ABSTRACT
Some trials administered antituberculosis agents for 5 of 7 days (5/7-day regimen) to optimize adherence. Since moxifloxacin has a longer half-life than rifampin, rifampin concentrations are <1% of the maximum concentration in serum (Cmax) on day 6 and nondetectable on day 7, while concentrations of moxifloxacin remain and are able to induce error-prone replication. We determined if functional moxifloxacin monotherapy for 24 h/week caused resistance. In in vitro pharmacodynamic experiments, Mycobacterium tuberculosis was treated with mean area under the concentration-time curve (AUC) exposures for moxifloxacin and rifampin of 400 and 600 mg/kg/day and exposures equal to 1 standard deviation (SD) above and below the mean values. The drugs were administered on schedules of 7/7 days and 5/7 days. Over the 28-day experiments, bacteria were plated onto antibiotic-free agar to determine the effects of exposure and schedule on the total population. MICs were checked for emergence of resistance. At days 7 and 14, there was a 0.56- to 1.22-log10-CFU/ml greater cell kill with the 7/7-day regimen versus the 5/7-day regimen (low exposure). This difference was not seen for the larger exposures at day 21. At day 23, the low-exposure 5/7-day arm had breakthrough resistance, with the total count increasing to >2 log10 CFU/ml above the low-exposure 7/7-day arm. Pharmacokinetic mismatching of drugs in the therapy of tuberculosis may result in emergence of resistance when a drug holiday is imposed during which there is functional monotherapy and where the remaining agent induces error-prone replication. This is particularly true for the portion of the population where the clearance is higher (1 SD above the mean).
IMPORTANCE
Directly observed therapy is a cornerstone of treatment of Mycobacterium tuberculosis. Patients are often given a drug holiday to facilitate the direct observation of therapy. With rifampin and moxifloxacin, there is a discordance between the half-lives of these agents (1.9 versus 6.5 h when employed in combination). In addition, moxifloxacin induces error-prone replication in Mycobacterium tuberculosis. In this experiment, we demonstrate that the drug holiday (5 of 7 days of therapy [5/7-day regimen]) allows the emergence of resistance to moxifloxacin, which was not seen with 7/7-day therapy. If drug holidays are used, it is imperative to better match pharmacokinetics to minimize the risk of emergence of resistance.
Introduction
A number of clinical and preclinical studies have administered therapy for Mycobacterium tuberculosis on a schedule of 5 days out of 7 days (5/7-day regimen), with the idea that this would improve regimen adherence. Most recently, these trials have included moxifloxacin and rifampin as part of the therapy (1, 2). Moxifloxacin and rifampin have somewhat discordant serum half-lives, and they have been demonstrated by Weiner et al. (3) to interact, causing moxifloxacin clearance to increase (and the moxifloxacin area under the concentration-time curve [AUC] to decrease) by 27% when administered in the presence of rifampin.
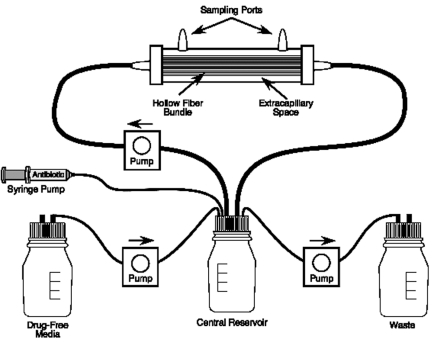
The discordance in serum half-lives of moxifloxacin and rifampin may have important implications. Over the 48 h in which both drugs are not administered, the concentration of rifampin will be less than 1% of the peak concentration at the end of the first day and undetectable on the second day, while moxifloxacin still retains considerable concentrations during this period because of its longer half-life. Furthermore, as a fluoroquinolone, moxifloxacin induces error-prone replication in M. tuberculosis (4). Consequently, it is reasonable to hypothesize that the discordance in half-lives may lead to emergence of resistance to one or both drugs. We decided to test this hypothesis with our hollow fiber infection model (HFIM) (Fig. 1). We compared the impacts of drug exposures on 7/7-day and a 5/7-day dosing schedules on breakthrough regrowth over 28 days and assessed whether regrowth was due to emergence of resistance to either moxifloxacin (expected) or rifampin. The ability to suppress resistance with combination chemotherapy is a key factor in the ultimate outcome of therapy for M. tuberculosis.
