Abstract
We have identified conserved orthologs in completely sequenced genomes of double-strand DNA phages and arranged them into evolutionary families (phage orthologous groups [POGs]). Using this resource to analyze the collection of known phage genomes, we find that most orthologs are unique in their genomes (having no diverged duplicates [paralogs]), and while many proteins contain multiple domains, the evolutionary recombination of these domains does not appear to be a major factor in evolution of these orthologous families. The number of POGs has been rapidly increasing over the past decade, the percentage of genes in phage genomes that have orthologs in other phages has also been increasing, and the percentage of unknown “ORFans” is decreasing as more proteins find homologs and establish a family. Other properties of phage genomes have remained relatively stable over time, most notably the high fraction of genes that are never or only rarely observed in their cellular hosts. This suggests that despite the renowned ability of phages to transduce cellular genes, these cellular “hitchhiker” genes do not dominate the phage genomic landscape, and a large fraction of the genes in phage genomes maintain an evolutionary trajectory that is distinct from that of the host genes.
INTRODUCTION
In recent years, it has become clear that virus particles are strikingly abundant in the biosphere, with 106 to 109 virus-like particles per milliliter of seawater being reported (7), similar estimated numbers in terrestrial populations (6), and possibly even higher abundances in some freshwater environments (63). This makes viruses of bacteria, archaea, and eukaryotes the most abundant organisms (and their genomes the most prevalent hereditary material) in the sea and, most likely, in the entire biosphere (27). Far from being mere parasites whose effect could be simplistically viewed as merely to reduce the host fitness, viruses are active participants in these ecosystems (11, 47, 52), playing crucial roles in host genome rearrangements (29, 58, 60), population dynamics (5, 12, 59), and geochemical and ecological processes (1, 23). Estimates of the frequency of encounters between bacteria and phages, whose particle count is about an order of magnitude more than the number of bacterial cells, are on the order of 1023 infections per second globally (52). Recent advances in sequencing technologies, leading to reporting of extensive metagenomics data (17, 19) and revealing breathtaking viral diversity (10, 24, 53), have enabled us to quantify the occurrence of viruses in the environment and to understand better these global roles (21). Sequence analysis of metagenomic samples reveals that, in addition to the genes that have homologs already sequenced and deposited into the databases, the substantial majority of genes packaged inside virus (or virus-like) capsids do not appear to be significantly similar to any known sequences (15, 17, 32, 64).
Most viruses detected in the environment are head-tail phages that infect prokaryotes (mostly bacteria, though archaeophages are being actively studied, and there is no reason to believe that they will be less diverse than bacteriophages [14, 44, 48]). There are almost 600 fully sequenced genomes of phages in sequence databases as of this writing. Phage genetic material may be represented by RNA or DNA, in single- or double-stranded form, and differ in size by 2 orders of magnitude (from <5 kb to >300 kb). Of these fully sequenced phages, 85% have a double-stranded DNA (dsDNA) genome, and it is this group of phages that is examined here. The number of protein-coding genes in dsDNA phages varies, from 4 in the circular DNA of Leuconostoc phage L5 to ∼400 in the linear genomes of Vibrio phage KVP40 and Pseudomonas phage 201phi2-1 (Bacillus phage G, with nearly 700 protein-coding genes [more than several small bacterial genomes have] appears to be the largest known bacteriophage thus far [43], though a fully sequenced and annotated genome is not yet publicly available).
The high diversity of dsDNA phage genomes, as well as fast evolution of their sequences (18, 42) and the genomic mosaicism (24), complicates the study of their evolutionary history, especially since the omnipresent phylogenetic molecular markers available for cellular organisms (such as rRNA and dozens of ubiquitous proteins [13, 45, 62]) are not found in phages; in fact, not a single gene is shared by all phages (39, 46), and even among phages that infect a common bacterium, the number of shared genes may be low (25).
Analysis of the extent of shared orthologs (i.e., homologs related by speciation rather than duplication) has many uses; it has been applied to studies of phage taxonomy and evolution (22, 34, 38, 46), allowing reconstruction of phage phylogeny (30, 45, 61). The identification of orthologous genes and their distribution in the genomes is a foundation of almost every comparative genomics study, be it in a viral or cellular context: orthologous groups (evolutionary families) provide information about sequence conservation, the tempo and mode of molecular evolution, and gene gain and loss, and they constitute a “parts list” for system-wide biological modeling. As a first step toward better understanding the evolutionary history and the repertoire of molecular functions in dsDNA phages, we have proposed the phage orthologous groups (POGs), a natural system of viral protein families that initially included 6,378 genes from 164 completely sequenced dsDNA bacteriophage genomes (39). Compared to that study, which included only genomes sequenced prior to 2004, the numbers of dsDNA phage genomes and proteins available in the NCBI database have both increased 3-fold and are expected to continue to increase at an even higher rate in the future, especially with the advent of assembled environmental sequences (2, 24, 36, 57).
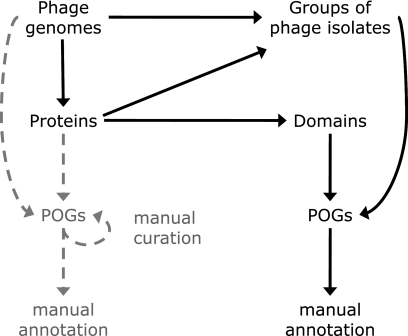
In this paper, we present an expanded POG framework (Fig. 1) that takes advantage of several computational improvements. Using this framework, we created two new sets of POGs (“annotated POGs-07” and “extended POGs-10”), which allow us to discern several evolutionary trends in dsDNA phages. We report that the majority of dsDNA phage genes have no duplicates (paralogs) in the same genome and there appears to be much less domain shuffling in phage proteins than in cellular proteins. With time, the number of POGs has been increasing, and we expect this to continue, as more proteins find homologs and create a family. The proportion of conserved proteins in any given phage genome will rise also, as more and more, and perhaps almost all, proteins will eventually find homologs in other genomes. We also observe a high proportion of phage genes that are never or rarely observed in cellular organisms, suggesting that despite their intimate contact, the proteomes of phages are quite distinct from those of their cellular hosts.
