Abstract
The tumor suppressor p53 is often inactivated in breast cancer cells because the overexpression of its repressors (e.g., MDM2 and MDMX). Restoration of p53 activity by small molecules through counteracting p53 repressors can lead to in vivo tumor regression and is therefore considered a promising strategy for treatments of cancer. Recent efforts in high-throughput drug screening and rational drug design have identified several structurally diverse small-molecule p53 activators, including a pseudourea derivative XI-011 (NSC146109). This small molecule strongly activates p53 while selectively inhibiting growth of transformed cells without inducing genotoxicity, indicating its potential as a drug lead for p53-targeted therapy. However, the mechanism(s) by which XI-011 activates p53 and the effects of XI-011 on growth of breast cancer cells are currently unknown. Here, we report that XI-011 promoted breast cancer cells to undergo apoptosis through activating p53 and inducing expression of proapoptotic genes. Importantly, we found that activation of p53 by this small molecule was achieved through a novel mechanism, that is, inhibition of MDMX expression. XI-011 repressed the MDMX promoter, resulting in down-regulation of MDMX messenger RNA level in MCF-7 cells. In line with these results, XI-011 decreased the viability of breast cancer cells expressing low levels of MDMX in a less-efficient manner. Interestingly, XI-011 acted additively with the MDM2 antagonist Nutlin-3a to inhibit growth of breast cancer cells. We conclude that XI-011 belongs to a novel class of small-molecule p53 activators that target MDMX and could be of value in treating breast cancer.
Introduction
Breast cancer is one of the leading causes of death due to cancer. A major factor contributing to the development of breast cancer is inactivation of the tumor suppressor p53. p53 is the guardian of the genome, and its main function is to maintain genetic stability on oncogenic challenges by inducing cell cycle arrest, apoptosis, or senescence [1,2]. Inactivation of p53 not only promotes tumorigenesis and cancer progression but also confers breast cancer cells with an ability to evade death induced by conventional therapeutic agents [3,4]. Not surprisingly, overexpression of major p53 repressors such as MDM2 and MDMX occurs in more than 25% of breast cancers [1]. Accordingly, restoration of p53 activity through counteracting MDM2 or MDMX has been considered a promising strategy for breast cancer treatments [5–7].
MDM2 inactivates p53 mainly through repressing its transcriptional activity [8] and promoting its proteasomal degradation [9,10]. Because such a regulation requires binding of MDM2 to p53, an effective strategy to restore p53 activity in cancer cells is to dissociate the MDM2-p53 complex. Indeed, it has been demonstrated that small molecules (e.g., RITA, Nutlin-3a, MI-219) capable of disrupting the MDM2-p53 interaction can activate p53, leading to in vivo tumor regression [11,12] while sensitizing cancer cells to conventional therapies [13–15]. Notably, these designed small-molecule p53 activators are advantageous over conventional chemotherapeutic agents because they do not exhibit genotoxicity [11,12]. However, these compounds seem largely ineffective in cancer cells (e.g., MCF-7) that overexpress MDMX, presumably because of their inability to prevent p53 from MDMX binding [16–18]. As a homolog to MDM2 [19], MDMX binds p53 and regulates p53 activity through repressing its transactivation activity [1] as well as promoting MDM2-mediated degradation [20–23]. It is thus likely that MDMX-targeted agents could be more effective in treating cancer cells expressing high levels of MDMX. Indeed, a recent report showed that an MDMX-binding small molecule activates p53 leading to death of MDMX-overexpressing retinoblastoma cells [24]. Similar results were obtained with two peptides that can interfere with the MDMX-p53 interaction [25,26]. Through a cell-based high-content drug screening, we recently identified a benzofuroxan derivative that can inhibit MDMX expression thereby activating p53, leading to cancer cell death [27]. These results thus indicate that MDMX-targeted agents could be of great value in treating cancer, particularly breast cancer, which often overexpresses MDMX rather than MDM2 [1].
NSC146109 ([10-methyl-9-anthryl]methyl imidothiocarbamate; referred to as XI-011 thereafter) (Figure 1A) is a pseudourea derivative, and it was recently found to be a potent p53 activator through a high-content drug screening [28]. XI-011 at 2 µM could increase the activity of a p53 reporter by more than five-fold and was one of the most potent p53-activating small molecules identified in that screening [28]. This small molecule was also shown to selectively decrease viability of cancer cells without genotoxicity [28]. Interestingly, XI-011 seemed to be an active hit in a screening designed to search for tumor-selective anticancer agents [29]. These results thus suggest that this small-molecule p53 activator could be of value in further development into a novel cancer therapeutic agent. However, the molecular mechanism underlying the anticancer effects of XI-011 is poorly understood. Here, we report that this small molecule could induce breast cancer cells to undergo apoptosis—an effect distinct from the MDM2 inhibitor Nutlin-3a. Importantly, we found that XI-011 activated p53 mainly through inhibiting MDMX expression. This small molecule thus belongs to a new class of p53 activator that can induce apoptosis of breast cancer cells through inhibiting MDMX expression.
Figure 1.
