Abstract
Various fatal neurodegenerative disorders are caused by altered metabolism of the prion protein (PrP). These diseases are typically transmissible by an unusual ‘protein-only’ mechanism in which a misfolded isomer, PrPSc, confers its aberrant conformation onto normal cellular PrP. An impressive range of studies has investigated nearly every aspect of this fascinating event; yet, our understanding of how PrPSc accumulation might lead to cellular dysfunction and neurodegeneration is trifling. Recent advances in our understanding of normal PrP biosynthesis and degradation might have unexpectedly shed new light on this complex problem. Indeed, our current understanding of normal PrP cell biology, coupled with a growing appreciation of its complex metabolism, is providing new hypotheses for PrP-mediated neurodegeneration.
Transmission versus neurodegeneration in prion diseases
Prion diseases are a class of invariably fatal protein misfolding diseases that cause neurodegeneration in various mammals including humans [1–3]. Examples include sheep scrapie, bovine spongiform encephalopathy (BSE), Kuru, Creutzfeldt-Jakob Disease (CJD) and Gerstmann-Straussler-Scheinker syndrome (GSS). The etiological agent of these pathologies is the prion protein (PrP), a widely expressed nonessential cell-surface glycoprotein of unclear function. Unique among protein-misfolding disorders, prion diseases are typically transmissible. The transmissible agent is composed primarily, if not exclusively, of a misfolded form of PrP termed PrPSc (Box 1). The most widely accepted model is that PrPSc is misfolded in such a manner that it is capable of interacting with and converting normal cellular PrP (termed PrPC) into the PrPSc conformation. Continued rounds of ‘replication’ concomitant with ongoing PrPC production by the host cell leads to PrPSc accumulation, thereby generating additional transmissible agent. The conformational conversions of PrP and analogous ‘infectious proteins’ in yeast [4] have been extensively studied, revealing a cogent framework for the mechanism of ‘protein-only’ disease transmission. In striking contrast, the downstream consequences of PrPSc production that lead to the observed neurodegenerative phenotype are very poorly understood.
Box 1. Nomenclature of PrP forms.
Classically, PrPC denoted normal cellular PrP, whereas PrPSc denoted the scrapie form associated with the transmissible agent, a prion. As the ‘protein-only’ hypothesis gained increasing experimental support, PrPSc was typically equated with the transmissible prion. By this definition, PrPSc has a conformation capable of converting PrPC to additional PrPSc molecules. The earliest studies correlated PrPSc with a high degree of protease resistance, relative insolubility, high β-sheet content and fibril-forming capacity. Over time, however, it has become clear that many PrP conformations (i.e. ‘strains’) with converting capacity merit the designation of PrPSc. Unfortunately, none of the biochemical features that initially characterized PrPSc are unique and, conversely, not all PrPSc ‘strains’ have all of these features. As a result, the literature contains considerable variation in nomenclature. For example, PrP-sen and PrP-res are used to denote forms that are sensitive or resistant to protease digestion. Yet, because there are countless ways for PrP (or any protein for that matter) to be sensitive or resistant to protease digestions, PrP-sen and PrP-res do not refer to specific forms of PrP; rather, they are biochemical descriptors. To minimize confusion, we use the following nomenclature and definitions:
PrPC: this is the major (most abundant) normal cellular form of PrP characterized by its glycosylated, GPI-anchored, cell-surface locale and trafficking through the secretory and endocytic pathways. Its normal function is poorly understood.
PrPSc: by definition this is the transmissible agent with a conformation capable of converting PrPC to additional PrPSc molecules. Its deposition and accumulation are not intrinsically toxic; instead, it causes pathology in only some cell types, and these must express PrPC.
CtmPrP: this refers to a transmembrane form in which the N terminus resides in the cytoplasm, the C terminus faces the exoplasmic environment and a central hydrophobic domain (residues ~112–135) spans the membrane. Its increased generation in mice causes neurodegeneration.
NtmPrP: this refers to a transmembrane form in which the C terminus resides in the cytoplasm, the N terminus faces the exoplasmic environment and a central hydrophobic domain (residues ~112–135) spans the membrane. It has been observed only in vitro; therefore, its role, if any, in neurodegeneration is unknown.
cyPrP: this refers to PrP molecules in the cytosol, irrespective of their origin. Forced generation of this form at higher than normal levels leads to neurodegeneration in mice.
PrP: this term will be used when not referring specifically to any of the forms above.
The two most obvious consequences of prion replication are the relative depletion of PrPC and the accumulation of PrPSc. However, numerous studies argue that neither an acute loss of PrPC nor direct cellular toxicity of PrPSc provides a fully satisfactory mechanism of neurodegeneration (Box 2). Indeed, it is now apparent that PrPSc is not intrinsically toxic to cells if they do not actively express PrPC, despite the fact that Prnp-null cells are not impaired in PrPSc internalization [5]. Conversely, several rare familial diseases caused by PRNP mutations are poorly or non-transmissible and cause little or no PrPSc accumulation [6–8].
Box 2. Relationships between PrPSc and neurotoxicity.
