Abstract
After initiation of transcription, a number of proteins participate during elongation and termination modifying the properties of the RNA polymerase (RNAP). Gre factors are one such group conserved across bacteria. They regulate transcription by projecting their N-terminal coiled-coil domain into the active center of RNAP through the secondary channel and stimulating hydrolysis of the newly synthesized RNA in backtracked elongation complexes. Rv1080c is a putative gre factor (MtbGre) in the genome of Mycobacterium tuberculosis. The protein enhanced the efficiency of promoter clearance by lowering abortive transcription and also rescued arrested and paused elongation complexes on the GC rich mycobacterial template. Although MtbGre is similar in domain organization and shares key residues for catalysis and RNAP interaction with the Gre factors of Escherichia coli, it could not complement an E. coli gre deficient strain. Moreover, MtbGre failed to rescue E. coli RNAP stalled elongation complexes, indicating the importance of specific protein-protein interactions for transcript cleavage. Decrease in the level of MtbGre reduced the bacterial survival by several fold indicating its essential role in mycobacteria. Another Gre homolog, Rv3788 was not functional in transcript cleavage activity indicating that a single Gre is sufficient for efficient transcription of the M. tuberculosis genome.
Introduction
Once the process of transcription is initiated by RNAP, it is important for the enzyme to carry out elongation and termination to ensure the full-length RNA synthesis. However, the movement of the RNAP along the template during the transcription elongation is not uniform and gets interrupted either accidentally or due to regulatory mechanisms [1]. Inadvertent disruption of the elongation complex would lead to the accumulation of non-functional RNA which can be potentially deleterious to the cell [2]. To overcome these interruptions, a number of transcription factors act during elongation and termination by modifying the properties of RNAP [1], [3], [4]. These factors deal with the accidental disruption of the elongation process and affect transcription processivity and fidelity by modulating pausing, arrest, termination or anti-termination of the enzyme [1], [5]. Prokaryotic transcript cleavage factors GreA and GreB [6], [7] and their eukaryotic analog, elongation factor TFIIS [8], stimulate intrinsic transcript cleavage activity of RNAP [9], [10] for removal of the aberrant RNA 3′ ends so that polymerization activity can be restored from the end of a cleaved RNA. They suppress the RNAP pausing to rescue arrested [7], [11] or road-blocked [12] transcription complexes, providing RNAP a second chance to resume elongation [13] by directly accessing the RNAP active center through the secondary channel [10], [14]. Although homologs of the Gre factors are found in most bacteria, they are well characterized only from a few species viz. E. coli [6], [7], Thermus thermophilus and Thermus aquaticus [15], [16], [17]. No information on the properties of the transcript cleavage factors is available from genus mycobacteria which harbors several pathogenic species. In this manuscript we describe the characteristics of M. tuberculosis Gre factor.
The genome of M. tuberculosis harbors a gre factor - Rv1080c [18], sharing 32% and 26% identity (48% and 43% similarity) with the E. coli GreA and GreB respectively. Other ORFs which show low degree of similarity with the E. coli Gre factors in the genome are Rv3788 which shares 16% identity and 33% similarity with the E. coli GreA (Figure S1A) and Rv2103 – a hypothetical protein, having much lower similarity (9% identity and 21% similarity with E. coli GreA). The former has Gre like domain organization while the latter lacks key acidic amino acids and the domains required for Gre like activity.
A number of molecular processes show significant differences in mycobacteria compared to the other well-studied bacterial systems [19]. Presence of a large number of sigma factors recognizing unique sequences of the promoters in their GC rich genomes [20], slow rates of transcription and macromolecular synthesis [21], [22] and occurrence of novel transcription activators [18] etc. point towards the differences in the transcription process. The GC rich genome of M. tuberculosis (65.6% G+C) may pose additional challenges to the transcribing RNAP and hence the role of Gre factor could be critical for high fidelity transcription. We demonstrate that Rv1080c, the primary Gre factor of the genome is essential for cell survival unlike the Gre factors characterized from other eubacteria. The protein is needed for efficient promoter escape by reducing the abortive initiation and anti-arrest action during transcription elongation. Although its properties resemble E. coli GreA in many respects, it does not appear to collaborate with E. coli RNAP during elongation process and much of its properties seem to be tailored for the mycobacterial transcription.
Results
Rv1080c has Gre factor like domain organization
Rv1080c encodes for a 164 amino-acid protein having sequence similarity with the E. coli transcript cleavage factors GreA and GreB (Figure S1). A homology model of the protein was generated by using the crystal structure of E. coli GreA (PDB code:1GRJ) [23] as a template (Figure S1B). GreA and GreB of E. coli have two distinct domains: an N-terminal coiled-coil (Gre-NTD) and a C-terminal globular domain (Gre-CTD) [14], [24], [25]. NTD is responsible for the stimulation of specific nucleolytic and anti-arrest activities, whereas the residues in Gre-CTD interact with RNAP-β′ subunit coiled-coil domain [26], [27]. From the model, it is evident that Rv1080c is more similar to the E. coli GreA than GreB in its surface charge distribution (Figure S1C). The homology model of Rv3788, the other Gre homolog in the M. tuberculosis genome, shows that most of the features of the Gre factor are conserved in the ORF (Figure S1A, S1B and S1C). The M. smegmatis Gre (MsGre) has 97% similarity with the M. tuberculosis protein in the amino acid sequence and shares similar domain architecture. To understand the function and the nature of transcript cleavage stimulatory activity of mycobacterial Gre factor and the Gre factor homolog Rv3788, the genes were cloned in pET20b for over-expression of the ∼18 kDa proteins in E. coli (Figure S2A, S2B and S2C). The identities of the expressed proteins were confirmed by peptide-mass-fingerprinting using MALDI-TOF (data not shown).
MtbGre stimulates the intrinsic cleavage activity of mycobacterial RNAP
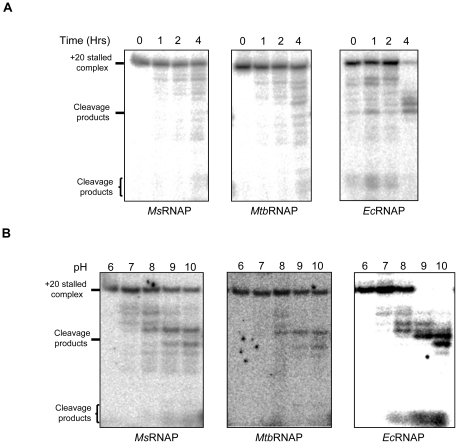
A stalled elongation complex comprising of 20 nt RNA was generated from the T7A1 promoter (T7A1-TEC) for studying transcript cleavage on the elongation complexes (Figure S3A). RNAP from both M. smegmatis (MsRNAP) and M. tuberculosis (MtbRNAP) were proficient in carrying out transcription from this template (Figure S3B). Transcript cleavage is an intrinsic property of the catalytic center of the RNAP [9] but is very slow and requires prolonged incubation. First, this intrinsic cleavage activity of the enzymes from E. coli, M. smegmatis and M. tuberculosis were compared. In all the three enzyme systems, RNA fragments of varied length were generated after incubation for a few hrs. Varied amount of short RNA fragments generated from the 3′ end of the stalled TEC could be detected at the bottom of the gels (Figure 1A). Both MtbRNAP and MsRNAP had lower intrinsic cleavage compared to E. coli RNAP (EcRNAP) (Figure 1A), but the cleavage activity was stimulated in alkaline pH similar to the E. coli enzyme (Figure 1B) indicating the conservation of the mechanism across different bacterial species. However, the cleavage of the TEC was not complete for the mycobacterial RNAPs even at alkaline pH. The slower nuclease activity seen above was inherent to the mycobacterial polymerases and not due to the co-purification of endogenous Gre factor (Figure S4).
Figure 1. (A) Intrinsic transcript cleavage property of RNAP.
Stalled elongation complexes bearing the 20 mer transcript were generated with M. smegmatis (Ms), M. tuberculosis (Mtb) and E. coli (Ec) RNAP respectively. The complexes were incubated for a prolonged time (1–4 hrs) in transcription buffer (pH 7.5), followed by resolving the cleavage products on 20% urea-PAGE. (B) pH-induced transcript-cleavage activity of RNAP. The gels show cleaved RNA generated from the 20 mer ternary complexes formed by Ms, Mtb and Ec RNAP in buffers of pH 6.0 to 10.0.
