Abstract
Cells, tissues and biological fluids contain a diverse repertoire of many tens of thousands of structurally distinct lipids that play multiple roles in cellular signaling, bioenergetics, and membrane structure and function. In an era where lipid-related disease states predominate, lipidomics has assumed a prominent role in Systems Biology through its unique ability to directly identify functional alterations in multiple lipid metabolic and signaling networks. The development of Shotgun Lipidomics has led to the facile accrual of high density information on alterations in the lipidome mediating physiologic cellular adaptation during health and pathologic alterations during disease. Through both targeted and non-targeted investigations, lipidomics has already revealed the chemical mechanisms underlying many lipid-related disease states.
The Multiple Roles of Lipids in Cellular Function
Lipids play multiple diverse roles in cellular function that must be effectively integrated into chemical and genetic networks to allow each cell to fulfill its specific biological function. By some recent estimates, cellular lipids encompass tens of thousands of structurally distinct compounds (van Meer, 2005; Shevchenko and Simons, 2010). On the most fundamental level, lipids are the main components of biological membranes where they serve to sequester, organize and distribute the molecular entities necessary for life processes. On a second level, lipids (e.g., fatty acids and triglycerides) are important fuel sources for many cell types. Lipids supply energy for cellular function through oxidation and facilitate metabolic flexibility through their ability to store chemical energy during times of caloric excess and harvest the energy stored in lipids (e.g., triglycerides) during energy depletion. Moreover, lipids play prominent roles in the regulation of cellular bioenergetics through integrating oxidative metabolic programs (Michalik et al., 2006), modulating systemic energy balance through eicosanoid and lysolipid production (Vegiopoulos et al., 2010; Skoura and Hla, 2009) and regulating mitochondrial electron transport chain flux and coupling efficiency (e.g., cardiolipin, fatty acids) (Zhang et al., 2002; Breen et al., 2005). On a third level, lipid membranes serve as molecular scaffolds that promote productive interactions among membrane-associated moieties that regulate cellular signaling to facilitate the transmission of biological information across cell membranes, between intracellular compartments or to other cells. Furthermore, the molecular dynamics and physical properties of membrane bilayers are critical determinants of the activities of transmembrane proteins such as ion channels and ion pumps (e.g., Schmidt and Mackinnon, 2008). Through the use of a diverse structural repertoire of lipids to regulate membrane surface charge, curvature and molecular dynamics, many biological functions of membranes can be fulfilled. Thus, the precise covalent nature of membrane molecular constituents within the lipid bilayers, including alterations in class, subclass and individual molecular species are important determinants of membrane structure that facilitate many specialized membrane-mediated cellular functions (Fig. 1). Finally, many classes of lipids serve as 2nd messengers of signal transduction (e.g., eicosanoids, lysolipids, phosphoinositides, endocannabinoids) that reside in biologic membranes in a latent state which can be activated either by hydrolysis and/or covalent transformation (e.g., Wolf and Gross 1985; Shaw and Cantley, 2006; Chiang et al., 2006; Simon and Cravatt, 2006). During evolution, adaptive alterations in membrane structure and function were selected by their integrated effects on cellular metabolism and signaling. This evolutionary process has led to the development of multiple membrane-delimited compartments (e.g., mitochondria, peroxisomes, endoplasmic reticulum, etc.) which perform integrated specialized processes that are essential for efficient cellular function and adaptation to external perturbations (Fig. 1).
Figure 1. The Pleiotropic Roles of Lipids in Cellular Function.
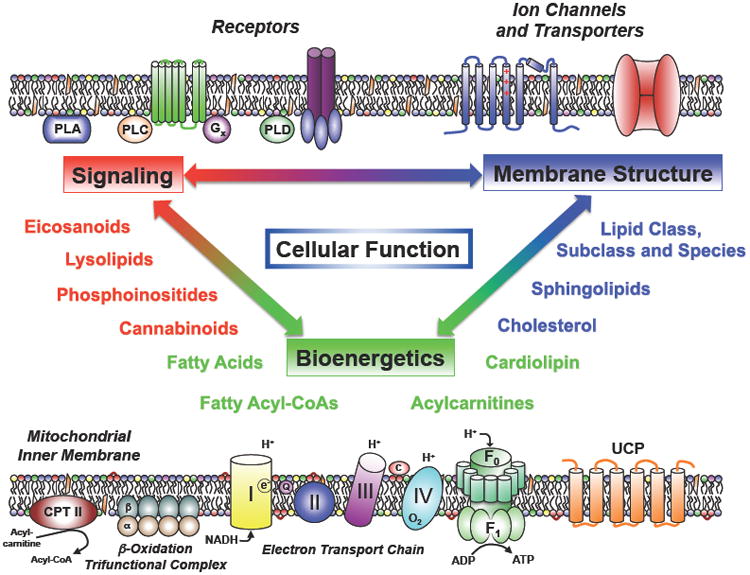
Lipids fulfill multiple roles in cellular function including cellular signaling (top left) through:1) harboring latent 2nd messengers of signal transduction that are released by phospholipases (PLA, PLC and PLD enzymes); 2) covalent transformation of membrane lipids into biologically active ligands by kinases (e.g., PI 3,4,5 triphosphate); 3) providing molecular scaffolds for the assembly of protein complexes mediating receptor/effector coupling (e.g., G-protein coupled receptors); and 4) coupling the vibrational, rotational and translational energies and dynamics of membrane lipids to transmembrane proteins such as ion channels and transporters (top right) thereby facilitating dynamic cooperative lipid–protein interactions that collectively regulate transmembrane protein function. Moreover, lipids play essential roles in mitochondrial cellular bioenergetics (bottom) through the use of fatty acids as substrates for mitochondrial β-oxidation (bottom left) that result in the production of reducing equivalents (e.g., NADH). The chemical energy in NADH is harvested through oxidative phosphorylation whose flux is tightly regulated by mitochondrial membrane constituents including cardiolipins which modulate electron transport chain (ETC) supercomplex formation. A second mechanism modulating mitochondrial energy production is the dissipation of the proton gradient by the transmembrane flip-flop of fatty acids in the mitochondrial inner membrane bilayer as well as the fatty acid-mediated regulation of uncoupling proteins (UCP).