Conversion of PrPC to PrPSc leads to neurodegeneration. However, neither PrPC depletion nor direct toxicity of PrPSc provides a fully satisfactory mechanism for neuronal death. No obvious neurodegenerative phenotypes have been observed in either germline or post-natal knockouts of the Prnp gene in mice [68–70]. Furthermore, depletion is unlikely to be complete during prion infection because PrPC is a requisite substrate for PrPSc replication. Thus, although PrPC depletion could be a (minor) contributing factor, it is usually accepted that a toxic ‘gain of function’ by PrPSc is the primary mechanism of pathogenesis. However, several elegant studies argue persuasively against PrPSc being intrinsically toxic to cells. First, brain-grafting studies showed that PrPSc produced at high levels by grafted normal brain tissue had no pathological effects on directly adjacent brain tissue derived from Prnp-null mice (despite considerable PrPSc deposition [71] and the ability of Prnp−/− cells to internalize PrPSc [5]). Second, Prnp+/− mice survive prion infection considerably longer than wild-type mice, despite accumulating PrPSc to the same high levels and with indistinguishable kinetics [72]. Third, an inducible knockout system was used to show that acute neuronal depletion of PrPC precluded and even reversed the pathologic effects of PrPSc deposition [73,74]. And, finally, infection of mice expressing ‘anchorless’ PrP led to PrPSc propogation and accumulation without overt neurotoxicity [75]. Thus, PrPSc is not intrinsically toxic to cells if they do not actively express PrPC.
Conversely, there seem to be many ways in which PrP can be neurotoxic in the absence of PrPSc. These include several familial PRNP mutations [6–8], artificial Prnp mutations that cause neurodegenerative syndromes in mouse models [18,37,38,44] and PrP constructs lacking the HD [76–78]. These non-transmissible disorders are perhaps better thought of as ‘proteinopathies’, analogous to numerous other diseases caused by the generation of a misfolded protein. Although their relationships to transmissible prion disease are unknown, these observations indicate that PrP species other than PrPSc can be neurotoxic and could have a role in disease. In addition to the pathways described here (involving altered PrP biosynthesis and mislocalization), other possible mechanisms involving aberrant signaling by PrP and putative toxic oligomeric PrP species have been reviewed elsewhere [2,79,80].
These non-transmissible inherited disorders are perhaps better thought of as ‘proteinopathies’, analogous to numerous other diseases caused by the generation of a misfolded protein. Although one might argue that neurodegeneration caused by such genetic variants are mechanistically unrelated to transmissible prion diseases, both types of disease, nonetheless, involve the same protein and share many pathological phenotypes [6,9]. Thus, apparently non-transmissible diseases caused by PRNP mutations could be related to PrPSc-mediated neurodegeneration. It is, therefore, reasonable to expect that the study of familial PrP-mediated ‘proteinopathies’ will provide some insight into the more complex transmissible prion diseases, in which high levels of biochemically intractable PrPSc accumulation complicates several analyses. By analogy, key insights into other complex diseases including Alzheimer’s, Parkinson’s, breast cancer and diabetes have come from the study of comparatively rare, seemingly unusual inherited variants that eventually proved to be relevant (sometimes indirectly) to the more common, but less tractable, sporadic disease.
PrP biosynthesis, trafficking and degradation
A common theme among almost all protein-misfolding diseases is the generation of an aberrant product owing to a problem in the normal biosynthesis, folding, processing, trafficking or quality control of either a specific protein or proteins in general [10]. Thus, understanding the proteinopathies caused by PrP almost certainly will require a detailed quantitative understanding of its normal metabolism. Deviations from these events that have detrimental effects can then be identified, studied and manipulated to determine their broader role in disease pathogenesis.
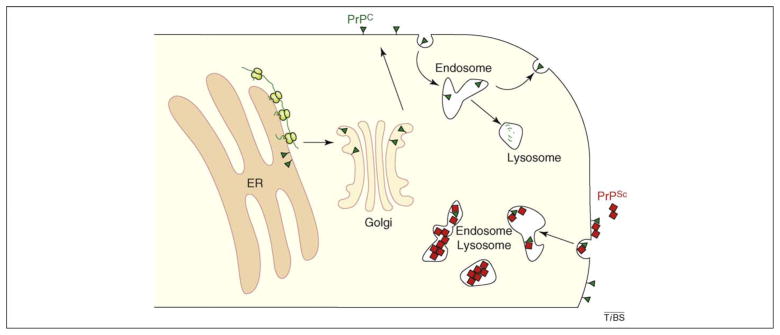
As a typical cell-surface glycoprotein, PrP is first imported into the endoplasmic reticulum (ER) [11], where nascent PrP is processed, glycosylated, modified by a C-terminal glycosylphosphatidyl inositol (GPI) anchor and properly folded before transport to the Golgi [12]. As it transits the Golgi stacks, PrP receives various additional modifications to its glycans and GPI anchor and is then exported to the cell surface. This mature cell-surface PrP can be endocytosed (possibly by multiple mechanisms including clathrin-coated pits and caveolae [13]) to internal endosomal compartments, from where it is either recycled to the cell surface or routed to lysosomes for degradation. Thus, the majority of PrP follows the traditional exocytic pathway to the cell surface and the endocytic pathway for turnover in the lysosome (Figure 1). In a typical cell, PrP biosynthesis and trafficking to the cell surface takes around 30 min, whereas it undergoes degradation with a half-life of ~3–6 h [14,15].
Figure 1.
Overview of PrPC and PrPSc metabolism. Nascent PrP (green line) is synthesized at the ER and imported into the ER lumen, where it is processed and folded into its final conformation (green triangle). Properly folded PrP is trafficked through the Golgi to the cell surface. Cell-surface PrPC recycles through endosomes, eventually being degraded in lysosomes with a half-life of ~3–6 h. The lower right shows the consequences of extrinsic PrPSc (red square). PrPC and PrPSc can interact, and both can be internalized into endosomes. Although the location is poorly defined, PrPSc converts PrPC into additional PrPSc. Because PrPSc has a longer half-life (~24 h) for lysosomal degradation, it can accumulate to relatively high levels in intracellular compartments of the endo-lysosomal system.
However, each of these biosynthetic, maturation, sorting, trafficking and degradation steps is presumably limited in their efficiency (after all, no biochemical process is 100% efficient). Thus, minor populations of PrP must follow non-canonical (and, in many cases, presumably aberrant) pathways. Such typically minor ‘aberrant’ species are usually not observed and are difficult to quantify, largely owing to robust and vigorous quality control systems that promptly recognize and dispose of them. Recently, an increasing appreciation of cellular quality control systems and the generation of precise tools for their manipulation and analysis have enabled various minor species to be uncovered.
