Abstract
Lung cancers harboring mutations in the epidermal growth factor receptor (EGFR) respond to EGFR tyrosine kinase inhibitors, but drug resistance invariably emerges. To elucidate mechanisms of acquired drug resistance, we performed systematic genetic and histological analyses of tumor biopsies from 37 patients with drug-resistant non–small cell lung cancers (NSCLCs) carrying EGFR mutations. All drug-resistant tumors retained their original activating EGFR mutations, and some acquired known mechanisms of resistance including the EGFR T790M mutation or MET gene amplification. Some resistant cancers showed unexpected genetic changes including EGFR amplification and mutations in the PIK3CA gene, whereas others underwent a pronounced epithelial-to-mesenchymal transition. Surprisingly, five resistant tumors (14%) transformed from NSCLC into small cell lung cancer (SCLC) and were sensitive to standard SCLC treatments. In three patients, serial biopsies revealed that genetic mechanisms of resistance were lost in the absence of the continued selective pressure of EGFR inhibitor treatment, and such cancers were sensitive to a second round of treatment with EGFR inhibitors. Collectively, these results deepen our understanding of resistance to EGFR inhibitors and underscore the importance of repeatedly assessing cancers throughout the course of the disease.
INTRODUCTION
Non–small cell lung cancer (NSCLC) is the leading cause of cancer death in the world, and traditional chemotherapeutic drugs are only modestly effective. Recent advances with targeted therapies have provided a marked benefit to subsets of patients whose tumors harbor specific genetic abnormalities. In particular, NSCLCs with mutations in the gene encoding the epidermal growth factor receptor (EGFR) are uniquely sensitive to EGFR blockade with specific tyrosine kinase inhibitors (TKIs) (1–3). Most cancers with EGFR mutations achieve marked and durable responses to treatment with the EGFR TKIs gefinitib or erlotinib. However, despite this initial response, patients with NSCLCs containing EGFR mutations acquire resistance to EGFR inhibitors, and the median time to disease progression is about 12 months (4, 5).
To date, two mechanisms of acquired drug resistance have been confirmed in patients. About half of cancers that acquire resistance to EGFR TKIs develop a secondary mutation in EGFR (T790M), which abrogates the inhibitory activity of the TKIs (6, 7). Another 15 to 20% undergo amplification of the MET receptor tyrosine kinase, which activates downstream intracellular signaling independent of EGFR (8, 9). Additionally, clinical experience has revealed that, after a drug-free interval, resistant cancers can respond again to EGFR TKIs (10, 11). However, the molecular basis for this phenomenon remains poorly understood. To increase our understanding of the full spectrum of acquired resistance by NSCLCs to EGFR TKIs, we rebiopsied recurrent disease sites in patients with EGFR mutations who developed resistance to EGFR TKIs. Molecular analyses were performed to assess the prevalence of known resistance mechanisms and to validate or refute potential mechanisms based on laboratory studies, with the aim of identifying new molecular mechanisms of resistance to EGFR TKIs. These investigations identified substantial histological and genetic changes in NSCLCs resistant to EGFR TKIs. In a few patients whose cancers were assessed at multiple points along their treatment course, we observed that genetic resistance mechanisms were “lost” without continued TKI treatment, thereby providing a molecular basis for the retreatment responses observed in the clinic. These results may provide a basis for developing new therapeutic strategies to overcome resistance and potentially to thwart its emergence. Additionally, our findings point to the value of repeat tumor biopsies throughout the course of a patient’s disease to determine the best treatment regimen.
RESULTS
Biopsies of resistant cancers
To identify how EGFR-mutant NSCLCs develop resistance to EGFR inhibitors, we performed biopsies on patients at the time that drug resistance was acquired. All patients had EGFR-mutant NSCLC and had achieved a clinical response to EGFR TKI therapy but subsequently developed progressive disease. They underwent repeat tumor tissue biopsies as part of routine clinical care. Clinical and pathological information was abstracted retrospectively under an Institutional Review Board (IRB)–approved protocol.
Thirty-seven patients had tumor tissue available both before and after TKI treatment. They included 15 men and 22 women (Table 1 and table S1). All patients had activating EGFR mutations; 20 (54%) had an exon 19 deletion mutation and 15 (41%) had the exon 21 point mutation L858R. All patients had responded clinically to either gefitinib (n = 5) or erlotinib (n = 32). Radiographs were obtained and robust treatment responses were confirmed with the Response Evaluation Criteria in Solid Tumors (RECIST) method in 14 of 17 patients with available scans (fig. S1) (12). The median duration of primary TKI therapy was 14.1 months (range, 4 to 69 months) and the 1- or 2-year progression-free rates were 64 or 30%, respectively. Most patients (78%) were still taking an EGFR TKI at the time of repeat biopsy, and biopsies were performed a median of 30 months (range,5 to 99 months) after original diagnosis. Only four patients received chemotherapy between the development of resistance and the repeat biopsy. Anatomic sites of repeat biopsy most commonly included lung lesions (38%), liver lesions (16%), and medi-astinal or cervical lymph nodes (16%). Most biopsies (68%) were percutaneous with either computed tomography or ultrasound guidance, but some were performed via bronchoscopy, mediastinoscopy, or another surgical procedure. There were no major biopsy-related complications, including no cases of clinically significant bleeding, pneumothorax, or unanticipated hospital admission.
Table 1.
Thirty-seven paired lung tumor biopsies resistant to EGFR inhibitors. EGFR, epidermal growth factor receptor; amp, amplification; del, deletion; Adeno, adenocarcinoma; Adenosquam, adenosquamous; NSCLC, non–small cell lung cancer not otherwise specified; SCLC, small cell lung cancer; CA, carcinoma; EMT, epithelial to mesenchymal transition; TKI, tyro-sine kinase inhibitor; Erlo, erlotinib; Gef, gefitinib.
| ID# | Age | Sex | EGFR mutation | Baseline histology | Summary of changes | Primary TKI (time on TKI) | TKI status at repeat biopsy |
|---|---|---|---|---|---|---|---|
| T790M | |||||||
| 1 | 66 | M | L858R | Adeno | T790M | Erlo (6 months) | Off (2 months) |
| 2 | 74 | F | Exon 19 del | Adeno | T790M | Erlo (12 months) | On |
| 3 | 47 | F | Exon 19 del | Adeno | T790M | Gef (15 months) | On |
| 4 | 60 | F | Exon 19 del | Adeno | T790M | Erlo (7 months) | On |
| 5 | 57 | M | L858R | Adeno | T790M | Gef (5+ years) | On |
| 6 | 47 | M | Exon 19 del | Adeno | T790M | Erlo (12 months) | Off (14 months) |
| 7 | 58 | F | Exon 19 del | Adeno | T790M | Erlo (3+ years) | On |
| 8 | 69 | M | L858R | Adeno | T790M* | Erlo (2 years) | On |
| 9 | 58 | F | G719C, S768I | Adeno | T790M | Erlo (2+ years) | On |
| 10 | 46 | F | Exon 19 del | Adeno | T790M | Erlo (3 years) | Off (3 months) |
| 11 | 53 | F | Exon 19 del | Adeno | T790M | Erlo (16 months) | On |
| 12 | 59 | F | L858R | Adeno | T790M | Erlo (8 months) | Off (5 months) |
| T790M + EGFR amp | |||||||
| 13 | 42 | M | Exon 19 del | Adeno | T790M, EGFR amp | Erlo (5 months) | On |
| 14 | 55 | M | Exon 19 del | Adeno | T790M, EGFR amp | Erlo (10 months) | On |
| 15 | 37 | F | Exon 19 del | Adeno | T790M, EGFR amp | Erlo (6 months) | On |
| T790M + new, additional mutations | |||||||
| 16 | 88 | F | Exon 19 del | Adeno | T790M, β-catenin | Erlo (2+ years) | On |
| 17 | 85 | M | Exon 19 del | Adeno | T790M, β-catenin | Erlo (22 months) | On |
| 18 | 75 | F | Exon 19 del | Adeno | T790M, APC† | Erlo (18 months) | On |
| MET amplification | |||||||
| 19 | 61 | M | L858R | Adenosquam | MET amp, loss EGFR amp | Erlo (15 months) | On |
| 20 | 76 | M | L858R | Adeno | MET amp | Erlo (13 months) | Off (5 months) |
| Acquired PIK3CA mutation | |||||||
| 21 | 65 | M | Exon 19 del | Adeno | PIK3CA acquisition | Erlo (21 months) | On |
| Histologic transformation (one with acquired PIK3CA mutation) | |||||||
| 22 | 67 | F | L858R | Adeno | SCLC transformation | Erlo (22 months) | On |
| 23 | 54 | F | Exon 19 del | Adeno | SCLC transformation | Erlo (3+ years) | On |
| 24 | 56 | F | L858R | Adeno | SCLC transformation, PIK3CA | Erlo (14 months) | On |
| 25 | 40 | F | Exon 19 del | Adeno | SCLC transformation | Erlo (2+ years) | Off (2 months) |
| 26 | 61 | F | L858R | Adeno | SCLC transformation | Erlo (18 months) | On |
| 27 | 66 | M | L858R | Adeno | EMT | Erlo (11 months) | On |
| 28 | 59 | M | Exon 20 ins‡ | Adeno | EMT | Gef (11 months) | On |
| 29 | 64 | M | L858R | Adeno | Sarcomatoid CA, loss of β-catenin | Erlo (11 months) | Off (2 weeks) |
| No histological or genetic changes identified | |||||||
| 30 | 62 | F | L858R | Adeno | None | Erlo (6 months) | On |
| 31 | 52 | F | Exon 19 del | Adeno | None | Gef (17 months) | On |
| 32 | 58 | F | Exon 19 del | Adeno | None | Erlo (14 months) | On |
| 33 | 61 | F | L858R | Adeno | None | Erlo (13 months) | On |
| 34 | 85 | F | Exon 19 del | Adeno | None | Erlo (6 months) | On |
| 35 | 62 | M | L858R | NSCLC | None | Gef (3+ years) | On |
| 36 | 56 | M | L858R | Adeno | None | Erlo (5 months) | Off (<2 weeks) |
| 37 | 51 | F | Exon 19 del | Adeno | None | Erlo (8 months) | On |
TP53 mutation suspected to be present, but not confirmed.