Fig. 1.
Method of POG construction. The earlier POGs (at left) were built from proteins in phages, whereas in this study we added (at right) the joining of closely related isolates (based on shared genes) and splitting proteins into their component domains.
MATERIALS AND METHODS
Phage genomes.
The list of fully sequenced phage genomes was obtained from NCBI at the URL http://www.ncbi.nlm.nih.gov/genomes/genlist.cgi?taxid=10239&type=6&name=Phages. Using taxid=35237 instead of taxid=10239 in the URL yields only “dsDNA viruses, no RNA stage.”
Gene prediction.
Protein-coding genes were predicted using GeneMarkS (8), starting with the codon frequency model of the best-known host organism and then updating on subsequent iterations with the model derived from already-detected phage genes, most of which have supporting evidence in the form of homologous sequences in the public databases. GeneMark.hmm (40) was used in cases where a host model was not available. Based on manual inspection of the resulting predicted genes and their correspondence to known genes in the well-studied T7, T4, and lambda phages, as well as 25 finished, expert-annotated phage genomes published on the websites of the Pittsburgh Bacteriophage Institute (PBI) (http://pbi.bio.pitt.edu/) and the J. Karam group at Tulane University (http://phage.bioc.tulane.edu/), these supplementary predicted genes were kept if they did not overlap with an existing gene by more than 50 bases and were not surrounded by ≥3 genes in each direction that are transcribed in the opposite direction. With the latter criteria helping to avoid overpredictions and with only a few underpredictions (mostly small or translationally recoded open reading frames [ORFs]), this approach provides high recall and precision (see Results).
Orthologous groups.
For easier comparison between methods of ortholog definition, when using the new version of COGtriangles based on the fast EdgeSearch algorithm (31), proteins were allowed to belong to only at most one POG. Otherwise, default parameters were used (specifically, an E-value cutoff of 10), except for the hit coverage threshold (percentage of protein length that must be matched in order to qualify as a best match) being set to 50%.
Domains.
We used the SMART, PFAM, LOAD, and CD subsets of the NCBI's Conserved Domain Database (41), as these entries are less likely to contain multiple domains. When multiple domains in a protein met certain criteria (probability, ≥90%; length, ≥30 amino acids; score, ≥40; alignment length, ≥40% of query length; E value, ≥10−3; P value, ≥10−3; and nonglobular and coiled coil regions excluded), first the domain matches were merged into clusters distributed along the length of the protein and then the protein was split evenly between these domains.
Phage isolates.
Shared genes were defined as those that are symmetric best matches between a pair of phage genomes. Phages sharing ≥90% of their genes (i.e., variants or isolates of essentially the same phage) were combined using single-linkage clustering, with the resulting group containing the union of all proteins from each member phage.
Phageness.
PQi is defined as log (frequency of occurrence of family i in phages/frequency of occurrence of family i in prokaryotes), where the frequency in each organism is calculated as the ratio of the observed number of organisms (phages or their hosts) with at least one match to the total number of organisms. Matches were defined using PSI-BLAST and the following criteria: E value of ≤0.001, aligned region 50% of the length of both query and target, and the match was found in the first or second iteration. Proteins in cellular genomes predicted to belong to integrated prophages were detected by using the ACLAME database (33), which uses the PROPHINDER tool (37) to predict viral and other mobile genetic elements. As of August 2008, data for 727 completely sequenced microbial genomes were available, of which 351 were found to contain no integrated prophages, while 376 contain at least one, with a total of 1,088 prophages and an average of 2.9 per genome.
Extending POGs.
The “extended POGs-10” were constructed using the same procedure as for the “annotated POGs-07,” except that the fully automated GeneMark.hmm was used to predict the supplementary genes and the choice of COGtriangles/EdgeSearch to allow multiple proteins per POG (31) was used (which affects 1.3% of proteins in 8% of POGs).
Growth of POGs.
Following the automated procedure used to build the extended POGs-10, sets of POGs were built for the genomes available as of the end of each successive year for the past decade, omitting only the steps of splitting proteins into their component domains and grouping together isolates from closely related genomes.
RESULTS AND DISCUSSION
The method of POG construction is outlined in Fig. 1. The original version of POGs, shown on the left and described previously (39), treated each phage genome as a separate evolutionary lineage and each protein as a discrete unit, while the updated framework joins together closely related isolates (groups of essentially the same phage) and identifies individual protein domains. POGs are a modification of the NCBI clusters of orthologous groups (COG) framework (28, 54, 55) that includes several new ingredients described below. Two data sets were used at different stages of this work: first we discuss a smaller set of “annotated POGs-07” that was constructed automatically; the performance of the construction method was verified by several computational approaches and by manual annotation. Next, this now-verified automated procedure was applied to construct a larger, updated set of “extended POGs-10” containing the additional dsDNA phage genomes that were made available during this investigation. Both sets and the relationship between them are described in more detail below.
Data set: genomes and proteins.
