon A, Muro-Pastor MI, Scazzocchio C, Gonzalez R. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol. Microbiol. 2000;35:223–233. doi: 10.1046/j.1365-2958.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 37.Rupp RA, Becker PB. Gene regulation by histone H1: new links to DNA methylation. Cell. 2005;123:1178–1179. doi: 10.1016/j.cell.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, El-Deiry WS. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res. 2006;66:6982–6989. doi: 10.1158/0008-5472.CAN-06-0511. [DOI] [PubMed] [Google Scholar]
- 39.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 40.Zahir F, et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J. Med. Genet. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakayama K, et al. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamura T, et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natsume T, et al. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 2002;74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- 45.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.