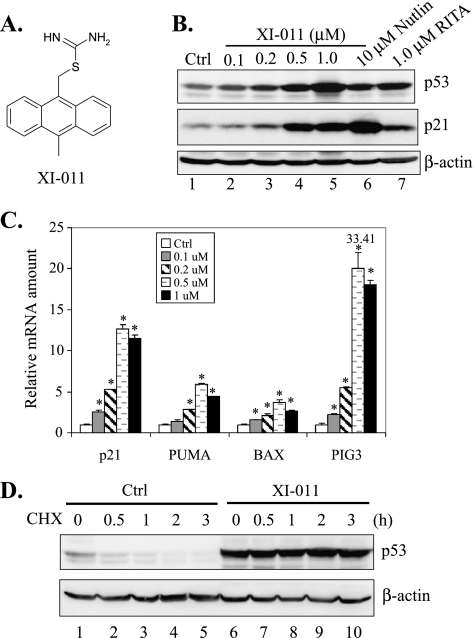
XI-011 activates p53 by increasing its stability. (A) The chemical structure of XI-011. (B) MCF-7 cells were treated with DMSO (Ctrl), 10 µM Nutlin-3a, 1.0 µM RITA, or indicated amounts of XI-011 for 16 hours and lysed for immunoblot assays to measure the expression levels of p53, p21, and β-actin. (C) MCF-7 cells treated with indicated amounts of XI-011 for 16 hours were lysed for RNA preparation. Total RNA was reverse-transcribed, and cDNA was subjected to qRT-PCR assays for measuring levels of p21, PUMA, BAX, and PIG3 mRNA. Data are depicted as mean ± SD values of three determinations. *P < .05 versus the DMSO control (Student's t test). (D) After treatment with 0.5 µM of XI-011 for 16 hours, MCF-7 cells were incubated with 100 µg/ml of cycloheximide. Cells were then lysed at indicated time points and subjected to immunoblot assays.
Materials and Methods
Cell Lines and Reagents
Breast cancer cell lines MCF-7, MDA-MB-175VII, ZR-75-1, ZR-75-30, HSS78T, BT474, and BT549 were cultured as recommended by the American Type Culture Collection (Manassas, VA). We obtained XI-011 (NSC146109) from the National Institutes of Health/National Cancer Institute Developmental Therapeutics Program. Nutlin-3a and RITA were purchased from Sigma-Aldrich (St Louis, MO) and Cayman Chemical (Ann Arbor, MI), respectively. These compounds were dissolved in dimethyl sulfoxide (DMSO) at 20 µM and diluted into working concentrations before use. Cell viability was measured with MTT assays.
Immunoblot Analysis and Cycloheximide Chase Assays
Immunoblot assays to determine protein levels were carried out as previously described [30]. The following antibodies were used: p53 (DO-1), MDM2 (SMP-14), and poly(ADP-ribose)polymerase (PARP; H-250) (from Santa Cruz Biotechnology, Santa Cruz, CA); MDMX (from Bethyl Laboratories, Montgomery, TX); p21 (from BD Biosciences, San Jose, CA); and β-actin (from Sigma-Aldrich). For cycloheximide chase assays, cells were treated with or without 100 µg/ml of cycloheximide (Sigma-Aldrich) for 0.5, 1, 2, and 3 hours and lysed for immunoblot analysis as described [31].
Quantitative Reverse Transcription-Polymerase Chain Reaction
Cells were lysed in TRIzol (Invitrogen, Carlsbad, CA). Total RNA was prepared, reverse-transcribed, and subjected to real-time polymerase chain reaction (PCR) assays using the ABI 7300 Real-time PCR System as described previously [32]. The sequences of primers are as follows: p21, forward 5′-CTGGAGACTCTCAGGGTCGAAA-3′ and reverse 5′-GATTAGGGCTTCCTCTTGGAGAA-3′; PUMA, forward 5′-CCTGGAGGGTCCTGTACAATCT-3′ and reverse 5′-GCACCTAATTGGGCTCCATCT-3′; BAX, forward 5′-TGGAGCTGCAGAGGATGATTG-3′ and reverse 5′-AAACATGTCAGCTGCCACTCG-3′; PIG3, forward 5′-TTGAGGCATCTGGACATGTG-3′ and reverse 5′-GGGTCAATCCCTCTGGGATAG-3′; and MDMX, forward 5′-GCCTTGAGGAAGGATTGGTA-3′ and reverse 5′-TCGACAATCAGGGACATCAT-3′.
Flow Cytometry and Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling Staining
Cells were fixed with cold 70% ethanol overnight and stained with the propidium iodide solution (50 µg/ml; Sigma-Aldrich) containing 20 µg/ml RNase A at 37°C for 20 minutes for flow cytometry analysis as described previously [33]. Data were analyzed using the FlowJo software. For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, cells fixed with 4% paraformaldehyde were subjected to dUTP labeling using In Situ Cell Death Detection Kit TMR Red (Roche, Indianapolis, IN) and observed under a fluorescence microscope. At least 300 cells were randomly chosen, and numbers of TUNEL-positive cells were counted.
Short Hairpin RNA Knockdown
Short hairpin RNA (shRNA)-mediated gene knockdown was carried out using a Lentivector-based system (pSIH-H1 shRNA Cloning and Lentivector Expression system; System Biosciences, Mountain View, CA) as described previously [34]. The targeted sequences for MDMX, p53, and MDM2 were 5′-GTG ATG ATA CCG ATG TAG A-3′, 5′-GAC TCC AGT GGT AAT CTA C-3′, and 5′-GGA ATT TAG ACA ACC TGA A-3′ based on publications [16,35,36], respectively. A firefly luciferase-targeted sequence (5′-CTT ACG CTG AGT ACT TCG A-3′) was also cloned into the Lentivector as a negative control.
