Abstract
The unique spectral properties of semiconductor quantum dots (QDs) enable long-term live-cell imaging and ultrasensitive detection of viral particles, which in turn can potentially provide a practical means for detailed analysis of the underlying molecular mechanisms of virus entry. In this study, we report a general method of labeling adeno-associated virus serotype 2 (AAV2) with QDs for enhanced visualization of the intracellular behavior of viruses in living target cells. It was found that the mild condition required for this QD conjugation reaction allowed for the retention of viral infectivity of AAV2. Furthermore, quantitative analysis of viral motility in living cells suggested that QD-labeling had no significant effect on the intracellular transport properties of AAV2 particles compared to those of conventional organic dye-labeled AAV2. Our imaging study demonstrated that QD-AAV2 was internalized mainly through a clathrin-dependent pathway and then trafficked through various endosomes. It was also observed that QD-AAV2 particles exploit the cytoskeleton network to facilitate their transport within cells, and the labeling study provided evidence that the ubiquitin-proteasome system was likely involved in the intracellular trafficking of AAV2, at least at the level of nuclear transport. Taken together, our findings reveal the potential of this QD-labeling method for monitoring the intracellular dynamics of virus-host cell interactions and interrogating the molecular mechanisms of viral infection in greater detail.
Keywords: quantum dot, adeno-associated virus, single virus tracking, intracellular trafficking, bionanotechnology, nanomedicine
During viral entry, viruses interact with various cellular structures and use them to optimize the delivery of the viral genome to the nucleus to promote efficient viral replication.1–4 Understanding virus trafficking requires elucidating the interactions between the virus and the target cell, including the cellular proteins involved and the viral infection routes, thus it can aid in the development of novel designs of therapeutic targets for controlling virus-triggered diseases, as well as crucial insights for improving virus-mediated gene delivery.5–6 Traditionally, to track single viruses in live cells, organic fluorophores are used to label and detect individual viruses.7–10 However, one key concern for organic dyes is their ability to fluorescently label viruses in a way so that they are bright enough to be detected and tracked. In addition, problems with metabolic degradation, or photobleaching of dyes, which is of great concern in confocal imaging, have limited their utility for long-term imaging of biological processes.6,11 The use of quantum dots (QDs), which can offer remarkable brightness and photostability and possess a wide absorption spectrum and a narrow emission spectrum,12–15 can potentially mitigate these concerns and allow for the development of improved techniques for detecting and studying viruses.
The adeno-associated virus (AAV), a small non-enveloped virus that belongs to the family of parvoviruses, has attracted considerable interest because it shows great promise for use in human gene therapy due to their non-pathogenicity and diverse tissue tropism with different AAV serotypes.16–18 However, the parameters governing the intracellular fate of AAV, even for the relatively well-characterized serotype 2 (AAV2), remain poorly understood. Although amine-reactive dyes have been employed to label AAVs,10,19–20 one challenge in single-virus tracking of AAV is the small size of these viral particles (~20 nm in diameter), which limits the number of fluorescent molecules that can be attached on the virus surface without causing a self-quenching effect or affecting viral infectivity.6,19 Utilization of probes with greater photostability and sensitivity can further facilitate detailed trafficking studies, which are characteristics that QDs can provide.14–15 As compared to the conventional organic dyes, labeling AAVs with the much brighter QDs can potentially offer a means to detect viral particles with a much lower load of the labeling molecules, which can ensure a minimal disturbance of virus-host interaction during virus trafficking.
We have previously developed a general method to label enveloped viruses with QDs.21 Enveloped viruses are surrounded by a lipid bilayer membrane, which is obtained from the host cell membrane during the budding process. Since circumstances that disturb membrane will also affect the envelope for these viruses, more gentle and non-disruptive labeling strategies are needed. Therefore, our strategy to label enveloped viruses with QDs was to incorporate a small tag onto the surface of viruses through the natural budding process. However, this labeling scheme is not applicable to non-enveloped viruses. One strategy to label non-enveloped viruses is through the genetic insertion of functional motifs, although this method can reduce viral production and infectivity.22–23
In this report, we investigated a general strategy for linking non-enveloped AAV2 viruses with QDs through a carbodiimide coupling reaction (Figure 1A). AAV2 virions are thought to be transported into the nucleus before viral uncoating occurs,19,24 and it is generally believed that non-enveloped viruses lacking a lipid bilayer membrane such as AAV2 are structurally more robust than enveloped viruses. Hence, it is conceivable that the capsid of purified AAV2 can be targeted for labeling by QDs to track the viral particle movements within the target cell. Our present study demonstrates the potential of this QD-labeling method for monitoring the dynamic interactions between AAV2 and the target cell structures in greater detail. This technique could also be readily adapted to label other types of non-enveloped viruses for use in virus trafficking studies.
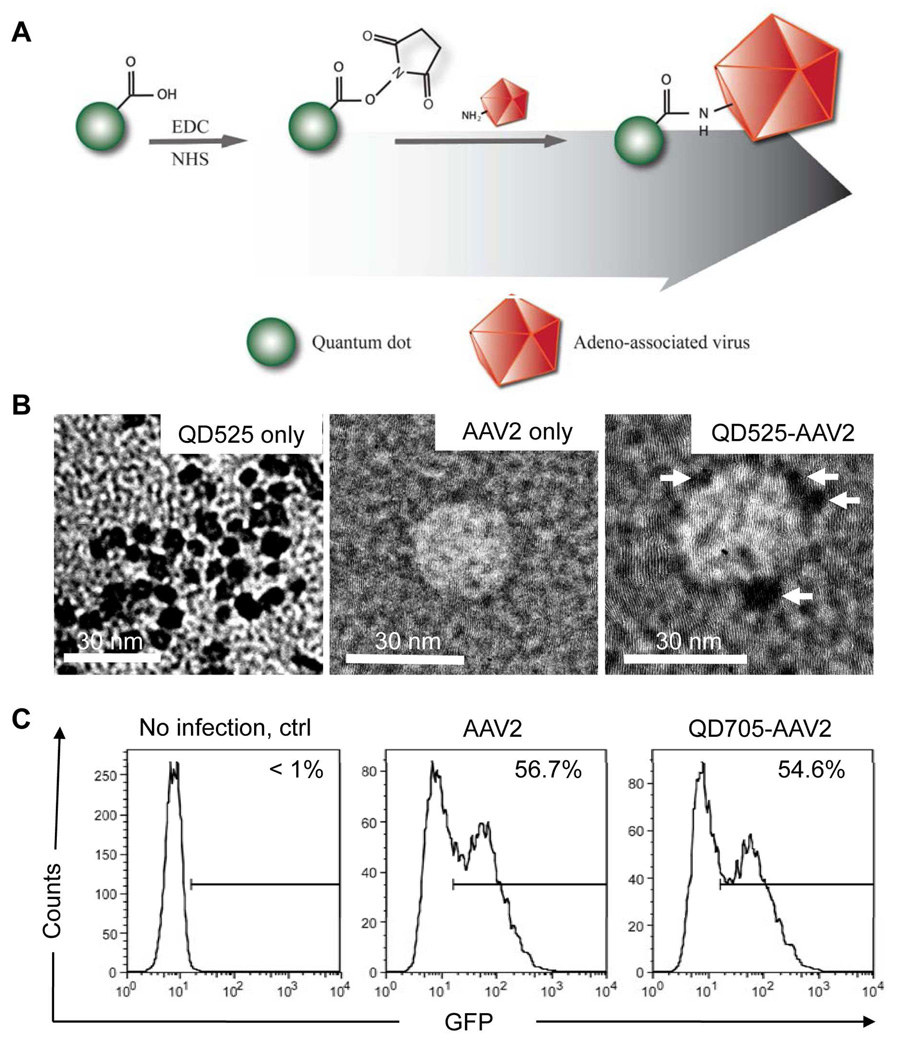
Figure 1.
Covalent attachment of QDs on AAV2 and characterization of QD-AAV2 conjugates. (A) QD-AAV2 networks are generated by an amide-bond formation between the carboxylic source on QDs and the primary amines from lysine residues on the AAV capsid via the carbodiimide chemistry. (B) Transmission electron microscope (TEM) images of unconjugated QD525 only (left), AAV2 only (middle) and QD525-labeled AAV2 (right). Arrows indicate QDs attached on AAV2. Scale bars represent 30 nm. (C) HeLa cells (1 × 105) were spin-infected with unlabeled (AAV2) or QD705-labeled viruses (QD705-AAV2). The resulting GFP expression was analyzed by flow cytometry.