FIG 1 .
Hollow fiber infection model system.
RESULTS
Organism rifampin and moxifloxacin MICs and mutation frequencies.
For M. tuberculosis strain H37Ra, the MIC for rifampin was 0.06 mg/liter and the mutation frequency to 2.0 mg/liter of rifampin was 1/6.921 log10 CFU. For moxifloxacin, the MIC was 0.25 mg/liter and the mutation frequency to resistance to 1.0 mg/liter was 1/6.524 log10 CFU.
M. tuberculosis cell kill for different drug exposures and schedules of administration.
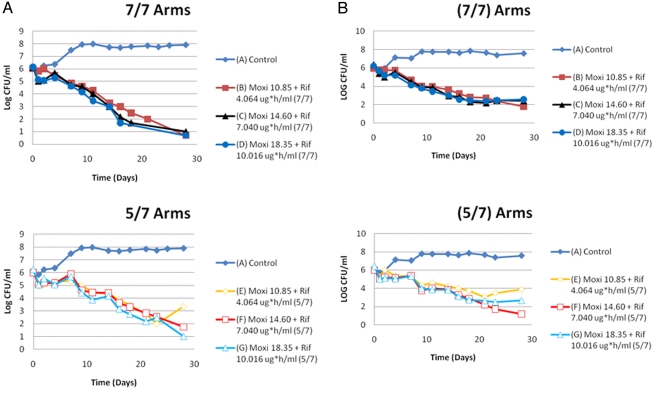
Examination of the top panel of Fig. 2A demonstrates that the administration of moxifloxacin plus rifampin provided excellent cell kill over time when the drugs were administered 7/7 days. The relatively small free 24-h AUC (AUC24 h) exposure range examined did not result in a reliable exposure response for the organism kill. At day 28, the cell kill results for all three regimens (mean AUC for both drugs from the paper by Weiner [3] ± 1 standard deviation [SD]) for this group were essentially identical. This was recapitulated in the second independent experiment (top panel of Fig. 2B).
FIG 2 .
(A) First trial of moxifloxacin-plus-rifampin 7/7-day versus 5/7-day regimens. The AUC exposures in the symbol keys are the free AUC24 hs that were infused into that particular hollow fiber system experimental arm on the days that the drugs were administered. (B) Second trial of moxifloxacin-plus-rifampin 7/7-day versus 5/7-day regimens. The AUC exposures in the figure legends are the free AUC24 hs that were infused into that particular hollow fiber system experimental arm on the days that the drugs were administered. There is re-growth between day 23 and day 28, specifically in the lowest exposure group (arm E) in the experimental arm where the drugs were administered 5/7days. There was a 2.25 Log10 (CFU/ml) difference between arms B and E, attributable to difference in administration schedule and weekly drug exposures. In arm E colonies recovered had a wild-type MIC for rifampin (0.03 mg/L), but increased 4-fold for moxifloxacin (0.25 to 1.0 mg/L).
Importantly, in the groups treated 5/7 days with moxifloxacin and rifampin, there was a general trend for the cell kill to be less than that in the group treated 7/7 days, but the antimicrobial effect was not statistically different until after day 21. At this juncture, there was clear discordance among the regimens. The highest-exposure regimens performed nearly identically at both 5/7 days and 7/7 days (top and bottom panels of Fig. 2A for the first experiment and top and bottom panels of Fig. 2B for the second experiment). However, in the lowest-exposure groups of the two administration schedules, there was a 2.25- to 2.5-log10-CFU/ml difference between the 7/7-day treatment group and the 5/7-day group in both experimental trials (top versus bottom panels of Fig. 2A and B).