Chromatin Immunoprecipitation Assays
Enrichment of RNA polymerase II (pol II) on the MDMX or ATF3 promoter was determined by chromatin immunoprecipitation assays as described previously [32] with modifications. Briefly, cells were cross-linked with 1% formaldehyde for 10 minutes, followed by lysis in a solution containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mMEDTA, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate. After sonication, lysates were incubated with 1 µg of the RNA pol II antibody (N-20; Santa Cruz) or 1 µg of normal rabbit IgG, at 4°C overnight. The chromatin-associated pol II was then precipitated with 30 µl of ssDNA-treated protein A agarose (Millipore, Billerica, MA). The agarose was washed, and the bound chromatin was eluted with a buffer containing 100 mM NaHCO3 and 1% SDS. After elution, chromatin solutions were treated with proteinase K and RNase A and incubated at 65°C overnight to reverse cross-links. Pol II-bound DNA were then purified using the QIAquick DNA purification kit (Qiagen, Valencia, CA) and subjected to real-time PCR for quantitation [32]. The primers used for amplifying the MDMX promoter (-96 to -31) were 5′-TGGTAGTGAGCTCAGCCTTTCTAG-3′ and 5′-GCCTCCGGAAACCATAGATG-3′. The primers for the ATF3 promoter (-75 to +42) were 5′-TGCCAACACGGAGTAAACGA-3′ and 5′-AGAGAAGAGAGCTGTGCAGTGC-3′ [37].
Results
XI-011 Activates p53 in Breast Cancer Cells
The pseudourea derivative XI-011 (Figure 1A) was identified as one of the most potent active hits in a reporter-based screening for small-molecule p53 activators [28]. We therefore sought to determine whether this small molecule could activate p53 in p53 wild-type MCF-7 breast cancer cells. Toward this end, we treated MCF-7 cells with XI-011 and measured expression levels of p53 and its downstream target gene p21 using immunoblot assays. As controls, we also treated cells with Nutlin-3a and RITA—two well-characterized small-molecule p53 activators. Interestingly, XI-011 increased the cellular p53 protein level as efficient as the two known p53 activators (Figure 1B). Moreover, XI-011 treatments resulted in a dose-dependent increase in the p21 protein level, suggesting that p53 was indeed activated by the small molecule (Figure 1B). To corroborate these results, we carried out quantitative reverse transcription-PCR (qRT-PCR) assays and found that XI-011 dramatically increase messenger RNA (mRNA) levels of p21 as well as p53-targeted proapoptotic genes including PUMA, BAX, and PIG3 (Figure 1C). These results indicated that XI-011 could strongly activate p53 in breast cancer cells. Moreover, XI-011 significantly extended the half-life of p53 (Figure 1D), consistent with a notion that this small-molecule p53 activator could increase the stability of p53 in breast cancer cells.
XI-011 Induces Apoptosis in Breast Cancer Cells
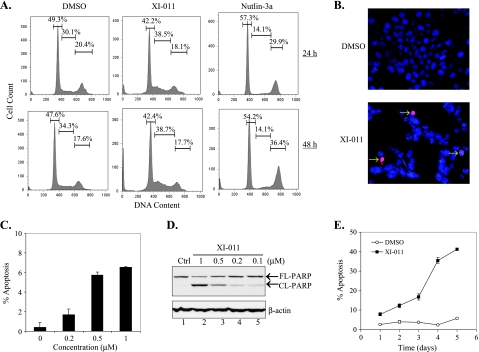
p53 activation can lead to cell cycle arrest or apoptotic cell death. To determine the consequence of p53 activation induced by XI-011, we treated MCF-7 cells with 0.5 µM of XI-011 and carried out flow cytometry analysis to examine cell cycle progression. It has been shown that activation of p53 by the MDM2 inhibitor Nutlin-3a results in only cell cycle arrest but not apoptosis in cancer cells overexpressing MDMX (e.g., MCF-7) [17]. Indeed, we found that Nutlin-3a induced a significant decrease in the number of S-phase cells (Figure 2A). In contrast, XI-011 did not appear to diminish the S-phase subpopulation (Figure 2A). However, XI-011 treatments resulted in apparent accumulation of cells in sub-G0/G1 phase (Figure 2A), suggesting that this small molecule could rather induce MCF-7 cells to undergo apoptosis. To confirm this important finding, we used TUNEL staining to specifically label apoptotic cells. In line with the notion that XI-011 could induce apoptosis, numbers of TUNEL-positive cells were largely increased after XI-011 treatments (Figure 2, B and C). This finding was further confirmed by the observations that cleavage of PARP—a biochemical marker for apoptosis—was induced by XI-011 in a dose-dependent manner (Figure 2D). Indeed, more than 40% of MCF-7 cells were apoptotic after treatments with 0.5 µMXI-011 for 5 days (Figure 2E). These results thus indicate that one of the major effects of XI-011 on breast cancer cells was to induce apoptosis. This effect was in sharp contrast to the effect conferred by Nutlin-3a, suggesting that XI-011 might activate p53 through a mechanism distinct from the MDM2 inhibitor.
Figure 2.
XI-011 induces apoptosis in breast cancer cells. (A) MCF-7 cells treated with 0.5 µM of XI-011, 5 µM of Nutlin-3a, or DMSO, for 24 and 48 hours were stained with propidium iodide and subjected to flow cytometry analysis. Numbers inserted in graphs indicate percentages of cells at different stages of cell cycle. (B, C) MCF-7 cells treated with XI-011 for 48 hours were subjected to TUNEL staining assays. At least 300 cells were randomly chosen, and the numbers of TUNEL-positive cells were counted. (D) MCF-7 cells were treated with indicated concentrations of XI-011 for 2 days and subjected to immunoblot assays to detect cleaved (CL-PARP) and full-length (FL-PARP) PARP. (E) MCF-7 cells treated with 0.5 µM of XI-011 or DMSO for different days were subjected to flow cytometry to determine percentages of apoptotic cells (sub-G0/G1 cells).