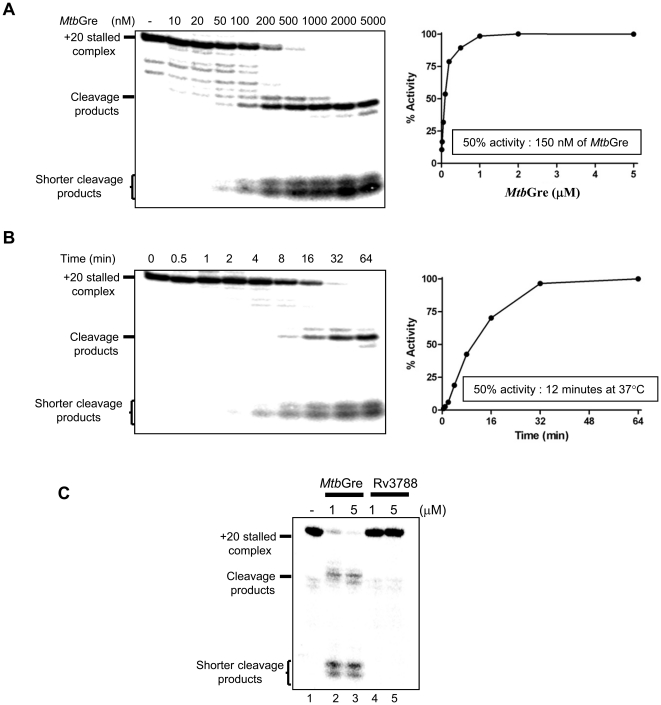
MtbGre factor stimulated the cleavage of short fragments (2–3 nt) from the 3′ end of the nascent RNA in 20-mer T7A1-TEC, and 50% of the cleavage could be achieved in less than 12 minutes (Figure 2A and 2B) indicating that Rv1080c indeed functions like a Gre factor. The pattern seen with MsGre was nearly identical mirroring their high degree of similarity (Figure S5A). However, its transcript cleavage activity appears to be higher compared to the MtbGre. In E. coli, GreA - induced hydrolysis generates mostly shorter di- and tri-nucleotides (type I cleavage), while GreB - induced hydrolysis generates variable length of fragments up to 18 nt in length (type II cleavage) depending on the extent of RNAP backtracking [6], [7], [28]. The pattern shown in Figure 2A and 2B and Figure S5A indicate that mycobacterial Gre factor follows type I cleavage.
Figure 2. MtbGre factor stimulates the cleavage of 20 mer transcript.
(A) Determination of the unit activity of MtbGre. Stalled TEC generated with M. smegmatis RNAP was incubated with different concentrations of MtbGre (10 nM to 5 µM) for 30 mins. Reactions were terminated and resolved on a 20% urea PAGE. (B) Time-course of MtbGre activity. Stalled TECs were incubated at 37°C with 1 µM MtbGre and aliquots were removed at different time points and quenched with urea gel loading dye followed by resolving on a 20% urea PAGE. The time required for 50% cleavage of the TEC was calculated from the plot. (C) The MtbGre homolog-Rv3788 does not induce transcript cleavage. MtbGre could induce the cleavage of +20 nt stalled elongation complex at T7A1 template (lanes 2 and 3). Rv3788 does not have detectable transcript cleavage stimulatory activity (lanes 4 and 5).
The MtbGre homolog – Rv3788 is a protein of 161 amino acids with a predicted coiled coil N-terminal domain and C-terminal globular domain (Figure S1A and S1B). The key acidic residues required for transcript cleavage activity of Gre factors and the hydrophobic residues in the C-terminal RNAP interaction region are conserved in Rv3788. However, the transcript cleavage assays presented in Figure 2C show that Rv3788 lacks the cleavage stimulatory activity on the stalled elongation complexes in assay conditions used for canonical Gre factor and hence not investigated further.
Gre factor knock-down results in growth retardation in mycobacteria
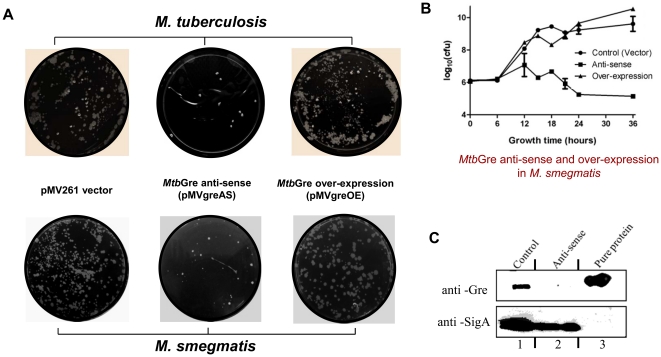
To check the importance of gre factor for cell growth, an anti-sense construct was generated by cloning the M. tuberculosis gre gene in reverse orientation under the control of the constitutive hsp60 promoter in pMV261 (Figure S5B). This strategy has been successfully employed to assess the physiological importance of several other mycobacterial genes [29], [30], [31]. The expression of M. tuberculosis gre anti-sense reduced the viability M. tuberculosis (Figure 3A) by several folds compared to the control cells transformed with only pMV261 vector. M. smegmatis cells transformed with the MtbGre anti-sense construct also showed reduced viability (Figure 3A) and were compromised in growth when compared to the cells transformed with vector or MtbGre over-expressing construct (Figure 3B). Western blots of the cell lysates probed with anti-Gre antibody showed highly reduced level of Gre protein in the cells with anti-sense construct, suggesting that the decreased survival could be due to the reduction in Gre concentration in the cells (Figure 3C). The M. smegmatis cells over-expressing MtbGre factor also showed an elongated phenotype (Figure S5C).
Figure 3. Knock-down of gre in mycobacteria is deleterious to the cell growth.
(A) The pMV261-vector control, MtbGre anti-sense (pMVgreAS) and MtbGre over-expressing (pMVgreOE) M. smegmatis and M. tuberculosis H37Ra cells were grown for 18 hrs or 7 days respectively in liquid cultures at 37°C under shaking condition, serially diluted (10−7 for vector control and over-expression and 10−5 for anti-sense) and plated to determine the cell viability. (B) Growth curve of M. smegmatis cells with vector control, MtbGre anti-sense and MtbGre over-expression constructs. Cultures were diluted in Middlebrook 7H9 broth to give an initial OD 600 of 0.02 to 0.04 and incubated for 36 hours. The growth curves were plotted by measuring cell viability by dilution plating at different time points. (C) Western blot of the M. smegmatis cell lysates from vector control (lane 1) and anti-sense MtbGre construct (lane 2) using a polyclonal antibody against MtbGre. Purified MtbGre has been used as a positive control (lane 3).
From the above data, it is apparent that the decrease in intracellular Gre levels could have caused the growth defects in both the organisms. This would also mean that a balanced pool of Gre may be required to sustain the cell viability. To measure the endogenous levels of the protein, semi-quantitative western blot analysis was carried out at different stages of cell growth. The expression level of the endogenous Gre was highest in mid-exponential phase, both in M. smegmatis and in M. tuberculosis (Figure S6A). The Gre concentration in M. smegmatis was ∼82 fmoles/µg total protein in early exponential stage cells and remained almost at the same level during late exponential phase, after which it declined slightly to 66 fmols/µg total protein in the stationary phase (Figure S6B). Gre levels in exponentially growing M. tuberculosis cells were also comparable to the levels seen with M. smegmatis cells (Figure S6A). Interestingly, the combined amount of GreA (∼53 fmol/µg of total protein) and GreB (∼13 fmol/µg of total protein) [32] in exponentially growing E. coli cells is comparable to the level of single Gre protein found in mycobacteria. The RNAP concentration also seems to be comparable between the two species (Gupta and Nagaraja, unpublished results). Next, the expression of Gre in response to different cellular stress conditions in M. smegmatis was determined by measuring the protein content, and was found to be mostly unperturbed (Figure S6C). RT-PCR experiments under various conditions also did not show significant alterations in the gre mRNA levels (data not shown). Together, these results indicate that a constant level of Gre is retained irrespective of growth phases or environmental conditions. Above findings are in contrast to the observations in several other organisms where under different stress conditions GreA level was found to be altered [33], [34]. Thus from all the results presented in Figure 3A to 3C (gre knock-down) and Figure S6A to S6C, we surmise that although amount of Gre in mycobacteria is found to be comparable to E. coli, maintaining the level is critical for cell survival.