APC mutation confirmed in resistant specimen, but not confirmed to be present in initial biopsy.
Exon 20 insertion assay added to SNaPshot for this patient given direct sequencing result from pretreatment sample.
Genotypic mechanisms of acquired drug resistance
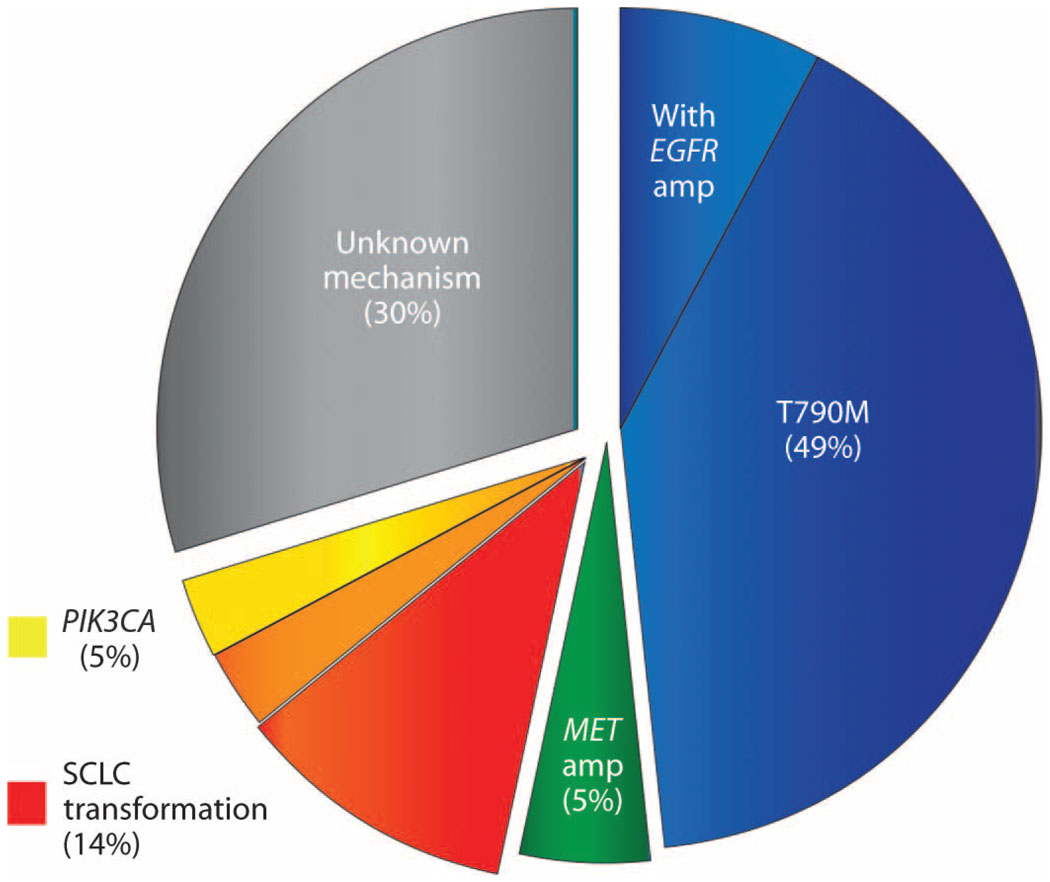
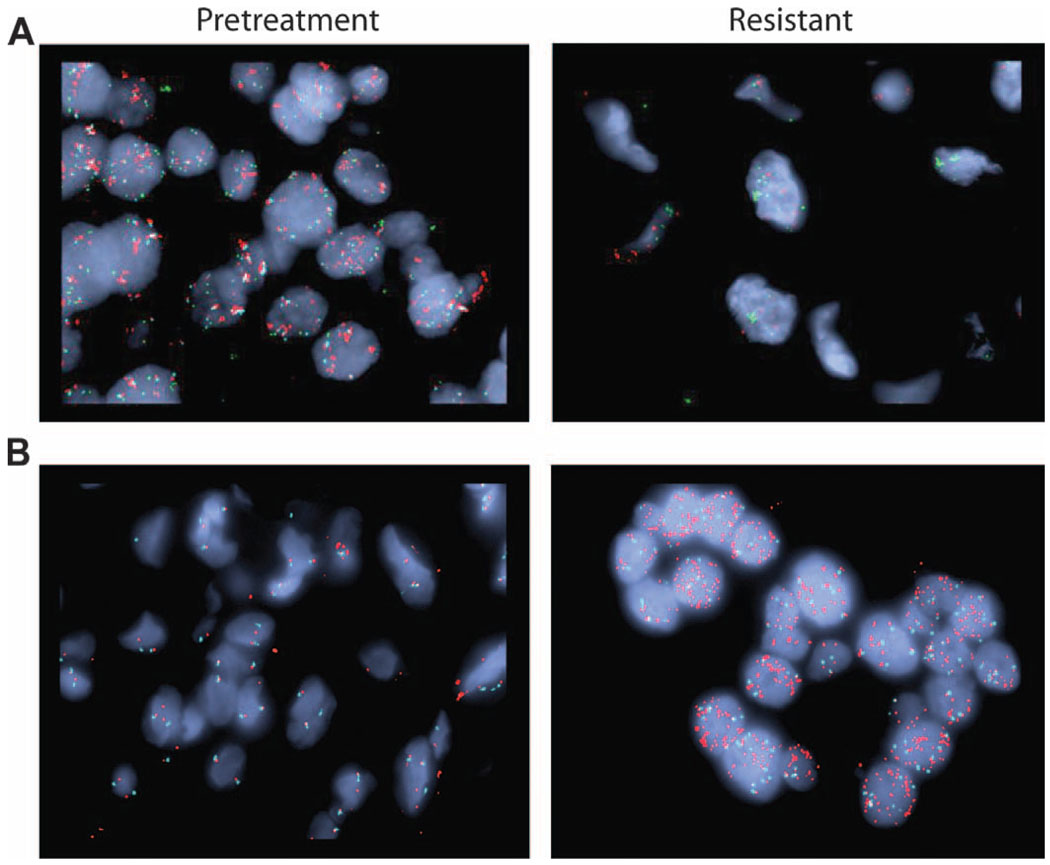
The 37 paired pre- and post-EGFR TKI tumor samples were analyzed for the presence of genetic alterations with our standard clinical geno-typing platform, the SNaPshot assay. SNaPshot is a multiplex platform that is used at Massachusetts General Hospital (MGH) to genotype cancers at specific genetic loci across 13 genes, as previously reported (table S2) (13). In addition, samples were analyzed for EGFR and MET amplification with fluorescence in situ hybridization (FISH). The pretreatment activating EGFR mutation was present in each drug-resistant specimen (Table 1 and table S1). As predicted, we observed mechanisms of TKI resistance that were previously validated in clinical specimens. Eighteen (49%) patients acquired the exon 20 EGFR mutation T790M, and two (5%) patients developed MET amplification (Fig. 1). In one case of an L858R EGFR-mutant cancer that subsequently developed MET amplification, the pretreatment specimen had marked EGFR amplification but no MET amplification (Fig. 2A). After resistance developed, MET amplification was abundant, but the EGFR amplification was lost (Fig. 2A). Given that the resistant lesion biopsied had initially responded to the TKI and harbored the same activating EGFR mutation as the treatment-naïve cancer, it seems most likely that the resistant tumor was derived from a distinct MET-amplified subpopulation of EGFR-mutant cells (that did not harbor EGFR amplification) that were selectively enriched during EGFR TKI administration, consistent with previous observations (14).
Fig. 1.
The frequency of observed drug resistance mechanisms. The pie chart depicts the prevalence of observed mechanisms of resistance to EGFR TKIs in 37 patients with NSCLC biopsied at the time that resistance was acquired. Pre- and posttreatment specimens were compared and only acquired mechanisms of resistance are depicted. The blue wedge represents resistant cancers that developed the EGFR T790M resistance mutation including a subset that developed concomitant EGFR amplification. The green wedge represents cancers that developed MET amplification, and the red wedge represents cancers that underwent transformation to SCLC. The yellow wedge represents cancers that developed PIK3CA mutations, and the orange wedge represents one patient who had both SCLC transformation and acquisition of a PIK3CA mutation.
Fig. 2.