Insights into PrP translocation from in vitro studies
Some of the earliest studies that showed heterogeneity in PrP biosynthesis involved in vitro systems that reconstituted the initial PrP translocation process at the ER. Oddly, subpopulations of PrP (varying from <10% to >50%), rather than being fully translocated into the lumen, were either retained in the cytosol or made as partially translocated transmembrane proteins [16–18]. Because none of these forms was readily observed in vivo, these minor species were considered to be in vitro artifacts. Although heterologous and intrinsically less efficient than in vivo, the in vitro system nonetheless pointed to potential alternative outcomes of PrP translocation. Thus, the in vitro results can be viewed as having grossly exaggerated the inefficiencies that would presumably also occur in vivo, but at markedly lower levels.
Subsequent studies analyzing the molecular basis of PrP translocation [19–23] revealed more precisely the nature and mechanisms of cytosolic and transmembrane PrP generation (Figure 2). As PrP begins synthesis, its N-terminal signal sequence is recognized by signal recognition particle and the entire ribosome–nascent chain complex is delivered to the ER. The next step, in which the signal interacts with the Sec61 translocation channel, proved to be incompletely efficient. Whereas model signal sequences, such as the one from the hormone prolactin, efficiently interacted with and gated open the Sec61 channel, the PrP signal was less effective. Consequently, a subpopulation of PrP slipped out of the Sec61 channel, either before or shortly after initiating translocation, and was exposed to the cytosol. Because these events occur co-translationally, continued protein synthesis releases the nascent PrP polypeptide further into the cytosol through a gap between the ribosome and Sec61.
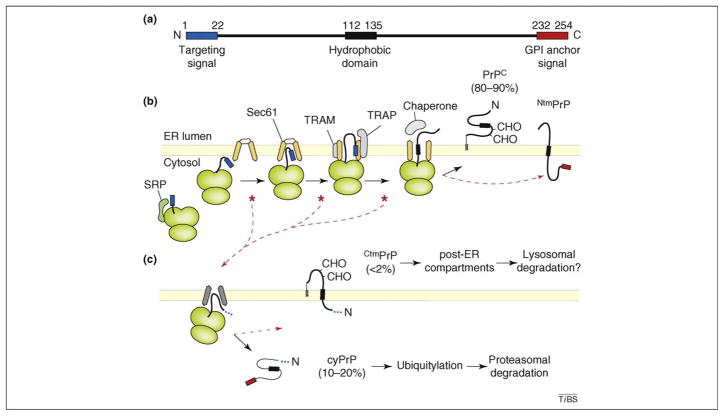
Figure 2.
Summary of the co-translational targeting and translocation of PrP into the ER, highlighting key steps that lead to the generation of its multiple isoforms. (a) Line diagram of PrP showing the location of its N-terminal signal sequence (blue), central hydrophobic domain (HD; black) and C-terminal GPI-anchoring sequence (red). (b) Important steps in PrP translocation taken by the majority (~80%) of molecules. As the N-terminal signal sequence emerges from the ribosome, it is recognized by the signal recognition particle (SRP) and targeted to the Sec61 translocon. The signal sequence then interacts with Sec61 and, with the aid of accessory factors such as TRAM and TRAP, gates open the Sec61 channel to initiate translocation. Forward transport into the lumen (or prevention of slippage back to the cytosol) might require chaperones. Sites of known or potential inefficiency of the signal sequence that lead to slipping of the N terminus into the cytosol are indicated by red asterisks. During translocation in vitro, some PrP molecules insert into the membrane to generate NtmPrP, a poorly studied form of which the relevance or existence in vivo remains to be studied. CHO denotes N-linked glycans. (c) Consequences of signal inefficiency. The ellipsis in place of the signal sequence indicates that both signal-containing and signal-cleaved molecules can be generated, depending on the precise step at which PrP slipped in part (b). Engagement of the translocon by the HD generates CtmPrP, whereas lack of engagement results in cyPrP. CtmPrP is typically a minor species, but it can be increased by mutations that raise HD hydrophobicity. Shown below each PrP species are their relative amounts thought to be generated in cells (not necessarily their steady state levels). Lysosomal degradation is presumed but has not been experimentally demonstrated yet.
Eventually a central hydrophobic domain (HD; residues ~112–135) emerges from the ribosome. For polypeptides that did not slip out of the Sec61 channel at the earlier step, this domain is predominantly translocated through the Sec61 channel, resulting in fully translocated PrP that subsequently becomes attached to the inner leaflet of the lipid bilayer via a GPI anchor [19,21,23]. However, translocation of the HD through the translocon is not completely efficient in vitro, and a population of it inserts into the membrane, generating NtmPrP, of which the N-terminus resides in the ER and the C-terminus resides in the cytosol [18–21]. Although this form also might be generated at very low levels in vivo, it has not been studied and, therefore, is not discussed further in this review.
For the minor fraction of PrP that slipped out of the translocon, the HD can potentially engage the Sec61 complex in a manner analogous to a signal sequence. This results in translocation of the C terminus and membrane insertion of the HD, generating a transmembrane form denoted CtmPrP [18–21]. If the HD does not engage the translocon, PrP is released in the cytosol to generate cytosolic PrP (cyPrP). Thus, inefficiency in the signal–Sec61 translocon interaction leads to a small proportion of PrP molecules being made in either the cytosol or, to a lesser extent, as CtmPrP [19]. As might be predicted, increasing the hydrophobicity of the HD will enable it to better engage the translocon, generating more CtmPrP [18,21,23]. Conversely, decreasing HD hydrophobicity decreases CtmPrP production. In addition, improving or weakening the signal–Sec61 interaction leads to less or more cyPrP and CtmPrP generation, respectively [19–23]. Thus, a combination of the signal and HD, and their relative efficacies in engaging Sec61, determines the amount of cyPrP and CtmPrP that are generated [21,23].
