ABSTRACT
This study demonstrates the prevalence, phylogenetic diversity, and physiology of nitrate-reducing microorganisms capable of utilizing reduced humic acids (HA) as electron donors in agricultural soils. Most probable number (MPN) enumeration of agricultural soils revealed large populations (104 to 106 cells g−1 soil) of microorganisms capable of reducing nitrate while oxidizing the reduced HA analog 2,6-anthrahydroquinone disulfonate (AH2DS) to its corresponding quinone. Nitrate-dependent HA-oxidizing organisms isolated from agricultural soils were phylogenetically diverse and included members of the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria. Advective up-flow columns inoculated with corn plot soil and amended with reduced HA and nitrate supported both HA oxidation and enhanced nitrate reduction relative to no-donor or oxidized HA controls. The additional electron donating capacity of reduced HA could reasonably be attributed to the oxidation of reduced functional groups. Subsequent 16S rRNA gene-based high-density oligonucleotide microarray (PhyloChip) indicated that reduced HA columns supported the development of a bacterial community enriched with members of the Acidobacteria, Firmicutes, and Betaproteobacteria relative to the no-donor control and initial inoculum. This study identifies a previously unrecognized role for HA in stimulating denitrification processes in saturated soil systems. Furthermore, this study indicates that reduced humic acids impact soil geochemistry and the indigenous bacterial community composition.
IMPORTANCE
This study identifies a new metabolic capacity in soil microbial communities that may be responsible for the mediation of significant nitrogen losses from soil systems. Nitrate-dependent humic acid (HA)-oxidizing organisms isolated from agricultural soils were phylogenetically diverse and included members of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria. Advective up-flow columns inoculated with corn plot soil and amended with reduced HA and nitrate supported both HA oxidation and enhanced nitrate reduction relative to no-donor or oxidized HA controls. The additional electron donating capacity of reduced HA could reasonably be attributed to the oxidation of reduced functional groups.
Introduction
Microbial respiration of nitrate to gaseous end products (denitrification) is estimated to account for as much as one-third of the loss of fertilizer or other nitrogen sources from soil containing plants (1–4). Therefore, understanding the respiratory activity of nitrate-reducing microorganisms indigenous to agricultural soils is important to modeling nitrogen flux within agricultural environments. The respiratory activity of nitrate-reducing bacteria in soils is controlled by several parameters, including pH, temperature, O2 concentrations, and redox state (3, 5–11). Additionally, the extent of nitrate respiration can be determined by the availability and chemical characteristics of potential electron donor compounds (1, 12–15).
Although most physiological studies of nitrate-respiring bacteria utilize labile organic compounds as electron donor sources, some nitrate-reducing organisms also oxidize inorganic compounds such as Fe(II) (16–19) and reduced hydroquinones within humic acids (HA) (20, 21). Of these compounds, HA may be of particular relevance to inherent soil denitrification. An operationally defined class of complex organic matter arising from the oxidative degradation, condensation, and transformation of biomass (22, 23), humic materials are ubiquitous in the environment and are prevalent in agricultural soils (24). Structurally diverse, HA contain numerous functional moieties, including carboxylic acid, ketone, quinone, and phenolic/alcoholic hydroxyl groups (23, 25, 26).
HA are recalcitrant and degrade relatively slowly. However, reduced redox-active functional groups within HA can be readily oxidized as electron donors for bacterial respiration (20, 21). The hydroquinone content of reduced HA are considered important humus-borne electron donors of this type, and microbial oxidation of hydroquinones to corresponding quinones has been demonstrated to support nitrate, perchlorate, arsenate, and selenate reduction (20, 21, 27–29). Microbial hydroquinone oxidation has been most clearly demonstrated with the model compound 2,6-anthrahydroquinone disulfonate (AH2DS), which can be oxidized to its quinone form 2,6-anthraquinone disulfonate (AQDS). To date, all tested AH2DS-oxidizing bacteria have alternatively utilized reduced HA, suggesting that AH2DS is a good model for investigating HA-oxidizing microbiology (21).
Although nitrate-dependent humic acid-oxidizing bacteria (NHOx) are common in the environment (21), their prevalence and metabolic activity in agricultural soils are unknown. Here we investigate the microbiology of HA oxidation in agricultural soils with both culture-dependent and culture-independent techniques. Our culture-dependent studies revealed the existence of a large and diverse population of NHOx and provided several novel isolates. NHOx populations coupled the oxidation of reduced HA to substantial nitrate reduction in advective flow columns. A 16S rRNA gene-based high-density oligonucleotide microarray (PhyloChip) (30) illustrated a phylogenetic response, with specific taxa enriched exlusively in HA-amended soils relative to control and background treatments. Together, the results presented in this article demonstrate that NHOx are prevalent in agricultural soils and may play an important role in nitrate respiration.
RESULTS
Most probable number analysis.
Most probable number (MPN) tubes were scored for microbial AH2DS oxidation based upon a visual color change from red (hydroquinone) to yellow (quinone) and microscopic examination. NHOx populations ranged from 3.3 × 104 cells g−1 in native grassland soil to over 106 cells g−1 in native woodland and corn plot soils (Fig. 1). When MPN tubes were left undisturbed for an extended period, gas production was evident as small bubbles on the tube walls in positive tubes. Uninoculated tubes and inoculated tubes without nitrate remained red with no visible bubble production.
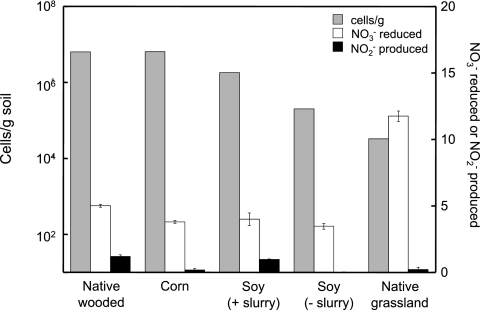
FIG 1 .
Populations of AH2DS-oxidizing, nitrate-reducing cells and degrees of nitrate reduction and nitrite production. Grey bars represent populations of AH2DS-oxidizing, nitrate-reducing cells in agricultural soils as measured by MPN analysis (6.4 × 106 cells g−1 in native wooded soil, 6.5 × 106 cells g−1 in soil from a plot of corn, 1.8 × 106 cells g−1 for soil from a soybean plot with a slurry added to the soil, 2.0 × 105 cells g−1 for soil from a soybean plot with no slurry added, and 3.3 × 104 cells g−1 for native grassland [bluegrass]). The degrees of nitrate reduction and nitrite production are also indicated for the 10−3 dilution of each MPN series.