In the NCBI list of all fully sequenced phage genomes (see Materials and Methods), 323 dsDNA phages were found as of November 2007, which is twice the number of genomes available for the first version of the POG resource (39). We froze this data set to develop and test the automated method for building POGs (and to build the annotated POGs-07) and to infer general trends in POG evolution, as described in this and five following sections. Afterwards, we used this automated method to update the POGs to include the genomes available through July 2010, as described in the “Extending POGs” below (extended POGs-10 set, which is a superset of annotated POGs-07). According to the NCBI taxonomy information, the 2007 freeze of 323 genomes included 169 Siphoviridae, 67 Myoviridae, 57 Podoviridae (and 8 more Caudovirales not assigned to a family), 8 Tectiviridae, 4 Fuselloviridae, 3 Lipothrixviridae, 3 Rudiviridae, 2 Bicaudaviridae, 1 Corticoviridae, and 1 Plasmaviridae. These phages infect a broad range of hosts: Staphylococcus aureus, Escherichia coli, Lactococcus lactis, and members of the genus Mycobacterium are each listed as the hosts of more than 20 distinct phages, but there are 100 other listed hosts that belong to a variety of phyla, including Proteobacteria, Firmicutes, Actinobacteria, Cyanobacteria, the Bacteroidetes/Chlorobi group, Tenericutes, the Deinococcus-Thermus group, Crenarchaeota, and Euryarchaeota. Most of these hosts are infected by only one or two phages with completely sequenced genomes from our data set. Some of the host information provided in GenBank entries could not be verified, and a complete set of natural hosts is not known for any phage; therefore, the information about host range was not used in constructing POGs.
The 323 completely sequenced genomes encoded 27,254 putative proteins. Of these, 25,675 (94%) were existing annotations in NCBI, while 1,579 (∼5 ORFs per genome) were additionally predicted by the GeneMark program. We used the proteomes of the well-studied T7, T4, and lambda phages, as well as 25 additional well-annotated genomes from curated databases as a gold standard (see Materials and Methods), and encountered few false negatives and false positives (average recall of 93% and precision of 95% among the 28 phages), mostly affecting the translationally recoded ORFs. Even as the list of unusual coding events in phage genomes is growing (6a), the absolute number of such events in any given genome tends to be low, at least for dsDNA phages, and thus the vast majority of all protein-coding genes in these genomes are expected to be included in the POGs even if they have been missed in the original annotations.
Orthologous groups.
A draft set of candidate POGs was constructed from the 27,254 proteins in 323 dsDNA phages following the same procedure that was used for the original POGs (as outlined at left in Fig. 1). The identification of orthologous groups was at first performed with the COGtriangles program used to build COGs in cellular organisms at NCBI (54), but in the process of this work, we developed and applied the efficient EdgeSearch algorithm, which scales much better to handle large numbers of genomes and has been described in detail previously (31). The process yields 2,015 candidate POGs containing 13,470 proteins, with 49% of all proteins in completely sequenced dsDNA phage genomes being conserved in three or more phages each. Of the 1,579 proteins that have been missed in the database annotation but detected by our gene-finding effort, 357 were included in POGs, with six POGs being composed entirely of such proteins. Another 1,017 of these proteins had at least one database match, not necessarily in a completed phage genome, indicating that many of them are conserved gene products that will join the future POGs after more phage genomes become completely sequenced.
The 2,015 candidate POGs include genes from all of the 323 genomes. Some large phages, such as enterobacterial phages RB32 and T4, have ≥200 conserved proteins (∼70% POG coverage in each), while others, such as Mycoplasma phage P1 and Pseudomonas phage 119X, have only a single one (representing 8% of the 12 genes in P1 and 2% of the 54 genes in 119X). On average, phages have about 42 (±35) proteins in POGs, representing an average coverage by POGs of 54%, albeit with large variance. An average POG contains about 7 proteins from 7 phages (varying from a minimum of 3 proteins from 3 organisms in a “minimal” POG up to 141 proteins in 136 organisms in a POG that consists of phage integrases). The proteins included in POGs tend to be 15% longer, overall, than those not in POGs (average lengths of 234 versus 204 amino acids, respectively), which is consistent with previous studies indicating that conserved genes tend toward larger size in the genome of Bacillus subtilis bacteriophage SPO1 (51) and that genes conserved in multiple clusters of mycobacteriophages are longer than those present only in a subset of them (26).
The vast majority of POGs do not contain paralogs, with only 7% containing even one paralog and 0.2% having more than one, in contrast with cellular proteins in the available release of the COG resource (ftp://ftp.ncbi.nih.gov/pub/COG/COG/), where 64% contain at least one paralog and 37% have even more. By design, COGs tend to include mostly in-paralogs (lineage-specific duplications) while excluding out-paralogs (55), and a low in-paralogy rate in small phage genomes is not unexpected. The maximum number of paralogs observed per phage genome is only 4, while in the cellular COGs it is 122. POGs that include multiple paralogs have such functions as Cro/cI repressors and Roi/ANT-type phage antirepressors that represent a substantial portion of the elements enabling phage gene regulation, as well as selected “selfish” genes such as DNA methylases and HNH homing endonucleases.
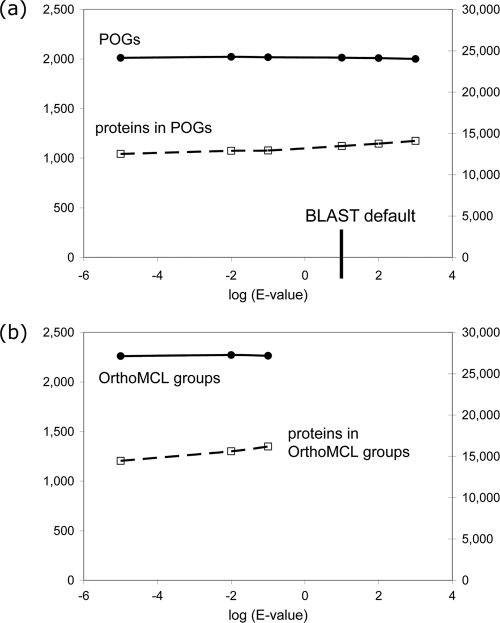
POGs are constructed from triplets of proteins that are each other's best-scoring matches (SymBeTs [55]). This ranking approach allows for the opportunity to detect fast-evolving orthologs in a way that is not possible when using thresholds based on similarity score (or statistics derived from it). We were interested in how much sensitivity is actually gained in our data set by using ranks instead of a score threshold, so we tested the effects of varying the BLAST E-value cutoff on the POG construction. Allowing the E value to reach 1,000 (2 orders of magnitude above the default cutoff of 10) (Fig. 2a, right) resulted in 5% more proteins being included in the POGs, but interestingly, these proteins grouped together into a slightly smaller (<1%) number of POGs with a larger average size. Conversely, restricting the E-value cutoff 6 orders of magnitude below the default, to 1e−5 (a number that is not strongly justified from either a statistical or biological point of view but nonetheless is frequently employed in the genome-scale identification of candidate orthologs) (Fig. 2a, left), reduced the number of proteins included in the POGs by 7%, though it again changed the number of POGs only slightly (0.1%). Across the 8 orders of magnitude of E values tested, the number of POG changes by ∼1% (solid line in Fig. 2a), whereas the number of proteins in POGs changes by 13%. Thus, BLAST cutoffs affect relatively little the definition of a POG, presumably because when the collection of genomes is very large, the chances that at least one triplet of phages will share a set of orthologs with relatively high similarity/low E value increase. On the other hand, the composition of a POG and overall coverage of a genome by POGs are much more dependent on the similarity cutoffs. Therefore, a more sensitive, rank-based detection of highly diverged orthologs may be important in gene annotation and phylogenetic analysis, especially when one wants to know whether a given POG is present or absent in a particular genome. Throughout the tested range, the number of phages contributing proteins to POGs shows virtually no change, the maximum number of paralogs per POG remains steady at 4, and the largest POGs, while changing in size, still remain the largest ones.