The Growth of Lipidomics in Systems Biology
Similar to the Systems Biology fields of genomics and proteomics, the field of lipidomics begins with the identification and quantitation of lipids that collectively comprise the lipidome (i.e., the collection of lipid molecular species) in each cell, tissue or biologic fluid of interest. Through identification and quantitation of alterations in lipid molecular species, a high-density matrix containing fundamental information on the metabolic state, nutritional history and functional status of each cell type can be obtained. Moreover, the field of lipidomics encompasses the roles of specific membrane lipid constituents in mediating membrane domain formation (e.g., rafts), in facilitating interactions among spatially interwoven networks of signaling proteins (e.g., G-protein receptor-effector coupling), and in providing dynamic highly specialized molecular scaffolds for the construction of microscopic and macroscopic chemical assemblies necessary for life processes (e.g., Klose et al., 2010).
The large majority of diseases in industrialized societies in the 21st century, such as diabetes, obesity, atherosclerosis, myocardial infarction and stroke, to name a few, are lipid-related disorders (e.g., Unger, 2002; Unger et al., 2010; Dixon, 2010). Thus, lipidomics offers a direct and unique perspective into the pathologic alterations in cellular regulatory networks that promote lipid-mediated disease states. Lipidomics has now matured into a field that has already identified biomarkers predictive of disease states, alterations in the profiles of lipids that reflect disease severity and can be used to determine treatment efficacy (Nomura et al., 2010; Porter et al., 2010; Bartz et al., 2007). Thus, through the comprehensive analysis of alterations in lipid molecular species and their abundance, the field of lipidomics produces a high-density array of diagnostic, therapeutic and mechanistic information which has greatly impacted our understanding of the complex roles of lipids in health and disease (e.g., Nomura et al., 2010; Mancuso et al., 2009; Liu et al., 2010).
The Challenges of Early Lipid Analytical Techniques
Historically, progress in lipid research has been hindered by the difficulties inherent in the identification of lipid structure (e.g., the chemical nature and regiospecificity of aliphatic chains in each lipid class and subclass) and quantitation of individual molecular species. Over many decades multiple approaches have been applied to the separation, quantitation and characterization of lipids including thin layer chromatography, gas chromatography, NMR and HPLC in conjunction with a variety of complementary procedures including chemical hydrolysis, regiospecific enzymatic cleavage and spectrophotometric assays in attempts to determine the diversity of lipid structures in biological systems and quantitate their abundance. However, these strategies required multiple sequential steps each of which possessed limited sensitivity and accuracy that collectively resulted in the propagation of errors.
Although the utility of GC-MS for the analysis of volatile non-polar lipids has provided a robust platform for analysis of volatile lipids for many years, the overwhelming majority of cellular membrane constituents are non-volatile charged moieties that are not accessible by GC-MS methods. Thus, progress in lipidomics was largely dependent on the development of a mass spectrometric approach for the identification and quantitation of charged non-volatile lipids.
The Emergence of Sequential HPLC and Mass Spectrometry as a Principal Strategy for Lipidomic Analyses
The development of fast atom bombardment-mass spectrometry (FAB-MS) in the early 1980’s allowed the relatively soft ionization and desorption of intact non-volatile charged lipids from a variety of chemical matrices resulting in the identification of the m/z of the molecular ion. Using HPLC separations (either straight phase, reversed phase or multidimensional orthogonal separations (e.g., sequential SCX and RPHPLC)) followed by FAB-MS, the molecular ions corresponding to individual lipid molecular species in mammalian tissues could be identified (Gross, 1984). Thus, in conjunction with regiospecific enzymatic hydrolysis, the identification and quantitation of non-volatile lipid molecular species was possible. This approach demonstrated that canine myocardial sarcolemmal membranes are comprised predominantly of plasmalogen molecular constituents, that the major storage depot for arachidonic acid in myocardium was plasmalogen molecular species and that specialized intracellular membrane compartments were comprised of discrete sets of phospholipid classes, subclasses and individual molecular species (Gross, 1984; Gross, 1985). Utilization of HPLC followed by FAB-MS provided abundant information on the specialized lipid compositions present in diverse cell types, subcellular compartments and many organ systems including the human heart (Hazen et al., 1993). The use of HPLC followed by direct ionization of intact non-volatile lipids and analysis by mass spectrometry heralded the beginning of a new era in understanding the pleiotropic roles of lipids in cellular function.