The ultimate difference, which largely explains the cell kill differences in the two schedules of administration, comes from the identification of emergence of resisitance in both performances of the experiment, solely in the low-exposure group in the 5/7-day administration schedule. The MIC for moxifloxacin increased from 0.25 mg/liter to 1.0 mg/liter, while the MIC for rifampin remained wild type.
Attainment of nominal concentration-time profiles.
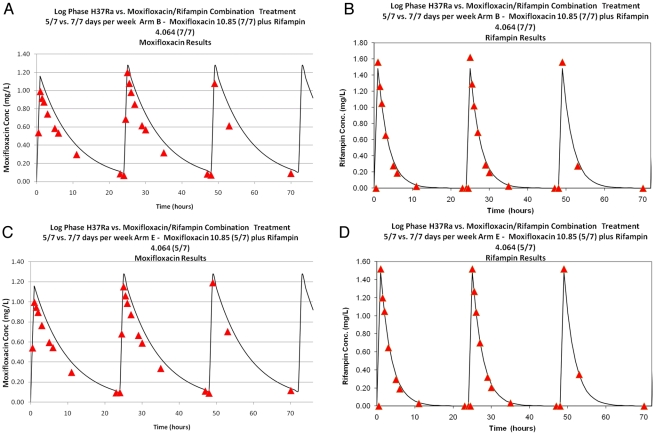
The simulated pharmacokinetic (PK) profiles for the first 72 h of the critical contrast arms (Fig. 3A, arm B—success versus arm E—failure) are displayed. The achieved drug concentrations for both agents were well matched between arms, indicating the difference in outcome is not attributable to difference in drug exposure, at least on a daily basis. For the other arms, the data indicate that the actual concentrations achieved were within approximately 10% of nominal. This was also true of the parallel experiment. For the lowest-exposure groups, the free AUC0-168 h values for moxifloxacin were 68.9 mg·h/liter for the 7/7-day schedule and 50.86 mg·h/liter for the 5/7-day group. For rifampin, the free AUC0-168 h values were 29.47 mg·h/liter and 22.12 mg·h/liter, respectively. On a per-day basis, the 5/7-day schedule group was marginally higher than the 7/7-day schedule group for both drugs (i.e., weekly exposure AUC0-168 h values divided by 7 and 5, respectively).
FIG 3 .
Moxifloxacin and rifampin concentrations (μg/ml) for the successful arm (A; arm B) versus failure arm (B; arm E). Solid curves are the targeted concentration-time profiles to be simulated in the hollow fiber systems. The red triangles are the measured concentrations of the respective drugs over the first 72 h of each experiment.
DISCUSSION
In the therapy of tuberculosis, maintenance of drug susceptibility is critical. Emergence of resistance has two main roots. The first is inadequate drug therapy. Getting the dose wrong may have consequences for rapid emergence of resistance, even in the combination chemotherapy setting. The second is regimen adherence. It may be argued that adherence is at least as important as drug regimen and perhaps may be a more important cause of emergence of resistance.
In order to minimize regimen nonadherence, directly observed therapy was instituted for use with treatment that includes the standard first-line antituberculosis drugs (rifampin, isoniazid, and pyrazinamide). Because of the practical difficulty of seeing each patient on 7/7 days, regimens have been designed with short drug holidays which, given the relatively long turnover half-time of M. tuberculosis (circa 20 h), should result in minimal regrowth over the standard gap of 2 days.