Reduction of abortive transcription initiation, and anti-arrest activity of MtbGre
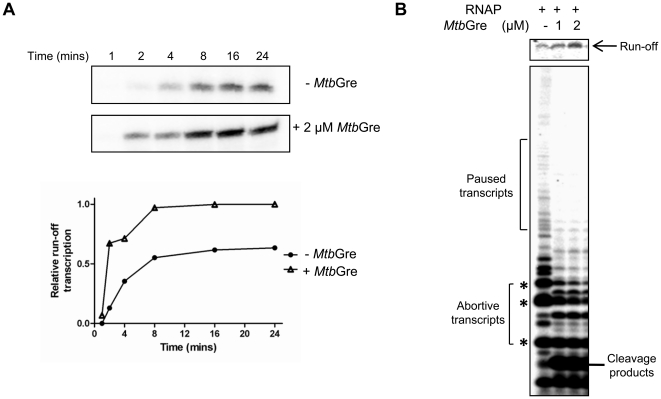
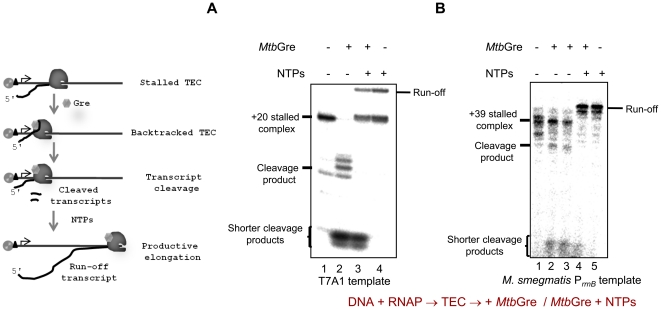
To determine the activity of MtbGre, in vitro transcriptions were carried out using M. smegmatis PrrnPCL1 as a template. The efficient open complex (RPO) formation is not effectively transmitted to the synthesis of full length transcripts in this promoter due to high abortive RNA synthesis [35]. One of the properties of the E. coli Gre factors is to reduce abortive RNA synthesis and enhance promoter clearance [36], [37]. MtbGre enhanced the full-length transcript synthesis from PrrnPCL1 by overcoming the abortive transcripts (Figure 4A). Notably, the intermittent pauses seen above the abortive transcripts in the transcription from PrrnPCL1 were also reduced in the presence of MtbGre (Figure 4B). After the cleavage of the transcript in the paused elongation complex, the trimmed TEC was capable of restarting the transcription in presence of all NTPs from both T7A1 promoter and mycobacterial PrrnB promoter templates (Figure 5A, 5B). However, the minor differences in the patterns in Figures 5A and 5B could be template specific effect. It is possible that some of the stalled elongation complexes generated on T7A1 template have entered an inactive arrested state which could not be elongated further. Taken together, data from these experiments indicate that MtbGre factor could function on pre-formed stalled elongation complexes and induce transcript cleavage-restart activity.
Figure 4. Effect of MtbGre factor on promoter clearance and abortive transcription.
(A) Promoter clearance assays were carried out in the absence (-•-) or presence (-Δ-) of 2 µM MtbGre. Transcripts were resolved on an 8% urea-PAGE and 109 nt long run-off transcripts were quantified using Image Guage (Fuji Film) and plotted (lower panel). The intensity of the bands was normalized against the amount of run-off transcript produced after 24 mins in presence of MtbGre. (B) MtbGre reduces abortive transcript level. In this assay, transcription reactions were carried out in the absence and presence of 1 µM and 2 µM MtbGre. The reactions were resolved on a 20% urea-PAGE to visualize abortive transcripts (marked by ‘*’).
Figure 5. RNAP can restart transcription elongation after transcript cleavage by MtbGre.
The starting materials in all the experiments were purified stalled complexes with radiolabeled RNA. (A) Cleavage-restart assay on T7A1-TEC. The scheme on the left depicts the reaction process. The TEC was incubated without (lane 1) or with MtbGre (lane 2 and 3), or NTPs (lane 3 and 4), as indicated. (B) Similar assay was carried out on M. smegmatis PrnB promoter template (the RNAP stalls at +39 position in the absence of UTP in the reaction mix). Transcripts were analyzed by resolving on 20% urea PAGE.
Structural features of Gre factors are conserved in MtbGre
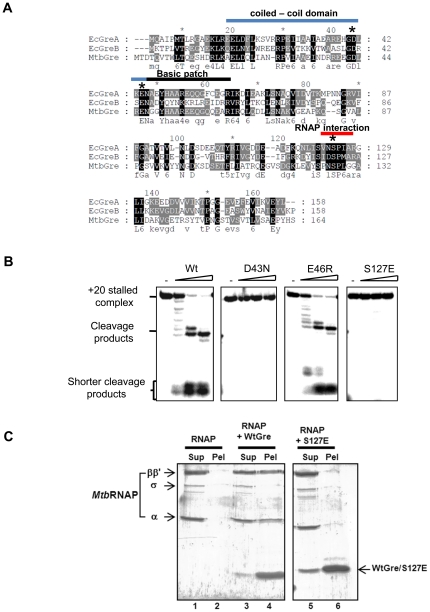
Alignment of the MtbGre with its E. coli counterparts revealed the following conserved features (Figure 6A). (i) Acidic amino acids at the tip of the predicted coiled-coil domain found in the N-terminus of the protein. In E. coli Gre factors, these residues are involved in Mg2+ co-ordination with the RNAP active center [10]. (ii) A short basic patch of residues on one side of a helix, which interacts with the 3′ end of RNA in E. coli [38]. (iii) A globular domain at the C-terminus of the protein. Residues in this domain of E. coli GreB interact with the carboxyl-terminal coiled-coil domain of RNAP β′ subunit [27]. The D43, E46 at the acidic tip of the coiled-coil domain (equivalent to the D36 and E39 of E. coli GreA) and S127 at the C-terminal globular domain of MtbGre factor (equivalent of E. coli GreA S119) (Figure 6A) were mutated to D43N, E46R, and S127E to address their function in MtbGre. The D43N and S127E mutations completely abolished the activity of MtbGre factor. On the other hand, E46R mutant retained the cleavage stimulation activity (Figure 6B). These results indicate that among the two acidic residues in the tip of N-terminal predicted coiled-coil domain, D43 is essential for the transcript cleavage activity. The loss of activity of the S127E mutant was probably due to its loss of interaction with the RNAP. Ni-NTA pull down assays were carried out to assess the direct interaction between purified MtbRNAP and histidine tagged MtbGre or its S127E variant. The MtbGre factor bound MtbRNAP (Lane 4 of Figure 6C), and as predicted S127E mutant did not interact with the RNAP (Lane 6 of Figure 6C).
Figure 6. Conserved residues of MtbGre factor are important for Mg++ co-ordination and RNAP binding.
(A) Multiple sequence alignment of MtbGren (164 aa) with E. coli GreA (158 aa) and GreB (158 aa). N-terminal coiled-coil domain is marked in blue and its basic patch in black. The C-terminus RNAP interaction domain is marked in red in the alignment. Conserved acidic residues at the tip of N- terminus coiled coil domain and S127 at the C-terminus, subjected to site directed mutagenesis are marked by an ‘*’.(B) Comparative activity of Wt with D43N, E46R and S127E mutants in T7A1 TEC. (C) Ni-NTA pull down of his-tagged Wt and S127E mutant with MtbRNAP. Lane-1 and 2 represent the supernatant and pellet fraction from the control reaction having only MtbRNAP. Lanes 3,4 represent the supernatant and pellet fraction of WtGre respectively and lanes 5 and 6 represents mutant S127E along with MtbRNAP respectively.