Electrospray Ionization as an Enabling Technology for Deep Penetrance into the Lipidome
Over the last decade, several different strategies for analysis of cellular lipidomes have been developed. Here, we will first focus on electrospray ionization-mass spectrometry (ESI-MS) and its robust contribution to the development of the field and subsequently discuss other emerging methodologies later in the text. Currently, the most commonly employed ionization method for lipidomics is ESI-MS which was developed by the late John Fenn (Fenn et al., 1989) in Nobel Prize winning work. Electrospray ionization is effected by passage of a solution containing analytes through a narrow orifice at a high electric potential (typically ~4 kV) resulting in the formation of charged droplets in the ionization chamber. The use of ESI improved ionization efficiencies for non-volatile charged lipids by two to three orders of magnitude in comparison to FAB-MS methods (Han and Gross, 1994). The ionization efficiency of most lipids during the electrospray process is largely dependent on the charge density present in each lipid. Thus, the charge state and magnitude of the dipole in the lipid analyte are the predominant contributors to its interactions with the electric field in the ion source and thus represent the primary determinants of the ionization efficiency of each analyte. Accordingly, amongst molecular classes compromised of similar charged or zwitterionic lipids, the ionization efficiencies of individual molecular species of lipids are largely independent of the aliphatic chain length within each lipid class analyzed in the low-concentration regime. In cases where analytes of interest do not carry an inherent charge, ESI can be affected through adduction with other anions or cations in solution to induce charge separation to facilitate ionization. The use of ESI and mass spectrometric analysis of molecular ions directly from lipid extracts led to many new insights into the complexity of lipids in mammalian cell membranes (Han and Gross, 1994), the roles of phospholipase-mediated cleavage of arachidonoyl containing lipids in modulating the gating currents of voltage-dependent ion channels (Gubitosi-Klug et al., 1995), the presence of chain elongation of acylcarnitine molecular species during myocardial ischemia (Ford et al., 1996), the identification of α-oxidation in adipocytes (Su et al., 2004) and dynamic alterations in cardiolipin content and molecular species distribution that occur during diabetes and obesity that lead to mitochondrial dysfunction (Han et al., 2007).
The dramatic increase in sensitivity resulting from ESI for mass spectrometric analysis of lipids clearly represented an enabling development in the growth and development of the field of lipidomics. Moreover, the robust increase in signal to noise during ESI readily led to application of tandem mass spectrometric approaches for lipid analysis (Han and Gross, 1995) allowing discrimination of the multiple isobaric and isomeric molecular species present at each m/z value. The assignment of the aliphatic chain composition and the regiospecificity of aliphatic chains could be directly determined by ESI MS/MS through the differential fragmentation kinetics of aliphatic moieties present at the sn-1 and sn-2 positions (Han and Gross, 1996). Two basic approaches for ESI MS/MS have been developed, each with context dependent strengths and limitations. In the traditional approach, lipids are separated by HPLC and directly sprayed into the ESI ion source for mass spectrometric analysis by molecular ion identification in conjunction with product ion analysis, selected reaction monitoring (SRM) or other fragmentation strategies. In a second approach, termed Shotgun Lipidomics, direct infusion of organic extracts of tissues, cells and biologic fluids into the mass spectrometer is performed. Direct infusion of organic extracts allows marked increases in the signal to noise ratio of molecular ions corresponding to individual molecular species. Concurrently, direct infusion facilitates the utilization of a wide variety of informative fragmentation strategies that are not limited by the transient elution of individual lipid molecular species during column chromatography.
Multi-Dimensional Mass Spectrometry-based Shotgun Lipidomics
The large majority of cellular lipids are comprised of linear combinations of aliphatic chains and polar head groups that are covalently linked to a glycerol or sphingosine backbone. Accordingly, determination of the head group structure as well as the nature and regiospecificity of the aliphatic chains represents a formal structural identification of the overwhelming majority of lipids. To exploit the limited degrees of freedom in lipid structure, in conjunction with chemically predictable fragmentation patterns during tandem mass spectrometry, we developed Multi-Dimensional Mass Spectrometry-based Shotgun Lipidomics (MDMS-SL) (Han and Gross, 2005) that has served as a critical technology to understand the multiple roles of lipids in biologic function. MDMS-SL is comprised of four component parts including: 1) multiplexed extractions and chemistries; 2) intrasource separation