Acquired genetic amplifications in drug-resistant lung tumors. Amplification of MET and EGFR genes was observed in biopsies of tumors from patients who had acquired resistance to EGFR TKIs. Shown are FISH analyses that detect the MET gene (green), EGFR gene (red), and the control CEP7 gene (aqua). (A) The pretreatment specimen from patient 19 (left panel) shows a normal MET copy number but significant EGFR amplification; the drug-resistant posttreatment specimen (right panel) from the same patient exhibits acquired MET amplification but normal EGFR copy number. (B) Patient 13 demonstrated an acquired EGFR amplification in the drug-resistant posttreatment specimen (right panel) compared to the pretreatment specimen (left panel).
We also observed acquired resistance mechanisms previously assessed only in in vitro studies and not previously identified in patients. These included two (5%) patients with acquired PIK3CA mutations (15). In addition, three (8%) patients acquired EGFR amplifications in their resistant specimens (Fig. 2B), all of which also acquired the classic T790M EGFR mutation. Moreover, in two cases with high-level EGFR amplification (>10-fold), it was clear by comparison of the peak heights on the SNaPshot chromatogram that the T790M allele was the amplified allele (fig. S2). In the third case, we were unable to make a definitive determination. Other cases with acquired mutations of uncertain significance included two (5%) cancers with β-catenin mutations, both of which occurred concomitantly with the EGFR T790M mutation. Fifteen (41%) posttreatment biopsies did not reveal any new mutations as assessed by the SNaPshot assay, nor MET or EGFR amplification. Two patients in this group had insufficient posttreatment tissue for EGFR and MET gene copy number analyses. Among the 15 patients without an identified genetic resistance mechanism, only 2 patients had stopped EGFR TKI therapy for more than 2 weeks at the time of biopsy.
Phenotypic changes in tumors with acquired resistance
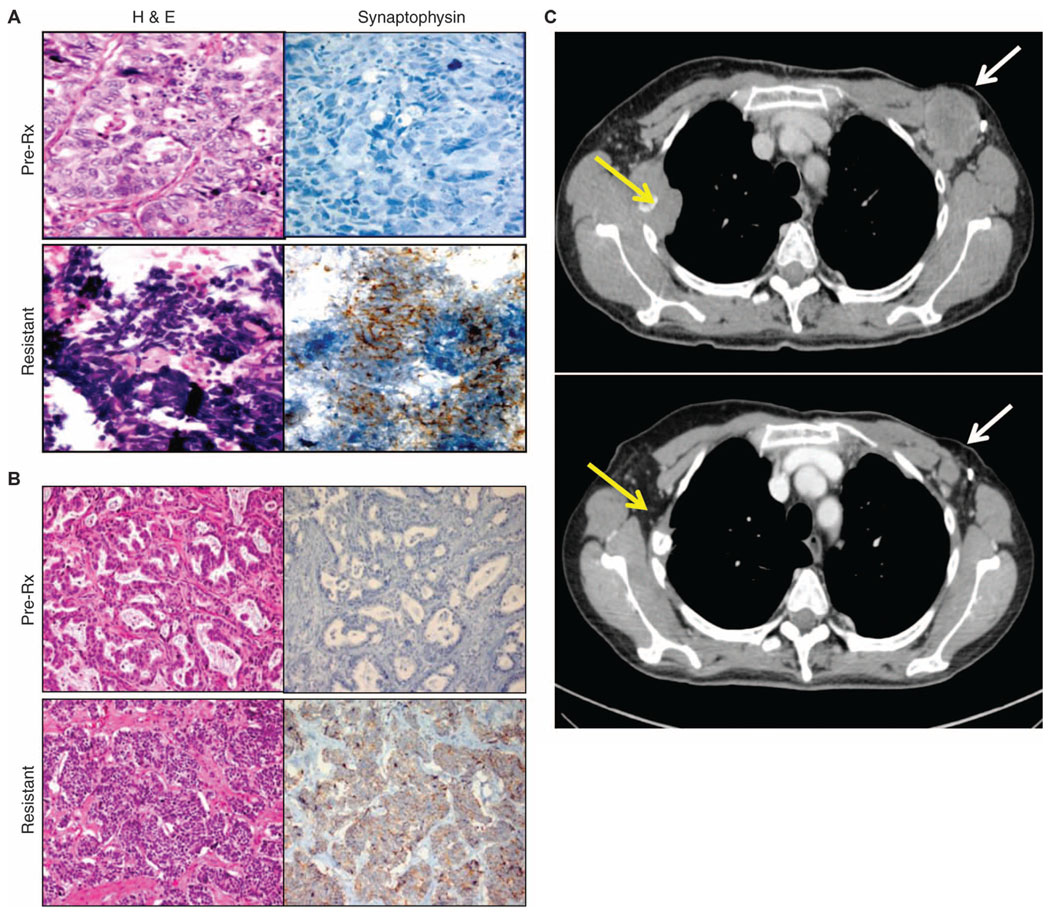
All of the drug-resistant tumor specimens underwent routine pathological analyses, and in some cases, significant alterations in the predominant histology of the resistant tumors were observed. To our surprise, five patients (14%) were found to have a diagnosis of small cell lung cancer (SCLC) in their drug-resistant tumor biopsies (Table 2). All of these cases were lung adenocarcinoma before EGFR TKI treatment. The transformation to SCLC at the time of clinical TKI resistance was validated by histological examination and confirmed by expression of neuroendocrine markers (Fig. 3, A and B). The original EGFR mutation was maintained during the histological transformation in all five cases. One patient also acquired a PIK3CA mutation accompanying the SCLC transformation. Clinically, these five patients ranged in their disease courses. Two patients had relatively indolent disease immediately after the SCLC transformation, whereas the other three patients showed a marked progression that was reminiscent of classic SCLC. Four patients were treated with a classic SCLC treatment, platinum-etoposide–based chemotherapy, which induced marked responses in three cases (Fig. 3C). The fourth treated patient had an initial response to radiation therapy, but declined quickly upon salvage chemotherapy. Autopsy of this case revealed extensive metastatic disease in the lung, thoracic lymph nodes, liver, and nodules along the diaphragm, all consisting entirely of SCLC and all maintaining the original EGFR L858R mutation with no additional mutations (table S3). However, brain metastases still retained the appearance of lung adenocarcinoma, consistent with the original diagnosis.
Table 2.
Patients with lung tumors showing an NSCLC to SCLC transformation. ID number refers to the patient number in Table 1. Pre-, pre-treatment; Post-, posttreatment/drug-resistant.
| Age (years) | Gender | ID no. | Biopsy | Histology | Synaptophysin | Chromogranin | CD56 | Genotype |
|---|---|---|---|---|---|---|---|---|
| 67 | Female | 22 | Pre- | Adenocarcinoma | − | − | − | L858R |
| Post- | SCLC | + | + | + | L858R | |||
| 54 | Female | 23 | Pre- | Adenocarcinoma | − | − | Weak+ | Exon 19 deletion |
| Post- | SCLC | Strong+ | Exon 19 deletion | |||||
| 56 | Female | 24 | Pre- | Adenocarcinoma | − | − | L858R | |
| Post- | SCLC | + | + | L858R, PIK3CA | ||||
| 40 | Female | 25 | Pre- | Adenocarcinoma | − | − | − | Exon 19 deletion |
| Post- | SCLC | + | − | Exon 19 deletion | ||||
| 61 | Female | 26 | Pre- | Adenocarcinoma | − | − | − | L858R |
| Post- | SCLC | + | + | L858R |
Fig. 3.
Drug resistance and transformation of NSCLC to SCLC. The SCLC histological phenotype was observed in five (14%) NSCLC patients who had acquired resistance. Two examples are shown. (A) Patient 23 had an exon 19 deletion in EGFR. (B) Patient 22 carried the L858R mutation in EGFR. The presence of the original activating mutation was confirmed in both pretreatment (pre-Rx) specimens (upper panels) and drug-resistant specimens (lower panels). Hematoxylin and eosin (H&E) (left) and synaptophysin (right) staining are shown for each case. Notably, the pretreatment biopsies (A and B, upper panels) demonstrate adenocarcinoma consisting of cellular growths of atypical glands with (A) or without (B) a cribriform pattern that are negative for synaptophysin. The post-resistance biopsies (A and B, lower panels) demonstrate a SCLC phenotype consisting of nests of small cells with a high nuclear-to-cytoplasmic ratio with (A) or without (B) the classic SCLC-associated finding of “crush artifact,” with positive immunohistochemical staining for synaptophysin. (C) Computed tomography scans of a representative patient (patient 25) with SCLC in the acquired resistance specimen before (above) and after (below) chemotherapy with cisplatin and etoposide (the standard regimen for treating SCLC). Yellow arrows denote right third rib metastases, and white arrows denote left axillary adenopathy.