Fig. 2.
Effect of BLAST E-value threshold on POGs (a) and OrthoMCL groups (b). Left axes, numbers of groups; right axes, numbers of proteins in groups.
Similar trends were observed when we defined the orthologs using another program, OrthoMCL (35), which is popular in the genomics community. In this case we only tested the effects of changing the E-value cutoff across 5 orders of magnitude from 10−5 to 10−1. Like POGs, the number of groups did not change much (<1% in this range) (Fig. 2b), and the size of the groups increased by 12%, although the maximum number of paralogs increased from 8 to 18. In short, in this range of E values, both the POGs and OrthoMCL families appear to be robust groupings that are fairly independent of an E-value cutoff, primarily adding new members to existing groups as this cutoff is progressively relaxed. We note, however, that the EdgeSearch/COGtriangles program includes fewer paralogs in POGs at any cutoff and has much lower worst-case complexity and runs much faster in practice than OrthoMCL (31).
Domains.
The evolutionary histories of individual protein domains within a multidomain protein may be different. Modular signaling domains in eukaryotes present an extreme example of this phenomenon, but even relatively short phage gene products may include multiple conserved domains (49, 56). To perform automatic detection of domains, we used a heuristic approach based on the sensitive hidden Markov model (HMM) matching method HHpred (50), which presents both the query and target as HMMs and uses as the search space the libraries of domains with at least partially curated information about domain ends. Once matches to known domains were found, we split the unmatched regions of the protein evenly between the closest domains if the space between them (or between a domain and one end of the protein) was shorter than 100 amino acids and otherwise treated the unmatched regions as separate domains (see Materials and Methods).
Applying this surrogate approach to the phage proteins prior to POG construction, we split 2,199 proteins into 5,128 candidate domains, which represent an average of seven multidomain proteins per genome. This number corresponds to 8% of the proteins in each phage genome, which, predictably, is much lower than the estimated 60% of multidomain proteins in unicellular organisms from the three kingdoms of life and more than 80% of multidomain proteins in metazoans (4). Among these predicted phage multidomain proteins, 74% have two domains and 20% have three, leaving only 6% with four or more domains per protein, which again represents a much lower percentage of proteins having three or more domains than in either prokaryotes or eukaryotes (20). When POGs are constructed with this approach, their total number increases by 11% to 2,227, and of the original 2,015 POGs, 86% remain unchanged while the other 14% are affected in some way (either adding or removing members). The overall number of proteins/domains that are represented in POGs increases by 14%, but as before, this corresponds to about half (51%) of the new data set. On average, each phage has about 48 (±38) such POG members, which is slightly higher than the 42 (±35) observed prior to splitting into domains, with each POG again on average having seven proteins/domains from seven phages.
In this new set of POGs built with individual domains, we searched for instances where a POG built with full-length proteins was split into multiple POGs that had different sets of “parent” proteins. This provides a rough estimate of the number of chimeric POGs that were improperly joined, with different parts of their member proteins possibly having different evolutionary histories. Such cases are actually quite rare in the phage data set (∼1%), and when they do occur, many appear to be due to different combinations of domains that typically occur together, such as variable N- and C-terminal domains surrounding a central catalytic domain. Examples include the Ig-like domains present in structural proteins, a variety of domains in several cell wall-associated lytic enzymes (for instance bacterial SH3, amidase, peptidase_M23 superfamily, and peptidoglycan-binding domains), several antirepressors having various combinations of ANT, Bro-N, HTH_XRE, KilA, Phage_pRha, and other domains, and the set of cI repressors that were artificially joined to other DNA-binding proteins due to the presence of a common XRE-type helix-turn-helix domain. In short, while the automated domain dissection method may provide an advantage in annotating COGs from cellular organisms, especially multicellular eukaryotes, the actual number of such cases affecting POGs in the dsDNA phages is modest.
Phage isolates.
