Abstract
Our previous studies revealed that a novel two-component signal transduction system, YhcSR, is essential for the survival of Staphylococcus aureus; however, the biological function of YhcSR remains unknown. In this study, we demonstrated that YhcSR plays an important role in the modulation of the nitrate respiratory pathway under anaerobic conditions. Specifically, we determined that nitrate induces yhcS transcription in the early log phase of growth under anaerobic conditions and that the downregulation of yhcSR expression eliminates the stimulatory effect of nitrate on bacterial growth. Using semiquantitative real-time reverse transcription-PCR (qPCR) and promoter-lux reporter fusions, we established that YhcSR positively modulates the transcription of the narG operon, which is involved in the nitrate respiratory pathway. Our gel shift assays revealed that YhcR binds to the promoter regions of narG and nreABC. Collectively, the above data indicate that the yhcSR system directly regulates the expression of both narG and nreABC operons, which in turn positively modulate the nitrate respiratory pathway of S. aureus under anaerobic conditions. These results provide a new insight into the biological functions of the essential two-component YhcSR system.
INTRODUCTION
The continuing increase of hospital- and community-associated methicillin-resistant Staphylococcus aureus infections highlights an urgent need for alternative potent antibacterial agents (8, 13, 25). The ability of this organism to resist antibiotics and cause infection is partially due to the coordinated regulation of gene expression, which allows the bacteria to survive under different stress conditions. Two-component signal (TCS) transduction systems play important roles in the adaptation of the microbial organisms within different niches, as well as in pathogenesis and biofilm formation for various bacterial species (7, 15, 22, 27). Our previous studies have demonstrated that a novel two-component signal transduction system, YhcSR, is required for the viability of S. aureus in vitro culture (29). However, the biological function of YhcSR is still unclear.
It is well known that oxygen is a contributing factor in the regulation of virulence gene expression in pathogens and enables bacteria to persist and survive in ecological niches similar to host conditions, which are required to facilitate the pathogenicity of pathogens. Although there are different oxygen tensions between different sites in the host, especially completely anaerobic conditions in abscesses (23), S. aureus is able to invade almost every kind of tissue. Thus, S. aureus must evolve mechanisms to sense the availability of oxygen and adapt to a dynamic host environment with a variety of oxygen limitations by employing either nitrate respiration with nitrate as the terminal electron acceptor (2) or carbohydrate fermentation (28). In Bacillus subtilis, it has been determined that a ResD/ResE system is involved in the regulation of genes required for anaerobic respiration; the ResD/ResE system is controlled by PhoP/PhoR, which responds to phosphate starvation (1). In S. aureus, SrrAB, which is a homolog of ResDE in B. subtilis, has been identified in the modulation of anaerobic gene expression, suggesting its sensitivity to oxygen tension (30, 33). It has been reported that anaerobic conditions and low carbon dioxide concentrations impede the production of toxic shock syndrome toxin 1 (34) and enhance bacterial adhesion and biofilm formation by inducing the expression of polysaccharide intracellular adhesin in S. aureus and Staphylococcus epidermidis (3). The recently characterized novel two-component system NreBC has been shown to control the nitrate reductase and nitrite reductase operons in S. aureus (26). In Escherichia coli, a global regulator, Fnr (fumarate and nitrate reductase regulation), controls gene expression in response to anaerobic environments by acting as a sensor and a regulator (31); however, in S. aureus no Fnr homolog seems to exist (30). The oxygen-liable iron-sulfur cluster of the NreBC system senses oxygen depletion and regulates anaerobic gene expression in S. aureus (12).