We felt that the combination of moxifloxacin plus rifampin might be a special case with regard to emergence of resistance, because the serum half-lives of the drugs that are active against M. tuberculosis in log-phase growth are mismatched. Weiner et al. (3) found that there was drug interaction in volunteers, such that at 2 weeks, the half-life of moxifloxacin of 10.4 ± 2.0 h when administered alone decreased to 6.5 ± 0.9 h when moxifloxacin was administered in combination with rifampin. Rifampin, studied only in combination, had a half-life of 1.9 ± 0.3 h. For a no-drug period of 48 h, rifampin will be essentially gone within the first 24 h (>12 terminal half-lives), while there will still be some moxifloxacin left (circa 3.7 terminal half-lives). O’Sullivan et al. (4) demonstrated that low concentrations of a fluoroquinolone caused rapid upregulation of some of the components of the SOS response in M. tuberculosis. Concentrations of ciprofloxacin of 1/4 and 1/2 the MIC resulted in upregulation of expression of lexA and recA within 4 h and dnaE2 within 24 h. Given the kinetics of moxifloxacin in the serum of patients, we hypothesized that the induction of error-prone replication would, after a number of rounds of replication, result in emergence of resistance.
We employed exposures at the mean of the free AUC24 h for moxifloxacin and rifampin (3) as well as the free AUC24 hs that were 1 SD above and below the mean exposure. Our previous publication examining this combination (5) demonstrated that, when administered on a daily basis, even small exposures to rifampin in combination with moxifloxacin still suppressed resistance.
We performed the experiment completely independently with two different teams at the same time. Examination of the top panels of Fig. 2A and B shows that, when administered 7/7 days, there was little evidence of an exposure response, and at day 28, there was no emergence of resistance.
In contrast, using the 5/7-day schedule, there was a divergence in the colony counts that became manifest after day 23 in both experiments, with a 2.25- to 2.5-log10-CFU/ml increase in colony counts when contrasting the lowest exposure arms of the 7/7- and 5/7-day experiments (arm B versus arm E in Fig. 2A and B). More importantly, the low-exposure arm (arm E) allowed emergence of resistance for moxifloxacin (4-fold increase in baseline MIC) in both experiments. It is important to note that in a normal distribution, 15.8% of the total population will have an AUC value at or below the value of the mean − 1 SD. This places a considerable fraction of the population at risk in this circumstance.
In conclusion, the results were able to be replicated, and in both instances, resistance emerged in the lowest-exposure arms, but not in the two higher-exposure arms when the drugs were administered 5/7 days. Administration of moxifloxacin and rifampin on a 7/7-day schedule suppressed resistance, even at the lowest exposure. This is concordant with our earlier publication (6). The emergence of resistance explained the increase in colony counts in the low-exposure arms in the 5/7-day administration schedule relative to the 7/7-day schedule (increase of 2.25 to 2.5 log10 CFU/ml in arm B versus arm E). The outcomes were not due to differences in achieved drug concentrations. Resistance occurred only to moxifloxacin.
When holiday schedules are designed to enhance regimen adherence, attention should be paid to whether or not error-prone replication is induced by one of the drugs, and there should be no discordance in the PK profiles so that both drugs are concurrently available to help suppress resistance. In the case of moxifloxacin plus rifamycin, it may be that rifapentine may be a better choice if a 5/7-day administration schedule is to be contemplated since rifapentine has a serum half-life in humans of 13 h (5) compared with a 1.9-h serum half-life for rifampin.
MATERIALS AND METHODS
Bacterial isolate.
M. tuberculosis H37Ra (ATCC 25177; American Type Culture Collection) was used in our studies. Bacterial cultures were stored at −80°C in Middlebrook 7H9 broth with 10% albumin-dextrose-catalase (ADC) and 0.025% Tween 80 (Becton Dickinson), and aliquots were thawed for study. The cultures were incubated in Middlebrook 7H9 broth with 10% ADC at 37°C with 5% CO2 for 4 days to achieve exponential-phase growth.
Antimicrobial agents.