RESULTS AND DISCUSSION
Our strategy to label adeno-associated virus (AAV) with QDs utilizes a coupling reaction that operates under mild conditions, as illustrated in Figure 1A. QD-AAV2 networks were produced by covalent amide bonds formed via carbodiimide chemistry between the carboxylic moieties on the QDs and the primary amines from the lysine residues on the viral capsid protein. The carboxyl QDs were incubated with 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) to modify the carboxyl group into an amine-reactive NHS ester. Next, the excess EDC and NHS were removed by a gel filtration column and AAV2 was added to provide the primary amine source to initiate the coupling reaction. We first performed assays to examine whether QDs could be conjugated to the surface of AAV2. Transmission electron microscope (TEM) analysis showed that the size of individual QDs was approximately 6–7 nm in diameter by TEM (Figure 1B, left). The TEM images also suggested that around 2 to 4 of QDs were detected onto a single AAV2 particle (Figure1B, right). In addition, the number of QDs linked to AAV2 was also determined by measuring the spectroscopic property of the purified QD-labeled AAV2. The measurement indicated an average dye-to-protein ratio (QDs/AAV2 molar ratio) of approximately 2.2. Furthermore, the hydrodynamic radius of QDs, AAV2, and QD-AAV2 in aqueous solutions were measured by the dynamic light scattering (DLS), and the results revealed that there was an increase in the mean radius of QD-AAV2 (69.9 nm) compared to that of AAV2 (34.4 nm), as expected, suggesting that significant aggregation is negligible in QD-AAV2 networks (See Supporting information, Figure S1). It is noteworthy that the value of the hydrodynamic radius is generally larger than the size detected by TEM due to the influence of the counterion cloud on the particle mobility25 and especially, the intrinsic aggregation tendency of AAV particles in aqueous solutions.26–27 A fluorescence measurement showed that there was a slight red-shift (1 nm) of the emission wavelength for QD-AAV2 in comparison with that of QDs alone (Figure S2B), which has also been observed in QD-encapsulated/attached plant viruses.28–29 We further confirmed that the QDs were coupled to AAV2 by overlaying the particle solution onto coverslips and immunostaining the adhered viral particles with an antibody specific for intact AAV2. The fluorescence signal of the QDs on the viral surface was readily detected and most of the QD signals (>75%) were colocalized with the signals generated by the anti-AAV2 antibody staining, which was confirmed by calculating centroid-to-centroid distance (approximated 9 ~ 30 nm) between red (AAV2) and green (QDs) signals (Figure S2A). Next, to examine whether the QD-labeled viruses remained infectious, unlabeled or QD705-labeled AAV2 carrying a GFP reporter gene were used to infect HeLa cells. It was also found that a similar level of transduction efficiency as measured by the expression of GFP reporter gene using flow cytometry (Figure 1C) and integrated mean fluorescence intensity (iMFI) that reflects the total intensity of GFP signals from the virus transduced cells (Figure S2C) was obtained for the labeled and unlabeled viruses, suggesting that this QD-labeling strategy had a minimal effect on the viral infectivity.
To examine the photostability of QD-labeled viruses compared to that of fluorescent dye-labeled viruses, QD705- or fluorescein isothiocyanate (FITC)-labeled AAV2 were co-incubated with HeLa cells for 30 min at 37°C. The cells were then fixed and illuminated continuously with a laser at 491 nm (50 mW) to excite the QD705 and FITC. As shown in Figures S3A and S3B and Movie S1 in supporting information, the dye-labeled AAV2 showed marked photobleaching, whereas the QD-labeled AAV2 retained its fluorescent signal during the imaging, confirming that the photostable QDs are better fluorescent probes for long-term and real-time imaging of single AAV2 viruses in living cells.
For a multicolor real-time imaging study, it is always desirable to acquire images with exposure times as short as possible to obtain sufficiently high time resolution to monitor the detailed dynamics of virus trafficking in living cells. However, AAV2 virions are very small, limiting the number of fluorescent dyes that can be attached on a single virus without affecting viral infectivity, which sets an upper limit to the exposure time needed to detect the fluorescent signal emitted from such a tiny virus. Therefore, we compared the detection sensitivity of the QD-labeled AAV2 particles with dye-labeled AAV2 by acquiring images of the same region of cells with different exposure times. As expected, the QD signal was much brighter and could be detected at a shorter exposure time (Figure S3C). Thus, the use of QDs can potentially present advantages for fast multicolor time-lapse imaging due to their remarkable brightness and sensitivity as compared with conventional organic fluorophores.
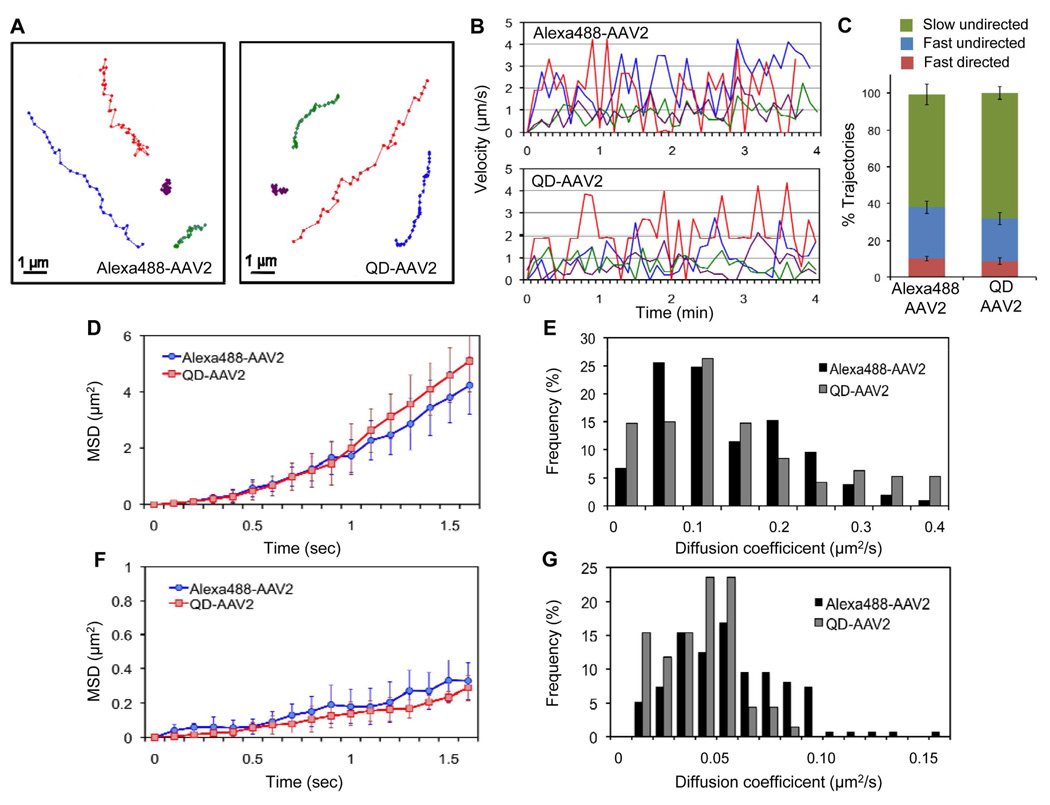
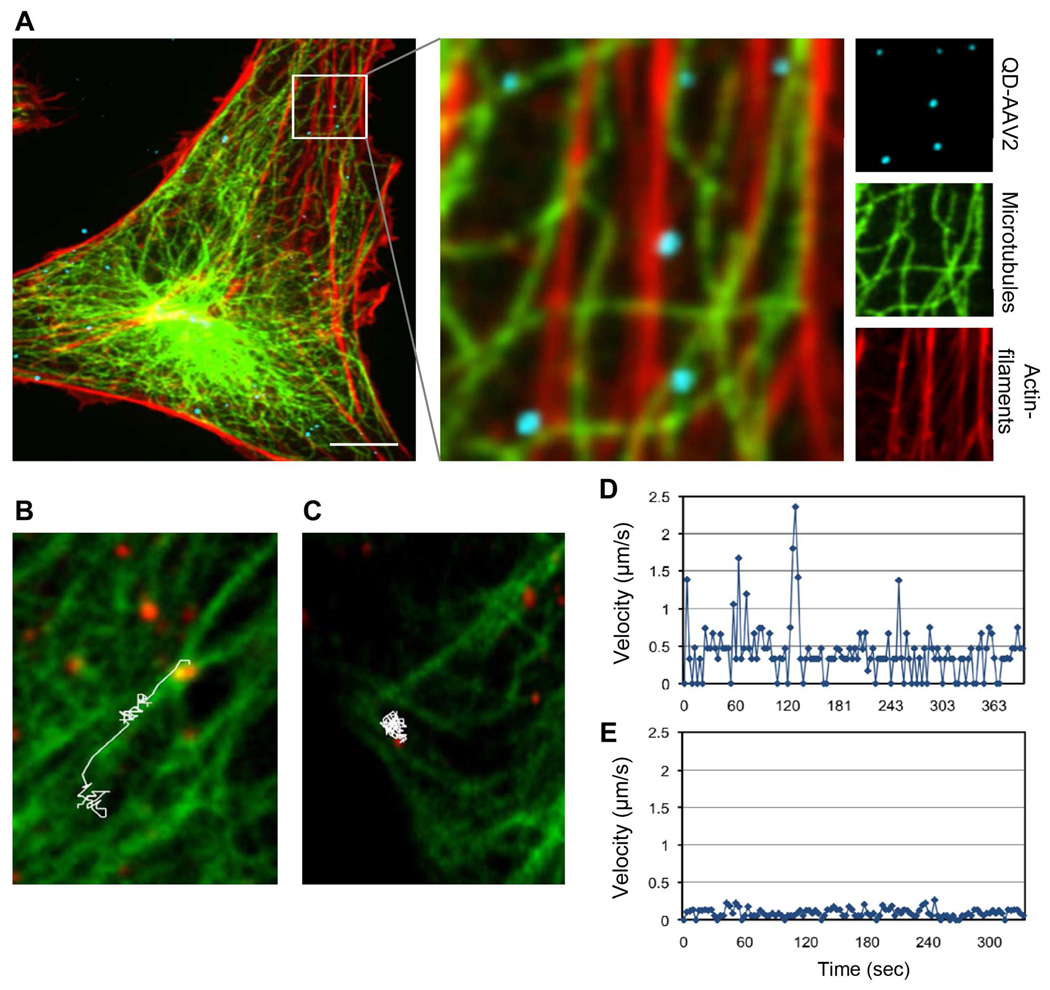
Although QDs hold several advantages over conventional organic dyes, it is important to determine whether QD conjugation to biomolecules and biological particles alters their intracellular transport properties.30–32 To investigate whether the intracellular viral motility can be affected by QD-labeling, the movements of individual labeled particles were monitored in living cells by real-time imaging, and viruses labeled with a fluorescent dye, which has been commonly used for labeling non-enveloped viruses, were used for comparison. Various types of intracellular movements of viral particles were detected during imaging, and representative trajectories of QD- or Alexa488-labeled AAV2 in HeLa cells are shown in Figure 2A. Based on the two dimensional data analysis of the three dimensional AAV2 particle trafficking, we found that many particles exhibited relatively slow movement (e.g. the purple and green trajectories in Figure 2A and 2B), while some particles showed fast and directed transport (e.g. the red and blue trajectories in Figure 2A and 2B). No significant difference in viral speed and movement patterns was observed between Alexa488- or QD-labeled AAV2. For a more detailed characterization of their intracellular motility, we tracked a large number of viral particles within the cytoplasm and analyzed their mean-square displacement (MSD) and diffusion coefficient. We observed diverse types of viral movements. In our analysis, the trajectories having a peak speed ≥ 2µm/s and containing uni-directional movement in more than 5 consecutive frames were defined as directed transport, trajectories with fast (≥ 2µm/s) but undirectional motions were defined as fast undirected transport, and trajectories with slow (≤ 2µm/s) and undirectional motions were defined as slow undirected transport. Using these definitions, 10.1% of Alexa-AAV2 trajectories and 8.7% of QD-AAV2 trajectories were fast and directed, 27.6% of Alexa-AAV2 trajectories and 22.8% of QD-AAV2 trajectories were fast but undirected, whereas the remaining were slow undirected transport (Figure 2C). The directed transport subgroup of Alexa488- and QD-labeled AAV2, which exhibited a parabolic shape of MSD curve with time and likely involved microtubule-dependent movement,19 showed similar patterns for MSD and diffusion coefficient distribution (Figure 2D, 2E, and S4A). On the other hand, the undirected subgroup including slow and fast motions showed a linear dependency of MSD with time indicating normal diffusion, and no significant distinction in MSD and diffusion coefficient distribution between Alexa488- or QD-labeled AAV2 (Figure 2F, 2G, and S4B). Thus, these experiments suggest that the intracellular transport properties of AAV2 are not significantly altered by the QD conjugation.