In this study, we identified that nitrate is able to induce the expression of YhcS and demonstrated that the YhcSR system directly regulates the nitrate reductase and the NreABC operons. These findings indicate that the essential YhcSR system also plays an important role in the regulation of nitrate respiratory metabolism pathways.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. aureus strains used in this study are listed in Table 1. All S. aureus strains were cultured at 37°C in BM broth (1% soy peptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose). Media were supplemented with erythromycin (5 μg/ml) as appropriate. Anaerobic cultures were incubated in screw-cap tubes containing chemically defined medium (CDM) covered with sterile mineral oil at 37°C with shaking at 100 rpm.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. aureus strains | ||
| WCUH29 | Clinical human methicillin-resistant S. aureus isolate, rsbU+ | NCIMB40771 |
| WCUH29/pYH3 | WCUH29 containing plasmid pYH3, Ermr | 29 |
| JSAS909 | WCUH29 containing plasmid pSAS909, Ermr | 29 |
| YJ106 | WCUH29 containing plasmid pCY106, Cmr | This study |
| YJ2185 | WCUH29 containing plasmid pMY2185, Cmr | This study |
| YJ606 | WCUH29 containing plasmid pCY606, Ermr | 32 |
| YJ2185-1 | WCUH29 containing plasmid pMY2185-1 | This study |
| Plasmids | ||
| pCY1006 | agr promoter-luxABCDE reporter system, Cmr | 16 |
| pCY106 | yhcSR promoter-luxABCDE reporter system, Cmr | This study |
| pMY2185 | narG promoter-luxABCDE reporter system, Cmr | This study |
| pCY606 | Shuttle vector, derived from pSAS909, carrying promoterless luxABCDE and yhcS antisense, Ermr | 32 |
| pMY2185-1 | Derived from pCY606 carrying narG promoter-luxABCDE reporter and yhcS antisense, Ermr | This study |
| Primers | ||
| yhcSNdefor | 5′-TATGGCTAGCATGGAAAAAGGACGCGAC-3′ | |
| yhcSXhorev | 5′-CGCACTCGAGTTTTATAGGAATTGTGAATTG-3′ | |
| yhcRforNdeI | 5′-GGAATTCCATATGAACAAAGTAATATTAGTAG-3′ | |
| yhcRrevXhoI | 5′-CCGCTCGAGAATCAACTTATTTTCCATTGC-3′ | |
| PyhcSforp | 5′-AATACACGTAAAAATGAATCCCG-3′ | |
| Pyhcrev | 5′-TACCCGGGATTCATTTTTACGTGTATT-3′ | |
| narGproEcoNotfor | 5′-ATGAATTCGCGGCCGCCAACTTCTAATCCGACTCA-3′ | |
| narGproNotrev | 5′-TAGTGCGGCCGCTATTTATATCCTCCTACGTATA-3′ | |
| narGproXmarev | 5′-TACCCGGGTATTTATATCCTCCTACGTATA-3′ | |
| narGRTfor | 5′-CACCTATTCCAGCGATGTCAATG-3′ | |
| narGRTrev | 5′-ATGTGCATCCGGAGTACGTGTTA-3′ | |
| narGprGSfor | 5′-AAAATAAATGAATAAGTAAGGTTTC-3′ | |
| narGprGSrev | 5′-CTTTCTAGGATCGACCAATTC-3′ | |
| Sa2180RTfor | 5′-CGCTTCTTTGGATGATCTAGG-3′ | |
| Sa2180RTrev | 5′-TCAACGCATTTAGAATAGCTTC-3′ |
RNA isolation and purification.
Overnight cultures of S. aureus were inoculated at 1% in tryptic soy broth (TSB) medium and grown to the mid-exponential phase (∼4 h) of growth. Total RNA was purified from the above cultures, as described previously (11). Briefly, bacterial cells were harvested by centrifugation at 4,000 × g, and the RNA was isolated using an SV total RNA isolation system (Promega). Contaminating DNA was removed with a Turbo DNA-free kit (Ambion), and the RNA yield was determined spectrophotometrically at 260 nm.
qPCR analysis.
In order to determine whether the downregulation of yhcSR expression has any impact on the expression of identified genes, we employed semiquantitative real-time reverse transcription (RT)-PCR (qPCR) to compare the RNA levels, as described previously (11, 16). The first-strand cDNA was synthesized using SuperScript III reverse transcriptase and random primers (Invitrogen). For each RNA sample, we performed duplicate reactions of reverse transcription, as well as a control without reverse transcriptase, in order to determine the levels of DNA contamination. PCRs were set up in triplicate by using a SYBR green PCR Master Mix (Stratagene). Real-time sequence-specific detection and relative quantitation were performed with the Stratagene Mx3000P real-time PCR system. Gene-specific primers were designed to yield 100 to 200 bp of specific products (Table 1). Relative quantification of the product was calculated using the comparative threshold cycle (CT) method, as described for the Stratagene Mx3000P system. The housekeeping gene 16S rRNA was used as an endogenous control (16). All samples were analyzed in triplicate and normalized against 16S rRNA gene expression. The experiments were repeated at least three times.
Construction of promoter-lux reporter fusions.