The positive scored 10−3 dilution tubes for each soil type were examined for extent of nitrate reduction and nitrite production relative to cell-free control medium. In all cases, nitrate reduction relative to control tubes was apparent (Fig. 1). No nitrate reduction or nitrite production occurred in uninoculated MPN tubes (data not shown). In most cases, low concentrations of nitrite were produced, but nitrate reduction was in stoichiometric excess of nitrite production in all tubes tested (Fig. 1). Along with the observation of gas bubbles, these results support active denitrification coupled to AH2DS oxidation within each inoculated MPN series.
Pure cultures.
Several NHOx isolates were obtained from the MPN series. Analyses of 16S rRNA gene sequences revealed that all isolates were phylogenetically distinct members of the Proteobacteria (see Fig. S1 in the supplemental material). In the presence of nitrate and acetate (100 µM), all AH2DS-respiring NHOx isolates grew to higher cell densities than in controls lacking AH2DS, demonstrating that hydroquinone oxidation was coupled to growth (data not shown). While none of the tested isolates degraded the anthraquinone, each culture supported a different pattern of respiratory nitrate reduction. For example, strain A7, a gammaproteobacterium of the Pseudomonas genus, produced a transient nitrite spike during nitrate-dependent AH2DS oxidation. In contrast, strain A5, a betaproteobacterium, produced nitrite as the end product of the metabolism, and strain D1, an alphaproteobacterium, reduced nitrate without observable nitrite accumulation.
Column studies.
In order to examine the effect of reduced HA electron donors on nitrate reduction in dynamic soil systems, advective up-flow column studies . Sealed anaerobic columns inoculated with corn plot soil were constructed using one column per treatment as described in Materials and Methods (see Fig. S2 in the supplemental material). During the initial 7-day equilibration period, limited nitrate reduction occurred in all columns in the absence of electron donor amendment, suggesting the presence of an electron donor intrinsic to the soil. After equilibration, the columns were continuously injected at port A with electron acceptor medium containing approximately 0.5 mM nitrate. Directly above this point at port B, electron donor treatments (donor-free medium, oxidized HA medium, or reduced HA medium), were continually injected at the same flow rate. Bromide tracers in electron donor treatments allowed for dilution factor calculation throughout each column.
(i) Column performance.
Average retention times were 164 ± 56 h throughout operation for the donor-free control column, 139 ± 30 h for the oxidized HA column, and 135 ± 9 h for the reduced HA column. Total flow was very similar between columns for each time point (data not shown), suggesting that cross-comparison of column treatments for extent of nitrate reduction could be reasonably achieved. Bromide tracer data indicated similar donor medium gradients between column treatments and an approximate 1:1 mixing of donor and acceptor media within each column (data not shown).
(ii) Donor-free control column.
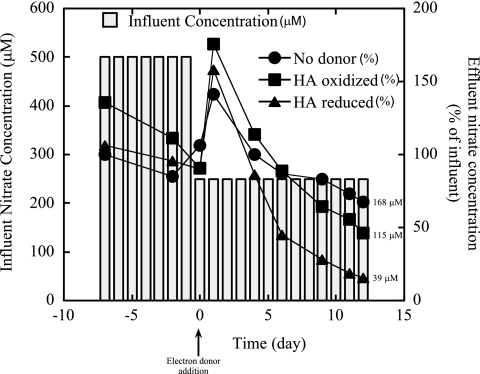
During the initial equilibration, each column hosted a limited amount of nitrate reduction (data not shown). The trend was sustained for the control column continually injected with donor-free medium (Fig. 2; see Fig. S3a in the supplemental material). By the end of the experimental period, samples collected from each port of the column contained similar nitrate concentrations (163, 161, 166, and 185 µM for ports 1, 2, 3, and 4, respectively), indicating a limited concentration gradient throughout the length of the column. This suggests that the majority of the microbial nitrate removal occurred between the injection ports and sampling port 1. Based upon the observed 1:1 mixing ratio of the two types of media, the donor-free column removed an average of 33% (82 µM) of the introduced nitrate by the end of the experimental time period. Nitrite production in the donor-free column was never detected at concentrations higher than 10 µM (data not shown), suggesting the production of more-reduced respiratory end products.
FIG 2 .
Effective influent nitrate concentration (accounting for dilution) and measured effluent nitrate concentration throughout operation in columns treated with no electron donor, oxidized HA, or reduced HA electron donor medium.
(iii) Oxidized HA column.
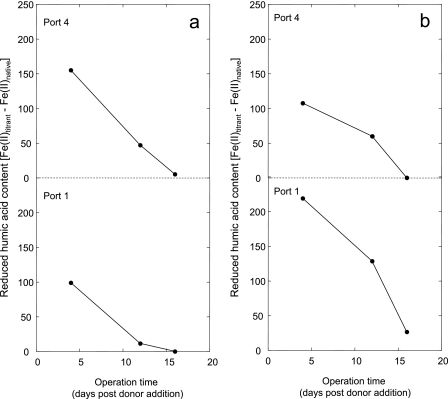
Relative to the no-donor control, the column treated with oxidized HA supported a greater extent of nitrate reduction (Fig. 2; see Fig. S3b in the supplemental material). By the end of the experimental period, the average nitrate concentration across the oxidized HA column was 115 µM, suggesting a 54% reduction of the introduced nitrate based upon the observed 1:1 mixing ratio of the media. Similarly to the donor-free column, there was a minimal nitrate gradient along the length of the column by the end of the operation (124, 105, 110, and 122 µM for ports 1, 2, 3, and 4, respectively), suggesting that the majority of the microbial nitrate reduction was limited to the area between the injection ports and sampling port 1. Nitrite production (0 µM to 21.9 µM) was limited and transient. Fe(III) back-titration revealed that the oxidized HA medium contained a moderate amount of reduced functionalities amounting to 448.7 ± 37.8 reducing microequivalents (µEq). Reducing equivalents were also detected in samples collected from the oxidized HA columns at ports 1 and 4 (Fig. 3a). Over time, the column community was capable of more rapidly oxidizing introduced HA microequivalents (Fig. 3a). By the end of the experimental time period, essentially all reduced microequivalents were oxidized immediately at port 1, suggesting development of a HA-oxidizing community colocalized with the most active zone of apparent nitrate reduction near the point of donor medium injection. Assuming a five-electron transfer to dinitrogen gas and accounting for 1:1 mixing of the media, oxidation of these reduced functionalities in donor medium (224 µEq) accounted for approximately 33% of the total observed nitrate reduction observed in acceptor medium (135 µM; 675 µEq) within the oxidized HA column. The remaining nitrate reduction may be accounted for by the intrinsic electron donating capacity of inoculum soil, degradation of introduced HA, or degradation of labile organics associated with HA.