Figure 2.
Quantitative analysis of the intracellular viral motility of QD-labeled and Alexa488-labeled AAV2. (A, B) Representative trajectories of Alexa488- and QD-labeled AAV2 trafficking in HeLa cells. HeLa cells were incubated with Alexa488-labeled or QD-labeled AAV2 for 10 min at 37°C, after which confocal time-lapse images were then recorded. Typical trajectories for Alexa488-AAV2 and QD-labeled AAV2 are presented (A), and the time trajectories of the viral velocity are shown (B). (C) The fraction of trajectories of Alexa488-AAV2 and QD-AAV2 that are categorized as “slow undirected”, “fast undirected”, and “fast directed” as defined in the text. Error bars represent the standard deviation of the mean from triplicate experiments (total 595 trajectories). (D to G) Mean-square displacement (MSD) and diffusion coefficient analysis of the directed transport subgroup and the undirected transport subgroup. A large number of viral particles within the cytoplasm (~50 trajectories, ~900 frames) were tracked and further analyzed. MSD versus time for the directed transport (D) and for the undirected transport subgroup (F) were plotted. The diffusion coefficients of the directed transport (E) and the undirected transport (G) subgroups were calculated from the slope of the MSD plot.
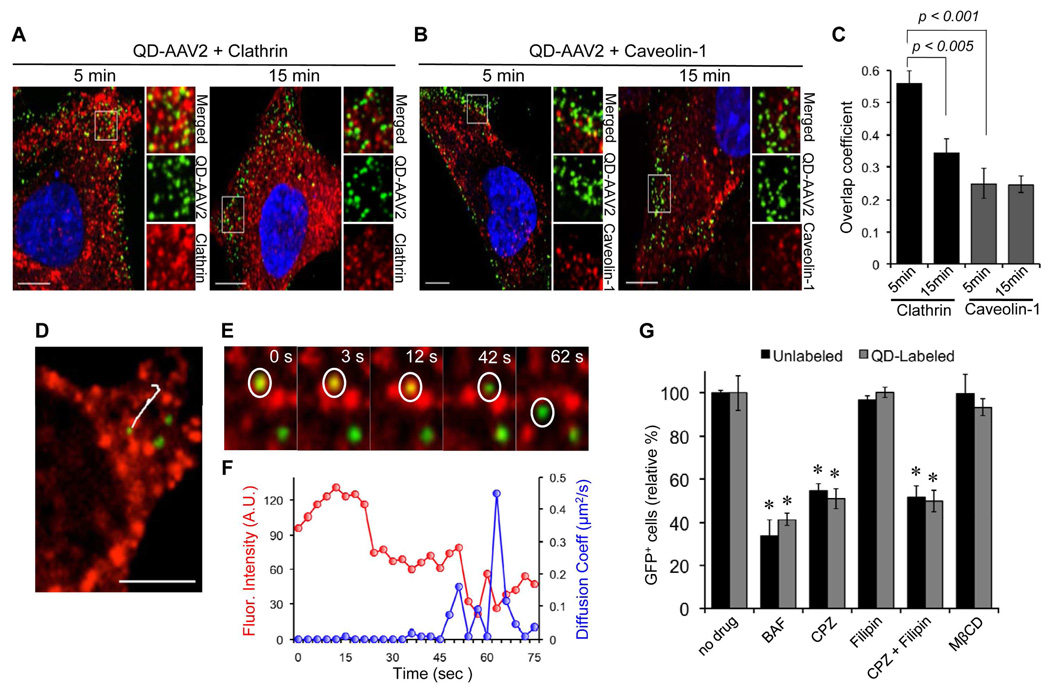
In the subsequent series of investigations, we conducted experiments to demonstrate the utility of this QD-labeling method for trafficking studies of AAV2. It has been generally believed that AAV2 enters cells through clathrin-coated pits in a dynamin-dependent manner.33 To confirm the role of clathrin- or caveolin-mediated endocytosis in the entry of AAV2, we visualized the individual viral particles and endocytic structures (clathrin or caveolin) in target cells after 5 or 15 min of incubation at 37°C. As shown in Figure 3A and 3C, a significant level of colocalization (56% ± 4.0%) of AAV2 particles with discrete clathrin structures was detected. After incubation for 15 min, less colocalization of AAV2 with the clathrin structures were observed (33.4% ± 4.4%), suggesting that many of the viruses had already been dissociated from uncoated clathrin vesicles and had likely been transported to early endosomes. Although some particles were overlaid with caveolin structures, a marked level of colocalization was not observed for both 5 and 15 min incubations (< 25%) (Figure 3B and 3C). These imaging results were further confirmed by drug-inhibition assays (Figure 3G). Chlorpromazine is a drug known to prevent clathrin polymerization and inhibit internalization mediated by clathrin-coated vesicles,34 while filipin is a cholesterol-binding reagent that blocks caveolin-dependent internalization.35 It was found that 10 µg/ml chlorpromazine could significantly inhibit AAV2 infection of 293T cells, whereas filipin (5 µg/ml) exhibited no inhibitory effect, confirming that the entry of AAV2 is mediated by clathrin-dependent endocytosis, and that caveolin might not be involved in the productive infection process. Another experiment revealed that AAV2 entry is dependent on low pH-associated endosomal processes, as a significant inhibition of infection was seen when the cells were treated with bafilomycin A1 (BAF) (Figure 3G), a specific inhibitor of vacuolar proton ATPases.36 It also appeared that cholesterol on the target cell membrane is not associated with the AAV2 infection pathway because the depletion of cholesterol with methyl-beta-cyclodextrin (MβCD) had little impact on the infectivity of AAV2 (Figure 3G).37–38 No significant difference in inhibitory effect of drugs on virus infection was observed between unlabeled and QD-labeled AAV2, suggesting that the infection pathway of AAV2 is not significantly altered by the QD conjugation.
Figure 3.
Clathrin/Caveolin-dependent entry of AAV2. (A, B) HeLa cells were incubated with QD-labeled AAV2 for 30 min at 4°C to synchronize infection. The cells were then shifted to 37°C for 5 or 15 min, fixed, permeabilized, and immunostained with anti-clathrin (A) or anti-caveolin-1 antibody (B). The boxed regions are enlarged in the right panels. Scale bar represents 5 µm. (C) Quantification of QD-AAV2 colocalized with clathrin or caveolin-1 signals after 5 or 15 min of incubation. Colocalization coefficients were calculated using the Manders' overlap coefficient by viewing more than 10 cells at each time point using the Nikon NIS-Elements software. Error bars represent the standard deviation of the mean from analysis of multiple images. (D to F) Real-time monitoring of QD-AAV2 internalization through the clathrin-coated pit. HeLa cells seeded on a glass bottom dish were transiently transfected with mRFP-clathrin (red). At 24 h post-transfection, the cells were incubated with QD-labeled AAV2 (green) for 30 min at 4°C, and were then warmed to 37°C to initiate virus internalization. Confocal time-lapse images were then recorded. A representative trajectory of QD-AAV2 in HeLa cells expressing mRFP-clathrin (D) and the selected frames of the real-time imaging (E) are presented. (F) Kinetics of the fluorescence intensity of mRFP-clathrin (red) and diffusion coefficients (blue) of AAV2 particle indicated by the white circle is shown. Scale bar represents 2 µm. (G) Inhibition of low pH-dependent endosomal processing by bafilomycin A1 (BAF, 50 nM), clathrin-dependent internalization by chlorpromazine (CPZ, 25 µg/ml), caveolin-dependent internalization by filipin (5 µg/ml), or depletion of cholesterol by methyl-beta-cyclodextrin (MβCD, 15 mM). HeLa cells were preincubated with the indicated drugs at 37°C for 30 min except for MβCD (15 min). The cells (1 × 105) were spin-infected with unlabeled or QD-labeled AAV2 in the presence of drugs (BAF, CPZ, and filipin) or in the absence of drug (MβCD). After an additional 3 h of incubation with drugs (BAF, CPZ, and filipin) at 37°C, the cells were washed with PBS and replenished with fresh media. The percentage of GFP-positive cells was analyzed by flow cytometry. Error bars represent the standard deviation of the mean from triplicate experiments. Asterisk indicates comparison to the no drug treatment group (* p<0.05).