In order to identify potential stimuli for the YhcSR system and to further confirm whether YhcSR regulates the transcription of narG and nreABC, we created different promoter-lux reporter fusions using pCY1006 (16) and pCY606, as described previously (32). The promoter regions of yhcSR and narG genes were obtained by PCR using the indicated primer pairs listed in Table 1 and ligated into the upstream segment of promoterless luxABCDE and resulted in recombinant plasmids pCY106 and pMY2185, respectively. The narG promoter region was also ligated into the upstream segment of promoterless luxABCDE in pCY606 and formed plasmid pMY2185-1. These resulting plasmids were transformed into E. coli DH10B competent cells and confirmed by PCR, restriction enzyme digestion, and DNA sequencing. The plasmids were then electroporated into RN4220, purified, and electroporated into S. aureus WCUH29, generating YJ106, YJ2185, and YJ2185-1 strains as described previously (10). The lux expression level was detected by measuring the bioluminescence intensity using a Chiron luminometer. The relative light units (RLU) were calculated (bioluminescence intensity/optical density at 600 nm [OD600]). To identify a potential stimulus, we examined the effects on yhcSR expression of calcium (1 mM CaCl2), magnesium (1 to 2 mM MgCl2), and pH (4.0 and 5.2) under aerobic conditions and of nitrate (20 mM NaNO3) and nitrite (5 mM NaNO2) under anaerobic conditions. Each experiment was repeated at least three times.
Cloning, expression, and purification of YhcS- and YhcR-His tag fusion proteins.
In order to differentiate which identified genes are directly regulated by the yhcSR regulator, we purified His-tagged YhcS and YhcR proteins, as described previously (29). Both yhcS and yhcR coding regions were obtained by PCR from S. aureus using corresponding primers (Table 1) and cloned into NdeI and XhoI sites of the E. coli expression vector pET24b. The recombinant DNA (pETyhcS or pETyhcR) was confirmed by PCR and DNA sequencing (data not shown) and transformed into E. coli strain BL21(DE3). The transformants were incubated until mid-log phase (OD600 of ∼0.6), and then yhcS and yhcR expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 1 mM). After 4 h of incubation, cells were harvested and lysed by sonication. The expression of YhcS and YhcR was confirmed by SDS-PAGE followed by Coomassie bright blue staining (data not shown).
To purify His-tagged YhcS and YhcR proteins, a 500-ml culture of BL21(DE3) containing pETyhcS or pETyhcR was induced. Next, the cell pellet was collected and lysed in buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 20 mM imidazole) containing 10 mg/ml lysozyme followed by sonication at 4°C. The supernatant was collected by centrifugation at 10,000 × g, and applied to the nickel-nitrilotriacetic acid (Ni-NTA) agarose column (Novagen). After being washed with washing buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 30 mM imidazole), the His-tagged YhcS or YhcR protein was eluted with elution buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 300 mM imidazole) and dialyzed against a dialysis buffer (10 mM NaH2PO4, pH 8, 300 mM NaCl, 10% glycerol). The purified His-tagged YhcS and YhcR proteins were confirmed by SDS-PAGE followed by Coomassie bright blue staining (data not shown). The concentrations of purified proteins were determined by the Bradford method.
Gel mobility shift DNA binding assay.
To determine which identified gene(s) is directly regulated by YhcR, we performed gel shift assays. DNA fragments of the upstream regions of narG and nreABC were obtained by PCR using the primers listed in Table 1. The amplified DNA fragments were purified and labeled with digoxigenin (DIG) using a DIG gel shift kit (Roche) according to the manufacturer's protocol. The DNA binding and electrophoresis were performed as described previously (17, 20). Briefly, the purified PCR products were labeled with digoxigenin using terminal transferase (Roche). The labeled DNA fragments were further purified to remove the redundant DIG-ddUTP and salts. The interaction of YhcR with DNA was conducted in a 20-μl reaction mixture containing 0.2 pmol DIG-labeled DNA, 1 μg of poly(dI-dC), 25 mM NaH2PO4 (pH 8.0), 50 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 10% glycerol, 0.1 mM EDTA, and different concentrations of YhcR protein (final concentrations of YhcR were 0.4, 1, 2, 4, and 8 μM, corresponding to 0.2, 0.5, 1, 2, and 4 μg, respectively). Unlabeled DNA fragments of the promoter region as a specific competitor were added into the reaction mixture with a 100-fold excess over labeled probe. Gene internal fragments were obtained by PCR, purified, and labeled as nonspecific controls. Bovine serum albumin (BSA) was used as a nonspecific protein binding control. The DNA binding reaction was initiated by the addition of YhcR, and the reaction mixture was incubated at room temperature for 25 min. Samples were then loaded directly onto a 5% native polyacrylamide gel (acrylamide-bisacrylamide [29:1] in 0.5× Tris-borate-EDTA [TBE] buffer). Electrophoresis was run for 2 h at 4°C with 7 V/cm, and the gels were transferred to a nylon membrane via electroblotting in 0.5× TBE at 300 mA for 90 min at 4°C. After cross-linking of DNA fragments using UV, the membranes were hybridized with anti-digoxigenin-alkaline phosphatase (AP) antibody and exposed to X-ray film for 4 h to achieve the desired signal.