In addition to the analysis of cis-acting sequences within PrP, parallel studies analyzed the machinery required to translocate PrP across the membrane. In addition to Sec61, reconstitution studies showed that the translocon-associated protein (TRAP) complex stimulates PrP translocation at the crucial step of signal-mediated initiation of transport [24]. Other factors, such as the translocating chain associating membrane protein (TRAM), might also maximize the functionality of certain signal sequences in initiating translocation [25]. Thus, the picture that has emerged of PrP translocation from these in vitro studies is one of relative inefficiency and unexpected dependence on additional factors not required by ‘model’ proteins.
In vivo analyses of PrP translocation and metabolism
Analyses of these same events using in vivo systems is challenging because of their relatively diminished manipulability. Nonetheless, slightly inefficient PrP translocation can indeed be detected if the degradation of the non-translocated (and, hence, non-glycosylated and non-processed) population is prevented by proteasome inhibitors [26–29]. That this cyPrP accumulation results from failed translocation (as opposed to retrotranslocation from the ER lumen) can be readily demonstrated by replacing the PrP signal sequence with signals from proteins predicted to be more efficiently translocated. In this instance, proteasome inhibition leads to little or no cyPrP accumulation [28].
Furthermore, sensitive assays to detect the non-translocated population (by fusion to a reporter that is only active in the cytosol) revealed that the PrP signal is indeed slightly inefficient compared with those from model proteins [30]. Thus, PrP signal inefficiency from in vitro studies can be verified in cells, but so too can the relative disparities among signal efficiencies. In addition, as predicted from in vitro studies, manipulating HD hydrophobicity and/or the signal sequence can influence the generation of CtmPrP in predictable ways [23,31]. Unlike cyPrP, CtmPrP is likely to be degraded in the lysosome in vivo, as is typical for membrane proteins that leave the ER (as CtmPrP apparently does [18,32]). These findings suggest that, although artifactually exaggerated in vitro, the basic model of PrP translocation for CtmPrP and cyPrP biogenesis is likely to be valid in vivo.
More puzzling, however, is the issue of why PrP (and, as it turns out, many other proteins) contains a less than perfect signal sequence, the proper function of which depends on accessory factors. One possibility is that it serves a regulatory purpose: by making a decisive step in translocation depend on multiple factors, the cell can selectively regulate the entry of proteins into the ER under different conditions [33]. Recent studies show that PrP translocation efficiency indeed does change under different conditions, being selectively reduced during ER stress [34,35]. Remarkably, the ER stress-induced changes in PrP translocation depend on its signal sequence [34]. Replacing the native PrP signal with a more efficient model signal permitted efficient translocation even during ER stress. This forced constitutive translocation caused PrP to misfold in the ER lumen during ongoing stress, in lieu of being degraded in the cytosol. Because misfolded PrP in the ER seems to be a poor substrate for ER-associated degradation, this misfolding decreases cell viability [34]. Interestingly, under a different type of stress (proteasome inhibition), an efficient signal sequence on PrP was beneficial because it precluded the accumulation of cyPrP aggregates [28]. Thus, it seems that PrP tolerates some inefficiency in its signal sequence under normal conditions (and the potential risk of toxicity during impaired proteasomal degradation) because of the advantage it provides during ER stress [34].
These analyses of PrP biosynthesis in vitro and in cells lead to several conclusions. First, in addition to the major population of properly translocated, glycosylated and processed PrP, minor cytosolic and transmembrane forms are generated. Second, production of cyPrP and CtmPrP results from intrinsic inefficiencies in normal biological reactions (i.e. the interaction between the signal sequence of PrP and the Sec61 translocon). Third, these minor species, owing to their low abundance and transient existence, are difficult to detect in vivo without additional manipulation. Fourth, the increased complexity of PrP biosynthesis instigated by low-level basal cellular inefficiencies enables cells to control PrP transloction. And fifth, regulatable PrP translocation is apparently important for avoiding excessive PrP misfolding during conditions such as ER stress, thus providing an explanation for why the weakness of the PrP signal sequence has been evolutionarily conserved [20,21].
Deviation from normal levels of PrP forms can cause neurodegeneration
Remarkably, CtmPrP and cyPrP, although apparently low-level normal byproducts of PrP biosynthesis, can be detrimental in larger amounts in vivo. Mutations that increase the hydrophobicity of the HD lead to increased CtmPrP in vitro, in cultured cells and in transgenic mice. Although CtmPrP levels in wild-type mice are very low (~1% of total), the levels in HD mutants can range from 5–20% of total PrP [36]. A strong correlation between CtmPrP levels and the severity of neurodegenerative disease suggests that this form of PrP can be harmful to a subset of cells in the central nervous system [18,36,37]. Subsequent studies that further exaggerated CtmPrP production by a combination of a weakened signal sequence and increased HD hydrophobicity led to an even more severe neurodegenerative phenotype in mice [37]. This finding not only further validated the model of PrP biosynthesis in vivo but also lent additional support to the idea that elevated CtmPrP levels are detrimental. Importantly, at least five naturally occurring mutations in human PRNP (resulting in Pro105-Leu, Gly114Val, Ala117Val, Gly131Val and Ala133Val) have been found that increase HD hydrophobicity, and one of these (Ala117Val) causes neurodegeneration when expressed in mice [36]. Thus, this normally minor species can have a role in at least some forms of naturally occurring PrP-mediated neurodegenerative disease.