To monitor the interactions between AAV2 and the clathrin structures in real-time, QD-labeled viruses were incubated with HeLa cells expressing clathrin fused with red fluorescent protein (mRFP-clathrin), followed by live-cell imaging was conducted using time-lapse spinning confocal microscopy (See Supporting information, Movie S2). A representative trajectory of QD-AAV2 and selected images obtained from the time series are shown in Figure 3D and 3E. The viral particle (green) was initially colocalized with the clathrin signal (red). This colocalization was maintained for ~45 s, and the clathrin signal then rapidly disappeared, suggesting that the virus was dissociated from the uncoated clathrin vesicle (Figure 3E and 3F). While the particle remained colocalized with the clathrin signal, the instantaneous diffusion coefficients of the viral particle were significantly lower compared to those after dissociation from the clathrin signal (Figure 3F), suggesting that the viral particle entered through the confined region of the clathrin-coated pit into the cell.
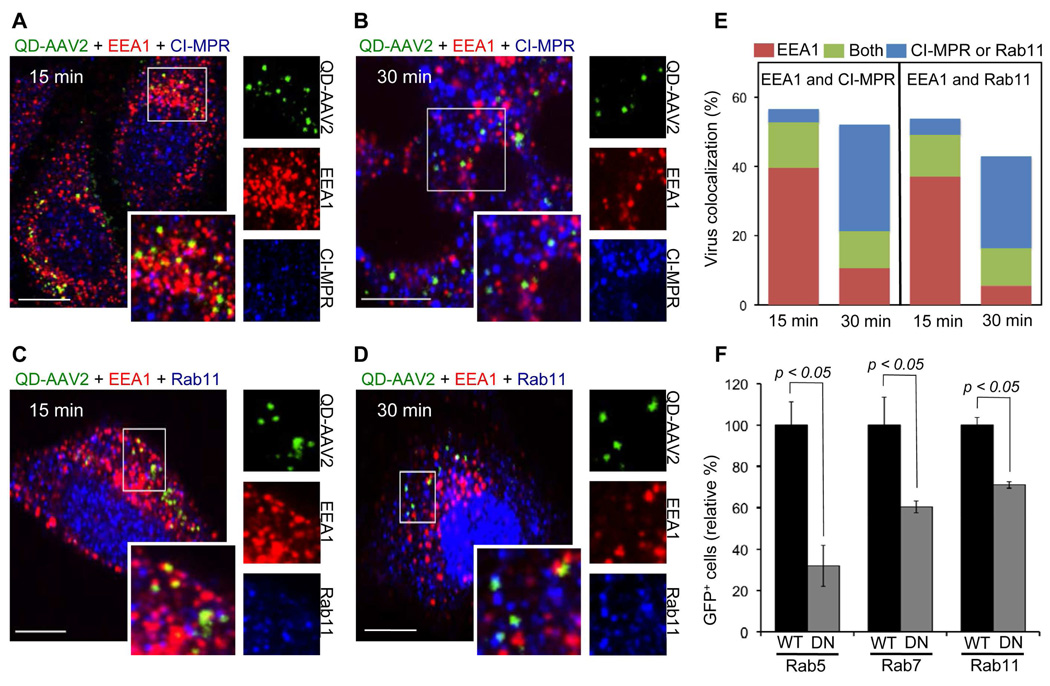
Next, we demonstrated whether AAV2 particles traffic through various endosomal compartments using a colocalization experiment with early endosome antigen 1 (EEA1),7,39–40 cation-independent mannose 6-phosphate receptor (CI-MPR),7,40–41 and Rab1142–43 as the early endosome, late endosome, and recycling endosome markers, respectively. After 15 min of incubation at 37°C, the acquired images showed that ~40% of the viral particles (n=320) were observed in endosomes positive for EEA1, but no significant colocalization of viruses with CI-MPR or Rab11 was detected (Figures 4A and 4C). At 30 min, a small fraction of the viral particles were colocalized with EEA1, but approximately 30.8% (n=159) of the particles were seen in late endosome (CI-MPR, Figure 4B) and ~26.4% (n=140) in the recycling endosome (Rab11, Figure 4D). Some viruses were observed in endosomes that were positive for both EEA1 and CI-MPR or EEA1 and Rab11, which might be interpreted to be intermediates transitioning into the late or recycling endosomes. These imaging results suggest that AAV2 is trafficked from early endosomes to both late and recycling endosomes. The quantification of viral particles colocalized with EEA1, CI-MPR, and Rab11 are shown in Figure 4E.
Figure 4.
The trafficking of QD-AAV2 through various endosomes. (A to D) HeLa cells were incubated with QD-AAV2 (green) for 30 min at 4°C to synchronize infection and were then shift to 37°C for 15 or 30 min. The cells were fixed, permeabilized, and immunostained with antibodies against EEA1 (red) and CI-MPR (blue) (A, B), or EEA1 and Rab11 (blue) (C, D). The boxed regions are enlarged in the right panels. Scale bar represents 5 µm. (E) Quantification of QD-AAV2 colocalized with EEA1+, CI-MPR+, Rab11+, EEA1+CI-MPR+, or EEA1+Rab11+ endosomes after 15 or 30 min of incubation. (F) Functional involvement of endosomes in the AAV2 transduction. 293T cells transiently transfected with the wild-type or dominant-negative mutant form of Rab5, Rab7, or Rab11 were infected with unlabeled AAV2. The percentage of GFP-positive cells was analyzed by flow cytometry. Error bars represent the standard deviation of the mean from triplicate experiments.
To further investigate whether viral trafficking into the early, late, and recycling endosomal compartments is functionally required for AAV2 transduction, the dominant-negative mutants of Rab proteins were used to disable either the early (Rab5),44 the late (Rab7),45 or the recycling (Rab11)46 endosome function. 293T cells transfected with either the wild-type or the dominant-negative form of Rab proteins (Rab5, Rab7, or Rab11) were incubated with unlabeled or QD-labeled AAV2. As shown in Figure 4F and Figure S6, expression of the Rab5 dominant-negative mutant reduced the transduction rate by ~70% as compared to the transduction of wild-type Rab5-expressing cells, suggesting that most of the AAV2 must travel to early endosomes for successful infection. The reduced transduction rate in 293T cells expressing the dominant-negative mutant form of Rab7 or Rab11 further indicated that the productive transduction pathways are associated with the functional transport of AAV2 to the late and recycling endosomes. These similar trafficking results for the functional involvement of endosomes in virus transduction between unlabeled and QD-labeled AAV2 further indicate that the endosomal transport of AAV2 is not significantly affected by the QD labeling.
It has been shown that the microtubule network and/or actin-filaments can facilitate the migration of AAV2 to the nucleus through a drug inhibition study.47 However, another recent study analyzed effect of microtubule-altering drugs and reported that AAV2 could transduce HeLa cells in a dynein- and microtubule-independent fashion.48 The reason for this discrepancy is not clear, but it is generally believed that the disruption of microtubule or actin-filaments by drugs may also inhibit the trafficking of other vesicles in the cell.24 To provide a better understanding of the functional involvement of the cytoskeletons in the intracellular transport of AAV2, we examined the interactions between QD-labeled AAV2 and microtubules/actin-filaments in cells. First, to examine whether cytoskeletons are involved in facilitating intracellular transport of AAV2 within cells, we conducted a colocalization experiment using antibodies specific to microtubules and actin-filaments in fixed cells. The results of the colocalization study using a tubulin-specific antibody and rhodamine-conjugated phalloidin suggested that approximately 41% of AAV2 particles were detected on microtubules and ~14.5% of particles on actin-filaments in fixed cells (total ~200 particles), implying that AAV2 particles could travel along microtubules and actin-filaments (Figure 5A). We further characterized the microtubule-mediated transport of AAV2 by real-time monitoring of QD-labeled AAV2 particles in HeLa cells expressing GFP-tubulin (See Supporting information, Movie S3). Analysis of the live-cell imaging experiment suggested that the microtubule-mediated movement of AAV2 was rapid and directional towards the nucleus (up to 2.3 µm/s), although this was only observed for a small fraction of the viral particles (< 10%) (Figures 5B and 5D, and Supporting information, Movie S4). Microtubule-independent movement of the viral particles was also observed, which was likely due to diffusion-associated transport (Figures 5C and 5E, and Supporting information, Movie S5). We also visualized the actin-mediated virus internalization into cells by live-cell imaging of QD-labeled AAV2 in HeLa cells expressing GFP-actin (See Supporting information, Movie S6). We observed that the QD-labeled AAV2 particles were moving toward the cell body along highly dynamic cell surface protrusions such as filopodia, which are actin-rich structures.49 However, it was difficult to clearly visualize actin-dependent transport of the viral particles inside the cell body because our current live-cell imaging procedure failed to resolve individual actin structures fluorescently labeled with GFP-actin in living cells, although actin structures were clearly visualized with rhodamine-conjugated phalloidin in fixed cells. We are developing new protocols aiming to improve the live-cell imaging capacity for visualizing actin-dependent movement of AAV2 within living cells.