It should come as no surprise that the complete phage genome data set has an uneven distribution, with some groups containing several very closely related phages while others are more sparsely sampled. For example, the three closely related Bordetella phage isolates, BPP-1, BMP-1, and BIP-1, have >99% sequence identity at the DNA level, and each of their 50 shared genes is therefore conserved in at least these three dsDNA phages, forming a POG in the procedure used so far (even though many of these genes are not seen anywhere outside these three Bordetella isolates). To deal with such instances of bias caused by uneven genomic sampling, we grouped together phage isolates by merging all genomes that share ≥90% of their genes (see Materials and Methods). In this way, strains of essentially the same phage with minor genetic rearrangements become merged into a single entity, for example, the three aforementioned Bordetella phage isolates, the four Staphylococcus phages 44AHJD, P68, 66, and SAP-2, the four Burkholderia phages Bcep781, Bcep1, Bcep43, and BcepNY3, the four Bacillus phages Cherry, Gamma, Wbeta, and Fah, and the six enterobacterial phages PRD1, L17, PR3, PR4, PR5, and PR772 (for a full listing, see “Availability” below). In constructing POGs as groups of orthologs shared by three or more evolutionary lineages, the use of lineages based on groups of isolates rather than individual phage genomes is designed not to assemble phages into high-level hierarchies (such as order, family, subfamily, or even genus) but only to alleviate the redundancy by grouping together closely related phages so that proteins must also be conserved outside this group (in at least two other lineages) to be considered a POG. Applying this process (as shown in Fig. 1) to the 323 individual phages yielded a total of 280 lineages, with most lineages (249, or 89%) consisting of only a single phage and the rest containing multiple (from two to six) phage isolates.
The use of lineages built from groups of isolates rather than single phages reduced the number of candidate POGs by 24%, and the group membership is affected in an additional 12% (with 97% of these representing a reduction in size). In the new set of 1,689 POGs, the coverage of phage genomes by POGs is reduced from 50% to 43%, with each lineage having at least one such conserved protein or domain, and on average each has about 47 (±40), with each POG contributing about eight proteins from seven lineages. The small but noticeable reduction in coverage indicates that without considering the bias presented by closely related phages isolates, the number of candidate POGs will be somewhat inflated, although the overall trend of about half of all phage proteins being conserved in the POGs remains clear.
Function annotation.
To complete the construction of the annotated POGs-07 using the procedure shown in Fig. 1, we applied PSI-BLAST searches, followed by the more sensitive HHpred searches (50) to find homologs with known structure and function for as many uncharacterized POGs as possible. At least some level of annotation could be provided for 905 (54%) of the 1,689 POGs (see “Availability” below). About one-third of these are functions related to virion structure and assembly (head, tail, scaffold, packaging, portal, etc.), another one-third are involved in genome maintenance and expression (replication, recombination, repair, transcription, regulation, etc.), and the remaining fraction includes other functions (host lysis, metabolism, host ribosomal control, etc.).
Phageness.
We are interested in the extent to which phages are exchanging genes with their cellular hosts. As one measure of the outcome of this exchange, we have defined the phageness quotient (PQ) of each protein family as the log-odds ratio of two hypotheses: H0 (a member of this protein family comes from a phage genome) versus the alternative H1 (a member of this protein family comes from a cellular genome) (39). Matches to the 323 dsDNA phage genomes used to construct the annotated POGs-07 were summed in the numerator of PQ, matches to the nonprophage regions of 727 completely sequenced microbial genomes were summed in the denominator, and matches to prophages did not count as either for the purpose of PQ determination (see Materials and Methods).
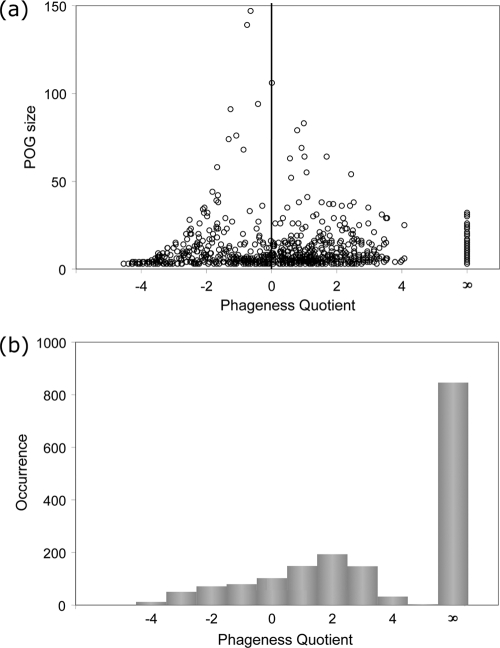
As might be expected from the wide but sparse distribution of viral proteins, the number of matches of these POG proteins to phage genomes is relatively low, with only one POG (a TMP repeat-containing tail protein) matching more than half of the genomes, and only 11 genomes matched per POG on average. This number is higher than the average number of genomes in the annotated POGs-07 due to the stricter requirement that a protein must form SymBeTs with at least two members of an existing POG in order to join it, whereas here unidirectional BLAST matches were also included. In comparison, POGs have a much wider distribution among their cellular hosts, with 45.5 microbial genomes matched on average. As shown in Fig. 3a, the relatively small number of very large POGs (>50 genes per POG) tend to have PQs near zero; this corresponds to ubiquitous, promiscuous genes, many of which, such as integrase genes, are shared by phages and cellular mobile elements. Among the remaining POGs, which span more than an order of magnitude in size (3 to 50 genes), there seems to be no close correlation between POG size and phageness quotient. The strongest trend, however, is the existence of 846 POGs consisting of 3 to 32 proteins (5.5 on average) that have a PQ of infinity, i.e., are never observed in the host genomes. This represents 50% of all annotated POGs-07 (Fig. 3b). Moreover, there are another 182 POGs (11%) that are highly phage specific, with 2 orders of magnitude more homologs in phages than in hosts (PQ ≥ 2). These numbers represent lower-bound estimates, since some POG matches in the host genomes are likely to be in the unrecognized prophage regions. Even so, the fact that more than half of the POGs have no or very few matches to proteins in microbial genomes, except to obvious prophages, suggests that dsDNA phages combine the ability to share and transduce the host genes with the propensity to maintain a large fraction of unique, phage-specific genes that may be “dsDNA phage hallmark genes.”
Fig. 3.
Distribution of phageness quotient according to the number of proteins in a POG (a) and the number of POGs (b).