of lipids based upon their intrinsic electrical propensities; 3) multidimensional mass spectrometry; and 4) bioinformatic array analysis. MDMS-SL begins with a multiplexed series of extractions and chemistries that are optimized to collectively result in the integrated analysis of over 30 lipid classes leading to the identification and quantitation of many hundreds to thousands of lipid molecular species directly from lipid extracts. Through alterations in the pH of the infusate solution, the highly selective ionization of groups of specific lipid classes based on their intrinsic electrical properties could be effectively achieved within the ion source (Han et al., 2004). This process, now known as intrasource separation, results in the resolution of lipid constituents in the ion source, thereby effectively replacing the need for chromatography prior to mass spectrometric analysis. Through construction of multidimensional arrays of full mass scans of molecular ions with precursor ion scanning or neutral loss scanning of aliphatic chains, polar head groups and other informative product ions, a comprehensive description of the diversity of lipid structures within biologic samples can be defined directly from their organic extracts. For example, if neutral loss scanning of aliphatic chains present in triglycerides for all major biologically occurring aliphatic chains is performed, then extending a perpendicular line from any m/z value of the full mass scan identifies the nature and relative abundance of fragment ions at each cross peak (Han and Gross, 2001) (Fig. 2). Bioinformatic array analysis of these cross peaks can then be used to identify the structure of each individual isobaric lipid molecular species present at each m/z value in the full mass scan. In this two dimensional array, the first dimension is comprised of the molecular ions (shown on the x-axis), while the second dimension corresponds to the abundance of individual aliphatic chains at each m/z of the molecular ion during neutral loss scanning of aliphatic chains. These two-dimensional mass spectrometric arrays have units of mass in each dimension and thus are analogous in both concept and principle to 2D-NMR plots which derive structural information from through-space or through-bond connectivities present in the targeted molecule, but have units of frequency on each axis. Thus, these two-dimensional maps of lipid structure have been referred to as two-dimensional mass spectrometry (2D-MS) since they are entirely analogous to 2D-NMR spectroscopy. Moreover, additional dimensions can be accessed through precursor ion or neutral loss scanning of multiple sets of informative fragment ions (e.g., polar head groups) thereby providing a high density matrix of structural information in n-dimensional space. These multidimensional spectra are ideally suited to bioinformatic array analysis allowing the comprehensive identification and quantitation of many hundreds to thousands of lipid molecular species. This strategy is also well-suited for quantitation of individual molecular species through utilization of ratiometric comparisons using a two-step approach. This two-step procedure optimally uses the advantages of both exogenously added internal standards as well as endogenous lipid constituents as standards for tandem mass spectrometry by using molecular ions of endogenous molecular species whose mass content is known with high precision through their intensities in the full mass scan. Through this tandem mass spectrometric approach, the dynamic range of quantitation is extended by over two orders of magnitude and is accomplished with greater precision through exploiting multiple additional comparisons using endogenous molecular species as standards. Finally, we point out the utility of alterations in the ionization conditions, ramping of collision energies, examination of fragmentation kinetics and n-dimensional scanning of informative fragment ions that each provide additional dimensions to substantiate structural identification and quantitation of the abundance of identified lipid molecular species.
Figure 2. Electrospray Ionization Mass Spectrometric Analysis of Extracts of Murine White Adipose Tissue by MDMS-SL.
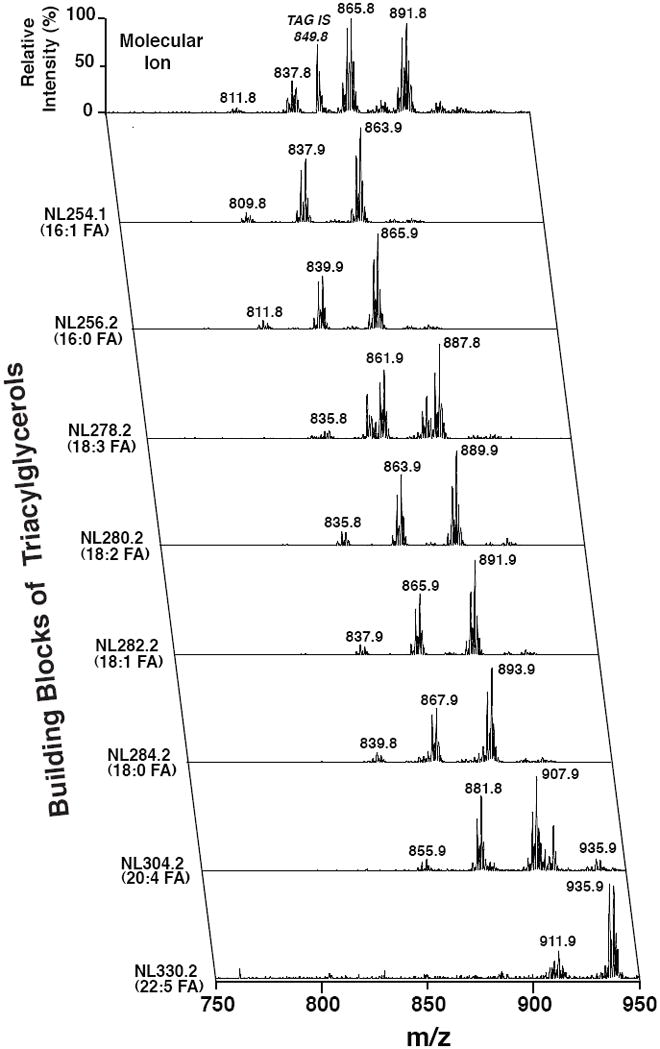
Bligh and Dyer extracts of white adipose tissue from mice fed a high fat diet were prepared as previously described (Han and Gross, 2005) and directly infused into the ESI ion source. Positive-ion ESI mass spectra identified multiple molecular ions in the full mass scan (top row; x-axis) that were quantitated through ratiometric comparisons with internal standard (tri 17:1 triacylglycerol, m/z 849.8) after corrections for isotope abundance and acyl chain length and unsaturation effects. Tandem mass spectrometric analysis was performed using neutral loss scanning for the indicated naturally occurring aliphatic chains. All scans were normalized to the base peak of the individual spectrum. Through bioinformatic analysis of the ion counts in each row (x-axis scan) and column (y-axis scans), the compositional identities of each individual molecular species can be determined and their relative abundance can be quantified.