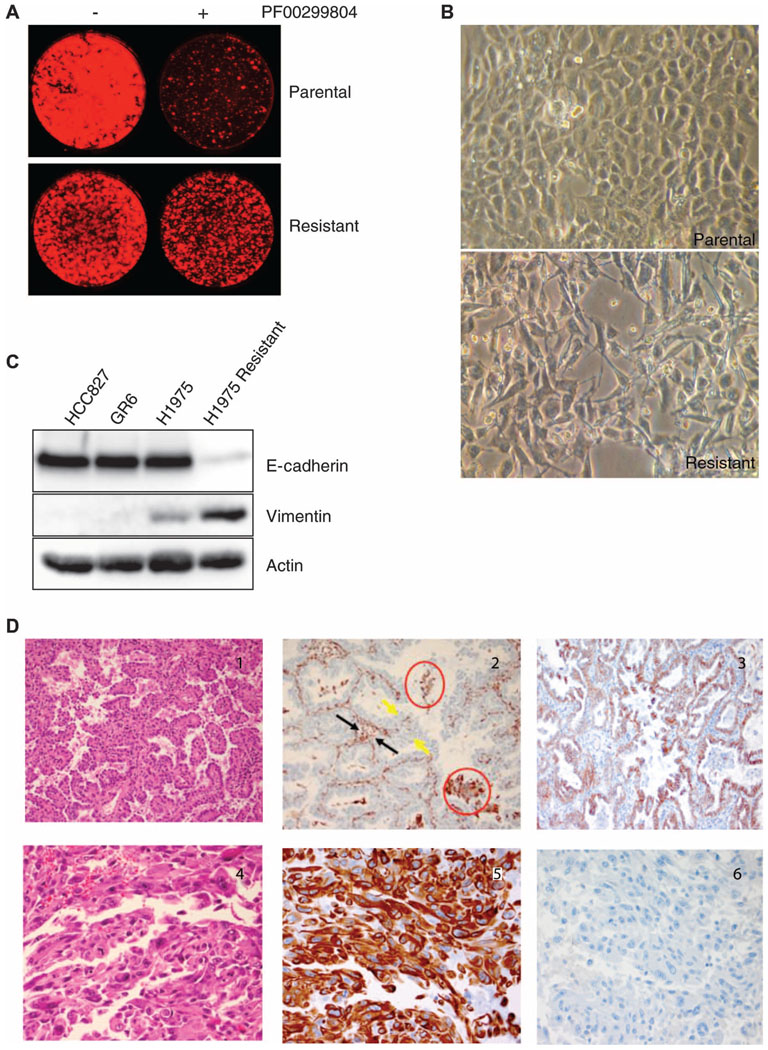
In the laboratory, we observed a different phenotypic transformation when using the H1975 (L858R/T790M) lung adenocarcinoma cell line to model acquired resistance to an EGFR inhibitor. The cell line was made resistant to the irreversible EGFR inhibitor, PF00299804, to which it was initially sensitive, as previously described (Fig. 4A) (8, 14–16). The resistant cell line (H1975 Resistant) did not acquire MET amplification, but did show an increased copy number of the EGFR T790M allele, consistent with previous reports (17). In addition, it underwent a marked histological change and developed a spindle-like morphology (Fig. 4B). Assessment of E-cadherin and vimentin expression confirmed that the resistant cell line had undergone an epithelial-to-mesenchymal transition (EMT) (Fig. 4C). EMT describes a cancer cell that loses its epithelial morphology and develops a more spindle-like mesenchymal morphology; this histological change is often associated with a shift in expression of specific proteins (for example, loss of E-cadherin and gain of vimentin) and a more invasive phenotype. In contrast, HCC827GR cells that had developed MET amplification upon resistance to an EGFR TKI (8) did not undergo an EMT (Fig. 4C).
Fig. 4.
EMT and acquired resistance. (A) H1975 cells were cultured in the presence of the irreversible EGFR inhibitor PF00299804 until drug resistance developed, as demonstrated by Syto60 viability assays. (B) Images of the parental and drug-resistant H1975 cells by bright-field microscopy demonstrate that the resistant cells have developed a spindle-like morphology. (C) Parental and resistant H1975 cells were lysed and probed with antibodies against E-cadherin, vimentin, and actin, revealing loss of E-cadherin expression and gain of vi-mentin expression among drug-resistant H1975 cells. For comparison, HCC827 cells and the derived HCC827 GR6 cell line (HCC827 cells that acquired resistance to gefitinib via MET amplification), which do not undergo an EMT, are shown. (D) Example of a case (patient 28) whose drug-resistant tumor shows evidence of an EMT (top, pretreatment specimens; bottom, drug-resistant posttreatment specimens). Left, H&E staining; middle, staining for vimentin; right, staining for E-cadherin. Notably, the pretreatment cancer had an adenocarcino-ma histology (panel 1), does not stain for vimentin (panel 2), and shows preserved membranous staining with E-cadherin (panel 3). The vimentin-positive areas in panel 2 include alveolar macrophages (red circles), inflammatory and stromal cells in fibro-vascular cores (black arrows), but not tumor cells lining papillary structures (yellow arrows). The drug-resistant posttreatment specimen has sarcomatoid histology (panel 4), is positive for vimentin (panel 5), and is negative for E-cadherin (panel 6), consistent with an EMT.
This finding supported previous observations that cancer cell lines undergoing an EMT have intrinsic resistance to EGFR inhibitors (18–22). This prompted us to analyze paired tissue samples from seven patients with unknown mechanisms of resistance and five patients with the T790M EGFR mutation (who had sufficient remaining tissue) for the development of mesenchymal features and changes in vimentin and E-cadherin expression. Three of the 12 resistant specimens had phenotypic changes consistent with a mesenchymal appearance at the time of TKI resistance; all 3 cases were among the 7 without another identified resistance mechanism. Further analyses confirmed that two of these three posttreatment specimens had acquired vimentin expression and lost E-cadherin expression compared to their pretreatment counterparts, supporting an EMT (Fig. 4D). Both cancers that underwent this transition retained their original EGFR mutation. Furthermore, one of these patients subsequently underwent autopsy, and phenotypic heterogeneity was observed among the differing sites of metastatic disease (table S4). A left bronchial lymph node exhibited adenocarcinoma and did not have immunohistochemical evidence of EMT. However, another specimen from the right lower lobe with sarcomatoid morphology had marked evidence of EMT (Fig. 4D and table S4). Both of these tissues retained the original EGFR mutation, an exon 20 insertion. Notably, although exon 20 insertions are not uniformly activating and have been associated with TKI resistance, this patient had achieved stable disease and symptom improvement on gefitinib treatment lasting 11 months, which is consistent with the clinical criteria of acquired resistance to EGFR TKIs (23). In contrast to these cases that underwent an EMT upon the development of resistance, we failed to observe this transition in all five cases examined that had developed T790M as their resistance mechanism.
It appears that an EMT and a histological change to SCLC may be enriched specifically in EGFR-mutant cancers acquiring resistance to TKI therapy, because we failed to observe EMT in 10 available biopsy specimens from EGFR wild-type tumors that developed resistance to chemotherapy. Additionally, we failed to identify a changeover to SCLC in these 10 samples and in an additional 69 cases of stage III NSCLC that were resected after preoperative chemotherapy and radiation. The overlap of the genotypic and phenotypic changes observed in the entire cohort of EGFR-mutant TKI-resistant specimens is shown in fig. S3.
Longitudinal genotypic and phenotypic changes in response to EGFR TKI
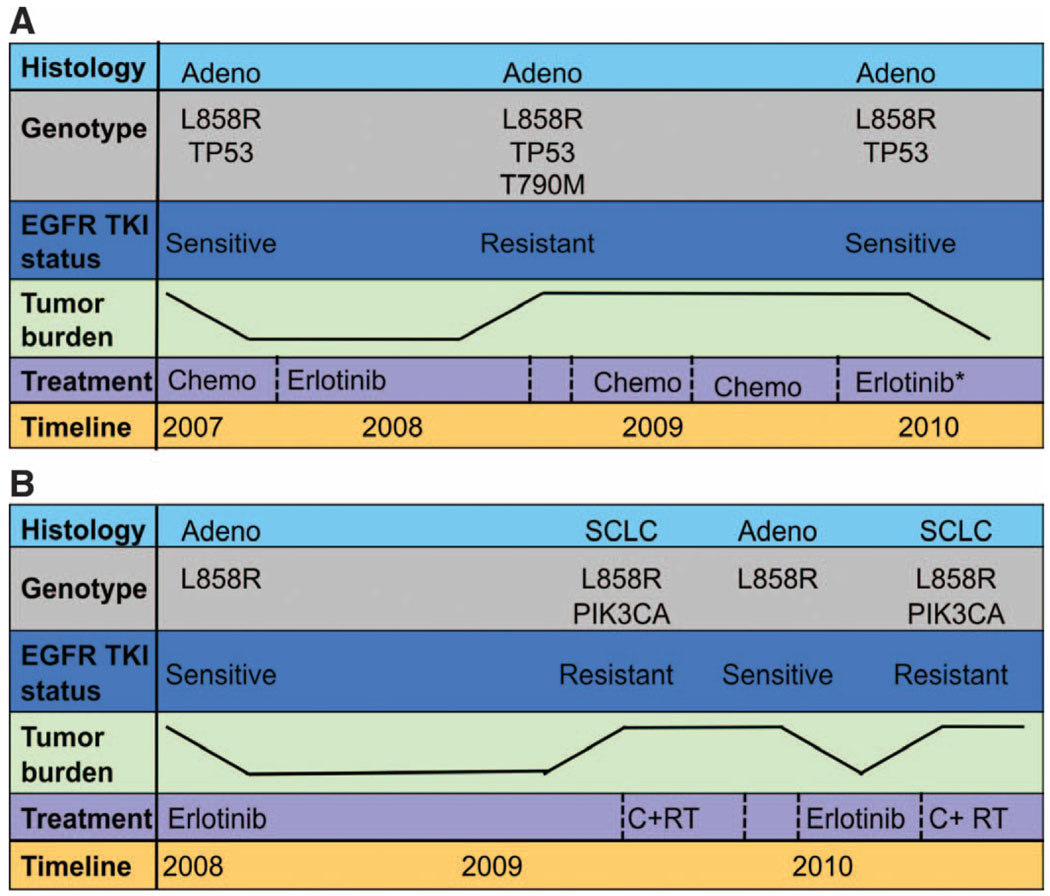
Three patients underwent multiple repeat biopsies over the course of their disease (Fig. 5). The first patient (Fig. 5A) had adenocarcinoma that harbored the L858R EGFR mutation and a mutation in the tumor suppressor TP53. As expected, this patient experienced a substantial initial response to erlotinib lasting 8 months, at which time a lung core biopsy revealed adenocarcinoma with the same L858R and p53 mutations, as well as an acquired T790M EGFR mutation. After a 10-month interval without any EGFR TKI exposure, a second repeat biopsy performed on the same lung lesion as the first repeat biopsy revealed that the T790M mutation could no longer be detected. The patient subsequently responded to treatment in a clinical trial of erlotinib plus an investigational agent that does not target T790M. A second patient with an exon 19 deletion had a similar clinical course involving gain and loss of the T790M mutation in multiple biopsies from the same anatomical location during periods of erlotinib and chemotherapy treatment, respectively.