Figure 5.
Cytoskeleton-mediated AAV2 transport. (A) QD-labeled AAV2 (blue) was incubated with HeLa cells for 30 min at 37°C. The cells were then fixed, permeabilized, and stained for microtubules (green) and actin-filaments (red) with the monoclonal antibody to α-tubulin and rhodamine-conjugated phalloidin, respectively. The boxed regions are enlarged in the right panels. (B to E) Live cell imaging of microtubule-dependent or microtubule-independent movement of QD-AAV2. HeLa cells were transiently transfected with GFP-α-tubulin. At 24 h post-transfection, the cells were incubated with QD-labeled AAV2 (green) for 30 min at 4°C, and were then warmed to 37°C for 15 min. Confocal time-lapse images were then recorded. A representative trajectory and viral velocity of microtubule-dependent (B, D) and microtubule-independent movement (C, E) of QD-AAV2 are shown. Scale bars represent 5 µm.
To further validate the result from the confocal imaging of QD-AAV2 trafficking through cytoskeletons, the effect of cytoskeleton-disrupting drugs on viral motility was evaluated (Figure S7). Treatment of cells with nocodazole, which depolymerizes microtubules, moderately reduced the motility of QD-AAV2, but remarkably, no directed transport was observed in nocodazole-treated cells (Figure S7B and S7E). The cytochalasin D (Cyto-D) treatment, which perturbs the actin cytoskeleton, resulted in greater reduction of the viral motility (Figure S7C and S7F). The drug inhibition results support the functional involvement of microtubules and actin-filaments in the intracellular transport of AAV2.
It is generally believed that AAV2 must escape from endosomes into the cytoplasm and then subsequently be trafficked to the nucleus.24,50 It has been suggested that the ubiquitin-proteasome pathway is likely to be involved in this intracellular processing of AAV2. Although it remains unclear which mechanism is triggered by the treatment with proteasome inhibitors, many studies have shown that the inhibition of proteasome activity resulted in an enhanced level of AAV transduction.51–53 Furthermore, it was demonstrated that both AAV2 and AAV5 capsid proteins are substrates for ubiquitination, and interestingly, the level of capsid ubiquitination was significantly increased in the presence of the proteasome inhibitor.51
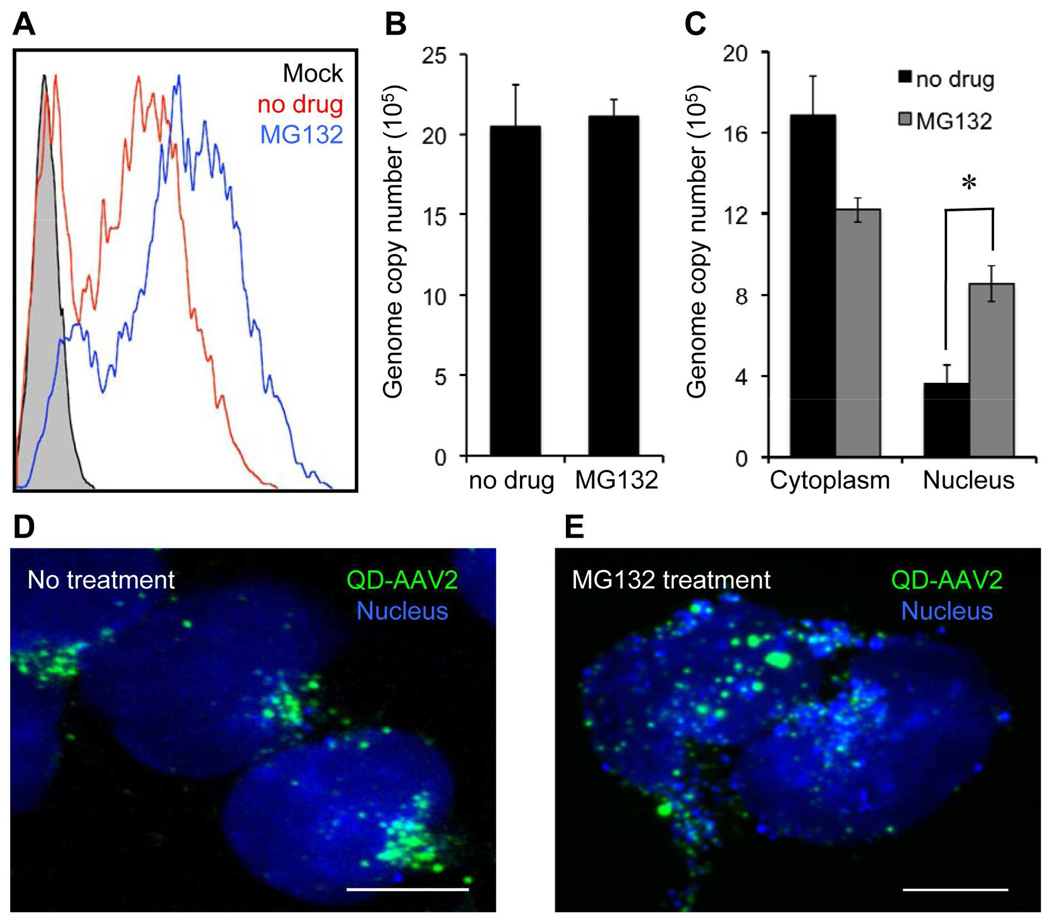
These prior studies prompted us to further investigate the functional involvement of the ubiquitin-proteasome system in the AAV2 entry pathway. As expected, treatment with the proteasome inhibitor (MG132) markedly augmented the transduction efficiency of AAV2 in HeLa cells (Figure 6A). However, the enhanced transduction does not seem to be a direct effect of blocking AAV genome degradation in the presence of the proteasome inhibitor; no significant change was observed in the intracellular viral genome copy number in infected cells treated with or without MG132 at 24 h post-infection (Figure 6B), which is consistent with the previous reports.51,54 Additionally, the enhanced transduction could not be a result of a modulation in the second-strand genome synthesis caused by the proteasome inhibitor.53,55 Instead, our confocal images showed that particle numbers accumulated in the nucleus were remarkably increased in MG132-treated cells (Figure 6D and 6E). Also, quantification of the viral genome copies isolated from the cytoplasmic and nuclear fractions of the infected cells confirmed that the accumulation of viral genomes in the nucleus was significantly enhanced in the presence of MG132 without a marked change in the total number of intracellular viral genomes (Figure 6C). Our imaging and genome quantification studies imply that modulation of the ubiquitin-proteasome system might alter the intracellular trafficking of AAV2, at least, in terms of facilitating the nuclear translocation of viruses. Although the detailed effects of proteasome inhibitor on the intracellular processing of AAV2 remains to be further studied, the enhanced nuclear uptake of AAV2 could possibly be correlated with the increased level of capsid ubiquitination observed with the use of the proteasome inhibitor,51 suggesting that the extent of capsid ubiquitination might be critical in modulating the intracellular fate of AAV2.
Figure 6.
Enhanced nuclear uptake of AAV in the presence of the proteasome inhibitor. (A) HeLa cells were pretreated with MG132 (25 µM) for 30 min at 37°C and then spin-infected with AAV2 in the presence of MG132. After an additional 3 h of incubation with MG132 at 37°C, the cells were washed with PBS and resupplied with fresh media. The resulting GFP expression was analyzed by flow cytometry at 3 days post-infection. (B) Quantification of the intracellular viral genome copy number in infected cells with or without MG132 at 24 h post-infection. (C) Quantification of intracellular viral genomes isolated from the cytoplasmic and nuclear fractions in infected cells with or without MG132 at 24 h post-infection. Error bars represent the standard deviation of the mean from triplicate experiments (* p<0.05). (D, E) Enhanced nuclear translocation of QD-AAV2. HeLa cells were incubated with QD-AAV2 (green) for 18 h at 37°C with (E) or without MG132 (D). The cells were then fixed, permeabilized, and counterstained with TO-PRO-3 (blue). Scale bars represent 5 µm.
To summarize, the goal of this study was to develop and evaluate a general method to label non-enveloped viruses with QDs via the carbodiimide chemistry. By the use of this method, AAV2 were successfully labeled with QDs, which subsequently enabled us to monitor viral behaviors in living cells in a more detailed manner. It was demonstrated that the use of QDs could potentially offer advantages for long-term imaging and fast and multicolor time-lapse imaging due to their remarkable photostability and brightness compared to conventional organic fluorophores. As a demonstration of the utility of the QD-labeled AAV2, we investigated the entry mechanism of AAV2 by visualizing the interactions of single viral particles with endocytic structures in living cells. This study suggested that AAV2 is internalized into cells mainly via a clathrin-dependent pathway and is trafficked through the early, late, and recycling endosomes. Ding et al. demonstrated that AAV2 traffics through both late and recycling endosomes in a dose-dependent manner.20 From our confocal imaging at the dose of ~1,000 genomes/cells, we observed that QD-AAV2 traffics through both late and recycling endosomes. However, at a lower dose of AAV2, we also found that QD-AAV2 particles were much less colocalized with the recycling endosomal markers.