Growth characterization of S. aureus strains under anaerobic conditions in the present of nitrate and nitrite.
Overnight cultures of S. aureus strains were diluted in fresh medium to an OD600 of 0.07. Bacterial cells covered with oil were cultured in Falcon tubes (50 ml) with moderate shaking (100 rpm). When applicable, 20 mM sodium nitrate (NaNO3) or 1 mM sodium nitrite (NaNO2) was added to the medium at final concentrations as mentioned elsewhere in the text. Both bioluminescence signals and cell growth were monitored at 37°C by measuring the light intensity with a Chiron luminometer and measuring the optical density at 600 nm with a SpectraMax Plus spectrophotometer. To eliminate the effect of bacterial growth, the relative light units (RLU) were calculated (light intensity/OD600) from triplicate readings at different times during growth.
RESULTS
Nitrate enhances expression of yhcSR in early log phase of growth under anaerobic conditions.
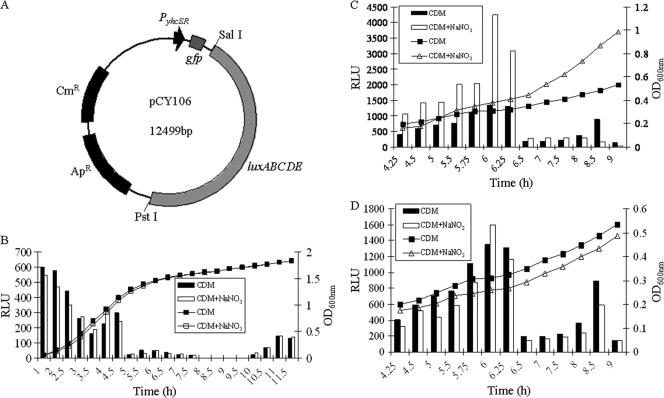
In order to identify potential stimuli and understand the base level of yhcSR expression during growth, we constructed a yhcSR promoter-lux reporter system, pCY106 (Fig. 1A), and detected the yhcSR expression levels by quantitatively measuring bioluminescence signal intensity during growth under different conditions. We found that under aerobic conditions yhcSR was highly expressed from the early log to the middle log phases of growth and then was dramatically repressed after the late log phase of growth (Fig. 1B). Similarly, under anaerobic conditions, yhcSR exhibited higher expression levels from the early log (OD, ∼0.2) to mid-log (OD, ∼0.5) phases of growth than in other stages of growth (Fig. 1C). Interestingly, we found that under anaerobic conditions, the addition of sodium nitrate in the chemically defined medium dramatically elevated the yhcSR expression (Fig. 1C), whereas the addition of sodium nitrite had no impact on yhcSR expression (Fig. 1D). Moreover, under aerobic conditions, the addition of nitrate had no influence on yhcSR expression (Fig. 1B). In addition, we found that neither calcium nor magnesium, nor pH, had any impact on yhcSR expression (data not shown). The above results suggest that the YhcSR system may be involved in the modulation of alternative respiratory functions under anaerobic conditions.
Fig. 1.
(A) Construction of yhcSR promoter-lux reporter fusion. (B) Examination of yhcSR expression under aerobic conditions in CDM. (C and D) Determination of the effects of nitrate (20 mM NaNO3) and nitrite (5 mM NaNO2) on yhcSR expression under anaerobic conditions in the CDM using the yhcSR promoter-lux reporter fusion. The results are representative of three independent experiments.
Downregulation of yhcSR expression eliminates the enhancive effect of nitrate on bacterial growth under anaerobic conditions.