Fig. 5.
Longitudinal evaluation of patients treated repeatedly with erlotinib. The color-coded boxes to the left of each panel describe the data displayed across the timeline. The tumor burden depicted is not quantitative but represents tumor growth and shrinkage. (A) Patient 12 with a lung adenocar-cinoma carrying the L858R EGFR mutation and a mutation in the tumor suppressor p53 had a modest response to first-line chemotherapy. The patient then achieved a more robust and durable response to second-line erlotinib, with near-complete resolution of her lung nodules. After 8 months on an EGFR TKI, she developed resistance with growing bilateral pulmonary nodules. A lung core biopsy revealed an acquired T790M mutation in EGFR. There was no response to chemotherapy and she subsequently developed bone and liver metastases. At that time (after not taking the EGFR inhibitor for 8 months), a second lung core biopsy revealed the L858R EGFR mutation, but no detectable T790M EGFR resistace mutation. The patient responded to erlotinib (in combination with an investigational agent that did not target T790M). (B) Patient 24 had an L858R EGFR-mutant adenocarcinoma that responded markedly to first-line erlotinib for 12 months with resolution of her pleural effusion and pulmonary nodules. After 1 year, there was progression of the largest nodule. Core biopsy of this lesion revealed histological transformation to SCLC that harbored the EGFR L858R mutation and acquired a PIK3CA mutation. She was treated with radiation and chemotherapy. After a 6-month break from all treatment, her pleural effusion reaccumulated and small bilateral pulmonary nodules reappeared. Pleural effusion analysis revealed adenocarcinoma with the L858R EGFR mutation only (no PIK3CA mutation). Her disease responded to a second-line course of erlotinib. After 6 months, bony metastases and an adrenal lesion developed. Assessment of a growing bone metastasis revealed SCLC with both the L858R EGFR and PIK3CA mutations.
The lung core biopsy from the drug-resistant tumor of a third patient (Fig. 5B) demonstrated SCLC with the original EGFR L858R mutation plus an acquired PIK3CA mutation (Table 2). This patient was treated with chemotherapy and radiation for SCLC and her cancer went into a partial remission. After a 7-month interval without any erlotinib exposure, she developed a symptomatic pleural effusion and a thoracentesis revealed adenocarcinoma (negative for neuroendocrine markers) with the L858R EGFR mutation only; the PIK3CA mutation was not detectable. Erlotinib was readministered with a second clinical response. When this patient developed resistance once again, a soft tissue metastasis originating from bone revealed SCLC with the EGFR L858R and the PIK3CA mutation. In total, these findings provide a molecular link to the clinical observation that patients with EGFR-mutant NSCLC tumors will often respond to erlotinib after a TKI-free interval (10, 11). Without the continued selective pressure of the TKI, the genetic resistance mechanisms and potentially the phenotypic resistance mechanisms are lost.
DISCUSSION
Here, we have performed in-depth genetic and histological analyses on cancers that acquired resistance to EGFR inhibitors. We observed both known molecular mechanisms of acquired resistance and also several genotypic and phenotypic changes that we believe broaden the conceptual model of acquired drug resistance. Notably, we observed a surprisingly high frequency of conversion of NSCLC to SCLC, marked EGFR amplification in a subset of cases with the T790M EGFR mutation, the development of PIK3CA mutations, EMT, and the loss of genetic resistance mechanisms in the absence of continuous TKI treatment. These findings provide new insights into our understanding of drug resistance and emphasize the need to perform tumor biopsies after the development of resistance to identify the best treatment options for patients.
The development of drug resistance that invariably occurs after about 12 months of initiating treatment (4, 5, 24–26) has spurred efforts to understand the biology underlying resistance and to identify therapeutic strategies to overcome or prevent it. These laboratory studies have primarily focused on exposing EGFR-mutant, TKI-sensitive cell lines to EGFR TKIs until resistance develops. They have identified several resistance mechanisms, two of which—EGFR mutation T790M (6, 7) and MET amplification (8, 9)—have been validated in the clinic. Other acquired resistance mechanisms identified by studying the development of resistance to EGFR TKIs in vitro include loss of PTEN (27, 28) and activation of the insulin growth factor receptor (in cell lines addicted to wild-type EGFR) (16). However, these resistance mechanisms have not yet been validated in the clinic. Activation of MET by hepatocyte growth factor (HGF) has been shown to drive resistance to EGFR TKIs, but these experiments were performed by adding exogenous HGF or HGF-secreting tumor-derived fibroblasts (14, 29–31), not by selecting cells after chronic exposure to TKIs. Analyses of resistant specimens support, but do not prove, that HGF may be a resistance mechanism in patients. To date, the various EGFR TKI resistance mechanisms share the same underlying concept: They enable the cancer cell to maintain its intracellular growth signaling pathways, especially the phosphatidylinositol 3-kinase (PI3K)–AKT pathway, in the presence of the EGFR TKI (32–37).
In our cohort of patients with EGFR mutation–positive NSCLC and acquired EGFR TKI resistance, we observed known mechanisms of resistance, the EGFR T790M mutation and MET amplification. Forty-nine percent developed the T790M mutation, consistent with the previously reported incidence of this mutation in patients with acquired resistance (8, 38–40). A subset of these patients also developed pronounced EGFR amplification, and it appears that the T790M allele is selectively amplified. To the best of our knowledge, amplification of EGFR T790M has not been previously appreciated in TKI-resistant specimens of NSCLC tumors. Balak et al. (40) reported one patient with about twofold increase in EGFR copy number in a drug-resistant specimen, but that case did not harbor the T790M mutation in EGFR. Despite the promising activity of newer, irreversible EGFR inhibitors in patients with EGFR mutations (41), their efficacy has been minimal in patients with acquired resistance to gefitinib and erlotinib (42, 43). The recent report by Ercan and colleagues (17) that amplified T790M mutations may promote resistance to irreversible EGFR inhibitors suggests that these patients may not respond to the current irreversible EGFR inhibitors and should be directed to other potential therapeutic strategies such as combined PI3K and MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase) inhibition (44), newer, more potent T790M–specific EGFR inhibitors (45), or combinations of anti-EGFR therapies (46). In addition, we observed that a subset of the T790M patients also acquired additional mutations, including two (11% of the T790M cohort) with acquired mutations in β-catenin. To our knowledge, β-catenin has not been postulated as an EGFR TKI resistance mechanism. Anecdotally, in our clinic, we have three patients with concurrent EGFR and β-catenin mutations at baseline, all of whom responded well to erlotinib without evidence of early-onset resistance.
MET amplification was identified in only two (5%) patients, which is less than the 15 to 20% frequency reported by our group and others (8, 9, 38). We cannot easily explain this lower than expected frequency. Possible contributing reasons include the lack of sufficient tissue for MET testing in two patients in the “unknown mechanism” category, the fairly conservative (high) threshold used for designating amplification used by our pathologists, and the sample size of our cohort. In addition, we failed to identify any acquired genetic resistance mechanism in several cases. Although we were unable to test for all potential resistance mechanisms because of tissue exhaustion and inadequate reagents, it does seem likely that further analyses with more sophisticated techniques such as deep sequencing will lead to the identification of new mechanisms of resistance to EGFR TKIs.