Rifampin was obtained from Sigma-Aldrich, Inc. (St. Louis, MO). Stock solutions were prepared in dimethyl sulfoxide (DMSO), and aliquots were stored at –80°C. Moxifloxacin was purchased from CuraScript SD (Orlando, FL). Stock solutions of this antibiotic were prepared in sterile water, and aliquots were stored at –80°C. For each study, a sample of each drug was thawed, diluted to the desired concentration in sterile water or Middlebrook 7H9 broth with ADC, and used immediately. The concentration of DMSO that was infused into the hollow fiber systems with drug administration did not affect the growth of the bacterium.
MIC determination.
The MICs for moxifloxacin and rifampin for the M. tuberculosis isolate were determined by plating 10 µl of 1 × 106 CFU/ml of log-phase-growth bacterium onto Middlebrook 7H10 plus 10% oleic acid, albumen, dextrose, and catalase (OADC) agar containing geometric 2-fold dilutions of rifampin or moxifloxacin. Plates were incubated at 37°C with 5% CO2 in ambient air. The results were read after 21 days of incubation. The MIC was defined as the lowest concentration that allowed growth of M. tuberculosis of ≤1% compared with untreated controls.
Mutation frequency determinations.
M. tuberculosis H37Ra was grown to exponential-phase growth in 7H9 broth plus ADC, as described above. On the fourth day, the mutation frequency (MF) in the cultures was determined by plating 5 ml of the bacterium onto plates containing Middlebrook 7H10 plus OADC containing either 32× MIC of rifampin or 4× MIC of moxifloxacin. The mutation frequency was determined after 21 days of incubation at 37°C with 5% CO2. This determination was performed aerobically.
Hollow fiber infection model.
The hollow fiber infection model (HFIM) permits us to simulate the concentration-time profile for any dose of antibiotic (Fig. 1). A computer-controlled syringe pump delivers the antibiotic into a central reservoir in the required amount at the desired schedule of administration. Fresh medium is pumped into the system while drug-containing medium is isovolumetrically removed from the system at rates programmed by the investigator to simulate the desired drug half-life.
In these experiments, 15 ml of M. tuberculosis H37Ra was inoculated into the peripheral compartments of hollow fiber cartridges at a concentration of 1 × 106 to 5 × 106 CFU/ml. Using human PK parameters from Weiner et al. (3), the free-drug serum concentration-time profiles for moxifloxacin and rifampin were simulated. The free mean values of AUC of both agents were used, but also regimens with free AUC exposures that were 1 SD above and below the mean exposure were simulated. There was a no-treatment control. The experiment lasted for 28 days. The experiment was conducted in duplicate.
As combination therapy was employed with drugs with significantly different half-lives, the correct and quite different half-lives for rifampin and moxifloxacin were developed using the approach of Blaser (7).
Bacterial samples (400 µl) were taken from each hollow fiber cartridge and were washed twice with sterile saline to prevent drug carryover. The samples were sonicated, vortexed, and then quantitatively plated onto 7H10-OADC agars to identify the effect of each drug exposure and schedule of administration on the total M. tuberculosis populations. After the plates were incubated at 37°C in 5% CO2 for 21 days, the colonies were counted. Multiple colonies were picked and retested for both rifampin and moxifloxacin MIC values to look for emergence of resistance.
To document the concentration-time profiles, the central reservoir was sampled 22 times over 60 h for determination of both moxifloxacin and rifampin concentrations by liquid chromatography-tandem mass spectroscopy (LC-MS/MS).
Determination of moxifloxacin and rifampin concentrations.
Middlebrook 7H9-ADC medium PK simulation samples (0.050 ml) were added to autosampler vials containing 1.00 ml high-performance liquid chromatography (HPLC) water and 0.050 ml d3-rifampin internal standard. They were analyzed by high-pressure liquid chromatography-tandem mass spectrometry (LC-MS/MS) simultaneously for moxifloxacin and rifampin concentrations. The LC-MS/MS system was comprised of a Shimadzu Prominence HPLC system and an ABSciex API5000 LC-MS/MS.