XI-011 Induces Apoptosis through Activating p53
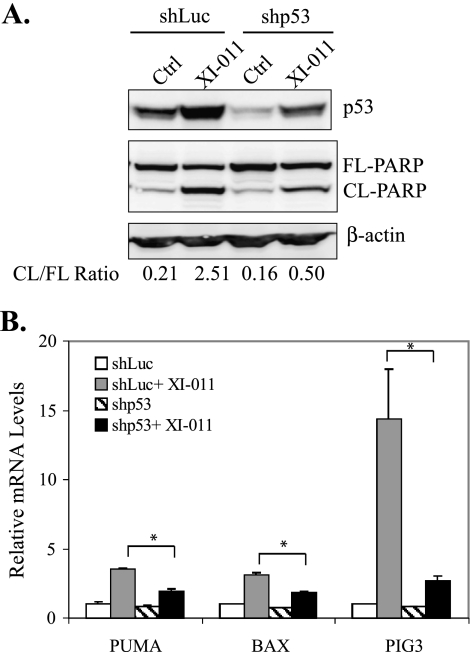
To demonstrate that the apoptosis-inducing activity of XI-011 was a consequence of p53 activation, we knocked down p53 expression in MCF-7 cells using a p53-specific shRNA [27] and determined PARP cleavage induced by XI-011. Indeed, XI-011-induced PARP cleavage was largely impaired in p53-downregulated cells (Figure 3A). Moreover, the XI-011-induced expression of proapoptotic genes (i.e., PUMA, BAX, and PIG3) was significantly diminished by knockdown of p53 expression (Figure 3B).
Figure 3.
XI-011 induces apoptosis and proapoptotic gene expression through activation of p53. (A) MCF-7 cells infected with Lenti-viruses expressing shRNA specific to p53 (shp53) or luciferase (shLuc) were treated with 0.5 µM of XI-011 for 2 days and lysed for immunoblot assays to detect cleaved (CL-PARP) and full-length (FL-PARP) PARP. Intensities of CL-PARP and FL-PARP bands were estimated by densitometry analysis and used for calculating the CL-PARP to FL-PARP ratio (CL/FL-PARP) (shown below the blots). (B) MCF-7 cells expressing shp53 or shLuc were treated with 0.5 µM of XI-011 for 16 hours and then subjected to qRT-PCR analysis to measure levels of indicated mRNA. Data are depicted as the mean ± SD values of three determinations. *P < .05 versus the corresponding shLuc cells (Student's t test).
XI-011 Activates p53 and Induces Apoptosis through Inhibiting MDMX Expression
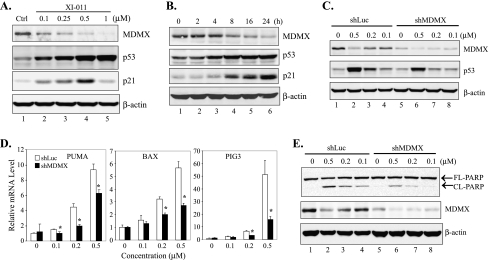
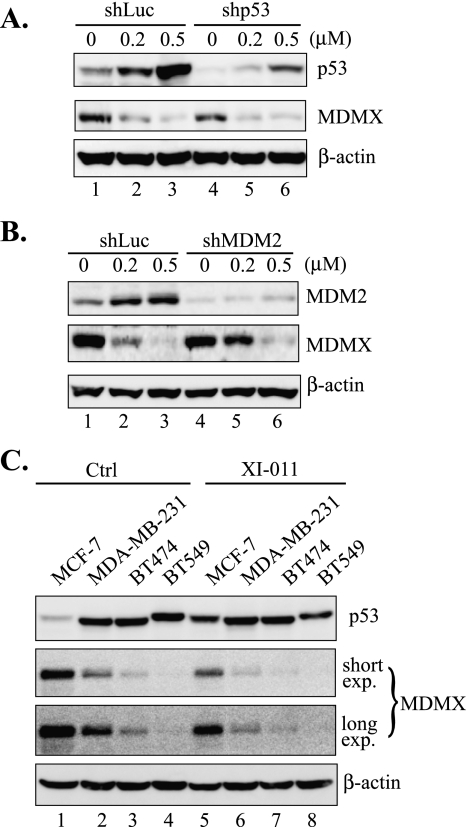
It has been suggested that the failure in induction of apoptosis by Nutlin-3a in MCF-7 cells (Figure 3A) was likely due to MDMX overexpression [17]. Because we have shown that XI-011 was distinct from Nutlin-3a and could induce MCF-7 cells to undergo apoptosis, we tested a hypothesis that XI-011 activates p53 through targeting MDMX. Interestingly, accompanied by p53 activation, the MDMX expression level was dramatically decreased after MCF-7 cells were treated with XI-011 (Figure 4A). Inhibition of MDMX expression by XI-011 occurred 4 to 8 hours after treatment when p53 activation started to become notable (Figures 4B and W1A). These results suggest a strong possibility that p53 activation by XI-011 could be a consequence of MDMX inhibition. To explore this possibility, we knocked down MDMX expression in MCF-7 cells with an MDMX-specific shRNA and determined p53 activation by immunoblot analysis and qRT-PCR assays. Indeed, activation of p53 (Figure 4C) and induction of expression of p53 target genes (i.e., PUMA, BAX, and PIG3) (Figure 4D) were largely impaired in cells expressing a low level of MDMX. Moreover, the cleavage of PARP induced by XI-011 was also impaired by knockdown of MDMX expression (Figure 4E). Furthermore, a small interfering RNA (siRNA) specific to MDMX but targeting a sequence different from the shRNA used above also impaired p53 activation induced by XI-011 (Figure W1B). These results thus indicate that XI-011 activated p53 and induced apoptosis through inhibiting MDMX expression. Of note, knockdown of MDMX expression by shRNA alone seemed insufficient to activate p53 and induce apoptosis (Figure 4, C and E, lane 5)—an observation in agreement with several [16,17,38] but in contrast to other reports [20,25]. The reason for this apparent discrepancy might be related to the fact that the p53 activity is regulated by a complicated network and thus effects of MDMX shRNA might be cell context-dependent [27].