In addition to these two well-described mechanisms of TKI resistance, we observed acquired PIK3CA mutations in two patients. To our knowledge, this represents the first documentation of PIK3CA mutations leading to drug resistance in cancer patients. This finding is supported by our previous laboratory findings that introduction of a PIK3CA mutation in EGFR-mutant HCC827 cells confers resistance to gefitinib (15). This has important therapeutic implications because there are several ongoing early-phase clinical trials combining EGFR and PI3K pathway inhibitors that are attractive targeted therapy strategies to overcome this mode of resistance. We also hypothesize that patients who have EGFR and PIK3CA mutations in the original primary tumor (baseline) might experience an abbreviated duration of benefit from EGFR TKI therapy compared with patients lacking PIK3CA mutations, and could be considered for enrollment in a first-line clinical trial combining an EGFR and PI3K inhibitor. Indeed, we have observed two patients with EGFR and PIK3CA mutations at baseline who both responded to first-line erlotinib therapy, but the responses lasted only 5 and 7 months. Neither of these cases is included in this cohort of patients who received repeat biopsies; one underwent a repeat biopsy but the tissue was nondiagnostic, and the other was not offered a repeat biopsy.
Perhaps, one of the more surprising findings from our study is the observation that 5 (14%) of the 37 patients experienced a fundamental histology transformation from NSCLC to SCLC at the time of TKI resistance. The original EGFR mutation was maintained in all five patients, disputing the rare possibility that these patients developed a second primary cancer. One patient also acquired a PIK3CA mutation in the SCLC specimen, but none of the patients demonstrated EGFR T790M or MET amplification. The pre- and posttreatment tissues were subjected to neuroendocrine immunohistochemical analyses including staining for synaptophysin, chromogranin, and/or CD56. Although the posttreatment (SCLC) specimens were all positive for neuroendocrine markers, most consistently synaptophysin, the pretreatment (NSCLC) samples were uniformly negative for neuroendocrine markers. We speculate that the high frequency of recognizing this unusual histological phenomenon may have been partly because of the implementation of thorough pathological evaluation of drug-resistant specimens as part of routine clinical care. These findings directly affected patient care decisions, and four of the five patients received SCLC chemotherapy regimens with a response obtained in three patients. This unequivocally suggests that the posttreatment biopsies provided useful clinical information in addition to research information, and that repeat biopsies at the time that clinical resistance to EGFR TKIs develops can directly benefit patients. The transition from NSCLC to SCLC appears to be specific for resistance to EGFR TKIs. We observed no evidence of SCLC in 10 cases of EGFR wild-type chemotherapy-resistant NSCLC and in 69 resected stage III lung cancers, where the patients had received chemotherapy and radiation.
Previous case reports have described patients with biopsy-proven SCLC and EGFR mutations (47–51). The individual cases reported by Zakowski et al. (47) and by Morinaga et al. (48) are most similar to our patients, and each describes a never-smoking female that presented with EGFR-mutant metastatic adenocarcinoma that transformed into SCLC after developing resistance. Okamoto et al. (49) describe a never-smoking female diagnosed with CD56-positive advanced SCLC harboring an exon 19 deletion in EGFR, who had a good partial response to first-line gefitinib. Fukui et al. (50) identified 6 patients with combined NSCLC-SCLC histology from a cohort of 64 SCLC patients undergoing surgical resection; one was a never-smoking female with an L858R EGFR mutation in both the SCLC and adenocarcinoma components. The final report is a case series arising from an analysis of 122 Asian patients with SCLC or mixed histology tumors that were screened for EGFR mutations, of which 5 (4%) samples were found to be mutation-positive (3 L858R, 1 exon 19 deletion, and 1 G719A) including a never-smoker and 4 smokers with tobacco histories ranging from 3 to 68 pack-years (51). In this series, only one patient had a pretreatment adenocarcinoma that transformed into a combined SCLC-adenocarcinoma after developing clinical resistance to an EGFR TKI. The other four patients had EGFR-mutant SCLC or mixed histology tumors at baseline.
The biological underpinnings of the SCLC transformation are unknown and are of great interest. The finding that the same EGFR-mutant cancer can manifest both as an adenocarcinoma and as a SCLC hints at the existence of a pluripotent population of EGFR-mutant cancer cells or cancer stem cells (52, 53) that are the source of resistance. The cause of the phenotypic switch to SCLC and concordant development of resistance remain to be determined. Perhaps, these patients developed drug resistance through a genetic or epigenetic event that concurrently led to a shift in phenotypic appearance. One of the marked molecular differences between NSCLC and SCLC is that most SCLCs exhibit loss of expression of the retinoblastoma protein (54–56), a tumor suppressor. We attempted to determine whether the resistant specimens had loss of retinoblastoma protein expression by immunohistochemistry, but staining was not of sufficient quality for interpretation.
In addition, we clearly observed the EMT in two cases of acquired TKI resistance. Neither case had another identified resistance mechanism, but more cases will be required to determine whether this mutual exclusivity can be generalized. Similarly, we observed an EMT in an EGFR-mutant cell line rendered resistant to an EGFR inhibitor in vitro. Several groups have noted that cell lines undergoing EMT are intrinsically resistant to EGFR inhibitors (18–22). However, those cancer models do not have EGFR mutations and many have KRAS mutations, so the relevance of those findings to acquired TKI resistance is less straightforward. Two case reports just published support our observation of an EMT in EGFR-mutant NSCLC at the time of TKI resistance (57, 58). The molecular mechanisms connecting the resistance of the cancer cells to the mesenchymal phenotype remain unknown. However, the recent findings that KRAS-mutant lung cancers with mesenchymal features are resistant to both KRAS knockdown (59) and combined PI3K and MEK inhibition (60) suggest that mesenchymal cells may have an intrinsic lack of sensitivity to the intracellular signaling pathway down-regulation that is normally the hallmark of sensitivity to EGFR TKIs.
Evidence from three patients with multiple biopsies over the course of their disease suggests that both tumor genotype and phenotype may evolve dynamically under the selective pressure of targeted therapies. Two patients developed T790M EGFR mutations at the time of TKI resistance and subsequently lost evidence of that resistance mutation in the same anatomic tumor after a period free from TKI treatment. These patients both responded to a challenge with an EGFR inhibitor subsequent to losing the T790M mutation. The third patient underwent a SCLC transformation with acquisition of a PIK3CA mutation at the time of resistance and, after a TKI-free interval, was found to have adenocarcinoma without a detectable PIK3CA mutation. This cycle was repeated when, after a second response to erlotinib, the cancer ultimately developed resistance again and the biopsy of the resistant cancer again revealed the SCLC phenotype with the EGFR L858R and PIK3CA mutations. The mechanisms underlying these fluctuations remain to be proven, but it is tempting to speculate that the baseline heterogeneity of the cancers may contribute to these findings. Indeed, it is possible that substantial populations of “sensitive” cancer cells may remain dormant while subjected to TKI treatment, as recently suggested by laboratory studies (61). Withdrawal of the TKI may permit their rapid expansion to a degree that overtakes the bulk of the tumor burden. Such a mechanism could also provide insight into the pronounced tumor flare that is often clinically observed when the TKI is removed from slowly progressing cancers (62). Indeed, these findings confirm that even “genetic” mechanisms of resistance are potentially reversible. Therefore, a static diagnostic biopsy may be insufficient to guide therapeutic decisionmaking throughout the course of a patient’s disease. Moreover, all of our patients experienced a second response to erlotinib when their resistance mechanism was no longer detectable, suggesting that repeat biopsies can provide molecular guidance about the likely benefit of a second treatment regimen with EGFR TKI therapy.
The primary limitations of our study are its retrospective nature and the heterogeneity among practice patterns that led to patients undergoing repeat biopsies at various times during their disease (Table 1). Although all of these treatment variations could have affected the resistance mechanisms observed, the most direct confounder is likely to be whether the patient was “on” or “off” of the primary TKI at the time of biopsy. All of our patients except one were on TKI at the time of biopsy, or had been off drug treatment for ≤5 months (table S1). Another limitation is that in many cases, because of safety and feasibility concerns or because of the predominant radiographic progression in one anatomic area over another, the repeat biopsies were obtained from different tumor locations compared to the original biopsies. Although distinct mechanisms of resistance in different anatomic locations within the same patient have been described (8), we observed that the primary resistance mechanism was often consistent throughout different metastatic sites both in our autopsy cases and in patients with multiple sites biopsied over time. Larger studies will be helpful in further clarifying the impact of these variables.
In conclusion, this study provides further impetus for the utility of reassessing cancers after they acquire resistance to targeted therapies. As our study shows, there is tremendous heterogeneity in resistance mechanisms, each of which may require its own therapeutic strategy. A recent report suggests that cancers with various resistance mechanisms may have distinct prognoses (63). Although invasive biopsies have associated risks, we did not encounter any significant complications. We anticipate that technologies to assess cancers via noninvasive measures such as circulating tumor cell analyses, plasma DNA analyses, or molecular radiology may eventually obviate the need for invasive procedures. The knowledge gained from our repeat biopsy program directly affected treatment decisions and outcomes, and we were better equipped to rationally treat patients (for example, those with SCLC) as their tumors evolved. Several patients in our cohort were enrolled in clinical trials specifically targeting T790M, MET, or the PI3K signaling pathway after biopsies of their drug-resistant tumors, and several had disease stabilization or response to those therapies. Indeed, it is becoming increasingly clear, from experiences with both chronic myelogenous leukemia treated with ABL kinase inhibitors and EGFR-mutant lung cancers treated with EGFR kinase inhibitors, that the era of targeted therapies will mandate continual assessment of each cancer’s evolution over the course of treatment to determine how it became resistant to therapy and to identify the optimal strategies to prevent or overcome it.