Advantages and Limitations of Multi-Dimensional Mass Spectrometry-based Shotgun Lipidomics
The obvious advantage of MDMS-SL is the direct acquisition of high-density information on lipid structure and abundance directly from biological extracts using the mass spectrometer as a separations device thereby avoiding the need for chromatography. A second advantage of MDMS-SL is through direct infusion large increases in S/N can be achieved in comparison to chromatographic approaches where limited amounts of time for signal acquisition and tandem mass spectrometric analyses are available. A third benefit of MDMS-SL is that it does not require previous knowledge of the compound(s) present in the mixture to identify transitions, but rather interrogates all mass space that contains naturally occurring aliphatic chains, head groups and signature fragmentation ions through precursor ion and neutral loss scanning, thereby allowing for the identification of previously unknown compounds. A fourth use of MDMS-SL is the benefit from analysis of peak contours in multidimensional space that lead to multiple bioinformatic advantages that facilitate refinements in quantitation through corrections for peak shape, mass offset and isotopologue analysis. A fifth advantage of MDMS-SL is that it is ideally suited to exploit the distinctive chemical characteristics of many lipid classes. Prominent examples include the use of the M+1/2 isotopologue approach for cardiolipin analyses (Han et al., 2006), the use of Fmoc derivatization for the analysis of ethanolamine containing constituents (Glaser and Gross, 1995), the use of specifically deuterated amine selective reagents for ratiometric quantitation through precursor ion scanning (Zemski Berry et al., 2009), alkaline hydrolysis to greatly enhance penetrance into the sphingolipidome (Jiang et al., 2007) and charge-switch derivatization methods to greatly increase the sensitivity for eicosanoid analyses (e.g., Bollinger et al., 2010) (Fig. 3).
Figure 3. Enhanced Shotgun Lipidomics Approaches.
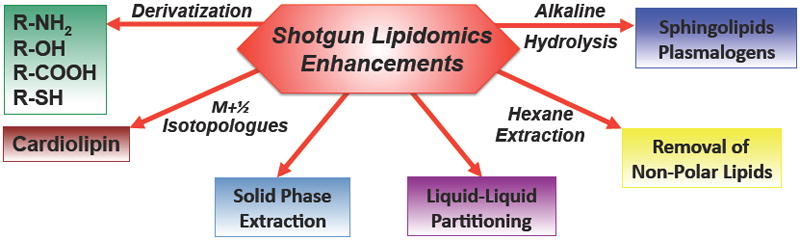
The analytic power of shotgun lipidomics can be extended through exploiting the unique chemical characteristics of specific lipid classes. These include the derivatization of nucleophilic lipid moieties to increase signal intensity and/or engender a mass shift to facilitate mass spectrometric analyses, the use of a M+1/2 isotopologue approach for doubly negatively charged cardiolipins, multiplexed extractions including liquid/liquid partitioning, liquid/solid partitioning (solid phase extraction) and/or alkaline hydrolysis to enrich for sphingolipids or ether lipids. In some tissues (e.g., adipose tissue), the removal of non-polar lipids by hexane extraction is judicious prior to the subsequent analysis of polar lipid constituents.
One limitation of MDMS-based SL is the limits of detection and quantitation that are feasible for low abundance species. Currently, the lower limits of detection for lipids in favorable cases using MDMS-SL are in the high attomole to low femtomole level. Although the MDMS-SL approach takes advantage of the 4 orders of magnitude dynamic range inherent in mass spectrometry, we explicitly point out that enrichment approaches are necessary for examination of extremely low abundance lipids which Shotgun Lipidomics is unable to access with the sensitivities of currently available mass spectrometers. Chromatographic separation, liquid-liquid partitioning, solid phase extraction and other enrichment methods can greatly extend the limits of detection for extremely low abundance molecular species. In our hands, chromatographic enrichment of many different classes of low abundance lipids is most effectively accomplished by orthogonal chromatographies employing an initial straight phase (or SCX) separation that resolves lipid classes followed by RPHPLC to affect the resolution of lipid subclasses and individual molecular species within each class. Even with orthogonal chromatographies, multiple molecular species are typically present at each m/z value that require analysis by tandem mass spectrometric approaches. A second limitation of MDMS-SL is the difficulty in the localization and the stereochemistry of the olefinic linkages in aliphatic chains which can be accessed through multistage fragmentation. A third limitation of MDMS-SL is the inability to discriminate enantiomeric lipids. Through chiral chromatography Blair et al. resolved eicosanoid enantiomers for direct mass spectrometric analysis (Lee and Blair, 2009). Whether direct infusion, chromatographic enrichment, partitioning or derivatization is used, many of the strategic advantages of multidimensional mass spectrometry can be readily accessed.
The Future Challenges of Lipidomics
The future challenges of lipidomics include the development of chemistries and technologies to increase the penetrance into the lipidome, identification of the subcellular compartments or domains where alterations are manifest during cellular adaptation or disease, and the development of bioinformatic approaches to define changes in lipid metabolic networks that can predict the onset of disease, identify disease progression and evaluate treatment efficacy. The development of increasingly sophisticated databases that contain the exact mass, fragmentation patterns and HPLC elution characteristics of each lipid molecular species is essential for the continued development of lipidomics and in its application to biomedical research. Currently there are three main databases that are routinely utilized for lipidomics research including METLIN (Smith et al., 2005), The Human Metabolome Database (Wishart et al., 2008), and LIPID MAPS (Sud et al., 2007). Through the use of ESI in conjunction with time of flight mass spectrometry, Shevchenko et al. used a Shotgun Lipidomics strategy in conjunction with custom software to identify over 250 lipids from yeast (Ejsing et al., 2009). Dennis et al identified over 400 lipids in RAW 267.2 cells using the LIPID MAPS database and demonstrated alterations in multiple lipid classes and molecular species after cellular stimulation (Dennis et al., 2010). Siuzdak et al. identified over 150 lipids in stem cells during differentiation and found increases in the unsaturation index of aliphatic chains that alter membrane structure and dynamics during differentiation (Yanes et al., 2010). Collectively, these studies demonstrate the dynamic role of alterations in lipid molecular species content and membrane composition that serve to facilitate cellular differentiation and cellular activation. Through orchestrating changes in membrane lipid composition, molecular structure and membrane dynamics the execution of intrinsic biologic programs (e.g., differentiation) or responses to external perturbations necessary for life processes are facilitated.