MtbGre factor is specific to the mycobacterial RNAP
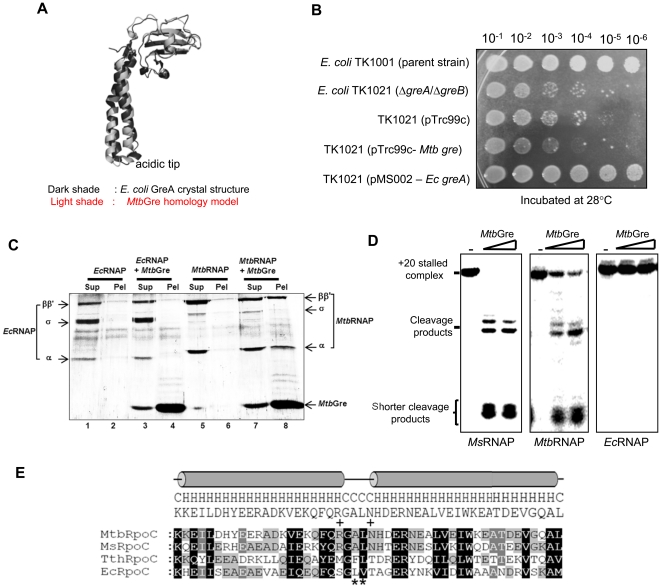
The MtbGre factor shares similar structural features (Figure 7A) with E. coli GreA and could rescue halted elongation complexes. Therefore, the ability of MtbGre to functionally complement the E. coli Gre factors was tested by using an E. coli ΔgreA/ΔgreB double knock-out strain [39], which shows a cold-sensitive phenotype. MtbGre factor expressed from a pTrc construct could not complement E. coli ΔgreA/ΔgreB grown at 28°C (Figure 7B) although the protein was expressed in E. coli (Figure S7A). The failure to complement could be due to the lack of interaction between E. coli RNAP and MtbGre (Figure 7C). In support of this, in vitro assays showed that MtbGre factor functions only on mycobacterial, i.e., M. smegmatis and M. tuberculosis TECs (Figure 7D). It did not stimulate transcript cleavage on E. coli RNAP containing TEC even at a very high concentration (>10 µM). Similarly, E. coli GreA was also not functional on the mycobacterial elongation complexes (Figure S7B).
Figure 7. MtbGre factor is specific to mycobacterial TEC.
(A) Homology modeling of MtbGre using E. coli GreA crystal structure (PDB code: 1GRJ) as template. (B) Complementation of E. coli ΔgreA/ΔgreB strain (TK1021) with Mtb gre. The gene was cloned into pTrc99c and the E. coli cells harboring different plasmids were grown till OD600 0.6 and spotted onto IPTG containing plates and incubated at 28°C. (C) Interaction of histidine tagged MtbGre with Ec and Mtb RNAPs by Ni-NTA pull-down. Lanes1 and 2 - supernatant and pellet of only EcRNAP and lanes 5 and 6 are MtbRNAP from the control reactions. Lane 4 and 8 represent the pellet fractions of the reactions with Ec and Mtb RNAP with MtbGre respectively (D) TECs prepared with Ms, Mtb and Ec RNAP were incubated with MtbGre and the resulting products were resolved on a 20% urea PAGE. (E) Comparison of coiled-coil domain present in the C-terminus of the RNAP β′ subunit from M. tuberculosis, M. smegmatis, T. thermophillus and E. coli. Residues marked with ‘+’ are either charged or polar amino acids present in the loop region of mycobacterial β′ subunit but absent in the other two.
Discussion
In this study, we describe the characterization of Rv1080c - the Gre factor present in the M. tuberculosis genome. The MtbGre increased the transcription efficiency both during initiation and elongation phase of the process. During initiation, it reduced the abortive transcripts and enhanced the promoter clearance. At elongation phase, the protein rescued RNAP from the transcription pauses by inducing the transcript cleavage. Knocking down of the gene resulted in growth retardation and cell death indicating its essentiality for cell survival.
In organisms where Gre factors have been analyzed so far, they show remarkably similar structural features. Functional characterization of the Gre factors from E. coli [6], [7], T. aquaticus and T. thermophilus [15], [17], [40] revealed the conserved nature of the transcript cleavage stimulation activity required for efficient transcription process. However, gre genes were found to be dispensable in E. coli; ΔgreA - ΔgreB double knock-out strain showed only a mild cold-sensitive phenotype [39]. In contrast, in M. tuberculosis, the protein appears to have a more pronounced and indispensable role. In the first glance our results appear to be contradicting the earlier transposon mutagenesis studies which led to the isolation of insertional mutation of M. tuberculosis gre (http://mylims2.cvmbs.colostate.edu/tnlist/). We have noticed that the point of insertion of the transposon is at the 493rd position out of the 495 bases in the Rv1080c. Thus it is likely that, the gene was not inactivated in the mutant strain. Also, with the decrease in intracellular Gre levels, the cell survival was affected. Notably, significant amount of the protein is present at all growth phases indicating its house-keeping function. Further, the Gre protein level was not altered to a great extent during different stress conditions, indicating that an optimum level of the protein may be required for cell survival.
MtbGre can rescue a pre-formed halted elongation complex to exert its anti-arrest activity similar to E. coli GreA and ensure efficient transcription elongation. The transcript cleavage pattern of MtbGre showed type I cleavage products i.e. predominantly 2–3 nt fragments similar to the activity of E. coli GreA. The longer transcript cleavage pattern (2–18 nt, type II) seen with E. coli GreB is mediated by a large stretch of positively charged residues in its N-terminal domain [38]. MtbGre does not have such a large stretch of basic amino acids and the surface charge distribution is similar to that of E. coli GreA (Figure S1C). In organisms having GreB, RNAP could backtrack farther to have a larger RNA 3′ end fragment to be processed. Indeed, in such conditions, high affinity interaction between RNAP and GreB results in transcript cleavage activity [16], [27]. Earlier studies have revealed lower transcription elongation rates in mycobacteria [41], [42]. Organisms such as E. coli with faster transcription rates seem to require two Gre factors to process shorter and longer RNA.
The action of the MtbGre seems to be restricted to mycobacterial transcription machinery as it did not rescue a halted elongation complex of E. coli RNAP. Lack of interaction between these heterologous partners could account for the observation. The interaction surface on E. coli RNAP for E. coli GreB was mapped to a conserved hydrophobic loop in the coiled-coil domain in the C-terminus of the β′ subunit [27]. The region is also conserved in the mycobacterial RNAP (Figure 7E) indicating the conserved architecture of transcription machinery. However, the C-terminal globular domain of Gre factors (GreA, GreB of E. coli and MtbGre), which interacts with the RNAP, shows considerable variation, although certain specific residues in the hydrophobic patch are conserved in all these proteins. Importance of specific interactions between RNAP and Gre is suggested from the studies in T. aquaticus. GreA of T. aquaticus failed to induce transcript cleavage in EcRNAP elongation complexes [15] similar to the present observation with MtbGre. Thus it appears that the transcript cleavage activity requires species-specific interactions, although both partners viz RNAP and Gre have conserved characteristics across species. Gre may have a more important function in mycobacteria to compensate for the low intrinsic cleavage activity of mycobacterial RNAP compared to its E. coli and themophilic counterparts. This deficiency could affect the recovery from arrest of backtracked MtbRNAP in the absence of MtbGre. The similar mechanism has been recently proposed to explain growth inhibition of the yeast strains expressing the cleavage deficient mutant of the eukaryotic Gre homolog, TFIIS [43]. The results presented here and the data emerged till date from a number of studies with Gre factors of diverse group of organisms emphasize the biological importance of these secondary channel binding proteins. The deletion of greA led to hypersensitivity phenotype under various stress conditions in E. coli [39], Sinorhizobium meliloti [44] and Rhizobium tropici [45] implicating the importance of Gre factors in the survival of the organism in the restrictive environment. In contrast, the decrease in Gre levels under normal cellular growth conditions itself reduced the viability of M. tuberculosis. The indispensability of the Gre factor in M. tuberculosis but not in E. coli [39] or T. thermophilus [17] indicates that the intracellular role of the factor is likely to be varied between different species of bacteria.