What about the cytosolic form of PrP? Here, too, analysis of transgenic mice and human disease mutants suggests that increased levels can be detrimental. Although non-physiologic, expression of a PrP mutant that lacks the signal sequence (and, hence, produces exclusively cyPrP) causes an atypical neurodegenerative disease in mice [38]. More recently, a mild neurodegenerative disease was observed in mice expressing a version of PrP that contains a less efficient signal sequence (designed to mimic the reduced PrP translocation seen during ER stress) [31]. In both cases, the steady-state cyPrP levels in the brain were very low (but detectable), which is consistent with its rapid proteasomal degradation as expected from cell-culture studies. In humans, two nonsense mutants in PRNP (at residues 145 and 160) that are associated with neurodegeneration seem to generate increased cyPrP owing to inefficient translocation into the ER [39,40].
Curiously, the adverse consequences of cyPrP seemed to be highly regional and cell-type specific despite widespread expression within and outside the central nervous system [31,38]. Indeed, cyPrP expression in cultured cells showed toxicity in some studies [38] but was inert or even protective in others [41,42]. Furthermore, there might be cell types that naturally express cyPrP in vivo [43], perhaps indicating a normal function in some contexts. Although the full role of cyPrP remains to be elucidated, it seems clear that, like CtmPrP, very low amounts are potentially cytotoxic in a tissue-specific (primarily the central nervous system) and cell-type-selective (specific subsets of neurons) manner in vivo. At least one of these forms is associated with some familial forms of neurodegeneration caused by PRNP mutations [36]. Importantly, neither cyPrP nor CtmPrP is transmissible [36,44]. They apparently elicit their effects by causing some type of proteinopathy, in which increased levels of an aberrant form of a normal protein cause cellular dysfunction (by downstream mechanisms that remain to be elucidated). What seems clear is that among the various naturally occurring forms of PrP, these two forms are most directly implicated in neurotoxicity and are causative of neurodegeneration.
Linking PrP metabolism to neurodegeneration in transmissible prion diseases
How then does our understanding of PrP biosynthesis and metabolism, combined with the (still limited) insight into non-transmissible neurodegenerative illnesses seen in certain transgenic mouse models, provide any insight into transmissible prion disease? One possibility is that the two are, in fact, completely unrelated; perhaps the phenotypes caused by cyPrP or CtmPrP share little or no mechanistic commonalities with transmissible (i.e. PrPSc-mediated) prion diseases. Certainly, it is abundantly clear that neither CtmPrP nor cyPrP favors spontaneous conversion to PrPSc, making a connection in this direction unlikely [36,44]. However, it remains a plausible hypothesis that PrPSc accumulation might, either directly or indirectly, lead to the increased generation or stabilization of cyPrP and/or CtmPrP (Figure 3). Given that both cyPrP and CtmPrP are known to be detrimental in larger than normal amounts, their accumulation could cause cellular dysfunction in some cell types.
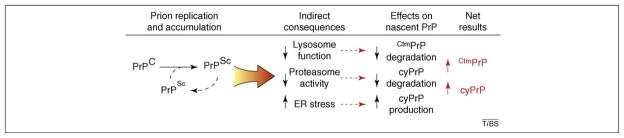
Figure 3.
Speculative working model of prion disease pathogenesis. Templated replication of PrPSc from PrPC leads to PrPSc accumulation. This has several indirect consequences for cellular function, three of which are indicated. The effect of each consequence on nascent PrP metabolism is listed, along with the net result of increased cyPrP and CtmPrP, which are both capable of causing neurodegeneration.
A key to this hypothesis is that PrPSc accumulation should have effects that impinge on cyPrP and/or CtmPrP metabolism. At least three possibilities have been described. First, it has long been known that PrPSc accumulates (and perhaps is generated) at endosomes and lysosomes [9,45–47]. Although the consequences of its accumulation here have been poorly studied, several lines of evidence suggest impairment of several lysosomal functions. For example, components normally degraded in the lysosome, such as lipofuscin, increase upon PrPSc accumulation. In addition, lysosomal hydrolases are upregulated in PrPSc-infected tissue, which is perhaps indicative of a cellular response to lysosomal insufficiency [46,48]. CtmPrP, which traffics to post-ER compartments of the secretory pathway (and, hence, acquires Golgi-specific modification [18,32]), probably relies on lysosomal function for its degradation. Second, PrPSc accumulation causes upregulation of various genes involved in ER function [49]. Such gene-expression patterns are often observed after activation of the unfolded protein response, and, indeed, studies in cell culture showed that PrPSc, perhaps via Ca+2 homeostasis dysregulation, causes ER stress [50]. Therefore, ER stress might have a role in transmissible prion disease [49]. PrP translocation into the ER is decreased during ER stress, resulting in increased cyPrP production [34,35]. Third, reduced proteasome system activity, a common feature of many neurodegenerative diseases, also occurs during the course of prion disease and PrPSc accumulation [51].
Thus, there seem to be at least three plausible ways to tie PrPSc to downstream cellular toxicity via cyPrP or CtmPrP. In one study, the role of ER-stress-mediated increases in cyPrP in neurodegeneration was investigated. Transgenic mice were produced that express, at modest levels, a version of PrP with reduced translocation at levels expected to occur during mild ER stress. This manipulation alone was sufficient to cause a mild neurodegenerative disease [31]. Of course, PrPSc accumulation has effects beyond just ER stress, including proteasome inhibition [51]. Thus, reduced translocation combined with partial proteasome impairment could cause a more severe phenotype. Another study observed a correlation between CtmPrP production and the apparent toxicity of PrPSc in mice [36]: an HD mutant that leads to a slight increase in CtmPrP production was highly sensitive to PrPSc accumulation, succumbing to infection by ~60 days (as compared with ~75 days for mice expressing wild-type PrP at comparable levels). By comparison, a PrP mutant that produces lower than normal levels of CtmPrP tolerated greater PrPSc accumulation before showing disease at over 300 days. Indirect assays showed that CtmPrP levels might increase concomitantly with PrPSc accumulation [36]. These results are consistent with the notion that PrPSc accumulation can stabilize (or possibly increase the production of) CtmPrP and that the resulting increase in CtmPrP levels leads to adverse downstream consequences.