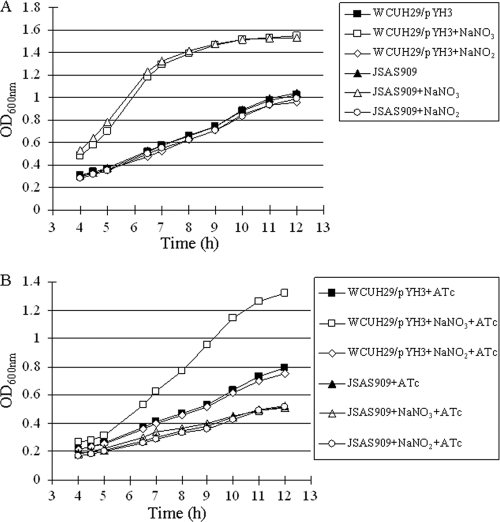
Bioinformatic analysis of the YhcR amino acid sequence revealed that YhcR has more than 30% identity with the NarL protein, suggesting that the YhcSR system may belong to the NarL family and be associated with regulation of the nitrate respiratory pathway in S. aureus (data not shown). A TetR-regulated antisense RNA technology has been successfully used to investigate gene function in S. aureus (10, 29). Our previous studies have demonstrated that induced yhcS antisense RNA can effectively downregulate endogenous yhcR expression (29). In order to test whether YhcSR is involved in regulation of the nitrate respiratory pathway, we utilized the same TetR-regulated yhcS antisense RNA expression system and examined the impact of downregulating yhcSR expression on growth in the presence or absence of nitrate under anaerobic conditions. S. aureus strain WCUH29 carrying parental plasmid pYH3 (WCUH29/pYH3) was used as a control. Without the inducer, anhydrotetracycline (ATc), the growth of both the yhcS antisense RNA mutant (JSAS909) and the control strain (WCUH29/pYH3) was strongly enhanced by the addition of nitrate in CDM (Fig. 2A). In contrast, in the presence of the inducer, ATc, only the control strain exhibited enhanced growth with the addition of nitrate, whereas no difference in growth was detected for the yhcS antisense mutant between cultures with and without the addition of nitrate (Fig. 2B). As a control, we also investigated the influence of nitrite on S. aureus growth under anaerobic conditions using both the yhcS antisense mutant and the control. We found that the addition of nitrite in CDM had no impact on the growth of both yhcS antisense and control strains in the absence and presence of inducer ATc (Fig. 2A and B). The above data suggest that the YhcSR system may specifically sense nitrate and participate in the nitrate respiratory pathway of S. aureus under anaerobic conditions.
Fig. 2.
Growth curves of S. aureus control (WCUH29/pYH3) and yhcS antisense isogenic mutant (JSAS909) in CDM in the absence of nitrate or in the presence of nitrate (20 mM NaNO3) or nitrite (5 mM NaNO2) without the inducer, ATc (A), and with ATc (500 ng/ml) (B) under anaerobic conditions. The results are representative of three independent experiments.
Downregulation of yhcSR expression dramatically decreased the transcription of genes involved in nitrate respiration.
Nitrate reductase (NarG) is a membrane-bound enzyme and involved in respiratory energy conservation (21). To elucidate the involvement of YhcSR in the regulation of the nitrate respiratory pathway, we examined the impact of YhcSR on the transcription of narG. The real-time PCR results showed that the downregulation of yhcSR apparently decreased the transcription of narG (52-fold).
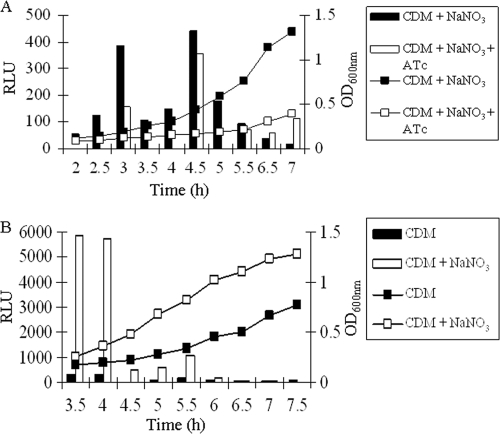
To further confirm the transcriptional regulation, we constructed a narG promoter-lux reporter fusion system. This system allows us to effectively downregulate yhcSR expression and simultaneously to monitor the reporter gene expression by measuring the bioluminescence intensity (32). The results showed that the bioluminescence intensity was apparently decreased from early log to late log phases of growth during yhcSR downregulation (Fig. 3A). To determine whether the regulatory pattern of YhcSR is correlated with the narG expression patterns, we constructed a narG promoter-lux reporter system. Consistent with the above results, in the presence of nitrate, the expression of narG was dramatically upregulated from early log to late log phases of growth (Fig. 3B). Taken together, these data indicate that the YhcSR system transcriptionally regulates the expression of narG.
Fig. 3.
(A) Analysis of transcriptional regulation of the narG gene by YhcSR using the narG promoter-lux reporter fusion. Solid bar and square, YJ2185 incubated in CDM with nitrate; open bar and square, YJ2185 incubated in CDM with nitrate in the presence of inducer, ATc (500 ng/ml). (B) Examination of narG expression pattern using the narG promoter-lux reporter fusion in CDM in the absence or presence of nitrate (20 mM NaNO3). The impact of the downregulation of yhcSR on narG expression was determined by monitoring the bioluminescence intensity during growth. The results are representative of three independent experiments.
YhcSR directly regulates the transcription of narG in S. aureus.