MtbGre seems to be the only transcription elongation factor in the genome possessing cleavage activity as the other ORF - Rv3788 found in the genome with lower degree of relatedness do not appear to participate in the process. The lack of transcript cleavage stimulatory activity in Rv3788 may be attributed to the absence of several key residues in the N-terminus which are found in Gre factors across different organisms. Although the two acidic residues needed for Mg2+ co-ordination are conserved in Rv3788 (Figure S1A), Asn47 and Tyr50 (present in MtbGre), required for binding to the backtracked protruding nascent RNA are absent. Nevertheless, Rv3788, has several features similar to the RNAP secondary channel binding proteins and hence may have some other intracellular role. It is also apparent that the RNAP secondary channel binding proteins are emerging to be the key regulators of different cellular functions apart from the transcript cleavage stimulatory functions [5].
In conclusion, Rv1080c functions like a bona fide Gre factor with transcript cleavage stimulatory activity in M. tuberculosis. Gre function is required for the optimal growth of the mycobacteria in contrast to its dispensability in E. coli. GC rich templates are known to impose blockage during transcription due to the formation of stable RNA-DNA hybrids [46]. Such strong barriers have to be overcome to ensure high fidelity RNA synthesis. Slower transcription rates in mycobacteria may lead to intermittent pauses and stalling at specific signals. Under these circumstances RNAP has to ensure completing the elongation process. Transcription factors like Gre, which maintain the efficiency by preventing premature pauses, appear to have a more profound role in maintaining the genomic integrity of M. tuberculosis.
Methods
Bacterial strains, plasmids and the growth conditions
M. smegmatis mc2155 [47] and M. smegmatis SM07sigA [48], [49] were cultured in Middlebrook 7H9 medium (Difco) containing 0.05% Tween-80 (Sigma) and 0.4% glucose (Sigma) under shaking conditions at 37°C. M. tuberculosis H37Ra [50] cells were cultured in Middlebrook 7H9 medium supplemented with ADC consisting of 0.2% glycerol (Sigma) and 0.05% Tween-80 at 37°C. To check the expression pattern of Gre at different growth phases in M. smegmatis and M. tuberculosis, cells were grown for 12, 18, 24, 30, 36 and 48 hrs (for M. smegmatis) or 3, 5, 7, 12 and 20 days (for M. tuberculosis), pelleted down by centrifugation, lysed by sonication and cell extracts were prepared.
Knock-down of gre expression in M. smegmatis mc2155 and M. tuberculosis H37Ra was carried out by generating the plasmid pMVgreAS (Mtbgre in anti-sense orientation) in pMV261 [51]. The coding sequence was amplified using primers with BamH1 site (Table 1) and cloned downstream of the hsp60 promoter at a BamH1 site of the vector pMV261 to generate plasmid pMVgreOE (Table 1) for over-expression of MtbGre in both M. smegmatis and M. tuberculosis. Comparison of the growth rates of different strains was carried out by inoculating (1% inoculum) 30 ml of Middlebrook 7H9 medium with 25 µg ml−1 kanamycin to obtain an initial OD600 of 0.02 to 0.04. Growth of the strains was monitored by dilution - plating from 8 day culture of M. tuberculosis or 20 hrs cultures of M. smegmatis grown at 37°C in shaking conditions. The cells were diluted in fresh media and plated into the middlebrook 7H10 agar plates to determine the cell viability by counting the cfu.
Table 1. Oligonucleotides, strains and plasmids used in this study.
| Name | Description | Reference |
| M. smegmatis SM07sigA | (HygR, his-rpoC, pJAM2mysA) | [46], [47] |
| M. smegmatis mc2 155 | (A high efficiency transformation strain of M. smegmatis) | [45] |
| M. tuberculosis H37Ra | (An attenuated strain of M. tuberculosis H37Ra) | [48] |
| E. coli BL21 | (hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | [51] |
| E. coli TK1001 | MC1061 zgj-203::Tn10 | [39] |
| E. coli TK1021 | MC1061 greA::kan, greB::cat, zgj-203::Tn10 | [39] |
| pMS002 | Derivative of pBR322 containing Mtb greA gene | [39] |
| pMV261 | E. coli-mycobacteria shuttle vector with a hsp60 promoter | [49] |
| pET20bgre | M. tuberculosis gre cloned between NdeI and HincII of pET20b | This study |
| pET20brv3788 | M. tuberculosis Rv3788 cloned between NdeI and HindIII of pET20b | This study |
| pET20bmsgre | M. smegmatis gre cloned between NdeI and HindIII of pET20b | This study |
| pET20bgre-his | M. tuberculosis gre cloned in pET20bwith C-terminus his-tag | This study |
| pET20bgre-his | M. tuberculosis gre S127E cloned in pET20b with C-terminus his-tag | This study |
| pET20bEcgreA-his | E. coli greA cloned in pET20b with C-terminus his-tag | This study |
| pMVgreAS | M. tuberculosis gre cloned in anti-sense orientation under hsp60 promoter | This study |
| pMVgreOE | M. tuberculosis gre coading sequence cloned under hsp60 promoter | This study |
| pTrc99gre | M. tuberculosis gre cloned under trc promoter | This study |
| Gre D43N Mut | 5′ GAAGAAGGCAACCTGCGCGAGAAC 3′ | This study |
| Gre E46R Mut | 5′ GAAGGCGACCTGCGCCGTAACGGCGGATACCAC 3′ | This study |
| Gre S127E Mut | 5′ TACTCGCCGAATGAACCGCTCGGTGGG 3′ | This study |
| greBamH1For | 5′ ACGGATCCCGACCATATGACGGATACTCAAGTC 3′ | This study |
| greBamH1Rev | 5′ ACGGATCCCGACCTGCTCGGAGATCTCGAACAG 3′ | This study |
| greNdeIFor | 5′ CGACCATATGACGGATACTCAAGTC 3′ | This study |
| greHindIIIRev | 5′ ATAAGCTTCGACCTGCTCGGAGATCTCGAACAG 3′ | This study |
| rv3788NdeIFor | 5′ ATGCGACATATGAGCGAGAAAGTCGAGTC 3′ | This study |
| rv3788HindIIIRev | 5′ ATAAGCTTTTCTGAGGGCAGCTTGACAG 3′ | This study |
| MsgreNdeIFor | 5′ ATGCGACATATGACCGATACCCAGGTCACC 3′ | This study |
| MsgreHindIIIRev | 5′ ATAAGCTTTCCGCCTTGATACGGCTCAGC 3′ | This study |
Western blots
To detect the protein level at different growth phases, cell lysates were probed for Gre factor with a polyclonal antibody raised in mice and anti-SigA antibody in rabbit. The primary antibodies were probed with the secondary antibody coupled with HRP and blots were developed using a chemiluminescence substrate (GE Health Care). Expression of Gre factor during different stress conditions were also checked by growing M. smegmatis cells till mid-log phase and subjecting them to varied stresses as described [52]. The amount of Gre protein present in the M. smegmatis cells was determined by western blot. Varying concentrations of the purified M. smegmatis Gre were loaded in the same gel as standards along with 120 µg of cell extracts from different growth phase cultures and subsequently probed with anti-Gre antibody.
Microscopy
M. smegmatis cells harboring pMV261 or pMVgreAS or pMVgreOE constructs were grown in Middlebrook 7H9 medium at 37°C to mid-exponential phase. Cells were pre-fixed in PBS, 1% (v/v) Triton X-100 (Sigma) and 2% (v/v) toluene (Merck) solution and incubated overnight at 4°C. Cells were stained with DAPI solution (4′,6-diamidino-2-phenylindole), which binds specifically to DNA. Microscopic observations were carried out by using a Carl Zeiss fluorescent microscope at 1000× magnification.