FIG 3 .
Reduced functional group content of oxidized (a) and reduced (b) HA treatments over time at ports 1 and 4 of each column. Time is measured as days after the addition of an electron donor and is shown on the x axes. Reduced functionality (in microequivalents) is given on the y axes.
(iv) Reduced HA column.
The greatest extent of nitrate reduction was observed in the reduced HA treatment (Fig. 2; see Fig. S3c in the supplemental material). Again, there was a limited nitrate gradient along the length of the column by the end of operation (40, 40, 23, and 53 µM for ports 1, 2, 3, and 4, respectively) with an average concentration of 39 µM, indicating 84% reduction of the theoretically present nitrate when accounting for mixing of the media (Fig. 2). Similarly to the other columns, nitrite concentrations were low, between 0 and 19 µM throughout the column matrix over the experimental time period (data not shown).
Reduced HA medium contained 1,053.7 ± 30.2 reducing µEq in reduced humic moieties, which was equivalent to 527 reducing µEq upon 1:1 dilution in the column. Essentially all of these reducing equivalents were oxidized at port 1, near the donor injection point, by the end of the experimental time period (Fig. 3b). Assuming complete reduction of nitrate to N2 (5-electron transfer), the extent of HA oxidation accounts for approximately 40% of the total nitrate reduction observed in the reduced HA column, suggesting that HA oxidation was a significant nitrate reduction driver in the experimental system.
(v) Electron balance.
As illustrated in the no-donor control column and calculated for HA-treated columns, donor species intrinsic to soil as well as potential degradation of HA or HA-associated organics, supported nitrate reduction in each column system. Nevertheless, the presence of reduced HA functionalities increased the nitrate reduction capacity of column systems, accounting for up to 40% of total observed respiratory activity. The difference in nitrate reduction between oxidized and reduced column treatments (211 µM to 135 µM) is approximately 76 µM, corresponding to 380 µEq of additional electron transfer, assuming production of dinitrogen gas. The additional reduced functionalities in the reduced HA treatment relative to the oxidized treatment (approximately 303 µEq accounting for dilution) therefore accounts for approximately 80% of the additional nitrate reduction observed. This correlation implies that the additional electron donating capacity of the reduced HA treatment can be attributed to the enhanced content of reduced functional groups.
Relative to the no-donor control, the oxidized HA treatment supported a greater extent of microbial nitrate reduction. The additional nitrate reduction was partially accounted for by the intrinsic reduced functionalities observed for HA.
(vi) Possible role for iron content of HA.
The amount of electron donating capacity attributable to the iron content [Fe(II)] of the HA is expected to be small relative to the electron donating capacity attributable to reduced organic functional groups. In undiluted donor media, the reduced HA treatment contained approximately 1,054 µEq of reduced functionalities as calculated by back-titration. Meanwhile, the total maximum iron content of reduced HA solution was 125 µM based upon technical specifications of the reagent. Even if all of the iron in the reduced HA treatment were in the ferrous form, Fe(II) would account for only 12% of the total electron donating capacity.
In general, concentrations of Fe(II) detected at ports 1 and 4 of the HA column treatments were low (39 to 102 µM) relative to the reduced organic functional group content of donor media. Additionally, Fe(II) concentrations detected at ports 1 and 4 were similar between the reduced HA treatment (39 to 102 µM) and the oxidized HA treatment (40 to 98 µM). Together with calculation of total iron content in the donor media above, these observations suggest that Fe(II) does not account for a substantial portion of the difference in respiratory activity observed between oxidized and reduced HA treatments.
PhyloChip microbial community analysis.
To discern which bacterial taxa were specifically associated with the reduced HA treatment, the microbial community from port 1, where most NHOx activity took place, was characterized across the three column treatments upon completion of the experiment. DNA was extracted in three replicate samples from the no-donor control column (control), oxidized HA column (oxidized), reduced HA column (reduced), as well as from the initial inoculum (inoculum). The isolated DNA was combined for each treatment and PCR amplified with archaeal and bacterial primers, and the community was characterized using PhyloChip as previously described (31).
The four samples contained a diverse microbial community, which varied from the initial inoculum in richness, composition, and distribution based on column amendment. In the HA-treated columns, the richness of the bacterial community decreased from the initial inoculum (1,559 operational taxonomic units [OTUs]) in the reduced HA column (−18%; 1,343 OTUs) and in the oxidized HA column (−15%; 1,394 OTUs), while richness slightly increased in the nondonor control (+5%; 1,638 taxa). In contrast to the bacterial community, the richness, composition, and distribution of the archaeal community after HA treatment did not differ from that of the initial inoculum.
To better characterize the response of HA treatment on specific bacterial taxa, a subtractive analysis using 16S rRNA gene hybridization intensity scores from each of the oxidized and reduced HA columns relative to the control column was performed. Twelve phyla of the Bacteria domain increased in relative abundance in both HA-treated columns compared to the control column (see Fig. S4 in the supplemental material). Of these phyla, the Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria were most enriched in response to HA treatment. Together, the decrease in bacterial OTU richness coupled with an increase of specific bacterial taxa implies that the HA impart a selective pressure on the bacterial community that increases the dominance of specific bacterial OTUs.