Figure 4.
Activation of p53 and induction of apoptosis by XI-011 is a consequence of inhibition of MDMX expression. (A, B) MCF-7 cells were treated with XI-011 as indicated and lysed for immunoblot assays for measuring MDMX, p53, and p21 expression levels. (C) MCF-7 cells infected with Lentiviruses expressing shLuc or shRNA specific to MDMX (shMDMX) were treated with varying amounts of XI-011 for 16 hours and subjected to immunoblot assays for p53 and MDMX expression. (D) MCF-7 cells expressing shMDMX or shLuc were treated 0.5 µM of XI-011 and subjected to qRT-PCR analysis to measure levels of indicated mRNA. Data are depicted as the average ± SD values of three determinations. *P < .05 versus the corresponding shLuc cells (Student's t test). (E) MCF-7 cells expressing shLuc or shMDMX were treated with indicated amounts of XI-011 for 2 days and subjected to immunoblot assays to detect cleaved and full-length PARP.
Inhibition of MDMX Expression by XI-011 Is Not a Consequence of p53 Activation
Having shown that XI-011 activated p53 through inhibiting MDMX expression, we attempted to elucidate the mechanism by which XI-011 downregulated MDMX expression. Because MDM2 can bind MDMX and drive its ubiquitination and proteolysis [39], p53 activated by DNA damage might induce MDM2 expression leading to subsequent decrease in the MDMX expression level. Therefore, down-regulation of MDMX expression by XI-011 might be a consequence of p53 activation induced by DNA damage. To explore this possibility, we determined the effects of XI-011 on MDMX expression in cells where p53 or MDM2 expression was knocked down by shRNAs. Interestingly, knockdown of p53 expression or MDM2 expression did not affect or only slightly affect XI-011-induced inhibition of MDMX expression (Figure 5, A and B). Consistent with these observations, XI-011 could downregulate MDMX expression in breast cancer cells (i.e., MDA-MB-231, BT474, BT549) harboring functionally inactive p53 mutants (Figure 5C). These results thus do not support the notion that XI-011 inhibited MDMX expression through activating p53 and inducing MDM2 expression. Indeed, XI-011 failed to prolong the half-life of MDMX (Figure W2)—a result arguing against the possibility that XI-011 inhibited MDMX expression through promoting MDM2-mediated proteolysis.
Figure 5.
Inhibition of MDMX expression by XI-011 is not caused by p53 activation or MDM2 induction. (A, B) MCF-7 cells infected with shLuc, shp53 (A) or shRNA specific to MDM2 (shMDM2) (B) were treated with indicated amounts of XI-011 for 16 hours and then lysed for immunoblot assays. (C) MCF-7 and indicated p53 mutant cells were treated with 0.5 µM of XI-011 for 16 hours and subjected to immunoblot assays. Both shorter exposure (short exp.) and longer exposure (long exp.) were shown for the MDMX blot.
XI-011 Inhibits MDMX Gene Transcription
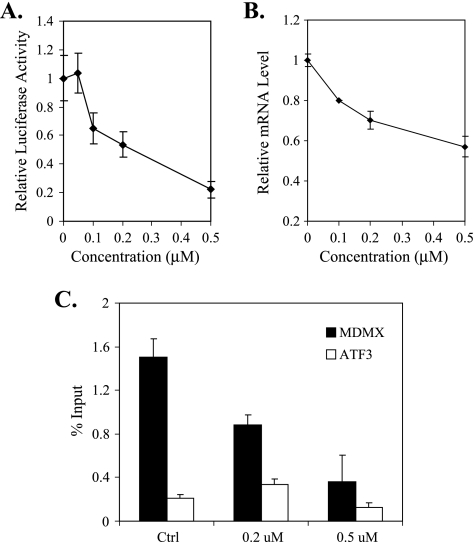
To explore the mechanism by which XI-011 inhibited MDMX expression, we determined whether this small molecule might regulate MDMX gene transcription. Toward this end, we first used a cell line that harbors a firefly luciferase reporter driven by a MDMX promoter [27] to determine the effects of XI-011 on MDMX promoter activity. Interestingly, XI-011 decreased the promoter activity in a dose-dependent manner (Figure 6A). Consistent with these results, the MDMX mRNA level was downregulated by XI-011 (Figure 6B). Because promoter occupancy of RNApol II reflects the transcription rate of a gene, we carried out chromatin immunoprecipitation assays using an anti-pol II antibody to confirm that XI-011 indeed intervened in MDMX gene transcription. Treatments of MCF-7 cells with XI-011 resulted in a dramatic reduction in the amounts of pol II associated with the MDMX promoter (Figure 6C). Such a reduction was unlikely an artifact due to loss of DNA mass in the experimental manipulations because amounts of pol II associated with the promoter of ATF3, a control gene whose expression was not inhibited by XI-011 (data not shown), was not dramatically reduced in the samples treated with the small molecule (Figure 6C). These results thus confirmed that XI-011 inhibited MDMX expression through repressing MDMX gene transcription.
Figure 6.
XI-011 inhibits MDMX gene transcription. (A) A reporter cell line carrying a stably integrated firefly luciferase gene under control of a MDMX promoter [27] were treated with various amounts of XI-011 for 16 hours and lysed for luciferase activity assays. (B) MCF-7 cells treated with indicated amounts of XI-011 were subjected to qRT-PCR assays to measure MDMX mRNA levels. (C) MCF-7 cells were treated with DMSO or indicated concentrations of XI-011 for 16 hours and then subjected to chromatin immunoprecipitation assays using an antibody against RNA pol II. The DNA associated with pol II were purified and quantified by real-time PCR.