Expression and purification of MtbGre, MsGre and Rv3788
gre (Rv1080c) and Rv3788 genes were PCR amplified from M. tuberculosis genomic DNA with specific primers (Table 1) and cloned between the NdeI and HindIII site of pET20b (pET20bgre and pET20brv3788). The M. smegmatis gre (MSMEG_5263) gene was PCR amplified from M. smegmatis mc2155 genomic DNA and cloned in pET20b (between NdeI and HindIII site). Site directed mutants of Mtbgre were generated using the mega-primer inverse PCR method with pET20bgre as a template (primer sequences are listed in Table 1). The purification of MtbGre, its mutants and MsGre was carried out as follows. E. coli BL21 cells [53] with pET20bgre or its mutants or pET20bmsgre were grown till OD600 0.6 at 37°C and induced with 0.3 mM IPTG. Cells were lysed by sonication and centrifuged at 100,000 g for 2 hrs. The supernatants were subjected to 0–65% ammonium sulfate precipitation and re-suspended in 3 ml of TGE buffer [10 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA] with 50 mM NaCl and subsequently resolved by a 120 ml Sephacryl S-100 gel filtration column. The fractions having Gre protein were further purified through DEAE - Sephacel chromatography by eluting with a linear NaCl gradient of 50 mM to 400 mM. The Rv3788 protein was purified from the E. coli BL21 cells harboring pET20brv3788. The purification involved a 45–60% ammonium sulfate precipitation of the cell lysate followed by DEAE - Sephacel chromatography. All the proteins purified were approximately 95% pure as judged by SDS-PAGE (Figure S2C). From 2 liters each of the cultures overexpressing the proteins (MtbGre, MsGre and Rv3788), about 5 mg of each of the protein were obtained. E. coli greA was cloned with a C-terminal His-tag in pET20b and the protein was purified from E. coli BL21 cells [53] over-expressing the protein using a Ni-NTA column. M. smegmatis RNAP was purified by following the method described earlier [49]. M. tuberculosis RNAP was purified from 2 liters of M. tuberculosis H37Ra cells grown for 8 days at 37°C in MB7H9 medium with ADC supplement (Difco). The purification involved gel filtration on Superdex S-200 matrix and subsequent heparin - Sepharose chromatography following the method described for native M. smegmatis RNAP purification [49].
Promoter clearance and abortive transcription
100 nM of RNAP and 20 nM of M. smegmatis PrrnPCL1 promoter containing template were incubated in transcription buffer [50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 100 µM DTT, 5% glycerol, 50 µg ml−1 BSA and 100 mM KCl] for 15 mins at 37°C to form the open complex. Subsequently 50 µg ml−1 heparin was added to the reactions and incubated for 1 min. Transcription was initiated by the addition of 100 µM NTPs and 2 µCi of α-32P[UTP]. Aliquots were withdrawn at different time intervals and reactions were incubated for indicated time. Reactions were analyzed in 22% urea-PAGE to resolve the abortive products.
Stalled TEC preparation
Transcription assays were carried out using T7A1 promoter and RNAPs from E. coli, M. smegmatis and M. tuberculosis. Ternary elongation complexes were generated on a 5′ biotinylated T7A1 promoter-containing DNA template (Figure S3A). The TECs for E. coli or the mycobacterial RNAPs were prepared by following the methods described for E. coli and T. thermophillus enzymes [15], [26]. RPO were formed by incubating 100 nM of M. smegmatis, M. tuberculosis or E. coli RNAP and 15 nM T7A1 promoter containing template DNA at 37°C for 15 min in transcription buffer. For multiple round transcription assays, 100 µM of NTPs were added to the reaction mix and incubated further for 15 mins at 37°C. Reactions were stopped with formamide dye and analyzed in Urea PAGE. For stalled complex formation assays, after RPO formation, 100 µM ATP, 100 µM GTP and 2 µCi [α-32P] ATP (300 Ci mmol−1, Perkin Elmer) were added. Reactions were carried out in the absence of CTP and UTP to generate stalled elongation complexes containing a 20 mer transcript. M. smegmatis PrrnB promoter template was used for the preparation of mycobacterial TEC. RNAP stalls at +39 position in the absence of UTP in this template. TECs were further purified by mixing 5 µl of Streptavidin-Sepharose beads (GE Healthcare) to each reaction and precipitated by centrifugation. Pellets containing the elongation complexes were washed thrice with transcription buffer supplemented with 200 mM KCl and 100 µg ml−1 heparin followed by washing twice with only transcription buffer. Indicated amounts of MtbGre or Rv3788 were added to the beads re-suspended in the transcription buffer followed by incubation at 37°C for 30 min. 150 nM MtbGre was found to be optimum for cleavage of 50% of the T7A1 TECs and was thus defined as the unit activity. Reactions were terminated with the addition of formamide dye [0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol FF, 0.08% amaranth (w/v), 10 mM EDTA, 0.025% SDS and 80% deionized formamide] and RNA cleavage products were analyzed by electrophoresis in a 20% denaturing PAGE. Amaranth dye included in the formamide stop mix served as a size marker. In a 22% urea PAGE the dye moves at a position corresponding to 2–3 nt short RNA fragments [54].
Intrinsic cleavage activity of RNAP
Intrinsic cleavage activity of the M. smegmatis, M. tuberculosis and E. coli RNAPs was detected by prolonged incubation (up to 4 hrs) of the TECs (prepared with 15 nM template and 100 nM RNAP) in transcription buffer (pH 7.5) at 37°C followed by resolving in a 20% urea - acrylamide gel. pH - induced transcript cleavage reactions were carried out in three different buffer systems at 37°C for 30 mins. (i) 40 mM PIPES adjusted to pH 6.0 by addition of 1 M NaOH; (ii) 40 mM Tris adjusted to pH 7.0, 8.0, and 9.0 by the addition of 1 M HCI; (iii) 40 mM CAPS adjusted to pH 10.0 by the addition of 1 M NaOH. All buffers contained 0.1 M KCl and 10 mM MgCl2.
Cleavage-restart activity of MtbGre
The 20 mer T7A1 TEC or the 39 mer M. smegmatis PrrnB TECs were prepared by using 15 nM biotinylated template and 100 nM RNAP. The RNAP was stalled at T7A1 template by using only 100 µM of ATP, GTP and 2 µCi of [α-32P] ATP (300 Ci mmol−1, Perkin Elmer) in each of the 10 µl reaction volume. For generating +39 stalled elongation complex at PrrnB promoter, 100 µM of ATP, GTP and CTP were used along with 2 µCi of [α-32P] ATP. To detect cleavage-restart activity of MtbGre, the TECs were incubated with the MtbGre factor in presence or absence all the four NTPs. Initially the complexes were incubated with 2 µM of MtbGre for 30 min followed by the addition of the NTPs and incubation was continued for another 10 min followed by resolving in a 20% urea PAGE.
MtbGre-RNAP interaction
C-terminal his-tagged MtbGre and its S127E mutant were cloned in pET20b and purified using a Ni-NTA column. 5 µg of both RNAP (Ec or Mtb) and Gre protein were used for analyzing direct interactions. Proteins were incubated together for 15 mins in 50 µl volume of incubation buffer containing 50 mM tris - HCl (pH 8.0), 100 mM potassium glutamate, 5% glycerol, and 20 mM imidazole at room temperature. 20 µl of Ni-NTA pre-equilibrated with incubation buffer was then added to the protein mixture and incubated for an additional 30 mins in a rotary mixer. The supernatant was separated and the pellet was washed thrice with 400 µl of the incubation buffer. Finally, the pellet was re-suspended in 50 µl of buffer mixed with SDS-gel loading buffer, boiled and loaded onto an 11% SDS-PAGE along with the supernatant fractions followed by silver staining of the gel.
Complementation of E. coli ΔgreA/ΔgreB strain with M. tuberculosis gre
The M. tuberculosis gre gene was cloned in pTrc99c vector to obtain pTrc99gre construct which was used for complementing the E. coli TK1021 strain (Table 1). The parental strain TK1001 was used as wild type E. coli control. E. coli greA expressing plasmid pMS002 was used as a positive control in these experiments [39]. The cells were grown in liquid culture and different dilutions were spotted on LB plates containing 0.3 mM IPTG and appropriate antibiotics (Table 1).