Although it has been reported that the NreBC system positively regulates the narG and nirR operons (26), it is possible that the YhcSR system also directly controls the expression of narG. To test this possibility, we expressed and purified His-tagged YhcR regulator protein and performed gel shift assays to examine whether YhcR binds to the promoter regions of narG. Negative controls included the labeled narG promoter region without protein and the labeled narG with BSA protein. The nonlabeled narG promoter region was used as a specific competitor, while an internal narG fragment probe was used as a nonspecific binding control. The results showed that the addition of YhcR in the reaction mixtures containing the narG promoter probe formed DNA-protein complexes and led to electrophoretic retardation of the complexes' mobility in a dose-dependent manner compared to the controls (Fig. 4). These data indicate that YhcR specifically binds to the narG promoter region.
Fig. 4.
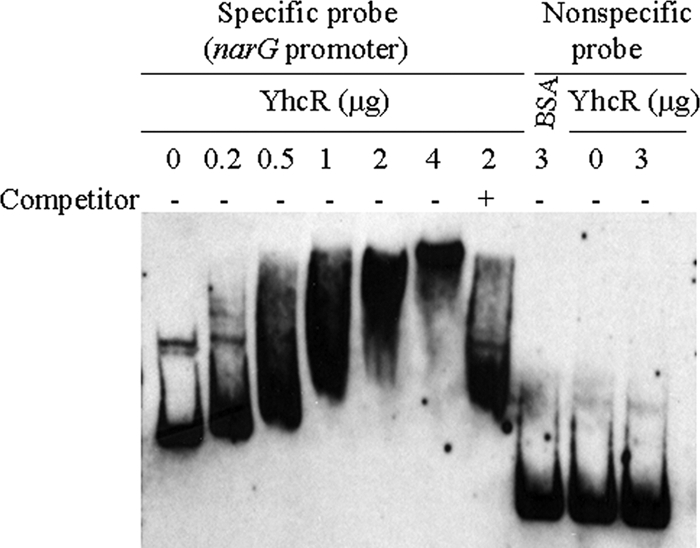
Gel shift mobility analysis of the narG gene regulated by YhcR. The promoter region of the narG gene was obtained as described in the text. The mobility of the labeled promoter fragment without the addition of YhcR is shown in the first lane. Different amounts of YhcR (0.2, 0.5, 1, 2, or 4 μg) were incubated with each DIG-labeled promoter probe narG in a 20-μl reaction volume. −, incubation without unlabeled specific competitor; +, incubation in the presence of 100-fold extra unlabeled specific competitor. BSA (3 μg) and a nonspecific internal gene probe were used as nonspecific binding controls. Approximately 0.2 pmol of DIG-labeled promoter DNA fragment was used in each reaction mixture.
The YhcSR system modulates the transcription of the oxygen-responsive NreABC two-component signal regulatory system in S. aureus.
It has been reported that a two-component system, NreBC, positively controls narG and nirR operons in both S. aureus (26) and Staphylococcus carnosus (4). This led us to hypothesize that the YhcSR system may directly or indirectly control the expression of NreBC. To test this possibility, we first performed quantitative RT-PCR analyses and revealed that the downregulation of yhcSR significantly reduced the nreB transcription (12-fold). Then, we employed gel shift assays to further determine whether the YhcR system directly or indirectly regulates nreABC. With the addition of YhcR, the DNA-YhcR complex was specifically formed in a dose-dependent manner, compared to controls (Fig. 5); this indicates that YhcR is likely to directly regulate the transcription of nreABC.
Fig. 5.
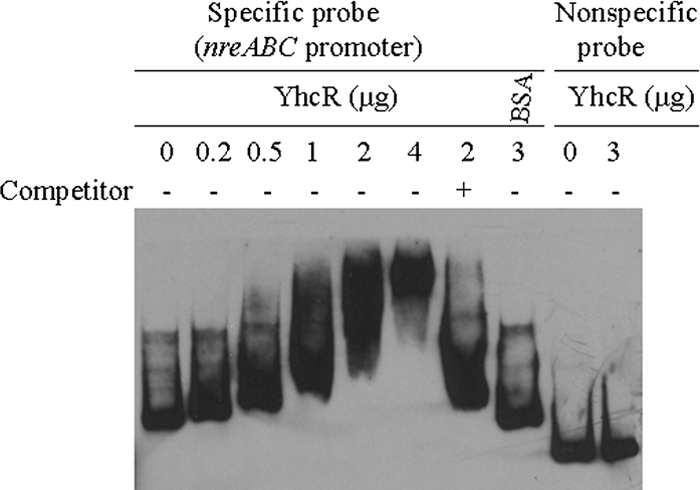
Gel shift mobility analysis of the nreABC gene regulated by YhcR. The promoter region of the nreABC gene was obtained as described in the text. The mobility of the labeled promoter fragment without addition of YhcR is shown in the first lane. Different amounts of YhcR (0.2, 0.5, 1, 2, or 4 μg) were incubated with each DIG-labeled promoter probe nreABC in a 20-μl reaction volume. −, incubation without unlabeled specific competitor; +, incubation in the presence of 100-fold extra unlabeled specific competitor. BSA (3 μg) and a nonspecific internal gene probe were used as nonspecific binding controls. Approximately 0.2 pmol of DIG-labeled promoter DNA fragment was used in each reaction mixture.