Not surprisingly, the POGs with a PQ of infinity tend to correspond to genes with essential functions, such as virion structure (tail, head, portal, components, and assembly factors), phage-specific components of DNA replication, repair, and recombination (single-stranded DNA-binding proteins and DNA primase-helicase), transcriptional control (transcriptional activator rinB and late promoter transcription accessory protein), translational control (sigma factor for T4 late transcription and RegA translational repressor), and virulence (cell wall hydrolase). Many proteins with unknown function also have high phageness, indicating that many still-uncharacterized phage-specific functions exist. The POGs with PQs of ≥2 add to this list several functions related to virulence, such as holins and lysis proteins, that also appear in their cellular hosts, although in far lower abundance than in phages. In contrast, enzymes of cellular metabolism tend to belong to POGs with low PQs, such as GTP cyclohydrolase I with a PQ of −4.5 (indicating its appearance in 4 to 5 orders of magnitude fewer phage genomes than microbial ones), although exceptions do occur, such as S-adenosyl-l-methionine hydrolase with a PQ of infinity, presumably used by the phage in restriction-modification warfare with the host to ensure the unmodified status of its own DNA, which would be useful, for example, when the phage encodes its own methylation-dependent restriction enzymes (9).
To assess the degree to which the cases of gene sharing between phages and their hosts involve conserved orthologs, the reverse search of cellular COGs to phage proteins was performed. In the publicly available release of the NCBI COG resource (ftp://ftp.ncbi.nih.gov/pub/COG/COG/), only 11% of cellular COGs match dsDNA phage proteins with E values of ≤0.01, and only 4% match a conserved protein in a POG. Thus, even though the absolute number of genes shared by phages and their hosts is high (and may grow for a given phage genome as more host genomes become sequenced), the relative number of such shared genes is low when expressed as the percentage of all phage genes, and orthologs broadly conserved in phages make up an even smaller subset of these cases.
Extending POGs.
The annotated POGs-07 set included proteins encoded by phage genomes deposited at NCBI as of November 2007. In order to include proteins from more recently obtained dsDNA phage genomes and to see whether periodic updates of the POG resources can be practically repeated in the future, we applied the automated procedure developed with the annotated POGs-07 set to the additional genomes sequenced in 2008 to 2010 to construct the larger, updated extended POGs-10 set. As of July 2010, NCBI's list of complete viral genomes contained 501 dsDNA phages, which includes 178 genomes not covered by the annotated POGs-07. These new members include 74 Siphoviridae, 39 Myoviridae, 35 Podoviridae (and 2 more Caudovirales not assigned to a family), 5 Lipothrixviridae, 5 Fuselloviridae, 1 Tectiviridae, 1 Rudiviridae, and 11 unclassified dsDNA phages; there are also representatives of three newly recognized virus groups, i.e., 2 Salterprovirus, 2 Globuloviridae, and 1 Ampullaviridae. Most of these phages have only one listed host, and most hosts are infected by only one or two of these new phages from our data set, with the exception of Escherichia coli and Pseudomonas aeruginosa, that are listed as the hosts for more than 15 phages each.
The genomes of these new phages encode 16,787 proteins. Compared to the 2007 data set used to construct the annotated POGs-07, these new genomes have larger average sizes (63 versus 54 kbp, encoding on average 91 versus 79 genes) and a lower number of “underpredicted” genes (∼3 versus ∼5 per genome additionally predicted by us using GeneMark). Adding these new proteins to the 2007 data set (where necessary, split into domains with statistics of domain content very similar to that in annotated POGs-07) results in an overall total of 48,850 proteins/domains from 501 genomes merged into 409 lineages. Using the fully automated portion of the procedure outlined in Fig. 1 (i.e., no inspection of each POG by human eye), this combined data set yielded a total of 2,371 extended POGs-10, a 40% increase over the 1,689 observed in the annotated-POGs-07 set, with each POG containing, on average, 10 proteins/domains from eight lineages. Only 15% of the extended POGs-10 remained unchanged compared to their annotated POGs-07, with 53% extended and 33% newly formed. Of the latter, 680 groups (29% of the total) were not in the 2007 data set and thus represent completely novel orthologous groups. Despite larger genome sizes in the new phages, their levels of paralogy are not significantly different from those of the earlier genomes and annotated POGs-07. The POG coverage in the new genomes is, however, significantly lower at 35%, whereas that of the genomes that were also in the 2007 data set increases to 54% (from 43% in the annotated POGs-07).
Growth of POGs.
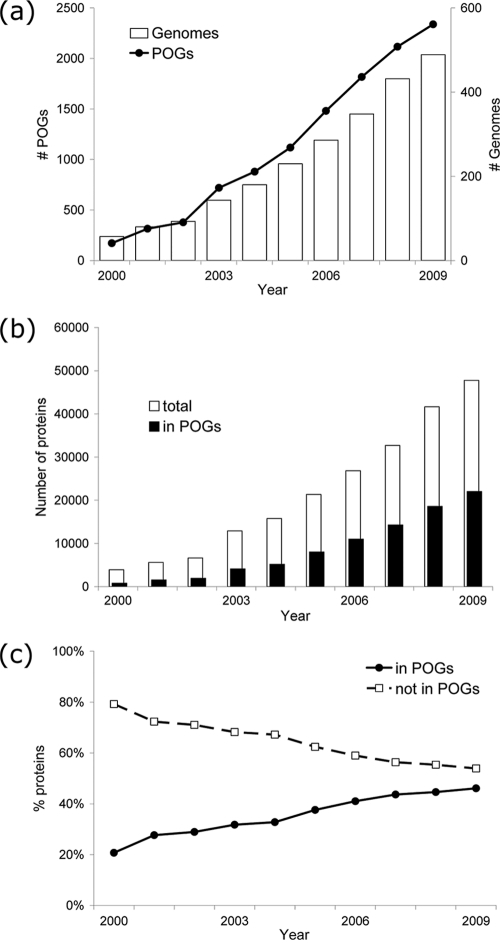
As shown in Fig. 4a, as the number of sequenced dsDNA phage genomes increases each year, the number of POGs also increases sharply. The automated procedure was used to build POGs for the set of genomes available at the end of each successive year (see Materials and Methods). The curve of genome count is far from saturation, which agrees with the earlier suggestions that a high fraction (perhaps 90%) of viral sequence diversity remains unknown (3, 16). As the number of POGs grows over time, both the number and percentage of proteins conserved in POGs also increase, while the percentage not in POGs correspondingly decreases, as shown in Fig. 4b and c. This occurs as new proteins are added to existing POGs and also by the creation of new POGs from singletons or pairs of proteins that had no mates to form the initial triangle before.