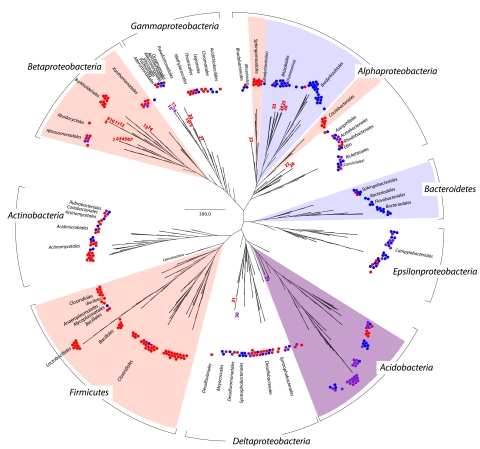
To visualize the bacterial response to HA treatment, we constructed a phylogenetic tree with 16S rRNA gene sequences from the five most enriched phyla (365 total OTUs) relative to the no-donor control (Fig. 4). The Acidobacteria phylum was nearly evenly enriched in both the oxidized and reduced HA columns (purple shading), while others like the Firmicutes (reduced; red shading) and Bacteroidetes (oxidized; blue shading) were enriched exclusively in one HA-treated column and not the other. Of the 66 Firmicutes sequences enriched by HA treatment, 97% (64 OTUs) were identified in the reduced HA column and only 3% (2 OTUs) were identified in the oxidized HA column. On the other hand, 20 of the 21 (95%) enriched Bacteroidetes sequences were identified in the oxidized column, while only 3 Bacteroidetes OTUs (14%) were enriched in the reduced column (Fig. 4).
FIG 4 .
A maximum likelihood tree constructed from 16S rRNA PhyloChip sequences and selected isolates illustrates the phylogenetic position of taxa enriched in abundance in columns treated with humic substance relative to the no-donor control column. To differentiate the response to HA treatment, PhyloChip-identified OTUs are shown by small purple circles if they were enriched in both the reduced and oxidized HA treatments, while the OTUs are shown by small red circles if they were enriched in the reduced HA treatments only and shown by small blue circles if they were enriched in the oxidized HA treatments only. A similar color scheme also identifies the 16S rRNA sequences of bacteria isolated as part of this study (numbered 8 to 14, 23, 24, 2 and 6), as well as bacterial isolates previously reported to be capable of HA redox reactions (remaining isolate numbers) (Materials and Methods). The colored pie-shaped wedges correspond to taxa with a collective HA response, with those increasing in reduced humic substance column (red wedge), oxidized humic substance column (blue wedge), or both columns (purple wedge) denoted. Bar, 300 inferred nucleotide changes per position rRNA gene sequence.
Unlike the Acidobacteria, Firmicutes, and Bacteroidetes, which had a phylum level response to HA treatment, the Actinobacteria and Proteobacteria response varied within each phylum. The Betaproteobacteria exhibited a class level response, with all of the sequences detected in the columns being enriched in the reduced HA column, indicating a functional role for these organisms in HA oxidation (Fig. 4). The Alphaproteobacteria demonstrated a discrete family level response to HA treatment, with certain orders like the Bradyrhizobiales and Rhizobiales predominantly enriched in the oxidized column, while the enrichment of Caulobacterales and Sphigomonadales taxa was found only in the reduced column. Although members of the Gammaproteobacteria, Actinobacteria, and, to an extent, Deltaproteobacteria did increase in proportional abundance in HA-treated columns relative to the control column, this response was stochastic and did not corroborate phylogenetic affiliation (Fig. 4). The Geobacteraceae, in the Deltaproteobacteria, were a notable exception to this trend, as they were exclusively enriched in the reduced HA column (Fig. 4) and decreased in the oxidized treatment relative to the control. Collectively, the PhyloChip results uncover much broader bacterial phylogenetic diversity associated with HA environments than has been previously observed.
DISCUSSION
These studies illustrate the prevalence and diversity of nitrate-reducing, HA-oxidizing bacteria in agricultural soils. Likewise, data presented here track the oxidation of reduced HA in complex, advective flow systems, as well as microbial community shifts associated with HA treatments. Taken together, these results suggest that a large, diverse, and potentially active population of nitrate-dependent humic acid-oxidizing bacteria exist in agricultural soils and that specific organisms may respond to the presence of HA.
Phylogenetically diverse NHOx microorganisms have already been shown to be common in nonagricultural soils, wetlands, and marine sediments (21). Similar ubiquity and diversity of NHOx populations were observed in this study which utilized source soils collected from a limited geographical area and differing only in plant cover or fertilizer treatment. The populations of NHOx observed in this study varied between 104 cells g−1 soil (native grassland soil) and 106 cells g−1 soil (forest and corn plot soil). Notably, MPN-based studies of heterotrophic nitrate-reducing bacteria in agricultural soils routinely reveal populations between 104 and 106 cells g−1 soil, in the same log range as the MPN analysis performed here (32, 33). Although total heterotrophic nitrate-reducing bacterial populations were not enumerated in this study, it is plausible that a significant portion of culturable nitrate-respiring organisms present in agricultural soils are capable of HA oxidation. Many of the isolates in this study were members of the Alcaligenes or Pseudomonas genera, which have routinely been isolated as denitrifying organisms from agricultural and rhizosphere soils (1, 32, 34). This observation highlights the potential culturing bias inherent to MPN and isolation analyses but suggests that previously described denitrifying bacteria in soil from plots containing plants may also prove capable of HA oxidation.
Native microbial populations in corn plot soil oxidized reduced functional groups within HA coupled to nitrate reduction in an advective system. The presence of reduced functional groups increased the electron donating capacity of HA, and the reduced functional group content roughly correlated with the difference in oxidation state between similar HA treatments. However, addition of either HA always supported a greater extent of nitrate reduction than would be expected based solely upon oxidation of the reduced functional group content alone. This discrepancy persists when electron donors intrinsic to the soil were accounted for in no-donor control systems. One potential explanation for this observation is microbial degradation of HA or HA-associated organics. Enhanced denitrification in response to humic amendments has been illustrated for soils and wetland or riverbed sediments (7, 14, 35, 36). These studies did not generally discriminate between degradative oxidation of humic materials and nondegradative oxidation of reduced functional groups. Although HA were potentially slowly degraded in column systems studied here, oxidation of reduced functional groups still accounted for a significant percentage of the nitrate respired in both oxidized and reduced HA columns (up to 40%).
There are few studies examining the influence of humic acids on bacterial communities in the literature. Here, a 16S rRNA gene-based phylogenetic microarray characterized the effect of oxidized and reduced HA treatment on the resident bacterial community in agricultural soil to better understand the role of reduced HA on bacterial communities. Specific bacterial sequences enriched in the reduced HA column included members of the Acidobacteria, Firmicutes, Betaproteobacteria, Caulobacterales, and Sphingomonadales of the Alphaproteobacteria and Geobacteraceae of the Deltaproteobacteria (Fig. 4).