Many other methods for ionization of lipids for mass spectrometry have been used with great success. In pioneering work, Marshall et al. demonstrated the utility of matrix assisted laser desorption/ionization (MALDI) for lipid analysis in conjunction with high mass accuracy using FT-ICR (Marto et al., 1995). The use of laser-based methods have the obvious advantage of allowing high throughput analyses with rapid repetition rate lasers that are unencumbered by the time constraints normally encountered using fluid-based methods for mass spectrometry. The use of MALDI/TOF-TOF mass spectrometry for lipidomic analysis of biologic tissues has recently been further developed for high throughput lipidomics where many dozens of samples can be directly analyzed within minutes facilitating the large scale application of lipidomics to human disease through this high throughput approach (Sun, et al., 2008). Moreover, laser or beam ionization methods provide a platform for innovative chemical strategies for matrix construction that can be exploited for selective ionization (e.g., the selective ionization/desorption of discrete lipid classes) or can be used for non-biased identification and quantitation of analytes. In elegant work, Siuzdak et al. engineered nanostructured surfaces to trap fluorinated siloxanes that desorb analytes upon laser-induced vaporization (e.g., Northen et al., 2007). Through multiplexing different surface-based technologies, doping with selected cavitands and the construction of engineered charge transfer complexes to facilitate class-specific ionization, the future growth of MALDI TOF/TOF mass spectrometry is assured. Moreover, recent success with MALDI imaging (Reyzer and Caprioli, 2005), desorption ESI (DESI) imaging (Takats et al., 2004) techniques, and nanostructured surfaces (Patti et al., 2010) has led to new insights into the spatial distribution of lipids in tissues, but at present these methods are limited by constraints on spatial resolution. A prominent issue in lipidomics is the identification of methods to delineate alterations in membrane composition in discrete subcellular loci or within specific membrane domains. Thus, the development of methods which precisely localize changes in specific compartments or domains in intact cells is a fundamental goal of future lipidomics research. The use of secondary ion mass spectrometry using ion beams has greatly enhanced spatial resolution to the order of 100 nm that can identify lipids in specific membrane domains (Winograd and Garrison, 2010). Additional technologic advances are anticipated in these fields that will define spatially privileged domains within cell membranes and subcellular compartments, identify unknown lipids and greatly facilitate our understanding of membrane structure and function.
Nuclear magnetic resonance (NMR) spectroscopy has been used to identify a wide variety of lipids in biological systems (e.g., Lindon and Nicholson, 2008 and references therein). Mountford and colleagues used 1H NMR to distinguish invasive breast cancer from benign lesions on the basis of increased total choline phospholipid species (Mackinnon et al., 1997). While NMR is a nondestructive and nonselective technique that provides unique information pertaining to molecular structure and dynamics, its modest sensitivity and resolving power to distinguish individual chemical species in comparison to mass spectrometry has limited utilization of NMR in lipidomics investigations. NMR interpretation is also complicated by the considerable number of spin-coupled multiplets that result in spectral crowding. Recently this problem has been addressed by using two-dimensional J-resolved NMR spectroscopy to visualize metabolite chemical shifts and J-couplings along different spectral dimensions and thereby increase peak dispersion (Ludwig and Viant, 2010). An advantage of magnetic resonance approaches is the ability to analyze lipids non-invasively in intact cells and tissues without losing chemical information about the analyte environment (Beckonert et al., 2010). A review of the methods used in high-resolution magic-angle spinning NMR, their application to lipidomics and their potential biomedical uses have been discussed recently (Fonville et al., 2010). Moreover, selective recoupling of dipolar and chemical-shift interactions removed by magic-angle spinning in the solid state allows for the characterization of regulatory interactions, dynamics, and ion channels within biological membranes (Guillion and Schaefer, 1989; deAzevedo et al., 1999; Kim et al., 2009; and Cady et al., 2010). Since NMR is a non-destructive technique it can be used to interrogate alterations in lipid structure and dynamics in biochemically functioning cells. Thus, it is anticipated that future developments will provide critical information on alterations in membrane structure and function through the synergistic application of a variety of solution state and solid state NMR approaches.
Summary
The past decades have witnessed the rapid growth and development of lipidomics, which has now become an essential part of Systems Biology. MultiDimensional Mass Spectrometry-based Shotgun Lipidomics has now been developed into a robust technology that can be used to provide rapid access to the mechanisms underlying diverse disease states. Continuing advances in ionization technologies, use of high mass accuracy lipidomics, novel fragmentation strategies, improvements in enrichment approaches and enhanced spatial resolution in conjunction with NMR-based approaches will collectively lead to the development of a comprehensive understanding of the pleiotropic roles of lipids in cellular function.