Fig. 4.
Properties of dsDNA phage genomes over the past decade. (a) Numbers of genomes and POGs; (b) total number of proteins in genomes and number that are in POGs; (c) percentage of those proteins in or not in POGs.
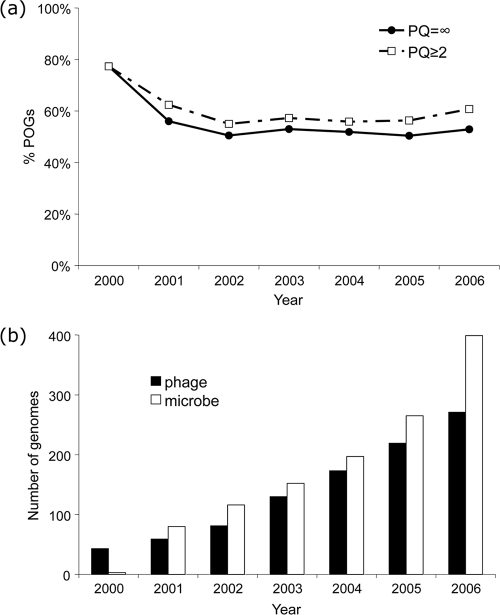
Despite the dramatic growth in the number of POGs over time, some measurements of phage protein properties already appear to have stabilized. Figure 4c shows that the lower bound of average phage genome POG coverage of almost 50% (47% in the extended POGs-10) may hold. As the number of completely sequenced phage and host genomes deposited into NCBI increases, the percentage of annotated POGs-07 with high phageness also remains stable, at about 50% for those with no cellular homologs and about 10% higher for those with ≥2 orders of magnitude more homologs among phages than in microbes (Fig. 5a). The higher phageness observed in 2000 must have been due to the insufficient sampling of the genome space at the turn of the century (Fig. 5b); this bias appears to have been reduced more recently.
Fig. 5.
(a) Percentage of POGs that are never or very rarely observed in their cellular hosts (PQ = infinity) or that have homologs in at least 2 orders of magnitude more phage species than host ones (PQ ≥ 2). (b) Overall number of phage and host (prokaryotic) genomes available at the end of each year.
We expect that as additional genomes and gene products become known, the POG resource will continue to expand, as new ortholog families are discovered and new proteins are added to existing families, but at the same time, POGs will start to provide more and more consistent data for estimating various parameters of phage protein evolution.
Availability.
The 1,689 annotated POGs-07 from the 323 dsDNA phage genomes and their annotations, as well as the 2,371 extended POGs-10 from the updated list of 501 genomes, are freely available at ftp://ftp.ncbi.nlm.nih.gov/pub/kristensen/ and mirrored at ftp://ftp.stowers.org/pub/dmk/. These data sets are available both in text flatfile database format and as BLAST-searchable databases, with the former containing 13,086 proteins (or domains) from 279 lineages and the latter containing 22,719 proteins/domains from 405 lineages (both ∼45% of the total proteins in the data set). Also available are BLAST-searchable databases for the subset that are observed only in phages and never in their cellular hosts (outside known prophage regions), which include 4,678 proteins (36% of those in POGs) from 846 (62%) of the annotated POGs-07. This repository of conserved phage proteins (including conserved motifs of each family) also provides data for future molecular systematics studies: with an average of 41 conserved proteins per phage, the annotated POGs-07 contain 2,824,817 amino acid characters (667,156 in the subset never observed in cellular genomes), and the extended POGs-10 contain nearly 5 million characters in 45 conserved proteins per phage.
ACKNOWLEDGMENT
This work was supported by the Stowers Institute for Medical Research.
Footnotes
Published ahead of print on 11 February 2011.