MATERIALS AND METHODS
Patients
All 43 consecutive EGFR-mutant NSCLC patients with acquired EGFR TKI resistance undergoing standard post-resistance biopsy of their tumor from January 2007 to May 2010 at the MGH were considered for inclusion in the study cohort. Patients included in the final analysis had to have both pre- and posttreatment tumor specimens available for testing at MGH. To ensure sufficient tissue for molecular analysis, we obtained core biopsies whenever possible, and all fine-needle aspiration samples (except one) undertook multiple passes, which were prospectively combined and spun down into a cell block. Six patients did not meet criteria and were excluded, including one whose repeat biopsy was nondiagnostic for malignancy, one bone biopsy with poor-quality DNA for molecular testing, one with a concomitant thyroid cancer in which the resistant biopsy showed malignant cells that were inconclusive regarding bronchogenic or thyroid origin, one fine-needle aspiration with insufficient DNA, one with a medical contraindication to biopsy, and one pretreatment biopsy that could not be located for molecular analysis. Thirty-seven patients were included in the study cohort; the feasibility of repeat biopsy and comparative molecular analysis in our clinic was therefore 37/43 or 86%. The electronic medical record was reviewed retrospectively to obtain all demographic and clinical information under an IRB-approved protocol.
Genetic analyses
Our group recently developed a multiplexed polymerase chain reaction (PCR)-based assay, based on the commercially available SNaPshot platform (Applied Biosystems), to detect mutations in tumor DNA from formalin-fixed, paraffin-embedded tissue (13). Our SNaPshot tumor genotyping assay detects multiple mutations in 13 key cancer genes including EGFR, KRAS, BRAF, PI3KCA, β-catenin, APC, and TP53 (table S2); these genes were selected on the basis of clinical relevance, with potential therapeutic agents either already available or with multiple pipeline drugs under development. The DNA of interest is amplified with multiplexed PCR. Genotypes are determined with a single-base extension sequencing reaction, in which allele-specific probes interrogate loci of interest and are extended by fluorescently labeled dideoxynucleotides. The allele-specific probes have different sizes and are subsequently resolved by electrophoresis and analyzed by an automated DNA sequencer. The sensitivity of the SNaPshot assay ranges from 94 to 99% per allele, with an average sensitivity of 95%. The average specificity is >95%. The SNaPshot assay has been validated for use in a Clinical Laboratory Improvement Act (CLIA)–certified lab and is performed as a clinical routine test, with results included in the medical record (13).
In our study, all pre- and posttreatment tumor specimens underwent genotyping with SNaPshot. Some pretreatment samples had also been analyzed via direct sequencing of EGFR at the time of diagnosis, as that was our standard clinical analysis up until 2009. Paired tumor samples also underwent FISH of both MET and EGFR using standard protocols (24). Tumor content by hematoxylin and eosin (H&E) was always confirmed before FISH slides were prepared. When tumor tissue was limited or at risk of becoming exhausted, the genetic tests were prioritized in the following order: (i) SNaPshot testing to confirm EGFR mutation, (ii) the remaining SNaPshot assays, (iii) MET FISH testing, and (iv) EGFR FISH testing.
Histological analyses
All biopsy specimens were reviewed at MGH to confirm diagnoses. Histology was confirmed by H&E staining, and tissue-specific markers such as TTF-1 (thyroid transcription factor 1) were included at the discretion of the pathologist. More tissue-specific markers were included for metastatic specimens when the primary site was in question. Neuroendocrine immunohistochemistry with synaptophysin, chromogranin, and/or CD56 was performed on both the pre- and posttreatment samples that were suggestive of SCLC transformation on H&E staining. Vimentin and E-cadherin immunohistochemistry was also performed on selected patient samples under an IRB-approved protocol. All immunohistochemical staining was performed on representative tissue sections from formalin-fixed and paraffin-embedded tissue blocks. A Ventana autostainer (Benchmark XT) and the company’s prediluted antibodies (Ventana) were used for synaptophysin, chromogranin, CD56, and vimentin immunostaining, following the manufacturer’s instructions. For E-cadherin immunohistochemistry, the antibody from a different vendor (M3612, dilution 1:50, Dako) was applied. HGF was not tested because of a lack of sufficient tissue in nearly all cases and is therefore not included in this article.
Analyses of H1975 cells made resistant to PF00299804
To generate a resistant cell line, we maintained H1975 cells (L858R/ T790M) in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and exposed them to increasing concentrations of PF00299804 similar to our previously described methods (8, 15). PF00299804 was provided by J. Christensen at Pfizer. PF00299804 concentrations were increased stepwise from 1 nM to 2 µM when the cells resumed growth kinetics similar to that of the untreated parental cells. The development of the resistant cell line took ~3 months. To confirm the emergence of a resistant clone, we performed survival assays after growth at each concentration after allowing the cells to grow in drug-free conditions for at least 4 days.
Western blots were performed as previously described (44). The E-cadherin antibody was from BD Biosciences, the vimentin antibody was from Cell Signaling, and the actin antibody was from Sigma.
Growth and inhibition of growth were assessed by Syto60 staining (Invitrogen). Cells were fixed with 4% formaldehyde for 20 min at 37°C and incubated with a 1:5000 dilution of Syto60 stain for 60 min. Cell density in each well was determined with an Odyssey Infrared Imager (LI-COR Biosciences), corrected for background fluorescence from empty wells, and normalized to untreated wells, as described previously (64).
Supplementary Material
REFERENCES AND NOTES
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sánchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M. Spanish Lung Cancer Group, Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 7.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Jänne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yano S, Nakataki E, Ohtsuka S, Inayama M, Tomimoto H, Edakuni N, Kakiuchi S, Nishikubo N, Muguruma H, Sone S. Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: A report of three cases. Oncol. Res. 2005;15:107–111. [PubMed] [Google Scholar]
- 11.Yoshimoto A, Inuzuka K, Kita T, Kawashima A, Kasahara K. Remarkable effect of gefitinib retreatment in a patient with nonsmall cell lung cancer who had a complete response to initial gefitinib. Am. J. Med. Sci. 2007;333:221–225. doi: 10.1097/MAJ.0b013e31803b8acb. [DOI] [PubMed] [Google Scholar]
- 12.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 13.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, Stubbs H, McDermott U, Settleman J, Kwak EL, Clark JW, Isakoff SJ, Sequist LV, Engelman JA, Lynch TJ, Haber DA, Louis DN, Ellisen LW, Borger DR, Iafrate AJ. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol. Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, Lindeman NI, Murphy C, Akhavanfard S, Yeap BY, Xiao Y, Capelletti M, Iafrate AJ, Lee C, Christensen JG, Engelman JA, Jänne PA. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borrás AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, Tracy S, Zhao X, Heymach JV, Johnson BE, Cantley LC, Jänne PA. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J. Clin. Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, Rinehart C, Seidel B, Yee D, Arteaga CL, Engelman JA. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J. Clin. Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, Lifshits E, Brown A, Lee C, Christensen JG, Kwiatkowski DJ, Engelman JA, Jänne PA. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, Iwata KK, Gibson N, Haley JD. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 19.Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr, Raben D. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol. Cancer Ther. 2007;6:1683–1691. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M, Tanabe KK. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391–2399. doi: 10.1158/0008-5472.CAN-07-2460. [DOI] [PubMed] [Google Scholar]
- 21.Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, Barón A, Franklin WA, Hirsch FR, Geraci MW, Bunn PA., Jr Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol. Cancer Res. 2006;4:521–528. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- 22.Rho JK, Choi YJ, Lee JK, Ryoo BY, Na II, Yang SH, Kim CH, Lee JC. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer. 2009;63:219–226. doi: 10.1016/j.lungcan.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Jänne PA, Lynch T, Johnson BE, Miller VA. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J. Clin. Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, Kuhlmann GL, Han M, Goldberg JS, Settleman J, Iafrate AJ, Engelman JA, Haber DA, Johnson BE, Lynch TJ. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J. Clin. Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 25.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M. West Japan Oncology Group, Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 26.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T. North-East Japan Study Group, Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 27.Yamasaki F, Johansen MJ, Zhang D, Krishnamurthy S, Felix E, Bartholomeusz C, Aguilar RJ, Kurisu K, Mills GB, Hortobagyi GN, Ueno NT. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res. 2007;67:5779–5788. doi: 10.1158/0008-5472.CAN-06-3020. [DOI] [PubMed] [Google Scholar]