Acknowledgments
This work was supported by the National Institutes of Health Grants PO1HL57278 (RWG), RO1HL41250 (RWG) and RO1-AG31675 (XH).
References
- Bartz R, Li W-H, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Beckonert O, Coe M, Keun HC, Wang Y, Ebbels TMD, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5:1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- Bollinger JG, Thompson W, Lai Y, Oslund RC, Hallstrand TS, Sadilek M, Turecek F, Gelb MH. Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Anal Chem. 2010;82:6790–6796. doi: 10.1021/ac100720p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen EP, Gouin SG, Murphy AF, Haines LR, Jackson AM, Pearson TW, Murphy PV, Porter RK. On the mechanism of mitochondrial uncoupling protein 1 function. J Biol Chem. 2005;281:2114–2119. doi: 10.1074/jbc.M511575200. [DOI] [PubMed] [Google Scholar]
- Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–693. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- de Azevedo ER, Hu W-G, Bonagamba TJ, Schmidt-Rohr K. Centerband-only detection of exchange: Efficient analysis of dynamics in solids by NMR. J Am Chem Soc. 1999;11:8411–8412. [Google Scholar]
- Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, Merrill AH, Jr, Sullards MC, Wang E, Murphy RC, Raetz CRH, Garrett TA, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy e, Subramaniam S. A mouse macrophage lipidome. J Biol Chem. 2010;285:39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JB. The effect of obesity on health outcomes. Mol Cell Endocrinol. 2010;316:104–108. doi: 10.1016/j.mce.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Nat Acad Sci USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Fonville JM, Maher AD, Coen M, Holmes E, Lindon JC, Nicholson JK. Evaluation of full-resolution J-resolved 1H NMR projections of biofluids for metabonomics information retrieval and biomarker identification. Anal Chem. 2010;82:1811–1821. doi: 10.1021/ac902443k. [DOI] [PubMed] [Google Scholar]
- Ford DA, Han X, Horner CC, Gross RW. Accumulation of unsaturated acylcarnitine molecular species during acute myocardial ischemia: metabolic compartmentalization of products of fatty acyl chain elongation in the acylcarnitine pool. Biochemistry. 1996;35:7903–7909. doi: 10.1021/bi960552n. [DOI] [PubMed] [Google Scholar]
- Glaser PE, Gross RW. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase: Discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry. 1995;34:12193–12203. doi: 10.1021/bi00038a013. [DOI] [PubMed] [Google Scholar]
- Gross RW. High plasmalogen and arachidonic acid content of canine myocardial sarcolemma: A fast atom bombardment mass spectroscopic and gas chromatography-mass spectroscopic characterization. Biochemistry. 1984;23:158–165. doi: 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- Gross RW. Identification of plasmalogen as the major phospholipid constituent of cardiac sarcoplasmic reticulum. Biochemistry. 1985;24:1662–1668. doi: 10.1021/bi00328a014. [DOI] [PubMed] [Google Scholar]
- Gubitosi-Klug R, Yu SP, Choi DW, Gross RW. Concomitant acceleration of the activation and inactivation kinetics of the human delayed rectifier K+ channel (Kv1.1) by Ca2+-independent phospholipase A2. J Biol Chem. 1995;270:2885–2888. doi: 10.1074/jbc.270.7.2885. [DOI] [PubMed] [Google Scholar]
- Guillion T, Schaefer J. Rotational-Echo Double-Resonance NMR. J Magn Reson. 1989;81:196. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Natl Acad Sci USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Gross RW. Structural determination of picomole amounts of phospholipids utilizing electrospray ionization tandem mass spectrometry. J Am Soc Mass Spec. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Structural determination of lysophospholipid regioisomers by electrospray ionization tandem mass spectroscopy. J Am Chem Soc. 1996;118:451–457. [Google Scholar]
- Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun Lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spec Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: A shotgun lipidomics study. Biochemistry. 2007;46:6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SL, Hall CR, Ford DA, Gross RW. Isolation of a human myocardial cytosolic phospholipase A2 isoform. Fast atom bombardment mass spectroscopic and reverse-phase high pressure liquid chromatography identification of choline and ethanolamine glycerophospholipid substrates. J Clin Invest. 1993;91:2513–2522. doi: 10.1172/JCI116487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Cheng H, Yang K, Gross RW, Han X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low abundance regime of cellular sphingolipids. Anal Biochem. 2007;371:135–145. doi: 10.1016/j.ab.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Singh M, Schaefer J. Oritavancin binds to isolated protoplast membranes but not intact protoplasts of staphylococcus aureus. J Mol Biol. 2009;391:414–425. doi: 10.1016/j.jmb.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Ejsing CS, Garcia-Saez AJ, Kalser HJ, Surma MA, Shevchenko A, Schwille P, Simons K. Yeast lipids can phase-separate into micrometer-scale membrane domains. J Biol Chem. 2010;285:30224–30232. doi: 10.1074/jbc.M110.123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Blair IA. Targeted chiral lipidomics analysis of bioactive eicosanoid lipids in cellular systems. BMB Reports. 2009;42:401–410. doi: 10.5483/bmbrep.2009.42.7.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon JC, Nicholson JK. Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Ann Rev Anal Chem. 2008;1:45–69. doi: 10.1146/annurev.anchem.1.031207.113026. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen Y, Momin A, Shaner R, Wang E, Bowen NJ, Matyunina LV, DeEtte Walker L, McDonald JF, Sullards MC, Merrill AH., Jr Elevation of sulfatides in ovarian cancer: An integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186–199. doi: 10.1186/1476-4598-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C, Viant MR. Two-dimensional J-resolved NMR spectroscopy: Review of a key methodology in the metabolomics toolbox. Phytochem Anal. 2010;21:22–32. doi: 10.1002/pca.1186. [DOI] [PubMed] [Google Scholar]