REFERENCES
- 1. Abedon S. T. 2009. Phage evolution and ecology. Adv. Appl. Microbiol. 67:1–45 [DOI] [PubMed] [Google Scholar]
- 2. Ackermann H. W., Kropinski A. M. 2007. Curated list of prokaryote viruses with fully sequenced genomes. Res. Microbiol. 158:555–566 [DOI] [PubMed] [Google Scholar]
- 3. Angly F. E., et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Apic G., Gough J., Teichmann S. A. 2001. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 310:311–325 [DOI] [PubMed] [Google Scholar]
- 5. Asadulghani M., et al. 2009. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ashelford K. E., Day M. J., Fry J. C. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a. Baranov P. V., Gurvich O. L., Hammer A. W., Gesteland R. F., Atkins J. F. 2003. Recode 2003. Nucleic Acids Res. 31:87–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergh O., Borsheim K. Y., Bratbak G., Heldal M. 1989. High abundance of viruses found in aquatic environments. Nature 340:467–468 [DOI] [PubMed] [Google Scholar]
- 8. Besemer J., Lomsadze A., Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bist P., et al. 2001. S-Adenosyl-l-methionine is required for DNA cleavage by type III restriction enzymes. J. Mol. Biol. 310:93–109 [DOI] [PubMed] [Google Scholar]
- 10. Breitbart M., Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278–284 [DOI] [PubMed] [Google Scholar]
- 11. Brussaard C. P., et al. 2008. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2:575–578 [DOI] [PubMed] [Google Scholar]
- 12. Brussow H., Canchaya C., Hardt W. D. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciccarelli F. D., et al. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287 [DOI] [PubMed] [Google Scholar]
- 14. Comeau A. M., et al. 2008. Exploring the prokaryotic virosphere. Res. Microbiol. 159:306–313 [DOI] [PubMed] [Google Scholar]
- 15. Cortez D., Forterre P., Gribaldo S. 2009. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 10:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desnues C., et al. 2008. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452:340–343 [DOI] [PubMed] [Google Scholar]
- 17. Dinsdale E. A., et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629–632 [DOI] [PubMed] [Google Scholar]
- 18. Drake J. W. 1999. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann. N. Y. Acad. Sci. 870:100–107 [DOI] [PubMed] [Google Scholar]
- 19. Edwards R. A., Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504–510 [DOI] [PubMed] [Google Scholar]
- 20. Ekman D., Bjorklund A. K., Frey-Skott J., Elofsson A. 2005. Multi-domain proteins in the three kingdoms of life: orphan domains and other unassigned regions. J. Mol. Biol. 348:231–243 [DOI] [PubMed] [Google Scholar]
- 21. Enquist L. W. 2009. Virology in the 21st century. J. Virol. 83:5296–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glazko G., Makarenkov V., Liu J., Mushegian A. 2007. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct. 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haaber J., Middelboe M. 2009. Viral lysis of Phaeocystis pouchetii: implications for algal population dynamics and heterotrophic C, N and P cycling. ISME J. 3:430–441 [DOI] [PubMed] [Google Scholar]
- 24. Hatfull G. F. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hatfull G. F. 2010. Mycobacteriophages: genes and genomes. Annu. Rev. Microbiol. 64:331–356 [DOI] [PubMed] [Google Scholar]
- 26. Hatfull G. F., et al. 2010. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hendrix R. W. 2002. Bacteriophages: evolution of the majority. Theor. Popul Biol. 61:471–480 [DOI] [PubMed] [Google Scholar]
- 28. Koonin E. V. 2005. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39:309–338 [DOI] [PubMed] [Google Scholar]
- 29. Koonin E. V., Wolf Y. I. 2009. The fundamental units, processes and patterns of evolution, and the tree of life conundrum. Biol. Direct. 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korbel J. O., Snel B., Huynen M. A., Bork P. 2002. SHOT: a web server for the construction of genome phylogenies. Trends Genet. 18:158–162 [DOI] [PubMed] [Google Scholar]
- 31. Kristensen D. M., et al. 2010. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26:1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kristensen D. M., Mushegian A. R., Dolja V. V., Koonin E. V. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leplae R., Hebrant A., Wodak S. J., Toussaint A. 2004. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 32:D45–D49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li J., Halgamuge S. K., Tang S. L. 2008. Genome classification by gene distribution: an overlapping subspace clustering approach. BMC Evol. Biol. 8:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L., Stoeckert C. J., Jr., Roos D. S. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lima-Mendez G., Toussaint A., Leplae R. 2007. Analysis of the phage sequence space: the benefit of structured information. Virology 365:241–249 [DOI] [PubMed] [Google Scholar]
- 37. Lima-Mendez G., Van Helden J., Toussaint A., Leplae R. 2008. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24:863–865 [DOI] [PubMed] [Google Scholar]
- 38. Lima-Mendez G., Van Helden J., Toussaint A., Leplae R. 2008. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25:762–777 [DOI] [PubMed] [Google Scholar]
- 39. Liu J., Glazko G., Mushegian A. 2006. Protein repertoire of double-stranded DNA bacteriophages. Virus Res. 117:68–80 [DOI] [PubMed] [Google Scholar]
- 40. Lukashin A. V., Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paterson S., et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464:275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pedulla M. L., et al. 2003. Bacteriophage G: analysis of a bacterium-sized phage genome, abstr. M-039. Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]
- 44. Prangishvili D., Garrett R. A., Koonin E. V. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52–67 [DOI] [PubMed] [Google Scholar]
- 45. Puigbo P., Wolf Y. I., Koonin E. V. 2009. Search for a ‘Tree of Life’ in the thicket of the phylogenetic forest. J. Biol. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rohwer F., Edwards R. 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohwer F., Thurber R. V. 2009. Viruses manipulate the marine environment. Nature 459:207–212 [DOI] [PubMed] [Google Scholar]
- 48. Santos F., et al. 2007. Metagenomic approach to the study of halophages: the environmental halophage 1. Environ. Microbiol. 9:1711–1723 [DOI] [PubMed] [Google Scholar]
- 49. Sekiguchi J., Shuman S. 1997. Domain structure of vaccinia DNA ligase. Nucleic Acids Res. 25:727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960 [DOI] [PubMed] [Google Scholar]
- 51. Stewart C. R., et al. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388:48–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suttle C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 53. Suttle C. A. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 54. Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tatusov R. L., Koonin E. V., Lipman D. J. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 56. Tetart F., Repoila F., Monod C., Krisch H. M. 1996. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 258:726–731 [DOI] [PubMed] [Google Scholar]
- 57. Toussaint A., Lima-Mendez G., Leplae R. 2007. PhiGO, a phage ontology associated with the ACLAME database. Res. Microbiol. 158:567–571 [DOI] [PubMed] [Google Scholar]
- 58. Vos M., Birkett P. J., Birch E., Griffiths R. I., Buckling A. 2009. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325:833. [DOI] [PubMed] [Google Scholar]
- 59. Wagner P. L., Waldor M. K. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weinbauer M. G., Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1–11 [DOI] [PubMed] [Google Scholar]
- 61. Wolf Y. I., Rogozin I. B., Grishin N. V., Koonin E. V. 2002. Genome trees and the tree of life. Trends Genet. 18:472–479 [DOI] [PubMed] [Google Scholar]
- 62. Wolf Y. I., Rogozin I. B., Grishin N. V., Tatusov R. L., Koonin E. V. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wommack K. E., Colwell R. R. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yin Y., Fischer D. 2008. Identification and investigation of ORFans in the viral world. BMC Genomics 9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]