Of these taxa, only the Acidobacteria contained a majority of sequences which were also enriched in the oxidized HA-treated column. A survey exploring the relative abundance of bacterial phyla within 71 soils from a range of ecosystems revealed a strong and ubiquitous inverse relationship between carbon availability and Acidobacteria abundance (37). Consequently, it is important to note that the enrichment of 16S rRNA gene sequences in this column study may not be a direct result of HA oxidation and reduction but an artifact of the microbial habitat created by the addition of HA (37). However, it is well documented that isolated members of the Acidobacteria are capable of both oxidation and reduction of quinone moieties (20), which suggests a physiological role in HA redox reactions for the Acidobacteria in these HA-amended columns. Unlike the Acidobacteria, Firmicutes sequences were identified only in reduced HA-treated columns. This selective enrichment of Firmicutes only in reduced HA columns suggests a functional role for these species in oxidation of reduced HA functional groups.
Within the Proteobacteria, each class reported a unique response to reduced HA. All 25 taxa that were identified as enriched relative to the control in the Betaproteobacteria were identified in the reduced HA column, with only five of these taxa increasing in the oxidized HA column. Correspondingly, a majority of the bacteria isolated as part of this study supported this finding, with seven of the NHOx isolates belonging to the Betaproteobacteria within the order Burkholderiales. Two of our isolates were closely related to Ralstonia sp., and five isolates were most closely related to Achromobacter sp. Further supporting our findings based on pure culture and community results, a recent study also identified the dominance of 16S rRNA gene sequences from the Burkholderiales, Ralstonia, and Cupriavidus spp. in reduced HA biofilms but not detected in controls not treated with humic acid (38).
In the Deltaproteobacteria, enrichment of members of the Geobacteraceae was consistent with previous studies demonstrating microbial oxidation of humic substances by several members of this family (20). However, it is also well documented that these bacteria are also capable of reduction of humic substances (39, 40), yet none of these members were enriched in the oxidized column relative to the control. The presence of excess nitrate in the system may have thermodynamically precluded (41, 42) or reduced the selective importance of HA-reducing respiratory processes by these organisms.
Like the Deltaproteobacteria, certain orders of the Alphaproteobacteria phylum (Caulobacterales and Sphigomonadales) were enriched only in reduced HA columns. These taxa are routinely isolated as members of the rhizosphere, but little is known regarding their role in humic substance redox reactions. It is plausible that the enrichment of these 16S rRNA gene sequences and our isolates reflect a previously unidentified role for alphaproteobacterial rhizosphere bacteria in HA oxidation coupled to denitrification.
Collectively, the results from our culture-dependent and -independent research suggest that a diverse set of microorganisms may be capable of oxidizing humic material coupled to nitrate reduction in agricultural soils. Furthermore, these results suggest that HA oxidation can potentially account for a significant percentage of denitrification observed in these soil types. Future studies examining the role of reduced humic materials in donating electrons for microbial respiration and the influence of these compounds on the structure and function of microbial communities are warranted.
MATERIALS AND METHODS
Medium techniques.
Basal, bicarbonate-buffered medium (pH 6.8 to 7.0) was utilized for MPN and isolate characterization studies. Media were prepared anaerobically under 80:20 N2-CO2 headspace as previously described (43). Where noted, quinone- or hydroquinone-containing media were prepared by adding AQDS at concentrations of 5 mM total anthraquinone. To prepare AH2DS, AQDS solutions were degassed and then bubbled with 80:20 H2-CO2 in the presence of palladium-coated aluminum chips (21, 44). When a red color developed, the medium was bubbled for 20 min with 80:20 N2-CO2 to remove dissolved H2 and subsequently decanted away from the palladium into pressure tubes or serum bottles before autoclaving. Acetate (0.1 mM) and nitrate (10.0 mM) were added as sodium salts from anoxic sterile stock solutions.
The medium for column experiments was comprised of 10 mM phosphate buffer (pH 7.2) amended with 5 ml liter−1 trace mineral solution (45). The medium was boiled and cooled under N2 gas and then sealed and autoclaved in 2-liter bottles. Electron acceptor medium contained nitrate (1.0 or 0.5 mM) added from stock solutions. Different electron donor media (donor-free, oxidized HA, and reduced HA media) were prepared for each column treatment. Donor-free medium consisted of phosphate-buffered water and mineral solution only. Oxidized HA treatments were prepared by adding 0.5 g liter−1 of Aldrich HA to previously degassed and cooled media prior to autoclaving. Reduced HA treatments were prepared by adding 0.5 g liter−1 of the same HA stock to degassed medium, chemically reducing the HA as previously described (44), and bubbling the medium for 20 minutes with N2 to remove H2 prior to autoclaving. Each donor bottle was supplemented with 200 µM bromide tracer from stock aqueous solutions of sodium salts.
Analytical techniques.
Anions were quantified by ion chromatography on a Dionex DX500 employing a CD20 conductivity detector suppressed with an ASRS-ULTRA II 4-mm system. Nitrate and nitrite concentrations were resolved on an IonPac AS9-HC anion-exchange column, with a 9 mM Na2CO3 mobile phase at a flow rate of 1 ml min−1. AH2DS and reduced humic acid concentrations were analyzed by ferric citrate back-titration or spectrophotometrically as previously described (21, 39, 40). Total anthraquinone concentrations were determined by aerating filtered samples and quantifying AQDS as previously described (29). Fe(II) was quantified with a ferrozine assay as previously described (46).
MPN analysis and isolation/characterization of pure cultures.
A family farm in the Platte River valley near Linwood, NE, was chosen as a sampling site. Soils were collected from the top 12 inches of plots containing corn or soy (either amended or unamended with swine manure in the previous growing season), as well as from native bluegrass grasslands and wooded areas containing ash, elm, oak, and cedar trees adjacent to the cultivated areas. The samples were sealed in autoclaved mason jars, brought to the laboratory, and subjected to most probable number (MPN) analysis for nitrate-reducing, AH2DS-oxidizing microorganisms as previously described, with an 87-day incubation period (21). Nitrate reduction and nitrite production in MPN tubes was measured via anion analysis of 0.2-µm-filtered subsamples from the 10−3 dilution series. Pure cultures of AH2DS-oxidizing, nitrate-reducing cells were obtained utilizing an agar shaking tube method previously described (21). Selected isolates were characterized for AH2DS-oxidizing, nitrate-reducing capabilities in the presence of 100 µM acetate as a suitable carbon source.
Phylogenetic analysis of isolates.