XI-011 Decreases Viability of Breast Cancer Cells
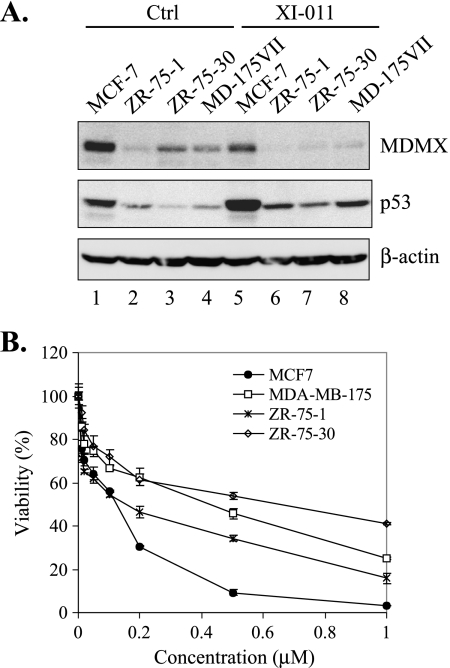
We have shown that XI-011 could activate p53 and induced apoptosis, which might in turn lead to a decrease in cell viability. Indeed, XI-011 dramatically decreased viability of MCF-7 cells (Figure 7B). An interesting question was surfaced as to whether efficacies of XI-011 to decrease cell viability were reduced in breast cancer cells expressing low levels of MDMX. To address this question, we treated three other breast cancer cell lines (i.e., ZR-75-1, ZR-75-30, and MD-175VII) that harbor a wild-type p53 gene but express a low level of MDMX with XI-011. As expected, XI-011 increased the p53 level in these cell lines, but the overall p53 level was much lower than MCF-7 cells after treatments (Figure 7A). Consistent with low p53 levels, XI-011 seemed less effective in decreasing viability of the three cell lines that express low levels of MDMX compared with MCF-7 cells (Figure 7B). These results are in support of the notion that XI-011 activated p53 and decreased cell viability mainly through inhibiting MDMX expression. Of note, the effects of XI-011 on cell viability did not correlate well with MDMX levels in the three cell lines expressing low levels of MDMX, probably because of the difference in cell contexts.
Figure 7.
XI-011 decreases the viability of breast cancer cells expressing a low level of MDMX in a less-efficient manner. (A) Indicated cells treated with DMSO or 0.5 µM of XI-011 were subjected to immunoblot assays for MDMX and p53 expression. (B) Indicated cells were treated with various amounts of XI-011 for 4 days and subjected to MTT assays for measuring cell viability.
XI-011 Acts Additively with Nutlin-3a to Inhibit Growth of Breast Cancer Cells
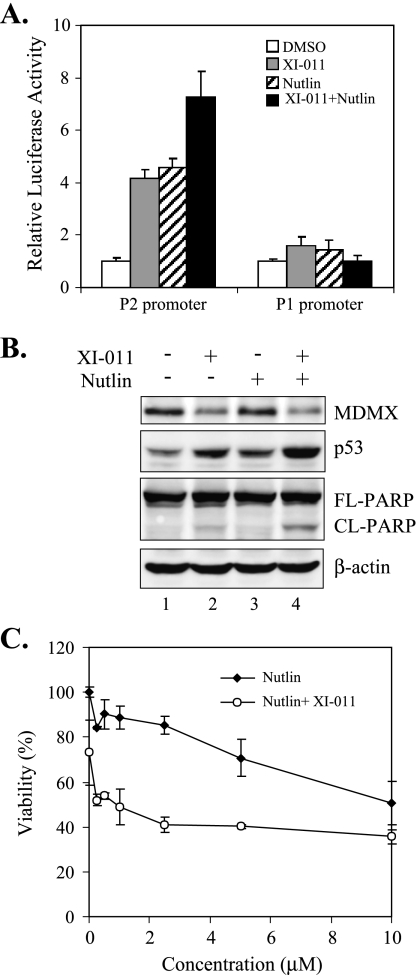
The MDM2 inhibitor Nutlin-3a can disrupt the p53-MDM2 interaction leading to p53 activation and cell death in many plural cancer cell lines [11] but fail to induce apoptosis in cancer cells (e.g., MCF-7) expressing high levels of MDMX [17–19]. Because we have shown that XI-011 could inhibit MDMX expression, we explored a possibility that XI-011 acts in concert with Nutlin-3a to inhibit growth of breast cancer cells. Previously, we have developed a cell line that expresses a luciferase reporter gene under control of a p53-responsive promoter (P2 promoter) and thus expression of the luciferase gene reflects the activity of p53 in the cells [27]. We therefore treated these cells with XI-011, Nutlin-3a, or both compounds, and performed luciferase activity assays to determine their effects on p53 activation. As expected, both XI-011 and Nutlin-3a increased the p53 reporter activity (Figure 8A). Interestingly, p53 activation was enhanced by coadministration of these two compounds (Figure 8A). As a control, expression of a luciferase reporter driven by a p53-nonresponsive promoter (P1 promoter) [27] was not affected by treatments with these agents (Figure 8A). These results, which were confirmed by immunoblot assays (Figure 8B), suggest that XI-011 acted additively, but not synergistically, with Nutlin-3a to activate p53. Consistent with these results, the viability of MCF-7 cells was decreased by XI-011 and Nutlin-3a in an additive manner (Figure 8C). These results are in line with our previous conclusion that an MDMX-targeting small molecule can cooperate with a MDM2 inhibitor to activate p53 and induce death of malignant cells [27].
Figure 8.