Furthermore, real-time imaging suggested that AAV2 exploits the cytoskeleton network to facilitate viral transport within cells. It was also demonstrated that the ubiquitin-proteasome system likely plays an important role in the intracellular trafficking of AAV2, at least at the level of the nuclear transport. This entry route of QD-labeled AAV2 is similar to that of organic dye-labeled AAV2 studied by us (Figure S5) and others.20 In addition, our quantitative analysis of the viral motility of QD- or dye-labeled AAV2 also revealed that the intracellular transport properties are not significantly different, suggesting that the QD-labeling procedure does not significantly affect the infection pathway and the intracellular trafficking properties of AAV2.
Recent studies have suggested that QD-labeling could possibly alter the uptake mechanism and intracellular trafficking of some ligands and TAT peptides.30–31 For example, one study showed that the QD-labeled TAT peptide is internalized into cells via lipid-raft-dependent macropinocytosis, which is different from clathrin-dependent pathway used by dye-labeled TAT peptides.31,56 However, it was also reported that the internalization process of TAT fused with green fluorescent protein (GFP) was different from that of dye-labeled or unconjugated TAT,57–58 suggesting that the different entry pathway of the QD-labeled TAT peptide might not be solely caused by the QD conjugation. In contrast, it was demonstrated that QD-labeled simian virus 40 (SV40) entered cells via the caveolae-mediated endocytosis and was transported to the endoplasmic reticulum (ER),59 which is a known entry route for the wild-type SV40 virus.60 A comparative study between organic dyes and QDs for the surface trafficking of a neurotransmitter receptor also revealed that the diffusion properties of individual receptors labeled with organic dyes or QDs were not significantly different.61 There are several studies in literature showing that QD-labeling is a great tool for revealing the intracellular trafficking of ligands and receptors.61–63 Thus, we predict that in the near future QDs can be also successfully utilized for single virus tracking studies to investigate the molecular mechanisms of viral infections. It is noteworthy that the proper labeling conditions (e.g. molar ratio, incubation time, etc.) are important for QD-labeling to avoid blocking of the functions of ligands or viruses. We found that under improper labeling conditions, we were able to label AAV2 with QDs, but the labeling resulted in significant inhibition of viral infectivity. Finally, it should be noted that our QD-labeling scheme reported in this study of AAV2 can be readily applied to labeling other non-enveloped viruses. Combined with our previous report on the QD-labeling of enveloped viruses,21 we have now established a complete tool set to utilize QDs to detect and monitor virus trafficking during infection. These labeling methods are expected to provide useful platforms for elucidating the molecular details of entry for many kinds of non-enveloped and enveloped viruses.
MATERIALS AND METHODS
Cell lines, Antibodies, and Reagents
HEK-293, HEK-293T, and HeLa cells were maintained in a 5% CO2 environment with Dulbecco’s modified Eagle medium (Mediatech, Inc., Manassas, VA) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO) and 2 mM L-glutamine (Hyclone Laboratories, Inc., Omaha, NE). The mouse monoclonal antibody against the intact AAV2 (clone: A20) was obtained from American Research Products, Inc. (Belmont, MA). The mouse monoclonal antibodies against clathrin and caveolin-1 and the rabbit polyclonal antibody specific to CI-MPR were purchased from Abcam (Cambridge, MA). The mouse monoclonal anti-EEA1 antibody and the rabbit polyclonal anti-Rab11 antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The mouse monoclonal antibody to α-tubulin was obtained from Sigma-Aldrich. The rhodamine-conjugated phalloidin, Texas red-conjugated or Alexa488-conjugated goat anti-mouse immunoglobulin G (IgG) antibody, and Alexa647-conjugated goat anti-rabbit IgG antibody were purchased from Invitrogen (Carlsbad, CA). Bafilomycin A1, chlorpromazine, filipin, MG132, and MβCD were obtained from Sigma-Aldrich. 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were purchased from Thermo Scientific (Rockford, IL).
Plasmids
The plasmid encoding the dominant-negative mutant of DsRed-Rab7 (Rab7T22N) was generated by site-directed mutagenesis as described.64 The constructs for mRFP-Clathrin, wild-type and dominant-negative forms of DsRed-Rab5, DeRed-Rab7 and DeRed-Rab11, and GFP-α-tubulin were purchased from Addgene (Cambridge, MA). The pAcGFP1-actin plasmid was obtained from Clontech (Palo Alto, CA).
Virus Production
Recombinant AAV2 vectors were produced in HEK-293 cells as previously described.65 Fifty 15-cm dishes of subconfluent 293 cells were triple-transfected with 650 µg each of AAV2 cis-plasmid carrying a GFP reporter gene under the control of the CMV promoter and AAV2 trans-plasmid containing the AAV2 rep and cap genes and 1300 µg of the adenovirus helper plasmid pΔF6 using the calcium phosphate precipitation method. After overnight incubation, the medium was replaced with fresh D10 media, and the cells were harvested at 3 days post-transfection, followed by three cycles of freeze and thaw. Viruses were then purified by cesium chloride gradient density centrifugation66 at 25,000 rpm and 15°C for 20 h (Optima L-90 K Ultracentrifuge, SW-28 rotor, Beckman Coulter, Brea, CA). Fractions containing AAV2 determined by refractive index were further desalted in PBS using an Amicon Ultra 100,000 MWCO centrifugal filter device (Millipore, Billerica, MA).
QD-Labeling of AAV2
To modify the carboxyl group to an amine-reactive NHS ester, 30 pmol of carboxyl QD525 or QD705 (~ 10 nm diameter, Invitrogen) were incubated with 100 nmol of EDC and 150 nmol of NHS for 15 min at room temperature. The excess EDC and NHS were then removed by a gel filtration column. The resultant QDs were incubated with purified AAV2 (1 ×1010 particles) for 2 h at room temperature. The reaction was then quenched by adding 50 mM Tris buffer. To remove unreacted QDs, the labeling mixture was further purified via the FPLC system (Bio-Rad BioLogic Duo Flow) using a HiTrap Heparin HP column (GE Healthcare). To label AAV2 with organic dyes, purified AAV2 (1 ×1011 particles) were incubated with 50 nmol of FITC-NHS (Pierce, Rockford, IL) or Alexa488-TFP ester (Invitrogen) for 2 h in 0.1 M sodium bicarbonate buffer (pH=9.3). Unbound dye molecules were removed via buffer exchange into PBS (pH=7.4) using a gel filtration column. For the spectroscopic measurement to determine the number of QDs bound to AAV2, the conjugate concentration was calculated by using the QD705 absorbance at 488 nm (ε=300,000 M−1 cm−1) of the purified QD705-AAV2 and assuming no loss of AAV2 during the column purification. The absorbance of the purified unlabeled AAV2 at 280 nm was experimentally determined, and the molar extinction coefficient of AAV2 (ε=6.61 × 106 M−1 cm−1) at 280 nm was used in calculation.67
Virus Transduction
HeLa cells (1 × 105 cells per well) were seeded in a 24-well culture dish and spin-infected with AAV2 (~1,000 viral genome copies per cell) at 2,500 rpm for 90 min at 30°C using a Sorval Legend centrifuge. After an additional 3 h of incubation, the medium was then removed and replaced with fresh medium and cultured for 3 days before flow cytometry analysis of GFP+ cells. For viral transduction with drug-treated cells, HeLa cells were preincubated with Bafilomycin A1 (BAF, 50 nM), Chlorpromazine (CPZ, 25 µg/ml), filipin (5 µg/ml), or MG132 (25 µM) for 30 min or with methyl-beta-cyclodextrin (MβCD, 15 mM) for 15 min at 37°C, and the cells (1 ×105 cells per well) were spin-infected with unlabeled or QD705-labeled AAV2 in the presence of drugs (BAF, CPZ, filipin, and MG132) or in the absence of drug (MβCD). After an additional 3 h of incubation with drugs (BAF, CPZ, filipin, and MG132) at 37°C, the cells were washed with PBS and replenished with fresh D10 media.
For the viral transduction with Rab protein-treated cells, 293T cells were seeded in a 24-well dish and grown at 37°C overnight. The cells were then transfected with DsRed-Rab5, -Rab7, or -Rab11 (either wild-type or dominant-negative mutants). After 4 h of incubation at 37°C, the media were then replaced with fresh D10 media. At 24 h post-transfection, unlabeled or QD705-labeled AAV2 viruses was added and incubated with the cells overnight, and the medium was then replaced with fresh media.
Confocal Imaging
Fluorescence images were acquired on a Yokogawa spinning-disk confocal scanner system (Solamere Technology Group, Salt Lake City, UT) using a Nikon eclipse Ti-E microscope equipped with a 60×/1.49 Apo TIRF oil objective and a Cascade II: 512 EMCCD camera (Photometrics, Tucson, AZ, USA). An AOTF (acousto-optical tunable filter) controlled laser-merge system (Solamere Technology Group Inc.) was used to provide illumination power at each of the following laser lines: 491 nm, 561 nm, and 640 nm solid state lasers (50mW for each laser). For the detection of individual viral particles, QD-labeled viruses were overlaid upon polylysine-coated glass bottom dishes (MatTek Corporation, Ashland, MA) for 60 min at 37°C. The dish was then rinsed, fixed with 4% formaldehyde and immunostained with the monoclonal antibody specific to intact AAV2 (A20).
For the photostability study of organic dye- and QD-labeled viral particles, FITC-labeled and QD705-labeled AAV2 were co-incubated with HeLa cells for 30 min at 37°C to initiate virus internalization. The cells were then fixed. The specimens were continuously illuminated by the laser at 491 nm over 10 min. Images were captured at ~3 s intervals. Fluorescent intensity versus time within the regions of interest was measured and analyzed by using the Nikon NIS-Elements software. To compare the detection sensitivity between QD705-labeled and FITC-labeled viral particles at different exposure times, FITC-labeled or QD705-labeled AAV2 were incubated with HeLa cells for 30 min at 37°C, and the cells were then fixed. Images of the same regions of cells were acquired with different exposure times.