- 28.Kokubo Y, Gemma A, Noro R, Seike M, Kataoka K, Matsuda K, Okano T, Minegishi Y, Yoshimura A, Shibuya M, Kudoh S. Reduction of PTEN protein and loss of epidermal growth factor receptor gene mutation in lung cancer with natural resistance to gefitinib (IRESSA) Br. J. Cancer. 2005;92:1711–1719. doi: 10.1038/sj.bjc.6602559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T, Matsumoto K, Wang W, Li Q, Nishioka Y, Sekido Y, Sone S, Yano S. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin. Cancer Res. 2010;16:174–183. doi: 10.1158/1078-0432.CCR-09-1204. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Li Q, Yamada T, Matsumoto K, Matsumoto I, Oda M, Watanabe G, Kayano Y, Nishioka Y, Sone S, Yano S. Crosstalk to stromal fibroblasts induces resistance of lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors. Clin. Cancer Res. 2009;15:6630–6638. doi: 10.1158/1078-0432.CCR-09-1001. [DOI] [PubMed] [Google Scholar]
- 31.Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, Ogino H, Kakiuchi S, Hanibuchi M, Nishioka Y, Uehara H, Mitsudomi T, Yatabe Y, Nakamura T, Sone S. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 32.Engelman JA, Jänne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. ErbB-3 mediates phosphoinositide3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 34.Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Jänne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 35.Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, Halmos B, Pearlberg J, Tsuchihashi Z, Cantley LC, Tenen DG, Johnson BE, Jänne PA. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J. Natl. Cancer Inst. 2005;97:1185–1194. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- 36.Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, Peyton M, Juroske D, Huang Y, Stuart Salmon J, Kim YH, Pollack JR, Yanagisawa K, Gazdar A, Minna JD, Kurie JM, Carbone DP. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. [PubMed] [Google Scholar]
- 37.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 38.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin. Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 39.Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, Mitsudomi T. Analysis of epidermal growth factor receptor gene mutation in patients with non–small cell lung cancer and acquired resistance to gefitinib. Clin. Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 40.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, Ladanyi M, Miller VA, Pao W. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor–mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin. Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 41.Mok T, Spigel DR, Park K, Socinski MA, Tung SY, Kim D, Borzillo G, Zhang H, O’Connell JP, Jänne PA. Efficacy and safety of PF-00299804 (PF299), an oral, irreversible, pan-human epidermal growth factor receptor (pan-HER) tyrosine kinase inhibitor (TKI), as first-line treatment (tx) of selected patients (pts) with advanced (adv) non-small cell lung cancer (NSCLC) J. Clin. Oncol. 2010;28 abstract 7537. [Google Scholar]
- 42.Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, Gitlitz B, Eaton K, Zacharchuk C, Freyman A, Powell C, Ananthakrishnan R, Quinn S, Soria JC. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: Results of a phase II trial in patients with advanced non–small-cell lung cancer. J. Clin. Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 43.Janne PA, Reckamp K, Koczywas M, Engelman JA, Camidge DR, Rajan A, Khuri F, Liang JQ, O’Connell J, Giaccone G. Efficacy and safety of PF-00299804 (PF299) in patients (pt) with advanced NSCLC after failure of at least one prior chemotherapy regimen and prior treatment with erlotinib (E): A two-arm, phase II trial. J. Clin. Oncol. 2009;27 abstract 8063. [Google Scholar]
- 44.Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, García-Echeverría C, Wong KK, Engelman JA. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Jänne PA. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Regales L, Gong Y, Shen R, de Stanchina E, Vivanco I, Goel A, Koutcher JA, Spassova M, Ouerfelli O, Mellinghoff IK, Zakowski MF, Politi KA, Pao W. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J. Clin. Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zakowski MF, Ladanyi M, Kris MG. Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group, EGFR mutations in small-cell lung cancers in patients who have never smoked. N. Engl. J. Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 48.Morinaga R, Okamoto I, Furuta K, Kawano Y, Sekijima M, Dote K, Satou T, Nishio K, Fukuoka M, Nakagawa K. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer. 2007;58:411–413. doi: 10.1016/j.lungcan.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Okamoto I, Araki J, Suto R, Shimada M, Nakagawa K, Fukuoka M. EGFR mutation in gefitinib-responsive small-cell lung cancer. Ann. Oncol. 2006;17:1028–1029. doi: 10.1093/annonc/mdj114. [DOI] [PubMed] [Google Scholar]
- 50.Fukui T, Tsuta K, Furuta K, Watanabe S, Asamura H, Ohe Y, Maeshima AM, Shibata T, Masuda N, Matsuno Y. Epidermal growth factor receptor mutation status and clinico-pathological features of combined small cell carcinoma with adenocarcinoma of the lung. Cancer Sci. 2007;98:1714–1719. doi: 10.1111/j.1349-7006.2007.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatematsu A, Shimizu J, Murakami Y, Horio Y, Nakamura S, Hida T, Mitsudomi T, Yatabe Y. Epidermal growth factor receptor mutations in small cell lung cancer. Clin. Cancer Res. 2008;14:6092–6096. doi: 10.1158/1078-0432.CCR-08-0332. [DOI] [PubMed] [Google Scholar]
- 52.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 53.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dosaka-Akita H, Cagle PT, Hiroumi H, Fujita M, Yamashita M, Sharma A, Kawakami Y, Benedict WF. Differential retinoblastoma and p16INK4A protein expression in neuroendocrine tumors of the lung. Cancer. 2000;88:550–556. [PubMed] [Google Scholar]
- 55.Yuan J, Knorr J, Altmannsberger M, Goeckenjan G, Ahr A, Scharl A, Strebhardt K. Expression of p16 and lack of pRB in primary small cell lung cancer. J. Pathol. 1999;189:358–362. doi: 10.1002/(SICI)1096-9896(199911)189:3<358::AID-PATH452>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 56.Beasley MB, Lantuejoul S, Abbondanzo S, Chu WS, Hasleton PS, Travis WD, Brambilla E. The P16/cyclin D1/Rb pathway in neuroendocrine tumors of the lung. Hum. Pathol. 2003;34:136–142. doi: 10.1053/hupa.2003.8. [DOI] [PubMed] [Google Scholar]
- 57.Chung JH, Rho JK, Xu X, Lee JS, Yoon HI, Lee CT, Choi YJ, Kim HR, Kim CH, Lee JC. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer. 2010 doi: 10.1016/j.lungcan.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 58.Uramoto H, Iwata T, Onitsuka T, Shimokawa H, Hanagiri T, Oyama T. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinom. Anticancer Res. 2010;30:2513–2517. [PubMed] [Google Scholar]
- 59.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, Buttarazzi MS, Sanofsky BA, McNamara KL, Brandstetter KA, Walton ZE, Gu TL, Silva JC, Crosby K, Shapiro GI, Maira SM, Ji H, Castrillon DH, Kim CF, García-Echeverría C, Bardeesy N, Sharpless NE, Hayes ND, Kim WY, Engelman JA, Wong KK. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riely GJ, Kris MG, Zhao B, Akhurst T, Milton DT, Moore E, Tyson L, Pao W, Rizvi NA, Schwartz LH, Miller VA. Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clin. Cancer Res. 2007;13:5150–5155. doi: 10.1158/1078-0432.CCR-07-0560. [DOI] [PubMed] [Google Scholar]
- 63.Oxnard GR, Arcila ME, Sima C, Riely GJ, Chmielecki J, Kris MG, Pao W, Ladanyi M, Miller VA. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin. Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothenberg SM, Engelman JA, Le S, Riese DJ, II, Haber DA, Settleman J. Modeling oncogene addiction using RNA interference. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12480–12484. doi: 10.1073/pnas.0803217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Acknowledgments: We thank A. Muzikansky and E. Bast for their help with data analysis. Funding: This study is supported by grants from Uniting Against Lung Cancer, sponsored by UALC: New England and the Marjorie E. Korff Fund (L.V.S.), the NIH Gastrointestinal Cancer SPORE P50 CA127003 (J.A.E. and M.M.-K.), K08 grant CA120060-01 (J.A.E.), R01CA137008-01 (J.A.E.), R01CA140594 (J.A.E.), 1U01CA141457-01 (J.A.E.), National Cancer Institute Lung SPORE P50CA090578 (J.A.E.), Dana-Farber/Harvard Cancer Center, the American Association for Cancer Research (J.A.E.), the V Foundation (J.A.E.), American Cancer Society RSG-06-102-01-CCE (J.A.E.), the Ellison Foundation Scholar (J.A.E.), and the Doris Duke Charitable Foundation (B.A.W.). This work was supported by a gift from the Daniel Crane Foundation. Author contributions: L.V.S., B.A.W., D.D.-S., A.J.I., M.M.-K., and J.A.E. were responsible for the concept and design of the study. L.V.S., P.F., A.T.S., S.G., R.S.H., J.T., J.C.W., T.J.L., M.L., and J.A.E. were responsible for identifying and providing patients. S.D., J.C.W., and M.L. were responsible for performing patient biopsies. D.D.-S., A.B.T., K.B., A.K.C., S.A., K.V., E.J.M., A.J.I., M.M.-K., and J.A.E. were responsible for laboratory analyses. S.D. was responsible for radiographic analyses. J.G.C. provided the PF00299804 compound. L.V.S., B.A.W., D.D.-S., M.M.-K., and J.A.E. were primarily responsible for writing the manuscript. All authors contributed to manuscript editing and approval. Competing interests: P.F. is an advisor to Genentech. D.D.-S. has a paid consulting relationship with Bio-Reference Laboratories Inc. D.D.-S. and A.J.I. are inventors on a patent filed for SNaPshot genotyping methods (U.S. patent number 12/799415). J.A.E. holds equity in Gatekeeper and is a co-inventor on a patent pertaining to the use of combined MET and EGFR inhibitors (U.S. patent number 12/450826). T.J.L. is an advisor to Boehringer-Ingelheim, Supergen, Merck, AstraZeneca, and Genentech; is on the Board of Directors of Infinity Pharmaceuticals; and is also an inventor on a patent held by Partners Healthcare for EGFR mutation testing (U.S. patent numbers 11/894135, 11/894159, and 11/894160). Other authors declare no competing interests.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.