Supporting Information
Sequence alignments and homology modeling of Gre. (A) Multiple sequence alignment of the E. coli GreA and GreB with MtbGre and the Rv3788 using ClustalW program (http://www.ebi.ac.uk/Tools/msa/clustalw2). The alignment figure is created using GenDoc – multiple sequence alignment editor (http://www.psc.edu/biomed/genedoc). The conserved amino acids are shaded in black and substitutions with similar amino acids in grey. The conserved acidic residues at the N-terminus are labeled as “♦” and hydrophobic residues in the C-terminus as “•”. (B) Homology model of the MtbGre and Rv3788 (using E. coli GreA crystal structure – 1GRJ as template). Models were generated using the comparative protein structure modeling program Modeller ver. 9.3. [Eswar, N et al. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement 15, 5.6.1–5.6.30, 2006]. (C) Surface charge distribution of MtbGre and Rv3788. Positively charged surface is shown in blue and the negatively charged region in red. E. coli GreA structure (Stebbins et al. [23]) is shown on left and GreB (Vassylyeva et al. [27]) second from the left and compared with the MtbGre (second from right) and Rv3788 (rightmost). Positively charged region on the surface of the coiled-coil domain is shown in the box.
(TIF)
Over-expression of Gre and Rv3788. (A) Gre factors of both M. tuberculosis and M. smegmatis were over-expressed and purified from E. coli BL21 cells. UN: un induced cell lysates and IN: induced cell lysate of MtbGre and MsGre over-expressing cells respectively. (B) Over-expression of M. tuberculosis Rv3788 in E. coli BL21 cells. Both un induced and IPTG induced samples of Rv3788 expressing cells show robust hyper-expression. (C) Purified proteins: MtbGre (17.8 kDa), Rv3788 (17.4 kDa) and MsGre (18 kDa). The yield of all the three proteins was ∼5 mg from 2 liters of culture. The proteins were >95% pure.
(TIF)
Transcription assays with T7A1 promoter templates using mycobacterial RNAPs. (A) A modified T7A1 promoter was used for generating the stalled complexes. Residues underlined are the ones replaced from the original residues showed above. Biotin tag is present at the 5′ end of the template. In presence of ATP and GTP, RNAP forms a stalled TEC with a 20mer RNA. (B) Multiple-round in vitro transcription with E. coli, M. smegmatis and M. tuberculosis RNAP from T7A1 promoter. Abortive transcripts are indicated in the lower panel.
(TIF)
Ms and Mtb RNAP are free from endogenous Gre factor contamination. 20 µg of both Ms and Mtb RNAP were probed with anti-Gre antibody. 100 ng of purified MtbGre was used as a control.
(TIF)
Activity of Ms Gre and the phenotypic effects of gre overexpression. (A) Transcript cleavage stimulatory activity of MsGre. (B) SDS-PAGE analysis of the cell lysates from M. smegmatis with pMV261, with the over-expression construct (pMVgreOE) and antisense (pMVgreAS) mediated knock-down construct. (C) Morphology of the M. smegmatis cells over-expressing MtbGre. Comparison of cellular morphology (left panel) and nucleoid (right panel) of M. smegmatis mc2155 cells harboring either the pMV261 vector or pMV-greAS or pMV-greOE constructs. Left panels- bright-field images; right panels-fluorescent images showing the DAPI-stained nucleoid.
(TIF)
Determination of the expression pattern of Gre and its amount in the cells. (A) Expression of gre in response to different stresses in M. smegmatis and M. tuberculosis determined by western blot. (B) Estimation of the level of Gre protein in the M. smegmatis cells. Purified M. smegmatis Gre protein was used as a standard. 120 µg of total cell lysate proteins were probed with the anti-Gre antibody to estimate the Gre protein level in cells at different growth phases. (C) Western blot analysis for Gre from cell lysate of M. smegmatis exposed to different stresses.
(TIF)
Expression of Gre from pTrc gre construct in E. coli TK1021. (A) The gel shows expression of MtbGre from the pTrcgre construct, induced with 0.3 mM IPTG. (B) Transcript cleavage assays using MtbGre and E. coli GreA. Only MtbGre shows cleavage of mycobacterial RNAP elongation complex.
(TIF)
Acknowledgments
We thank Nobuo Shimamoto, of Kyoto Sangyo University, for the strains TK1000, TK1021 and plasmid pMS002, O. Krishnadev from MBU, IISc, for generating the homology model of MtbGreA, G. Swapna for E. coli RNAP and AstraZeneca, Bangalore, for M. tuberculosis σA antibodies. Sergei Borukhov, of University of Medicine and Dentistry, New Jersey, USA, is acknowledged for helpful suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The Phosphorimager Facility of IISc is supported by the Department of Biotechnology, Government of India, and is acknowledged. AC was a recipient of Senior Research Fellowship from Council of Scientific and Industrial Research, Government of India. VN is a recipient of J.C. Bose Fellowship from the Department of Science and Technology and a Center for Excellence in Tuberculosis Research grant from Department of Biotechnology, Government of India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol. 2005;55:1315–1324. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 4.Vassylyev DG. Elongation by RNA polymerase: a race through roadblocks. Curr Opin Struct Biolol. 2009;19:691–700. doi: 10.1016/j.sbi.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci U S A. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 8.Reines D, Chamberlin MJ, Kane CM. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
- 9.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci U S A. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marr MT, Roberts JW. Function of Transcription Cleavage Factors GreA and GreB at a Regulatory Pause Site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 12.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 14.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 15.Hogan BP, Hartsch T, Erie DA. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J Biol Chem. 2002;277:967–975. doi: 10.1074/jbc.M108737200. [DOI] [PubMed] [Google Scholar]
- 16.Laptenko O, Borukhov S. Biochemical assays of Gre factors of Thermus thermophilus. Methods Enzymol. 2003;371:219–232. doi: 10.1016/S0076-6879(03)71016-7. [DOI] [PubMed] [Google Scholar]
- 17.Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, et al. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006;25:2131–2141. doi: 10.1038/sj.emboj.7601094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 19.Smith I, Bishai WR, Nagaraja V. Control of mycobacterial transcription. In: Cole ST, editor. Tuberculosis and the Tubercle Bacillus. Washington, DC: ASM Press; 2005. pp. 219–231. [Google Scholar]
- 20.Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 21.Harshey RM, Ramakrishnan T. Rate of ribonucleic acid chain growth in Mycobacterium tuberculosis H37Rv. J Bacteriol. 1977;129:616–622. doi: 10.1128/jb.129.2.616-622.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia MJ, Nunez MC, Cox RA. Measurement of the rates of synthesis of three components of ribosomes of Mycobacterium fortuitum: a theoretical approach to qRT-PCR experimentation. PLoS One. 2010;5:e11575. doi: 10.1371/journal.pone.0011575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stebbins CE, Borukhov S, Orlova M, Polyakov A, Goldfarb A, et al. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 24.Koulich D, Orlova M, Malhotra A, Sali A, Darst SA, et al. Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 1997;272:7201–7210. doi: 10.1074/jbc.272.11.7201. [DOI] [PubMed] [Google Scholar]
- 25.Koulich D, Nikiforov V, Borukhov S. Distinct functions of N and C-terminal domains of GreA, an Escherichia coli transcript cleavage factor. J Mol Biol. 1998;276:379–389. doi: 10.1006/jmbi.1997.1545. [DOI] [PubMed] [Google Scholar]
- 26.Loizos N, Darst SA. Mapping interactions of Escherichia coli GreB with RNA polymerase and ternary elongation complexes. J Biol Chem. 1999;274:23378–23386. doi: 10.1074/jbc.274.33.23378. [DOI] [PubMed] [Google Scholar]
- 27.Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I, et al. The carboxy-terminal coiled-coil of the RNA polymerase beta′-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borukhov S, Laptenko O, Lee J. Escherichia coli transcript cleavage factors GreA and GreB: functions and mechanisms of action. Methods Enzymol. 2001;342:64–76. doi: 10.1016/s0076-6879(01)42536-5. [DOI] [PubMed] [Google Scholar]