A testable model for prion disease pathogenesis
A potentially unifying model of prion disease pathogenesis can, therefore, be proposed (Figure 3). In this model, transmissible prion diseases are characterized by PrPSc, which upon interaction with host PrPC causes its conversion to additional PrPSc [1–3]. This conversion occurs in myriad different ways depending on subtle differences in PrPSc folding (i.e. ‘strains’ of prions), thereby influencing their location of accumulation (i.e. which regions of the brain are affected). The accumulated PrPSc, again in a strain-dependent manner, would influence several cellular processes to varying extents including ER stress [49], proteasome activity [51] and lysosomal function [46]. The severity of each perturbation is likely to be cell-type dependent given that the capacities and susceptibility to perturbation of each of these pathways vary widely. Consequently, the normally minor cyPrP and CtmPrP isoforms experience increased generation and/or decreased degradation. These isoforms, the transgenic expression of which elicits cell-type-selective effects [18,31,36–38], could have several downstream consequences. Because each of these events affects different cell types to different extents, it is possible that the most severe pathology will only be seen where multiple consequences intersect. Such a requirement might explain the regional pathology often observed in these diseases.
An important feature of this model is that it is eminently testable. As additional mechanistic insights are gained into PrP biosynthesis and degradation, specific events hypothesized to have a role in disease can be manipulated and tested in vivo. For example, in vitro and cell-culture studies indicate that CtmPrP generated by disease-causing HD mutants requires the slightly inefficient PrP signal sequence [19–21]. The utilization of more efficient signal sequences [20,30] should make it possible to reduce CtmPrP generation [19]. It will then be possible to test in transgenic mice whether the effects of the HD mutant stem solely from CtmPrP production or through other mechanisms (e.g. on PrP folding or some unknown activity). Similarly, the contribution of reduced PrP translocation during ER stress to PrPSc-mediated neurodegeneration can be tested by generating mice that express a version of PrP of which the translocation is constitutively efficient. Thus, one of the most practical consequences of a mechanistic understanding of PrP cell biology will be the tools it provides for testing various hypotheses with very high specificity.
Similarly, other minor forms of PrP (generated by inefficiencies in other cellular processes) could remain to be elucidated. These could include misfolded (but normally translocated) molecules of PrP that, although normally constituting a very small population, could be increased in the case of inherited disease-causing mutants or adverse cellular conditions. Such a hypothesis has often been invoked to explain altered metabolism, trafficking or biochemical behavior of various natural and disease-causing mutants [27,52–59]. Although most studies attempt to determine whether a given misfolded population shares ‘PrPSc-like’ features, these assays are typically nonspecific (e.g. relative protease-resistance and insolubility) and cannot be equated to infectivity. Instead, the connection between the misfolded PrPs (e.g. as caused by any of various inherited PrP mutants) and PrPSc might be more indirect. Thus, it would be a worthwhile endeavor to fully characterize the molecular events involved in misfolded PrP generation, trafficking and degradation so that they can be manipulated selectively to test their role in disease.
Concluding remarks and future perspectives
A common theme that emerges from this model of neurodegeneration is that inappropriate PrP localization can be detrimental. In considering CtmPrP, PrP is mislocalized in two ways: (i) it is embedded in the membrane and (ii) it might reside in different cellular locales than PrPC. Cytosolic PrP localizes to an entirely different cellular compartment than normal. In both cases, at least part of PrP (the N terminus) is exposed to the cytosol, an environment that typically does not see PrP at all. Why might this mislocalization be detrimental in a dominant manner? The simplest mechanism is that a protein in an incorrect environment makes inappropriate interactions that disrupt or otherwise alter the functionality of the interacting partners. For example, it could titrate both protein and non-protein cellular factor(s), change the biochemical activity of key factors, initiate inappropriate signaling cascades or disrupt cellular structures [60–62]. Intriguingly, it was recently demonstrated that mice lacking HSF1, a key regulator of the cytosolic stress response, are more susceptible to the adverse consequences of PrPSc [63], perhaps pointing to the cytosol as a site of dysfunction during disease.
But are such dramatic effects really plausible from such minor species of PrP? The answer to this question depends not only on the proportion of PrP in these minor forms but also on their absolute amounts. Protein abundances in a typical cell span around five orders of magnitude between the least and most abundant. PrP, at least in the central nervous system, is present at the higher end of abundance, estimated at between 70 and 400 micrograms per gram of total protein [64,65] (thus, ~0.01% to 0.04% by mass; perhaps higher in molar amounts). Because there are several crucial cellular factors, the levels of which are more than two orders of magnitude lower than that of of PrP, even 1% of normal PrP levels (i.e. ~1 microgram per gram of total protein) would exceed their abundance. Thus, seemingly low levels of cyPrP or CtmPrP (as judged by proportion) are not trivial and can easily have substantial consequences including physical titration of cellular factors. Precisely such mechanisms are beginning to emerge in other examples of protein-misfolding diseases.