- Mackinnon WB, Barry PA, Malycha PL, Gillett DJ, Russell P, Lean CL, Doran ST, Barraclough BH, Bilous M, Mountford CE. Fine-needle biopsy specimens of benign breast lesions distinguished from invasive cancer ex vivo with proton MR spectroscopy. Radiology. 1997;204:661–666. doi: 10.1148/radiology.204.3.9280241. [DOI] [PubMed] [Google Scholar]
- Mancuso DJ, Kotzbauer P, Wozniak DF, Sims HF, Jenkins CM, Guan S, Han X, Yang K, Sun G, Malik I, Conyers S, Green KG, Schmidt RE, Gross RW. Genetic ablation of calcium-independent phospholipase A2γ leads to alterations in hippocampal cardiolipin content and molecular species distribution, mitochondrial degeneration, autophagy, and cognitive dysfunction. J Biol Chem. 2009;284:35632–35644. doi: 10.1074/jbc.M109.055194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marto JA, White FM, Seldomridge S, Marshall AG. Structural characterization of phospholipids by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 1995;67:3978–3984. doi: 10.1021/ac00117a025. [DOI] [PubMed] [Google Scholar]
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O’Rahilly S, Palmer CN, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W. International Union of Pharnacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng S-W, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Suizdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Patti GJ, Shriver LP, Wassif CA, Woo HK, Uritboonthal W, Apon J, Manchester M, Porter FD, Siuzdak G. Nanostructure-initiator mass spectrometry (NIMS) imaging of brain cholesterol metabolites in Smith-Lemli-Opitz syndrome. Neuroscience. 2010 doi: 10.1016/j.neuroscience.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD, Scherrer DE, Lanier MH, Langmade SJ, Molugu V, Gale SE, Olzeski D, Sidhu R, Dietzen DJ, Fu R, Wassif CA, Yanjanin NM, Marso SP, House J, Vite C, Schaffer JE, Ory DS. Cholesterol oxidation products are sensitive and specific blood-based biomarkers for Niemann-Pick C1 disease. Sci Transl Med. 2010;2:56ra81. doi: 10.1126/scitranslmed.3001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyzer ML, Caprioli RM. MALDI mass spectrometry for direct tissue analysis: a new tool for biomarker discovery. J Proteome Res. 2005;4:1138–1142. doi: 10.1021/pr050095+. [DOI] [PubMed] [Google Scholar]
- Schmidt D, MacKinnon R. Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc Natl Acad Sci USA. 2008;105:19276–19281. doi: 10.1073/pnas.0810187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JS, Cantley LC. Ras, PI(3)K and m TOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for α/β-hydrolase 4 in this pathway. J Biol Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- Skoura A, Hla T. Lysophospholipid receptors in vertebrate development, physiology, and pathology. J Lipid Res. 2009;50:S293–S298. doi: 10.1194/jlr.R800047-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: A metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. http://metlin.scripps.edu. [DOI] [PubMed]
- Su X, Han X, Yang J, Mancuso DJ, Chen J, Bickel PE, Gross RW. Sequential ordered fatty acid α oxidation and Δ9 desaturation are major determinants of lipid storage and utilization in differentiating adipocytes. Biochemistry. 2004;43:5033–5044. doi: 10.1021/bi035867z. [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CRH, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. http://www.lipidmaps.org/ [DOI] [PMC free article] [PubMed]
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal Chem. 2008;80:7576–7585. doi: 10.1021/ac801200w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Unger RH. Lipotoxic diseases. Annu Rev Med. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- Unger RH, Clark GO, Scherer PE, Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim Biophys Acta. 2010;1801:209–214. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegiopoulos A, Muller-Decker K, Strzoda D, Schmitt I, Chichelnitskiy E, Ostertag A, Berriel Diaz M, Rozman J, Hrabe de Angelis M, Nusing RM, Meyer CW, Wahli W, Klingenspor M, Herzig S. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science. 2010;328:1158–1161. doi: 10.1126/science.1186034. [DOI] [PubMed] [Google Scholar]
- Winograd N, Garrison BJ. Biological cluster mass spectrometry. Annu Rev Phys Chem. 2010;61:305–322. doi: 10.1146/annurev.physchem.040808.090249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2008;37:D603–D610. doi: 10.1093/nar/gkn810. http://www.hmdb.ca/ [DOI] [PMC free article] [PubMed]
- Wolf RA, Gross RW. Identification of neutral active phospholipase C which hydrolyzes choline glycerophospholipids and plasmalogen selective phospholipase A2 in canine myocardium. J Biol Chem. 1985;260:7295–7303. [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemski Berry KA, Turner WW, VanNieuwenhze MS, Murphy RC. Stable isotope labeled 4-(Dimethylamino)benzoic acid derivatives of glycerophosphoethanolamine lipids. Anal Chem. 2009;81:6633–6640. doi: 10.1021/ac900583a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]