DISCUSSION
Two-component signal transduction systems play important roles in the ability of bacteria to adapt to various environments by sensing changes in their environment and subsequently altering gene expression levels (27). With the availability of S. aureus genomes, at least 16 pairs of two-component signal transduction systems have been revealed (14), with at least one being essential (19). Previous studies in our laboratory have demonstrated that a novel two-component signal system, YhcSR, is also required for bacterial viability in S. aureus (29); however, the biological functions of the YhcSR system remain poorly elucidated. This study provides the earliest direct evidence that the novel essential system, YhcSR, may sense nitrate and contributes to the modulation of the nitrate respiratory pathway by way of direct transcriptional regulation of the narG and nreABC operons under anaerobic conditions.
We have unveiled the different effects of nitrate on yhcSR expression during different stages of growth. Our yhcSR promoter-lux reporter results support the idea that YhcSR plays an important role in regulation of the nitrate respiratory pathway in the log phase of growth under anaerobic conditions. This finding is not surprising, since it has been previously demonstrated that a well-studied agr system differentially regulates the expression of both cell wall-associated proteins and exported toxins in different phases of cell growth (22). It is well known that nitrate reductase, a membrane-bound enzyme, is involved in respiratory energy conservation (21). The nitrate reductase operon consists of narG, narH, narJ, and narI genes, which are involved in the nitrogen metabolic pathway. Under anaerobic conditions, the transcription of genes, including narHJI and nasD (nirB), was upregulated (6). Our semiquantitative RT-PCR results have revealed that YhcSR positively controls the transcription of the narG operon. Our promoter-lux reporter experiments showed that under anaerobic conditions, both yhcSR and narG genes are highly expressed between early log and log phases of growth, which is consistent with the findings that YhcSR effectively regulates narG expression between early log and log phases of growth. These compelling data indicate the early regulatory function of YhcSR in respiratory energy conservation in oxygen-limited environments for S. aureus.
It has been reported that a two-component system, NreBC, positively controls narG and nirR operons in both S. aureus (26) and S. carnosus (4). Our results also reveal the regulatory function of YhcSR for narG expression. Our gel mobility shift DNA binding assays showed that YhcR bound to the promoter regions of narG and nreABC in a dose-dependent manner, whereas unlabeled competitors totally eliminated the shifted band, and internal control regions of narG and nreB failed to form a complex with YhcR. These data demonstrate that YhcR specifically binds to both the nar and nre promoter regions and indicate that YhcSR controls the production of nitrate reductase directly, by regulating the transcription of narG, and indirectly, by regulating the expression of NreABC, a narG regulator.
We also found that the upstream promoter regions of narG and nreABC showed multiple shifted bands with a low concentration of YhcR, suggesting that these promoter regions may bind YhcR as a dimer or at multiple sites. This phenomenon has been revealed in different regulators, including OmpR, SarA, and SrrA (9, 18, 24). In order to identify a potential consensus sequence of YhcR binding sites, it is necessary to define the YhcR binding sites in the promoter regions of narG and nreABC, which is beyond the scope of this report. To date, the TCS transduction systems, NreABC and YhcSR, have been shown to modulate the expression of genes involved in the nitrate respiratory pathway under oxygen-limited conditions. It remains to be determined which regulator is more important for the regulation of the nitrate respiratory pathway. Although our current studies have demonstrated that this novel YhcSR system regulates the expression of narG and nreABC genes at the transcriptional level, we cannot rule out the possible existence of posttranscriptional regulatory mechanisms. Despite the importance of narGHJI and nreABC as fitness factors under anaerobic conditions in the presence of nitrate as previously reported (26), neither operon is essential for cell viability (5, 26). Studies to determine the hierarchical regulation of nitrate respiration, as well as studies to elucidate the mechanism of YhcSR's essentiality, are ongoing in our laboratory.