- 29.Wu S, Howard ST, Lakey DL, Kipnis A, Samten B, et al. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol Microbiol. 2004;51:1551–1562. doi: 10.1111/j.1365-2958.2003.03922.x. [DOI] [PubMed] [Google Scholar]
- 30.Sharbati S, Schramm K, Rempel S, Wang H, Andrich R, et al. Characterisation of porin genes from Mycobacterium fortuitum and their impact on growth. BMC Microbiol. 2009;9:31. doi: 10.1186/1471-2180-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Wang X, Lau A, Liao TY, Bucci C, et al. Mycobacterial nucleoside diphosphate kinase blocks phagosome maturation in murine RAW 264.7 macrophages. PLoS One. 2010;5:e8769. doi: 10.1371/journal.pone.0008769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, et al. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh VK, Jayaswal RK, Wilkinson BJ. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiol Lett. 2001;199:79–84. doi: 10.1111/j.1574-6968.2001.tb10654.x. [DOI] [PubMed] [Google Scholar]
- 34.Len ACL, Harty DWS, Jacques NA. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology. 2004;150:1339–1351. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- 35.China A, Tare P, Nagaraja V. Comparison of promoter-specific events during transcription initiation in mycobacteria. Microbiology. 2010;156:1942–1952. doi: 10.1099/mic.0.038620-0. [DOI] [PubMed] [Google Scholar]
- 36.Hsu LM, Vo NV, Chamberlin MJ. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci U S A. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen R, Nagai H, Shimamoto N. Conformational switching of Escherichia coli RNA polymerase-promoter binary complex is facilitated by elongation factor GreA and GreB. Genes Cells. 2001;6:389–401. doi: 10.1046/j.1365-2443.2001.00436.x. [DOI] [PubMed] [Google Scholar]
- 38.Kulish D, Lee J, Lomakin I, Nowicka B, Das A, et al. The functional role of basic patch, a structural element of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 2000;275:12789–12798. doi: 10.1074/jbc.275.17.12789. [DOI] [PubMed] [Google Scholar]
- 39.Susa M, Kubori T, Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol Microbiol. 2006;59:1807–1817. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamour V, Hogan BP, Erie DA, Darst SA. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J Mol Biol. 2006;356:179–188. doi: 10.1016/j.jmb.2005.10.083. [DOI] [PubMed] [Google Scholar]
- 41.Harshey RM, Ramakrishnan T. Rate of ribonucleic acid chain growth in Mycobacterium tuberculosis H37Rv. J Bacteriol. 1977;129:616–622. doi: 10.1128/jb.129.2.616-622.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia MJ, Nunez MC, Cox RA. Measurement of the rates of synthesis of three components of ribosomes of Mycobacterium fortuitum: a theoretical approach to qRT-PCR experimentation. PLoS One. 2010;5:e11575. doi: 10.1371/journal.pone.0011575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell GRO, Sharypova LA, Scheidle H, Jones KM, Niehaus K, et al. Striking Complexity of Lipopolysaccharide Defects in a Collection of Sinorhizobium meliloti Mutants. J Bacteriol. 2003;185:3853–3862. doi: 10.1128/JB.185.13.3853-3862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nogales J, Campos R, BenAbdelkhalek H, Olivares J, Lluch C, et al. Rhizobium tropici Genes Involved in Free-Living Salt Tolerance are Required for the Establishment of Efficient Nitrogen-Fixing Symbiosis with Phaseolus vulgaris. Molecular Plant-Microbe Interactions. 2002;15:225–232. doi: 10.1094/MPMI.2002.15.3.225. [DOI] [PubMed] [Google Scholar]
- 46.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci U S A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 48.Mukherjee R, Chatterji D. Stationary phase induced alterations in mycobacterial RNA polymerase assembly: A cue to its phenotypic resistance towards rifampicin. Biochem Biophys Res Commun. 2008;369:899–904. doi: 10.1016/j.bbrc.2008.02.118. [DOI] [PubMed] [Google Scholar]
- 49.China A, Nagaraja V. Purification of RNA polymerase from mycobacteria for optimized promoter-polymerase interactions. Protein Expr Purif. 2010;69:235–242. doi: 10.1016/j.pep.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 50.Zheng H, Lu L, Wang B, Pu S, Zhang X, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 52.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 53.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 54.Hsu LM. Monitoring abortive initiation. Methods. 2009;47:25–36. doi: 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignments and homology modeling of Gre. (A) Multiple sequence alignment of the E. coli GreA and GreB with MtbGre and the Rv3788 using ClustalW program (http://www.ebi.ac.uk/Tools/msa/clustalw2). The alignment figure is created using GenDoc – multiple sequence alignment editor (http://www.psc.edu/biomed/genedoc). The conserved amino acids are shaded in black and substitutions with similar amino acids in grey. The conserved acidic residues at the N-terminus are labeled as “♦” and hydrophobic residues in the C-terminus as “•”. (B) Homology model of the MtbGre and Rv3788 (using E. coli GreA crystal structure – 1GRJ as template). Models were generated using the comparative protein structure modeling program Modeller ver. 9.3. [Eswar, N et al. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement 15, 5.6.1–5.6.30, 2006]. (C) Surface charge distribution of MtbGre and Rv3788. Positively charged surface is shown in blue and the negatively charged region in red. E. coli GreA structure (Stebbins et al. [23]) is shown on left and GreB (Vassylyeva et al. [27]) second from the left and compared with the MtbGre (second from right) and Rv3788 (rightmost). Positively charged region on the surface of the coiled-coil domain is shown in the box.
(TIF)
Over-expression of Gre and Rv3788. (A) Gre factors of both M. tuberculosis and M. smegmatis were over-expressed and purified from E. coli BL21 cells. UN: un induced cell lysates and IN: induced cell lysate of MtbGre and MsGre over-expressing cells respectively. (B) Over-expression of M. tuberculosis Rv3788 in E. coli BL21 cells. Both un induced and IPTG induced samples of Rv3788 expressing cells show robust hyper-expression. (C) Purified proteins: MtbGre (17.8 kDa), Rv3788 (17.4 kDa) and MsGre (18 kDa). The yield of all the three proteins was ∼5 mg from 2 liters of culture. The proteins were >95% pure.
(TIF)
Transcription assays with T7A1 promoter templates using mycobacterial RNAPs. (A) A modified T7A1 promoter was used for generating the stalled complexes. Residues underlined are the ones replaced from the original residues showed above. Biotin tag is present at the 5′ end of the template. In presence of ATP and GTP, RNAP forms a stalled TEC with a 20mer RNA. (B) Multiple-round in vitro transcription with E. coli, M. smegmatis and M. tuberculosis RNAP from T7A1 promoter. Abortive transcripts are indicated in the lower panel.
(TIF)
Ms and Mtb RNAP are free from endogenous Gre factor contamination. 20 µg of both Ms and Mtb RNAP were probed with anti-Gre antibody. 100 ng of purified MtbGre was used as a control.
(TIF)
Activity of Ms Gre and the phenotypic effects of gre overexpression. (A) Transcript cleavage stimulatory activity of MsGre. (B) SDS-PAGE analysis of the cell lysates from M. smegmatis with pMV261, with the over-expression construct (pMVgreOE) and antisense (pMVgreAS) mediated knock-down construct. (C) Morphology of the M. smegmatis cells over-expressing MtbGre. Comparison of cellular morphology (left panel) and nucleoid (right panel) of M. smegmatis mc2155 cells harboring either the pMV261 vector or pMV-greAS or pMV-greOE constructs. Left panels- bright-field images; right panels-fluorescent images showing the DAPI-stained nucleoid.
(TIF)
Determination of the expression pattern of Gre and its amount in the cells. (A) Expression of gre in response to different stresses in M. smegmatis and M. tuberculosis determined by western blot. (B) Estimation of the level of Gre protein in the M. smegmatis cells. Purified M. smegmatis Gre protein was used as a standard. 120 µg of total cell lysate proteins were probed with the anti-Gre antibody to estimate the Gre protein level in cells at different growth phases. (C) Western blot analysis for Gre from cell lysate of M. smegmatis exposed to different stresses.
(TIF)
Expression of Gre from pTrc gre construct in E. coli TK1021. (A) The gel shows expression of MtbGre from the pTrcgre construct, induced with 0.3 mM IPTG. (B) Transcript cleavage assays using MtbGre and E. coli GreA. Only MtbGre shows cleavage of mycobacterial RNAP elongation complex.
(TIF)