XI-011 decreases the viability of breast cancer cells in collaboration with Nutlin-3a. (A) Cells carrying a luciferase gene driven by a p53-responsive (P2) or p53-nonresponsive (P1) promoter were treated with 0.2 µM of XI-011, 5 µM of Nutlin-3a, or both agents, for 6 hours, and then lysed for luciferase activity assays. (B) MCF-7 cells were treated with 0.2 µM of XI-011, 5 µM of Nutlin-3a, or both agents, for 24 hours, and lysed for immunoblot assays to measure levels of MDMX, p53, and PARP. (C) MCF-7 cells were treated for 4 days with various amounts of Nutlin-3a in the absence or presence of 0.2 µM of XI-011 and then subjected to MTT assays for measuring cell viability. Data are depicted as mean ± SD values of three determinations.
Discussion
The pseudourea derivative XI-011 was previously found to activate p53 and specifically inhibit growth of transformed cells through unbiased, high-content drug screening [28,29]. In this study, we reported for the first time that this small molecule could activate p53 through inhibiting MDMX expression in various breast cancer cell lines. This notion was supported by our observations that both XI-011-induced p53 activation and subsequent decrease in cell viability were largely impaired in cells expressing low levels of MDMX. Our findings thus add XI-011 to a short list of small molecules that can activate p53 through counteracting MDMX functions in cancer cells. Previously, only two small molecules have been found to target MDMX, that is, SJ-172250, a compound that binds MDMX and blocks the p53-MDMX interaction [24], and XI-006, a benzofuroxan derivative that inhibits MDMX expression [27]. Given that MDMX is a major p53 repressor and overexpressed in nearly 20% of breast cancers [38], the results from this study are in strong support of a notion that therapeutic agents targeting MDMX overexpression in malignant cells could be of benefit to millions of patients with cancer [27].
Distinct from MDM2, MDMX contains a RING domain but lacks E3 ubiquitin ligase activity [1]. It is thus often assumed that binding of MDMX to p53 does not affect the p53 stability [1,40]. However, recent evidence indicates that MDMX can dimerize with MDM2 through the RING domains [7], thereby promoting efficient ubiquitination and degradation of p53 mediated by MDM2 [21–23,41]. This most recent model predicts that down-regulation of MDMX expression would lead to stabilization of the tumor suppressor. Indeed, we found that XI-011, like another MDMX inhibitor characterized in our previous report [27], dramatically increased the stability of p53 (Figure 1D). This effect was unlikely caused by an activity aside from inhibition of MDMX expression because knockdown of MDMX expression by a specific shRNA or siRNA could compromise XI-011-induced increase in the p53 level in breast cancer cells (Figure 4C). However, SJ-172250, a small molecule that counteracts MDMX through disrupting the p53-MDMX interaction, only slightly increases the cellular p53 level in retinoblastoma cells [24]. Because SJ-172250 intervenes in neither the p53-MDM2 interaction nor the MDMX-MDM2 interaction [24], MDMX might bind to MDM2 thereby retaining its capability to promote MDM2-mediated p53 degradation after SJ-172250 treatments. Conversely, down-regulation of the cellular MDMX level by XI-011 could limit the availability of MDMX to both p53 and MDM2, resulting in efficient stabilization of p53. In this regard, XI-011 and other small molecules inhibitory for MDMX expression (e.g., XI-006) [27] might be more potent in activating p53 and thus could lead to better therapeutic outcomes than small molecules targeting the p53-MDMX interaction (e.g., SJ-172250). It is important to note that XI-011, like XI-006 reported previously [27], could efficiently decrease the viability of breast cancer cells harboring mutant p53 (data not shown). These effects were not unexpected, and likely achieved through targeting MDMX for its functions independent of p53. Indeed, it has been documented that MDMX regulates other members of the p53 family including p63 and p73 [42–44], as well as Smad transactivation [45].
DNA damage induced by small molecules can activate p53, which may in turn induce MDM2 expression leading to subsequent degradation of MDMX [39]. Although XI-011 contains an anthracene ring and structurally resembles a DNA intercalator [29,46], this small molecule does not induce DNA damage at concentrations (20 µM) much higher than the concentrations (0.2–1 µM) at which p53 was strongly activated [28], arguing against the possibility that inhibition of MDMX expression by XI-011 was a consequence of p53 activation due to DNA damage. Indeed, although XI-011 did not alter the half-life of MDMX, this small-molecule p53 activator could downregulate MDMX expression in breast cancer cells harboring mutant p53 genes or in cells where p53 or MDM2 expression was knocked down by shRNA (Figure 5). Our results strongly suggest that XI-011 inhibited MDMX expression rather through inhibiting MDMX gene transcription. In support of this notion, XI-011 not only decreased the MDMX mRNA level but also reduced the amounts of pol II associated with the MDMX promoter (Figure 6). Given that XI-011 may bind DNA [46], it might be that this small molecule binds to the MDMX promoter thereby either directly preventing pol II from binding to the promoter or blocking recognition of cis-regulatory elements by transcription factors. However, XI-011 is not a general transcription inhibitor because expression of p53 target genes was induced, rather than inhibited, by the small molecule. Further exploration of the mechanism(s) by which XI-011 inhibits MDMX transcription is difficult, given that the cis-/trans-regulatory elements regulating MDMX transcription remain unclear.
Interestingly, pseudourea (NSC56054), the parent compound of XI-011, was found to be cytotoxic to malignant cells and entered clinical trials [47]. This compound was soon withdrawn from clinical testing because of severe toxicity [47]. Because XI-011 does not induce DNA damage at concentrations sufficient to activate p53 [28], and because XI-011 seems to selectively inhibit growth of transformed cells [29], this novel MDMX inhibitor might be more tolerant than pseudourea and thus could serve as a lead compound for further development into a therapeutic agent for breast cancer treatments.