Chromatographic separation was performed using a Thermo Scientific Hypersil gold C18 column (5 µm, 1,500 by 4.6 mm) employing a gradient using mobile phases of 0.1% formic acid in water and methanol at a flow rate of 1.00 ml/min.
Moxifloxacin and rifampin concentrations were obtained concurrently using LC-MS/MS monitoring the MS/MS transitions m/z 402 → m/z 384 for moxifloxacin, m/z 823 → m/z 791 for rifampin, and m/z 826 → m/z 794 for d3-rifampin. The analysis run time was 7.0 min.
The assay was linear over a range of 0.0050 to 5.00 µg/ml (r2 > 0.993) for both moxifloxacin and rifampin. The interday coefficients of variation (CVs) for the quality control samples, containing both moxifloxacin and rifampin, analyzed in replicates of four at three concentrations (0.0100, 0.100, and 1.00 µg/ml) on each analysis day ranged from to 4.94 to 8.82% for moxifloxacin and 5.91 to 9.66% for rifampin. Accuracies (% recovery) for the same quality control samples ranged between 97.4% and 107% for moxifloxacin and between 98.7 and 103% for rifampin.
Pharmacokinetic parameter identification and AUC calculation.
The pharmacokinetics of moxifloxacin and rifampin were estimated from the concentration-time profiles of both drugs using the ADAPT II package of programs of D’Argenio and Schumitzky (8). As the computer-controlled pumps were set to produce a single exponential decline, no model finding was performed and a 1-compartment model was fit to the data with maximum likelihood estimation.
ACKNOWLEDGMENTS
This work was supported by the Bill and Melinda Gates Foundation through their TB Drug Accelerator project.
The authors have no conflicts of interest to divulge.
Footnotes
Citation Drusano, GL, et al. 2011. Effect of administration of moxifloxacin plus rifampin against Mycobacterium tuberculosis for 7 of 7 days versus 5 of 7 days in an in vitro pharmacodynamic system. mBio 2(4):e00108-11. doi:10.1128/mBio.00108-11.
REFERENCES
- 1. Dorman SE, et al. 2009. Substitution of moxifloxacin for isoniazid during intensive phase treatment of pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 180:273–280 [DOI] [PubMed] [Google Scholar]
- 2. Conde MB, et al. 2009. Moxifloxacin versus ethambutol in the initial treatment of tuberculosis: a double-blind, randomised, controlled phase II trial. Lancet 373:1183–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weiner M, et al. 2007. Effects of rifampin and multidrug resistance gene polymorphism on concentrations of moxifloxacin. Antimicrob. Agents Chemother. 51:2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O’Sullivan DM, Hinds J, Butcher PD, Gillespie SH, McHugh TD. 2008. Mycobacterium tuberculosis DNA repair in response to subinhibitory concentrations of ciprofloxacin. J. Antimicrob. Chemother. 62:1199–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiner M, et al. 2004. Pharmacokinetics of rifapentine at 600, 900, and 1,200 mg during once-weekly tuberculosis therapy. Am. J. Respir. Crit. Care Med. 169:1191–1197 [DOI] [PubMed] [Google Scholar]
- 6. Drusano GL, et al. 2010. The combination of rifampin plus moxifloxacin is synergistic for resistance suppression, but is antagonistic for cell kill for Mycobacterium tuberculosis as determined in a hollow fiber infection model. mBio 1:e00139-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blaser J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J. Antimicrob. Chemother. 15(Suppl. A):125–130 [DOI] [PubMed] [Google Scholar]
- 8. D’Argenio DZ, Schumitzky A. 1997. ADAPT II. A program for simulation, identification, and optimal experimental design. User manual. Biomedical Simulations Resource, University of; Southern California, Los Angeles, CA: http://bmsr.usc.edu/ [Google Scholar]