For the colocalization study with endocytic markers, HeLa cells were incubated with QD-AAV2 (~1000 viral genome copies per cell) for 30 min at 4°C to synchronize infection. After being washed with PBS, the treated cells were then warmed to 37°C to initiate virus internalization for the indicated time periods. The cells were fixed, permeabilized with 0.1% Triton X-100, and then immunostained with the corresponding antibodies specific to clathrin, caveolin-1, EEA1, CI-MPR, Rab11, or microtubules or counterstained with TO-PRO-3 (Invitrogen).
Live-Cell Imaging
For real-time imaging of the dynamic interactions between QD-AAV2 and clathrin structures or microtubules, HeLa cells were seeded on glass bottom dishes and grown at 37°C overnight. The cells were then transiently transfected with mRFP-clathrin, GFP-α-tubulin, or pAcGFP1-actin. After 4 h of incubation at 37°C, the media was then replaced with fresh D10 media. At 24 h post-transfection, the cells were incubated with QD-labeled AAV2 for 30 min at 4°C, and were then warmed to 37°C to induce viral internalization. Confocal time-lapse images were then acquired.
Image Analysis
Image processing and data analysis were carried out using the Nikon NIS-Elements software. To quantify the extent of colocalization for a 2-color comparison, colocalization coefficients were calculated using the Manders' overlap coefficient by viewing more than 10 cells at each time point. The Manders overlap coefficient was generated by the Nikon NIS-Elements software. For the colocalization assay with endosome markers (a 3-color comparison), the level of colocalization was quantified by viewing more than 100 viral particles in more than 10 cells at each time points and counting the total number of viral particles and the number of particles positive for the endosome marker of interest. For single viral particle tracking, the trajectories, mean-square displacement (MSD), and diffusion coefficients were obtained and analyzed using the 2D tracking module of the Nikon NIS-Elements software. The diffusion coefficients were calculated from the slope of the MSD versus time plot, according to MSD = 4DΔt + ν2Δt2, where ν is the mean velocity, for directed transport, or MSD = 4DΔt for undirected transport.
Nuclear and Cytoplasmic Fractionation
To study the distribution of viral genomes in the cytoplasm and nucleus of infected cells, the Nuclear/Cytosol Fractionation Kit (BioVision, Inc., Mountain View, CA) was employed to separate the cytoplasmic and nuclear extracts from infected cells in the presence or absence of MG132. Fractions were obtained at 24 h post-infection according to the manufacturer’s instruction. All procedures were performed at 4°C. Extracts were stored at −80°C until the qPCR assay was performed.
Quantification of genome copies by PCR
DNA was extracted using the QIAamp MinElute Virus Spin Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Quantitative PCR was performed on the Bio-Rad MyiQ real-time system using a primer pair specific for the GFP transgene: 5'-GACATCATGAAGCCCCTTGAG-3' (forward) and 5'-GGTGGTCGAAATTCAGATCAAC-3' (backward).
Instruments
For transmission electron microscope (TEM) imaging, either unconjugated QDs were not stained, or unlabeled AAV2 and QD-AAV2 particles were negatively stained for 10 min with 2% uranyl acetate in alcoholic solution (50% ethanol) and were then deposited on the carbon-coated electron microscopy grids (Ted Pella, Inc., Redding, CA) and images were obtained using the Philips EM-120 TEM operated at 80 kV. The hydrodynamic size of AAV2, QDs, and QD-labeled AAV2 were measured by dynamic light scattering (Wyatt Technology, Santa Barbara, CA). Fluorescence spectra of QD525-AAV2 and unconjugated QD525 were obtained with the QuantaMaster QM-4SE Spectrofluorometer (Photon Technology International, Birmingham, NJ) using the PTI FeliX32 software.
Supplementary Material
ACKNOWLEDGEMENT
We thank the USC NanoBiophysics Core Facility. This work was supported by a National Institute of Health grant R01-AI068978.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Hydrodynamic size of QD-AAV2, characterization of QD-AAV2, comparison of the photostability and detection sensitivity of QD-AAV2, MSD of the directed and undirected transport of QD-AAV2, entry pathway of organic dye-labeled AAV2, functional involvement of endosomes in QD-AAV2 transduction, time trajectories of QD-AAV2 in the presence of cytoskeleton disrupting drugs, and details for supporting movies 1–6. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Anderson JL, Hope TJ. Intracellular Trafficking of Retroviral Vectors:Obstacles and Advances. Gene Ther. 2005;12:1667–1678. doi: 10.1038/sj.gt.3302591. [DOI] [PubMed] [Google Scholar]
- 2.Dimitrov DS. Virus Entry: Molecular Mechanisms and Biomedical Applications. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greber UF, Way M. A Superhighway to Virus Infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Marsh M, Helenius A. Virus Entry: Open Sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelkmans L, Helenius A. Insider Information: What Viruses Tell Us About Endocytosis. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/s0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 6.Brandenburg B, Zhuang X. Virus Trafficking - Learning from Single-Virus Tracking. Nat. Rev. Microbiol. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joo KI, Wang P. Visualization of Targeted Transduction by Engineered Lentiviral Vectors. Gene Ther. 2008;15:1384–1396. doi: 10.1038/gt.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing Infection of Individual Influenza Viruses. Proc. Natl. Acad. Sci. USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the Intracellular Behavior of Hiv in Living Cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartlett JS, Wilcher R, Samulski RJ. Infectious Entry Pathway of Adeno-Associated Virus and Adeno-Associated Virus Vectors. J. Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Immunofluorescent Labeling of Cancer Marker Her2 and Other Cellular Targets with Semiconductor Quantum Dots. Nat. Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 12.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 13.Chan WC, Nie S. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 14.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinaud F, Clarke S, Sittner A, Dahan M. Probing Cellular Events, One Quantum Dot at a Time. Nat. Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- 16.Mueller C, Flotte TR. Clinical Gene Therapy Using Recombinant Adeno-Associated Virus Vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 17.Wu ZJ, Asokan A, Samulski RJ. Adeno-Associated Virus Serotypes: Vector Toolkit for Human Gene Therapy. Mol. Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Choi VW, McCarty DM, Samulski RJ. AAV Hybrid Serotypes: Improved Vectors for Gene Delivery. Curr. Gene Ther. 2005;5:299–310. doi: 10.2174/1566523054064968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seisenberger G, Ried MU, Endress T, Buning H, Hallek M, Brauchle C. Real-Time Single-Molecule Imaging of the Infection Pathway of an Adeno-Associated Virus. Science. 2001;294:1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 20.Ding W, Zhang LN, Yeaman C, Engelhardt JF. rAAV2 Traffics through Both the Late and the Recycling Endosomes in a Dose-Dependent Fashion. Mol. Ther. 2006;13:671–682. doi: 10.1016/j.ymthe.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joo KI, Lei Y, Lee C-L, Lo J, Xie J, Hamm-Alvarez SF, Wang P. Site-Specific Labeling of Enveloped Viruses with Quantum Dots for Single Virus Tracking. ACS Nano. 2008;2:1553–1562. doi: 10.1021/nn8002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicklin SA, Buening H, Dishart KL, de Alwis M, Girod A, Hacker U, Thrasher AJ, Ali RR, Hallek M, Baker AH. Efficient and Selective AAV2-Mediated Gene Transfer Directed to Human Vascular Endothelial Cells. Mol. Ther. 2001;4:174–181. doi: 10.1006/mthe.2001.0424. [DOI] [PubMed] [Google Scholar]
- 23.Grifman M, Trepel M, Speece P, Gilbert LB, Arap W, Pasqualini R, Weitzman MD. Incorporation of Tumor-Targeting Peptides into Recombinant Adeno-Associated Virus Capsids. Mol. Ther. 2001;3:964–975. doi: 10.1006/mthe.2001.0345. [DOI] [PubMed] [Google Scholar]
- 24.Ding W, Zhang L, Yan Z, Engelhardt JF. Intracellular Trafficking of Adeno-Associated Viral Vectors. Gene Ther. 2005;12:873–880. doi: 10.1038/sj.gt.3302527. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Nieves A, Fernandez-Barbero A, de las Nieves FJ. Particle-Counterion Clustering in Highly Charge-Asymmetric Complex Fluids. Phys. Rev. E. 2001;63:041404. doi: 10.1103/PhysRevE.63.041404. [DOI] [PubMed] [Google Scholar]
- 26.Wright JF, Le T, Prado J, Bahr-Davidson J, Smith PH, Zhen Z, Sommer JM, Pierce GF, Qu G. Identification of Factors That Contribute to Recombinant AAV2 Particle Aggregation and Methods to Prevent Its Occurrence During Vector Purification and Formulation. Mol. Ther. 2005;12:171–178. doi: 10.1016/j.ymthe.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Li H, Yang HJ, Wang HB, Lu JH, Zhang Y, Hu J. Glycerol Facilitates the Disaggregation of Recombinant Adeno-Associated Virus Serotype 2 on Mica Surface. Colloids Surf B: Biointerfaces. 2007;60:264–267. doi: 10.1016/j.colsurfb.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Dixit SK, Goicochea NL, Daniel MC, Murali A, Bronstein L, De M, Stein B, Rotello VM, Kao CC, Dragnea B. Quantum Dot Encapsulation in Viral Capsids. Nano Lett. 2006;6:1993–1999. doi: 10.1021/nl061165u. [DOI] [PubMed] [Google Scholar]