Genomic DNA (gDNA) was extracted from isolate growth cultures oxidizing AH2DS with a MoBio Power Soil DNA kit (MoBio Laboratories Inc., Solana Beach, CA) per the manufacturer’s protocol. The 16S rRNA gene was amplified from gDNA extracts with universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG) and 1492R (5′-GGTTACCTTGTTACGACTT) (F stands for forward and R stands for reverse in the primer names). 16S rRNA gene amplification was performed with Takara Ex Taq HS according to the manufacturer’s protocol. Isolation of genomic DNA, 16S rRNA gene-specific PCR, and sequencing of PCR products were conducted as described previously (27, 31). 16S rRNA gene sequences were aligned with MUSCLE 3.6 (47), and Bayesian analysis of 16S rRNA gene phylogeny was completed with MrBayes 3.2 (48, 49). The program was run with four chains until the standard deviation of the split frequencies was stabilized below 0.01, in this case for 450,000 generations, with a sample frequency of 1,000. The first 25% of the samples were discarded for accurate estimation of the posterior probability distribution of the summary tree.
Humic column experiments.
Up-flow glass column reactors were constructed as diagrammed in Fig. S1 in the supplemental material. The columns were filled with a matrix containing 1 N HCl-washed, autoclaved 3-mm-diameter glass beads (Walter Stern no. 100C) inoculated 10% (wt/wt) by manual mixing with corn plot soil (~750 ml total matrix volume/column). The columns were sparged with He, sealed with rubber stoppers, and covered with aluminum foil. The columns were incubated for 2 days with approximately 370 ml of medium containing 500 µM nitrate to allow for microbial adherence and then equilibrated for flow rate and nitrate breakthrough over 7 days by continually injecting port A with electron acceptor medium (1.0 mM nitrate) and port B with donor-free medium (average total flow rate of 3.1 ± 0.2 ml/h). After 7 days, the nitrate concentrations in continually injected acceptor medium were reduced to 0.5 mM, and donor treatments commenced through continual injection of donor medium at port B. Donor treatments were as follows: donor-free medium, 0.5 g/liter oxidized Aldrich HA medium, and 0.5 g/liter reduced Aldrich HA medium. Column performance was assessed by frequent retention time and total medium flowthrough measurements. Nitrate, nitrite, bromide, and reduced HA content were analyzed at time points along the length of each column at ports 1, 2, 3, and 4. Reduced HA content was defined as the difference between Fe(II) generated in back-titration reactions and Fe(II) present in the native initial sample prior to Fe(III) citrate addition and was calculated as follows: reduced HA = Fe(II)back-titration – Fe(II)initial.
PhyloChip.
To characterize changes in the bacterial community after humic acid treatment, DNA was extracted from the initial inoculum and from port 1 of each of three columns (no donor, oxidized HA, and reduced HA). Due to the high humic content of the soil, the previously described extraction protocol (31) was modified to increase DNA quality and yields. Specifically, 50 µl of 0.1 M aluminum ammonium sulfate was added as flocculent to the cetyltrimethylammonium bromide (CTAB) extraction buffer to minimize coextraction of humic acid material, and nucleic acids were precipitated for two hours at room temperature with polyethylene glycol (PEG) rather than isopropanol (31). Extractions three times for each soil sample with pellets from each sample combined and resuspended in 100 µl of Tris-EDTA (TE) buffer. Coextraction of humic material in samples from the HA-treated columns resulted in brown resuspended DNA that failed to amplify by PCR. To remove humic substance contamination from the DNA, all the resuspended DNA samples were purified using the MoBio Power Soil DNA kit according to the manufacturer’s protocol apart from the fact that the samples were gently mixed rather than beadbeated or vortexed. DNA quality and quantification were assessed by gel visualization. PCR amplification was conducted as previously described (Wrighton et al. [31]); the amplifications over a gradient using 8 different annealing temperatures and nondegenerate primers and were restricted to 25 cycles to minimize PCR bias. The amount of PCR product loaded onto the chips, target fragmentation, biotin labeling, PhyloChip hybridization, scanning, and staining, as well as background subtraction, noise calculation, and detection and quantification criteria were based on the protocol given in reference 30.
PhyloChip arrays from the initial inoculum, no-donor control column, oxidized humic substance column, and reduced humic substance column. For bacterial richness, a taxon was considered present in the sample when 90% or more of its assigned probe pairs for its corresponding probe set were positive (positive fraction of ≥0.90). To determine the differences in bacterial community composition, nonmetric multidimensional scaling (NMDS) was performed on PhyloChip hybridization data using the statistical package Primer V (Plymouth Marine Laboratory, Plymouth, United Kingdom). To discriminate the bacterial populations enriched in the humic acid-treated columns, a subtractive analysis between the controls and humic acid-treated columns was performed.
With the intention of illustrating the phylogenetic response to HA across the 5 most enriched phyla, a maximum likelihood tree was constructed using the 365 16S rRNA sequences from the five dominant phyla demonstrated to be enriched relative to the control in either or both columns treated with humic acid. The sequences were aligned with MUSCLE 3.6 (Edgar), and phylogenetic analysis was performed with RAxML (Stamatakis). 16S rRNA sequences of previously isolated humic substance-oxidizing and -reducing bacteria and the bacteria characterized as part of this study (numbered isolates 8 to 14, 23, 24, and 26) are also included in the phylogenetic analyses and resulting tree. Isolate sequences are numbered as follows (accession numbers are given in the parentheses; all accession numbers are for the GenBank databank, except for accession numbers beginning with NC_, which are for the NCBI databank): 1, Dechloromonas sp. strain MissR (AF170357); 2, Dechloromonas aromatica (CP000089); 3, Dechloromonas sp. strain JJ (AY032611); 4, Dechloromonas agitata (AF047462); 5, Azospira (Dechlorosoma) suillum (AF170348); 6, Azoarcus evansii (X77679); 7, Azoarcus sp. strain HA (AF482683); 8, strain C7b; 9, strain C7a; 10, strain B7; 11, strain B2; 12, strain Bcol1; 13, strain A4; 14, strain A5; 15, Shewanella oneidensis MR-1 (NC_004347); 16, Shewanella algae isolate 62 (DQ883817); 17, Escherichia coli K-12 (NC_010473); 18, Pseudomonas sp. strain NMX (AF482685); 19, strain A7, 20, Pseudomonas sp. strain BU (AF482684); 21, Marinobacter sp. strain SBS (AF482686); 22, Paracoccus denitrificans (Y16927); 23, Agrobacterium sp. strain PB (AF482682); 24, strain D5; 25, strain D1; 26, strain A1; 27, Dechlorospirillum sp. strain VDY (EF405824); 28, Dechlorospirillum sp. strain WD (AF170352); 29, Geothrix fermentans (EF405824); 30, Geobacter metallirreducens (AF170352); and 31, Myxobacterium sp. strain KC (AF482687). The isolate sequences are color coded based on the reported humic substance response. A phylogenetic response is defined when >85% of the OTUs in a group respond uniformly to HA treatment. Taxon level responses are denoted with shading with colors similar to those used for the OTU designations.