Supplementary Material
Abbreviations
- PARP
poly (ADP-ribose)polymerase
- pol II
RNA polymerase II
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- shRNA
short hairpin RNA
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
Footnotes
This work was supported in part by a National Institutes of Health grant R01CA139107 and a Department of Defense grant PC061106 (W81XWH-07-1-0095) to C.Y. The authors thank Hua Lu, Jiandong Chen, and Guillermina Lozano for providing reagents and comments. The authors declare no potential conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.O'Conner PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, Weinstein JN, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 4.Scata KA, El-Deiry WS. p53, BRCA1 and breast cancer chemoresistance. Adv Exp Med Biol. 2007;608:70–86. doi: 10.1007/978-0-387-74039-3_5. [DOI] [PubMed] [Google Scholar]
- 5.Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 6.Vassilev LT. p53 activation by small molecules: application in oncology. J Med Chem. 2007;48:4491–4499. doi: 10.1021/jm058174k. [DOI] [PubMed] [Google Scholar]
- 7.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev LT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammolott U, Lukacs C, Klein C, Fotouhi N, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 12.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Qing K, Wang G, Chen J, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao C, Shinohara ET, Subhawong TK, Geng L, Woon Kim K, Albert JM, Hallahan DE, Lu B. Radiosensitization of lung cancer by nutlin, an inhibitor of murine double minute 2. Mol Cancer Ther. 2006;5:411–417. doi: 10.1158/1535-7163.MCT-05-0356. [DOI] [PubMed] [Google Scholar]
- 14.Barbieri E, Mehta P, Chen Z, Zhang L, Slack A, Berg S, Shohet J. MDM2 inhibition sensitizes neuroblastoma to chemotherapy-induced apoptotic cell death. Mol Cancer Ther. 2006;5:2358–2365. doi: 10.1158/1535-7163.MCT-06-0305. [DOI] [PubMed] [Google Scholar]
- 15.Coll-Mulet L, Lglesias-Serret D, Santidrian AF, Cosialls AM, de Frias M, Castano E, Campas C, Barragan M, de Sevilla AF, Domingo A, et al. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood. 2006;107:4109–4114. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- 16.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 17.Wade M, Ee TW, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 18.Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 19.Shvarts A, Steegenga WT, Riteco N, Laar V, Dekker P, Bazuine M, Van Ham RCA, Van Der Houven Van Oordt W, Hateboer G, van der Eb AJ, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J, Kawai H, Nie L, Kitao H, Widerschain D, Jochemsen AG, Parant J, Lozano G, Yuan Z-M. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- 21.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poyurovsky M, Priest C, Kentsis A, Borden K, Pan Z-Q, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uldrijan S, Pannekoek W-J, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786–10796. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 26.Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Ma X, Ren S, Buolamwini J, Yan C. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther. 2011;10:69–79. doi: 10.1158/1535-7163.MCT-10-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berkson RG, Hollick JJ, Westwood NJ, Wood JA, Lane DP, Lain S. Pilot screening programme for small molecule activators of p53. Int J Cancer. 2005;115:701–710. doi: 10.1002/ijc.20968. [DOI] [PubMed] [Google Scholar]
- 29.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 30.Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J. 2005;24:2425–2435. doi: 10.1038/sj.emboj.7600712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mo P, Wang H, Lu H, Boyd DD, Yan C. MDM2 mediates ubiquitination and degradation of activating transcription factor 3. J Biol Chem. 2010;285:26908–26915. doi: 10.1074/jbc.M110.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan C, Wang H, Boyd DD. ATF3 represses 72-kDa type IV collagenase (MMP-2) expression by antagonizing p53-dependent trans-activation of the collagenase promoter. J Biol Chem. 2002;277:10804–10812. doi: 10.1074/jbc.M112069200. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Mo P, Ren S, Yan C. Activating transcription factor 3 activates p53 by preventing E6-associated protein from binding to E6. J Biol Chem. 2010;285:13201–13210. doi: 10.1074/jbc.M109.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 36.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 37.Yan C, Jamaluddin M, Aggarwal B, Myers J, Boyd DD. Gene expression profiling identifies activating transcription factor 3 as a novel contributor to the proapoptotic effect of curcumin. Mol Cancer Ther. 2005;4:233–241. [PubMed] [Google Scholar]
- 38.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol. 2007;39:1476–1482. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 42.Ongkeko WM, Wang XQ, Siu WY, Lau AWS, Yamashita K, Harris AL, Cox LS, Poon RYC. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol. 1999;9:829–832. doi: 10.1016/s0960-9822(99)80367-4. [DOI] [PubMed] [Google Scholar]
- 43.Kadakia M, Slader C, Berberich SJ. Regulation of p63 function by Mdm2 and MdmX. DNA Cell Biol. 2010;20:321–330. doi: 10.1089/10445490152122433. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Arooz T, Siu WY, Chiu CHS, Lau A, Yamashita K, Poon PYC. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 2001;490:202–208. doi: 10.1016/s0014-5793(01)02124-x. [DOI] [PubMed] [Google Scholar]
- 45.Kadakia M, Brown T, McGorry M, Berberich S. MdmX inhibits Smad transactivation. Oncogene. 2002;21:8776–8785. doi: 10.1038/sj.onc.1205993. [DOI] [PubMed] [Google Scholar]
- 46.Wunz TP, Dorr RT, Alberts DS, Tunget CL, Einspahr J, Milton S, Remers WA. New antitumor agents containing the anthracene nucleus. J Med Chem. 1987;30:1313–1321. doi: 10.1021/jm00391a009. [DOI] [PubMed] [Google Scholar]
- 47.Frei E, Luce J, Loo T. Phase I and phototoxicity studies of pseudourea (NSC-56054) Cancer Chemother Rep. 1971;55:91–97. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.