For example, animal models of polyglutamine diseases suggest that misfolded-protein aggregates can sequester specific transcription factors, cytoskeletal proteins, autophagy factors, degradative proteins and molecular chaperones [66]. Even a transient association, as is observed between heat shock protein 70 and polyglutamine aggregates, could have downstream consequences including decreased fidelity of protein folding or quality control [67]. Toxic sequestration models have also been suggested for familial amyotrophic lateral sclerosis, Alzheimer disease and Parkinson disease, although the basis for these heterologous molecular interactions is less well established. Thus, elucidating putatively inappropriate interactions of a misfolded, misprocessed or mislocalized protein is likely to be essential in understanding the mechanisms underlying dominant neurodegenerative diseases. Unraveling such protein–protein interactions for cytosolically exposed PrP could be an important crucial step in further understanding the actual pathway for the onset of neuropathogenesis in prion diseases. A comprehensive and quantitative understanding of normal PrP cell biology, detailing its sorting, trafficking, interactions and degradation, will continue to yield new insights into how its derangement can lead to disease.
Acknowledgments
Work in our laboratory is supported by the Intramural Research Program of the National Institute of Child Health and Human Development (www.nichd.nih.gov) at the National Institutes of Health.
References
- 1.Aguzzi A, et al. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- 2.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB, et al. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 4.Wickner RB, et al. Prion genetics: new rules for a new kind of gene. Annu Rev Genet. 2004;38:681–707. doi: 10.1146/annurev.genet.38.072902.092200. [DOI] [PubMed] [Google Scholar]
- 5.Greil CS, et al. Acute cellular uptake of abnormal prion protein is cell type and scrapie-strain independent. Virology. 2008;379:284–293. doi: 10.1016/j.virol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovacs GG, et al. Mutations of the prion protein gene phenotypic spectrum. J Neurol. 2002;249:1567–1582. doi: 10.1007/s00415-002-0896-9. [DOI] [PubMed] [Google Scholar]
- 7.Tateishi J, Kitamoto T. Inherited prion diseases and transmission to rodents. Brain Pathol. 1995;5:53–59. doi: 10.1111/j.1750-3639.1995.tb00577.x. [DOI] [PubMed] [Google Scholar]
- 8.Tateishi J, et al. Experimental transmission of Creutzfeldt-Jakob disease and related diseases to rodents. Neurology. 1996;46:532–537. doi: 10.1212/wnl.46.2.532. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs GG, Budka H. Prion diseases: from protein to cell pathology. Am J Pathol. 2008;172:555–565. doi: 10.2353/ajpath.2008.070442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 12.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 13.Taylor DR, Hooper NM. The prion protein and lipid rafts. Mol Membr Biol. 2006;23:89–99. doi: 10.1080/09687860500449994. [DOI] [PubMed] [Google Scholar]
- 14.Borchelt DR, et al. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caughey B, et al. Prion protein biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J Virol. 1989;63:175–181. doi: 10.1128/jvi.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay B, et al. Biogenesis and transmembrane orientation of the cellular isoform of the scrapie prion protein. Mol Cell Biol. 1987;7:914–920. doi: 10.1128/mcb.7.2.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay B, et al. Evidence for a secretory form of the cellular prion protein. Biochemistry. 1987;26:8110–8115. doi: 10.1021/bi00399a014. [DOI] [PubMed] [Google Scholar]
- 18.Hegde RS, et al. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 19.Kim SJ, Hegde RS. Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol Biol Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SJ, et al. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- 21.Kim SJ, et al. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J Biol Chem. 2001;276:26132–26140. doi: 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- 22.Rutkowski DT, et al. Substrate-specific regulation of the ribosome- translocon junction by N-terminal signal sequences. Proc Natl Acad Sci U S A. 2001;98:7823–7828. doi: 10.1073/pnas.141125098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart RS, Harris DA. Mutational analysis of topological determinants in prion protein (PrP) and measurement of transmembrane and cytosolic PrP during prion infection. J Biol Chem. 2003;278:45960–45968. doi: 10.1074/jbc.M307833200. [DOI] [PubMed] [Google Scholar]
- 24.Fons RD, et al. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voigt S, et al. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drisaldi B, et al. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J Biol Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- 27.Ma J, Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci U S A. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rane NS, et al. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yedidia Y, et al. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 2001;20:5383–5391. doi: 10.1093/emboj/20.19.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine CG, et al. The efficiency of protein compartmentalization into the secretory pathway. Mol Biol Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rane NS, et al. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell. 2008;15:359–37. doi: 10.1016/j.devcel.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart RS, Harris DA. A transmembrane form of the prion protein is localized in the Golgi apparatus of neurons. J Biol Chem. 2005;280:15855–15864. doi: 10.1074/jbc.M412298200. [DOI] [PubMed] [Google Scholar]
- 33.Hegde RS, Kang SW. The concept of translocational regulation. J Cell Biol. 2008;182:225–232. doi: 10.1083/jcb.200804157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang SW, et al. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orsi A, et al. Conditions of endoplasmic reticulum stress favor the accumulation of cytosolic prion protein. J Biol Chem. 2006;281:30431–30438. doi: 10.1074/jbc.M605320200. [DOI] [PubMed] [Google Scholar]
- 36.Hegde RS, et al. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- 37.Stewart RS, et al. Neurodegenerative illness in transgenic mice expressing a transmembrane form of the prion protein. J Neurosci. 2005;25:3469–3477. doi: 10.1523/JNEUROSCI.0105-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, et al. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 39.Zanusso G, et al. Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J Biol Chem. 1999;274:23396–23404. doi: 10.1074/jbc.274.33.23396. [DOI] [PubMed] [Google Scholar]