SUPPLEMENTAL MATERIAL
Phylogenetic placement of humic substance-oxidizing bacteria among the Proteobacteria according to 16S rRNA gene analysis. Posterior probability values are indicated at nodes. The outgroup consists of Acidobacterium capsulatum and Geothrix fermentans in the Acidobacteria. Bar, 0.06 expected changes per site. Download Figure S1, DOC file, 0.408 MB.
Diagram of column systems amended with either no-donor, oxidized HA, or reduced HA electron donor medium. Electron acceptor medium, containing either 1 mM or 0.5 mM nitrate, was continually injected at port A as described in Materials and Methods. The various donor treatments were continually injected at port B. Nitrate and nitrite concentrations were sampled at ports 1, 2, 3, and 4, while Fe(II) and hydroquinone content were assayed at ports 1 and 4. Download Figure S2, DOC file, 0.220 MB.
Nitrate influent and effluent concentrations and percent nitrate removal in columns treated with either no-donor (a), oxidized HA (b), or reduced HA (c) electron donor medium throughout the experiment or operation. Download Figure S3, DOC file, 0.673 MB.
(a) Nonmetric multidimensional scaling (NMDS) output of PhyloChip hybridization intensity data. A stress value of 0.01 indicates little distortion from Bray-Curtis similarity matrix. (b) Contribution of major phyla enriched in reduced HA column (a) and oxidized HA column (b) relative to the control column. A total of 303 and 257 bacterial OTUs were enriched in the reduced and oxidized HA columns, respectively. Download Figure S4, DOC file, 0.704 MB.
ACKNOWLEDGMENTS
We thank Lonnie Spies for allowing access to his farm and surrounding properties.
Funding for K.C.W. is through the UC Berkeley Sustainable Products & Solutions (SPS) Program, and the Chang Tien Fellowship. Research on microbial humic substance oxidation in the laboratory of J.D.C. is supported through grant 2005-35107-16237 by the Natural Research Initiative of the USDA.
Footnotes
Citation Van Trump JI, et al. 2011. Humic acid-oxidizing, nitrate-reducing bacteria in agricultural soils. mBio 2(4):e00044-11. doi:10.1128/mBio.00044-11.
REFERENCES
- 1. Knowles R. 1982. Denitrification. Microbiol. Rev. 46:43–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hauck RD. 1981. Nitrogen fertilizer effects on nitrogen cycle processes. Ecol. Bull. 33:551–562 [Google Scholar]
- 3. Firestone MK. 1982. Biological denitrification, p. 289–326. In Stevenson FJ, Nitrogen in agricultural soils, vol. 22 American Society of Agronomy, Madison, WI [Google Scholar]
- 4. Weier KL, McEwan CW, Vallis I, Catchpole VR, Myers RJ. 1996. Potential for biological denitrification of fertilizer nitrogen in sugercane soils. Aust. J. Agr. Res. 47:67–79 [Google Scholar]
- 5. Wijler J, Delwiche CC. 1954. Investigations on the denitrifying process in soil. Plant Soil 5:155–169 [Google Scholar]
- 6. Stevens RJ, Laughlin RJ, Malone JP. 1998. Soil pH affects the processes reducing nitrate to nitrous oxide and di-nitrogen. Soil Biol. Biochem. 30:1119–1126 [Google Scholar]
- 7. Pfenning KS, McMahon PB. 1996. Effect of nitrate, organic carbon, and temperature on potential denitrification rates in nitrate-rich riverbed sediments. J. Hydrol. 187:283–295 [Google Scholar]
- 8. Herrman KS, Bouchard V, Moore RH. 2008. Factors affecting denitrification in agricultural headwater streams in northeast Ohio, USA. Hydrobiologia 598:305–314 [Google Scholar]
- 9. Sexstone AJ, Parkin TB, Tiedje JM. 1985. Temporal response of soil denitrification rates to rainfall and irrigation. Soil Sci. Soc. Am. J. 49:99–103 [Google Scholar]
- 10. Pilot L, Patrick WH. 1972. Nitrate reduction in soils: effect of soil moisture tension. Soil Sci. 114:312–316 [Google Scholar]
- 11. Rysgaard S, Risgaard-Peterson N, Sloth NP, Jensen K, Nielsen LP. 1994. Oxygen regulation of nitrification and denitrification in sediments. Limnol. Oceanogr. 39:1643–1652 [Google Scholar]
- 12. Beauchamp EG, Trevors JT, Paul JW. 1989. Carbon sources for bacterial denitrification. Adv. Soil Sci. 10:113–134 [Google Scholar]
- 13. Stanford G, Vander Pol RA, Dzienia S. 1975. Denitrification rates in relation to total and extractable soil carbon. Soil Sci. Soc. Am. J. 39:284–289 [Google Scholar]
- 14. Katz R, Hagin J, Kurtz LT. 1985. Participation of soluble and oxidizable soil organic compounds in denitrification. Biol. Fertil. Soils 1:209–213 [Google Scholar]
- 15. Burford JR, Bremner JM. 1975. Relationships between the denitrification capacities of soils and total, water-soluble, and readily decomposable soil organic matter. Soil Biol. Biochem. 7:389–394 [Google Scholar]
- 16. Weber KA, Picardal FW, Roden EE. 2001. Microbially catalyzed nitrate-dependent oxidation of biogenic solid-phase Fe(II) compounds. Environ. Sci. Technol. 35:1644–1650 [DOI] [PubMed] [Google Scholar]
- 17. Weber KA, Achenbach LA, Coates JD. 2006. Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4:752–764 [DOI] [PubMed] [Google Scholar]
- 18. Lack JG, Chaudhuri SK, Chakraborty R, Achenbach LA, Coates JD. 2002. Anaerobic biooxidation of Fe(II) by Dechlorosoma suillum. Microb. Ecol. 43:424–431 [DOI] [PubMed] [Google Scholar]
- 19. Chaudhuri SK, Lack JG, Coates JD. 2001. Biogenic magnetite formation through anaerobic biooxidation of Fe(II). Appl. Environ. Microbiol. 67:2844–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lovley DR, Fraga JL, Coates JD, Blunt-Harris EL. 1999. Humics as an electron donor for anaerobic respiration. Environ. Microbiol. 1:89–98 [DOI] [PubMed] [Google Scholar]