- 29.Blum AS, Soto CM, Wilson CD, Whitley JL, Moore MH, Sapsford KE, Lin TW, Chatterji A, Johnson JE, Ratna BR. Templated Self-Assembly of Quantum Dots from Aqueous Solution Using Protein Scaffolds. Nanotechnology. 2006;17:5073–5079. [Google Scholar]
- 30.Tekle C, Deurs Bv, Sandvig K, Iversen T-G. Cellular Trafficking of Quantum Dot-Ligand Bioconjugates and Their Induction of Changes in Normal Routing of Unconjugated Ligands. Nano Lett. 2008;8:1858–1865. doi: 10.1021/nl0803848. [DOI] [PubMed] [Google Scholar]
- 31.Chen B, Liu Q, Zhang Y, Xu L, Fang X. Transmembrane Delivery of the Cell-Penetrating Peptide Conjugated Semiconductor Quantum Dots. Langmuir. 2008;24:11866–11871. doi: 10.1021/la802048s. [DOI] [PubMed] [Google Scholar]
- 32.Delehanty J, Mattoussi H, Medintz I. Delivering Quantum Dots into Cells: Strategies, Progress and Remaining Issues. Anal. Bioanal. Chem. 2009;393:1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- 33.Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. Dynamin Is Required for Recombinant Adeno-Associated Virus Type 2 Infection. J. Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang LH, Rothberg KG, Anderson RG. Mis-Assembly of Clathrin Lattices on Endosomes Reveals a Regulatory Switch for Coated Pit Formation. J. Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlandi PA, Fishman PH. Filipin-Dependent Inhibition of Cholera Toxin: Evidence for Toxin Internalization and Activation through Caveolae-Like Domains. J. Cell Biol. 1998;141:905–915. doi: 10.1083/jcb.141.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowman E, Siebers A, Altendort K. Bafilomycins: A Class of Inhibitors of Membrane ATPases from Microorganisms, Animal Cells, and Plant Cells. Proc. Natl. Acad. Sci. USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imelli N, Meier O, Boucke K, Hemmi S, Greber UF. Cholesterol Is Required for Endocytosis and Endosomal Escape of Adenovirus Type 2. J. Virol. 2004;78:3089–3098. doi: 10.1128/JVI.78.6.3089-3098.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun X, Whittaker GR. Role for Influenza Virus Envelope Cholesterol in Virus Entry and Infection. J. Virol. 2003;77:12543–12551. doi: 10.1128/JVI.77.23.12543-12551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakadamyali M, Rust MJ, Zhuang X. Ligands for Clathrin-Mediated Endocytosis Are Differentially Sorted into Distinct Populations of Early Endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vonderheit A, Helenius A. Rab7 Associates with Early Endosomes to Mediate Sorting and Transport of Semliki Forest Virus to Late Endosomes. PLoS Biol. 2005;3:1225–1238. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urayama A, Grubb JH, Sly WS, Banks WA. Developmentally Regulated Mannose 6-Phosphate Receptor-Mediated Transport of a Lysosomal Enzyme across the Blood-Brain Barrier. Proc. Natl. Acad. Sci. USA. 2004;101:12658–12663. doi: 10.1073/pnas.0405042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheff DR, Daro EA, Hull M, Mellman I. The Receptor Recycling Pathway Contains Two Distinct Populations of Early Endosomes with Different Sorting Functions. J Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wikstrom K, Reid HM, Hill M, English KA, O'Keeffe MB, Kimbembe CC, Kinsella BT. Recycling of the Human Prostacyclin Receptor Is Regulated through a Direct Interaction with Rab11a Gtpase. Cell Signal. 2008;20:2332–2346. doi: 10.1016/j.cellsig.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Grunberg J, Zerial M. Inhibition of Rab5 Gtpase Activity Stimulates Membrane Fusion in Endosomes. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 Causes the Accumulation of Cathepsin D and Cation-Independent Mannose 6-Phosphate Receptor in an Early Endocytic Compartment. J. Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. Rab Proteins Mediate Golgi Transport of Caveola-Internalized Glycosphingolipids and Correct Lipid Trafficking in Niemann-Pick C Cells. J. Clin. Invest. 2002;109:1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanlioglu S, Benson PK, Yang J, Atkinson EM, Reynolds T, Engelhardt JF. Endocytosis and Nuclear Trafficking of Adeno-Associated Virus Type 2 Are Controlled by Rac1 and Phosphatidylinositol-3 Kinase Activation. J. Virol. 2000;74:9184–9196. doi: 10.1128/jvi.74.19.9184-9196.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirosue S, Senn K, ClÈment N, Nonnenmacher M, Gigout L, Linden RM, Weber T. Effect of Inhibition of Dynein Function and Microtubule-Altering Drugs on AAV2 Transduction. Virology. 2007;367:10–18. doi: 10.1016/j.virol.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and Myosin-Driven Movement of Viruses Along Filopodia Precedes Their Entry into Cells. J. Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harbison CE, Chiorini JA, Parrish CR. The Parvovirus Capsid Odyssey: From the Cell Surface to the Nucleus. Trends Microbiol. 2008;16:208–214. doi: 10.1016/j.tim.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 51.Yan Z, Zak R, Luxton GWG, Ritchie TC, Bantel-Schaal U, Engelhardt JF. Ubiquitination of Both Adeno-Associated Virus Type 2 and 5 Capsid Proteins Affects the Transduction Efficiency of Recombinant Vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douar A-M, Poulard K, Stockholm D, Danos O. Intracellular Trafficking of Adeno-Associated Virus Vectors: Routing to the Late Endosomal Compartment and Proteasome Degradation. J. Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan Z, Zak R, Zhang Y, Ding W, Godwin S, Munson K, Peluso R. Engelhardt, J. F., Distinct Classes of Proteasome-Modulating Agents Cooperatively Augment Recombinant Adeno-Associated Virus Type 2 and Type 5-Mediated Transduction from the Apical Surfaces of Human Airway Epithelia. J. Virol. 2004;78:2863–2874. doi: 10.1128/JVI.78.6.2863-2874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. Endosomal Processing Limits Gene Transfer to Polarized Airway Epithelia by Adeno-Associated Virus. J. Clin. Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding W, Yan Z, Zak R, Saavedra M, Rodman DM, Engelhardt JF. Second-Strand Genome Conversion of Adeno-Associated Virus Type 2 (AAV-2) and AAV-5 Is Not Rate Limiting Following Apical Infection of Polarized Human Airway Epithelia. J. Virol. 2003;77:7361–7366. doi: 10.1128/JVI.77.13.7361-7366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular Uptake of Unconjugated Tat Peptide Involves Clathrin-Dependent Endocytosis and Heparan Sulfate Receptors. J. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 57.Ferrari A, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Caveolae-Mediated Internalization of Extracellular HIV-1 Tat Fusion Proteins Visualized in Real Time. Mol. Ther. 2003;8:284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 58.Fittipaldi A, Ferrari A, Zoppe M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. Cell Membrane Lipid Rafts Mediate Caveolar Endocytosis of HIV-1 Tat Fusion Proteins. J. Biol. Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 59.Li F, Zhang ZP, Peng J, Cui ZQ, Pang DW, Li K, Wei HP, Zhou YF, Wen JK, Zhang XE. Imaging Viral Behavior in Mammalian Cells with Self-Assembled Capsid-Quantum Dot Hybrid Particles. Small. 2009;5:718–726. doi: 10.1002/smll.200801303. [DOI] [PubMed] [Google Scholar]
- 60.Pelkmans L, Kartenbeck J, Helenius A. Caveolar Endocytosis of Simian Virus 40 Reveals a New Two-Step Vesicular-Transport Pathway to the ER. Nat. Cell Biol. 2001;3:473–483. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- 61.Groc L, Lafourcade M, Heine M, Renner M, Racine V, Sibarita JB, Lounis B, Choquet D, Cognet L. Surface Trafficking of Neurotransmitter Receptor: Comparison between Single-Molecule/Quantum Dot Strategies. J. Neurosci. 2007;27:12433–12437. doi: 10.1523/JNEUROSCI.3349-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrini EM, Lu J, Cognet L, Lounis B, Ehlers MD, Choquet D. Endocytic Trafficking and Recycling Maintain a Pool of Mobile Surface AMPA Receptors Required for Synaptic Potentiation. Neuron. 2009;63:92–105. doi: 10.1016/j.neuron.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cambi A, Lidke DS, Arndt-Jovin DJ, Figdor CG, Jovin TM. Ligand-Conjugated Quantum Dots Monitor Antigen Uptake and Processing by Dendritic Cells. Nano Lett. 2007;7:970–977. doi: 10.1021/nl0700503. [DOI] [PubMed] [Google Scholar]
- 64.Joo KI, Tai A, Lee CL, Wong C, Wang P. Imaging Multiple Intermediates of Single-Virus Membrane Fusion Mediated by Distinct Fusion Proteins. Microsc. Res. Tech. 2010;73:886–900. doi: 10.1002/jemt.20841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auricchio A, Hildinger M, O'Connor E, Gao GP, Wilson JM. Isolation of Highly Infectious and Pure Adeno-Associated Virus Type 2 Vectors with a Single-Step Gravity-Flow Column. Hum. Gene Ther. 2001;12:71–76. doi: 10.1089/104303401450988. [DOI] [PubMed] [Google Scholar]
- 66.Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski RJ, Muzyczka N. Recombinant Adeno-Associated Virus Purification Using Novel Methods Improves Infectious Titer and Yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 67.Sommer JM, Smith PH, Parthasarathy S, Isaacs J, Vijay S, Kieran J, Powell SK, McClelland A, Wright JF. Quantification of Adeno-Associated Virus Particles and Empty Capsids by Optical Density Measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.