- 40.Heske J, et al. The C-terminal globular domain of the prion protein is necessary and sufficient for import into the endoplasmic reticulum. J Biol Chem. 2004;279:5435–5443. doi: 10.1074/jbc.M309570200. [DOI] [PubMed] [Google Scholar]
- 41.Roucou X, et al. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J Biol Chem. 2003;278:40877–40881. doi: 10.1074/jbc.M306177200. [DOI] [PubMed] [Google Scholar]
- 42.Fioriti L, et al. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J Biol Chem. 2005;280:11320–11328. doi: 10.1074/jbc.M412441200. [DOI] [PubMed] [Google Scholar]
- 43.Mironov A, Jr, et al. Cytosolic prion protein in neurons. J Neurosci. 2003;23:7183–7193. doi: 10.1523/JNEUROSCI.23-18-07183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norstrom EM, et al. Cytosolic prion protein toxicity is independent of cellular prion protein expression and prion propagation. J Virol. 2007;81:2831–2837. doi: 10.1128/JVI.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold JE, et al. The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. J Pathol. 1995;176:403–411. doi: 10.1002/path.1711760412. [DOI] [PubMed] [Google Scholar]
- 46.Kovacs GG, et al. Involvement of the endosomal-lysosomal system correlates with regional pathology in Creutzfeldt-Jakob disease. J Neuropathol Exp Neurol. 2007;66:628–636. doi: 10.1097/nen.0b013e318093ecc7. [DOI] [PubMed] [Google Scholar]
- 47.Laszlo L, et al. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J Pathol. 1992;166:333–341. doi: 10.1002/path.1711660404. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Up-regulation of cathepsin B and cathepsin L activities in scrapie-infected mouse Neuro2a cells. J Gen Virol. 2003;84:2279–2283. doi: 10.1099/vir.0.19153-0. [DOI] [PubMed] [Google Scholar]
- 49.Hetz CA, Soto C. Stressing out the ER: a role of the unfolded protein response in prion-related disorders. Curr Mol Med. 2006;6:37–43. doi: 10.2174/156652406775574578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hetz C, et al. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J. 2003;22:5435–5445. doi: 10.1093/emboj/cdg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kristiansen M, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Capellari S, et al. Effect of the E200K mutation on prion protein metabolism. Comparative study of a cell model and human brain. Am J Pathol. 2000;157:613–622. doi: 10.1016/S0002-9440(10)64572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen E, Taraboulos A. Scrapie-like prion protein accumulates in aggresomes of cyclosporin A-treated cells. EMBO J. 2003;22:404–417. doi: 10.1093/emboj/cdg045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanova L, et al. Mutant prion proteins are partially retained in the endoplasmic reticulum. J Biol Chem. 2001;276:42409–42421. doi: 10.1074/jbc.M106928200. [DOI] [PubMed] [Google Scholar]
- 55.Jin T, et al. The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J Biol Chem. 2000;275:38699–38704. doi: 10.1074/jbc.M005543200. [DOI] [PubMed] [Google Scholar]
- 56.Kiachopoulos S, et al. Pathogenic mutations located in the hydrophobic core of the prion protein interfere with folding and attachment of the glycosylphosphatidylinositol anchor. J Biol Chem. 2005;280:9320–9329. doi: 10.1074/jbc.M412525200. [DOI] [PubMed] [Google Scholar]
- 57.Lehmann S, Harris DA. Two mutant prion proteins expressed in cultured cells acquire biochemical properties reminiscent of the scrapie isoform. Proc Natl Acad Sci U S A. 1996;93:5610–5614. doi: 10.1073/pnas.93.11.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorenz H, et al. Cellular phenotyping of secretory and nuclear prion proteins associated with inherited prion diseases. J Biol Chem. 2002;277:8508–8516. doi: 10.1074/jbc.M110197200. [DOI] [PubMed] [Google Scholar]
- 59.Yin S, et al. Human prion proteins with pathogenic mutations share common conformational changes resulting in enhanced binding to glycosaminoglycans. Proc Natl Acad Sci U S A. 2007;104:7546–7551. doi: 10.1073/pnas.0610827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambold AS, et al. Association of Bcl-2 with misfolded prion protein is linked to the toxic potential of cytosolic PrP. Mol Biol Cell. 2006;17:3356–3368. doi: 10.1091/mbc.E06-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, et al. The interaction between cytoplasmic prion protein and the hydrophobic lipid core of membrane correlates with neurotoxicity. J Biol Chem. 2006;281:13559–13565. doi: 10.1074/jbc.M512306200. [DOI] [PubMed] [Google Scholar]
- 62.Spielhaupter C, Schatzl HM. PrPC directly interacts with proteins involved in signaling pathways. J Biol Chem. 2001;276:44604–44612. doi: 10.1074/jbc.M103289200. [DOI] [PubMed] [Google Scholar]
- 63.Steele AD, et al. Heat shock factor 1 regulates lifespan as distinct from disease onset in prion disease. Proc Natl Acad Sci U S A. 2008;105:13626–13631. doi: 10.1073/pnas.0806319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bendheim PE, et al. Purification and partial characterization of the normal cellular homologue of the scrapie agent protein. J Infect Dis. 1988;158:1198–1208. doi: 10.1093/infdis/158.6.1198. [DOI] [PubMed] [Google Scholar]
- 65.Pan KM, et al. Purification and properties of the cellular prion protein from Syrian hamster brain. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 67.Gidalevitz T, et al. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 68.Bueler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 69.Mallucci GR, et al. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 2002;21:202–210. doi: 10.1093/emboj/21.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prusiner SB, et al. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci U S A. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brandner S, et al. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 72.Bueler H, et al. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- 73.Mallucci G, et al. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 74.Mallucci GR, et al. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Chesebro B, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 76.Shmerling D, et al. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- 77.Li A, et al. Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105–125. EMBO J. 2007;26:548–558. doi: 10.1038/sj.emboj.7601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baumann F, et al. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J. 2007;26:538–547. doi: 10.1038/sj.emboj.7601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakaguchi S. Antagonistic roles of the N-terminal domain of prion protein to doppel. Prion. 2008;2:107–111. doi: 10.4161/pri.2.3.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]