- 21. Coates JD, Cole KA, Chakraborty R, O’Connor SM, Achenbach LA. 2002. Diversity and ubiquity of bacteria capable of utilizing humic substances as electron donors for anaerobic respiration. Appl. Environ. Microbiol. 68:2445–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Trump JI, Sun Y, Coates JD. 2006. Microbial interactions with humic substances. Adv. Appl. Microbiol. 60:55–96 [DOI] [PubMed] [Google Scholar]
- 23. Stevenson FJ. 1994. Humus chemistry: genesis, composition, reactions. John Wiley and Sons, New York, NY. [Google Scholar]
- 24. Boyer JN, Groffman PM. 1996. Bioavailability of water extractable organic carbon fractions in forest and agricultural soil profiles. Soil Biol. Biochem. 28:783–790 [Google Scholar]
- 25. Gaffney JS, Marley NA, Clark SB. 1996. Humic and fulvic acids and organic colloidal materials in the environment, p. 2–16. In Gaffney JS, Marley NA, Clark SB, Humic and fulvic acids—isolation, structure, and environmental role, vol. 651 American Chemical Society, Washington, DC [Google Scholar]
- 26. Schulten HR, Plage B, Schnitzer M. 1991. A chemical structure of humic substances. Naturwissenschaften 78:311–312 [DOI] [PubMed] [Google Scholar]
- 27. Thrash JC, et al. 2007. Electrochemical stimulation of microbial perchlorate reduction. Environ. Sci. Technol. 41:1740–1746 [DOI] [PubMed] [Google Scholar]
- 28. Bruce RA, Achenbach LA, Coates JD. 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319–331 [DOI] [PubMed] [Google Scholar]
- 29. Van Trump JI, Coates JD. 2009. Thermodynamic targeting of microbial perchlorate reduction by selective electron donors. ISME J. 3:466–476 [DOI] [PubMed] [Google Scholar]
- 30. Brodie EL, et al. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wrighton KC, et al. 2008. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2:1146–1156 [DOI] [PubMed] [Google Scholar]
- 32. Gamble TN, Betlach MR, Tiedje JM. 1977. Numerically dominant denitrifying bacteria from world soils. Appl. Environ. Microbiol. 33:926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chèneby D, Philippot L, Hartmann A, Hénault C, Germon J. 2000. 16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microb. Biol. 34:121–128 [DOI] [PubMed] [Google Scholar]
- 34. Weier KL, MacRae IC. 1992. Denitrifying bacteria in the profile of a brigalow clay soil beneath a permanent pasture and a cultivated crop. Soil Biol. Biochem. 24:919–923 [Google Scholar]
- 35. Sirivedhin T, Gray KA. 2006. Factors affecting denitrification rates in experimental wetlands: field and laboratory studies. Ecol. Eng. 26:167–181 [Google Scholar]
- 36. Hernandez ME, Mitsch WJ. 2007. Denitrification potential and organic matter as affected by vegetation community, wetland age, and plant introduction in created wetlands. J. Environ. Qual. 36:333–342 [DOI] [PubMed] [Google Scholar]
- 37. Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 38. Rodrigues AL, et al. 2008. Characterization of biofilm formation on a humic material. J. Ind. Microbiol. Biotechnol. 35:1269–1276 [DOI] [PubMed] [Google Scholar]
- 39. Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445–448 [Google Scholar]
- 40. Lovley DR, et al. 1998. Humic substances as a mediator for microbially catalyzed metal reduction. Acta Hydrochim. Hydrobiol. 26:152–157 [Google Scholar]
- 41. Champ DR, Gulens J, Jackson RE. 1979. Oxidation-reduction sequences in ground water flow systems. Can. J. Earth Sci. 16:12–23 [Google Scholar]
- 42. Lovley DR, Goodwin S. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim. Cosmochim. Acta 52:2993–3003 [Google Scholar]
- 43. Achenbach LA, Bruce RA, Michaelidou U, Coates JD. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527–533 [DOI] [PubMed] [Google Scholar]
- 44. Coates JD, Chakraborty R, Connor O, Schmidt C, Thieme J. 2001. The geochemical effects of microbial humic substances reduction. Acta Hydrochim. Hydrobiol. 28:420–427 [Google Scholar]
- 45. Lovley DR, Greening RC, Ferry JG. 1984. Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl. Environ. Microbiol. 48:81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779–781 [Google Scholar]
- 47. Edgar RC. 2004. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 49. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic placement of humic substance-oxidizing bacteria among the Proteobacteria according to 16S rRNA gene analysis. Posterior probability values are indicated at nodes. The outgroup consists of Acidobacterium capsulatum and Geothrix fermentans in the Acidobacteria. Bar, 0.06 expected changes per site. Download Figure S1, DOC file, 0.408 MB.
Diagram of column systems amended with either no-donor, oxidized HA, or reduced HA electron donor medium. Electron acceptor medium, containing either 1 mM or 0.5 mM nitrate, was continually injected at port A as described in Materials and Methods. The various donor treatments were continually injected at port B. Nitrate and nitrite concentrations were sampled at ports 1, 2, 3, and 4, while Fe(II) and hydroquinone content were assayed at ports 1 and 4. Download Figure S2, DOC file, 0.220 MB.
Nitrate influent and effluent concentrations and percent nitrate removal in columns treated with either no-donor (a), oxidized HA (b), or reduced HA (c) electron donor medium throughout the experiment or operation. Download Figure S3, DOC file, 0.673 MB.
(a) Nonmetric multidimensional scaling (NMDS) output of PhyloChip hybridization intensity data. A stress value of 0.01 indicates little distortion from Bray-Curtis similarity matrix. (b) Contribution of major phyla enriched in reduced HA column (a) and oxidized HA column (b) relative to the control column. A total of 303 and 257 bacterial OTUs were enriched in the reduced and oxidized HA columns, respectively. Download Figure S4, DOC file, 0.704 MB.