ACKNOWLEDGMENTS
We thank Jeffrey W. Hall and Valerie Neff for critical reading of the manuscript.
This project was supported by grant AI057451 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Birkey S. M., Liu W., Zhang X., Duggan M. F., Hulett F. M. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of the ResD. Mol. Microbiol. 30:943–953 [DOI] [PubMed] [Google Scholar]
- 2. Burke K. A., Lascelles J. 1975. Nitrate reductase system in Staphylococcus aureus wild type and mutants. J. Bacteriol. 123:308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cramton S., Ulrich M., Gotz F., Doring G. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fedtke I., Kamps A., Krismer B., Gotz F. 2002. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J. Bacteriol. 184:6624–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forsyth R. A., et al. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387–1400 [DOI] [PubMed] [Google Scholar]
- 6. Fuchs S., Pane-Farre J., Kohler C., Hecker M., Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189:4275–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gross R. 1993. Signal transduction and virulence regulation in human and animal pathogens. FEMS Microbiol. 10:301–326 [DOI] [PubMed] [Google Scholar]
- 8. Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885 [DOI] [PubMed] [Google Scholar]
- 9. Huang K. J., Lan C. Y., Igo M. M. 1997. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. U. S. A. 94:2828–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ji Y., Woodnutt G., Rosenberg M., Burnham M. K. R. 2002. Identification of essential genes in Staphylococcus aureus using inducible antisense RNA. Methods Enzymol. 358:123–128 [DOI] [PubMed] [Google Scholar]
- 11. Ji Y., Yu C., Liang X. 2007. Transcriptomic analysis of ArlSR two-component signaling operon in Staphylococcus aureus. Methods Enzymol. 423:502–513 [DOI] [PubMed] [Google Scholar]
- 12. Kamps A., Achebach S., Fedtke I., Unden G., Götz F. 2004. Staphylococcal NreB: an O(2)-sensing histidine protein kinase with an O(2)-labile iron-sulphur cluster of the FNR type. Mol. Microbiol. 52:713–723 [DOI] [PubMed] [Google Scholar]
- 13. Kazakova S. V., et al. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468–475 [DOI] [PubMed] [Google Scholar]
- 14. Kuroda M., et al. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 19:807–821 [DOI] [PubMed] [Google Scholar]
- 15. Li Y. H., et al. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacterol. 184:6333–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang X., et al. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manna A., Ingavale S., Maloney M., van Wamel W., Cheung A. L. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186:5267–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manna A., Cheung A. L. 2006. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbiol. 60:1289–1301 [DOI] [PubMed] [Google Scholar]
- 19. Martin P. K., Li T., Sun D., Biek D. P., Schmid M. B. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller A. A., Engleberg N. C., DiRita V. J. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976–990 [DOI] [PubMed] [Google Scholar]
- 21. Neubauer H., Gotz F. 1996. Physiology and interaction of nitrate and nitrite reduction in Staphylococcus carnosus. J. Bacteriol. 178:2005–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Novick R. 2006. Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation, p. 496–516 In Fischetti V., et al. (ed.), Gram-positive pathogens. ASM Press, Washington, DC [Google Scholar]
- 23. Park M., Myers R., Marzella L. 1992. Oxygen tensions and infections: modulation of microbial growth, activity of antibacterial agents, and immunologic responses. Clin. Infect. Dis. 14:720–740 [DOI] [PubMed] [Google Scholar]
- 24. Pragman A., Yarwood J., Tripp T., Schlievert P. M. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rice L. 2006. Antimicrobial resistance in Gram-positive bacteria. Am. J. Med. 119:S11–S19 [DOI] [PubMed] [Google Scholar]
- 26. Schlag S., et al. 2008. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 190:7847–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 28. Strasters K. C., Winkler K. C. 1963. Carbohydrate metabolism of Staphylococcus aureus. J. Gen. Microbiol. 33:213–229 [DOI] [PubMed] [Google Scholar]
- 29. Sun J., Zheng L., Landwehr C., Yang J., Ji Y. 2005. Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J. Bacteriol. 187:7876–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Throup J. P., et al. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401 [DOI] [PubMed] [Google Scholar]
- 31. Unden G., Schirawski J. 1997. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol. Microbiol. 25:205–210 [DOI] [PubMed] [Google Scholar]
- 32. Yan M., Yu C., Yang J., Ji Y. 2009. Development of shuttle vectors for evaluation of essential regulator regulated gene expression in Staphylococcus aureus. Plasmid. 61:188–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yarwood J. M., McCormick J., Schlievert P. M. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yarwood J. M., Schlievert P. M. 2000. Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J. Clin. Microbiol. 38:1797–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]