Abstract
The genome of Burkholderia cenocepacia contains two genes encoding closely related LysR-type transcriptional regulators, CysB and SsuR, involved in control of sulfur assimilation processes. In this study we show that the function of SsuR is essential for the utilization of a number of organic sulfur sources of either environmental or human origin. Among the genes upregulated by SsuR identified here are the tauABC operon encoding a predicted taurine transporter, three tauD-type genes encoding putative taurine dioxygenases, and atsA encoding a putative arylsulfatase. The role of SsuR in expression of these genes/operons was characterized through (i) construction of transcriptional reporter fusions to candidate promoter regions and analysis of their expression in the presence/absence of SsuR and (ii) testing the ability of SsuR to bind SsuR-responsive promoter regions. We also demonstrate that expression of SsuR-activated genes is not repressed in the presence of inorganic sulfate. A more detailed analysis of four SsuR-responsive promoter regions indicated that ∼44 bp of the DNA sequence preceding and/or overlapping the predicted −35 element of such promoters is sufficient for SsuR binding. The DNA sequence homology among SsuR “recognition motifs” at different responsive promoters appears to be limited.
INTRODUCTION
Members of the betaproteobacterial genus Burkholderia are widespread in diverse natural environments, including soil, the plant rhizosphere, and freshwater and urban settings (8). Some species, e.g., members of Burkholderia cepacia complex (BCC) are also well recognized as opportunistic human pathogens (6, 7, 22, 23, 24, 33, 34). Among them, B. cenocepacia is associated with severe infections of the respiratory tract in immunocompromised individuals, in particular, patients with cystic fibrosis (CF).
Connections between sulfur metabolism and virulence traits of various bacterial pathogens have been observed previously, e.g., the role of cysteine in repression of toxin synthesis in Bordetella and Clostridium species; the induction of sulfur metabolism-related genes upon interaction of host cells with Mycobacterium species or meningococci; a decreased virulence of mutants impaired in various steps of sulfur metabolism in Mycobacterium, Salmonella, and Brucella species; the importance of the master regulator of cysteine metabolism, CymR, for survival of Staphylococcus aureus in the host; and the role of the CysB regulator in expression of the alternative sigma factor PvdS- and PvdS-dependent virulence phenotypes of Pseudomonas aeruginosa (3, 4, 10, 12, 13, 16, 19, 21, 30). In B. cenocepacia, sulfate transport has been reported to be crucial for the biosynthesis of the siderophore pyochelin, one of the putative virulence factors of this organism (11, 28, 29).
The assimilation of sulfur from inorganic sulfate through the cysteine biosynthetic pathway and its regulation in Gram-negative bacteria are often assumed to follow the paradigm of the well-characterized “cysteine regulon” of Salmonella enterica serovar Typhimurium and Escherichia coli (20). Over 20 genes have been assigned to the cys regulon of Salmonella species. These genes are coordinately controlled by the LysR-type transcriptional activator CysB, and the molecular mechanisms underlying CysB-mediated transcriptional control in Salmonella species have been thoroughly described (20). Although the cysB gene is ubiquitous among genomes of Gram-negative bacteria, E. coli and several other species (but not all) possess an additional open reading frame (ORF) encoding a protein sharing significant amino acid sequence similarity with CysB (17). Studies on the function of this CysB-like regulator (Cbl) of E. coli disclosed its connection with the expression of genes involved in utilization of sulfur sources alternative to inorganic sulfate. In E. coli, the sulfate starvation-induced (ssi) genes whose transcription is dependent on Cbl are represented by the tauABCD and ssuEADCB operons, encoding transport and desulfonation functions for taurine and aliphatic sulfonates, respectively (35, 36, 37). Cbl activates transcription from the tauAP and ssuEP promoters when bound to loosely conserved 44-bp sequences preceding the promoter −35 region (5). An additional element of Cbl-mediated control is delivered by adenosine 5′ phosphosulfate (APS) which directly interacts with Cbl and abolishes its activating function (5, 31). The distinction between functions of CysB and CysB-like regulators in other bacteria (containing two cysB paralogs) has not been investigated.
In our previous study (18), we started a systematic experimental characterization of sulfur assimilation-related processes and their regulation in B. cenocepacia. Using the B. cenocepacia strain 715j (an isolate from a CF patient), we have identified two cysB-like genes encoding LysR-type transcription factors sharing homology with CysB and Cbl of E. coli as well as significant mutual homology. By studying the roles of both regulators in expression of some “signature” genes/operons involved in the utilization of sulfate and aliphatic sulfonates for cysteine biosynthesis, we proposed to designate these proteins CysBBc and SsuRBc. We noticed that the functions of B. cenocepacia CysB and SsuR may partially replace each other in the regulation of some target promoters, e.g., sbpP and cysIP (controlling expression of the sbp cysTWA operon encoding the sulfate transporter and the cysIXHD1NG operon encoding sulfate activation and reduction functions, respectively). One example of a B. cenocepacia promoter upregulated specifically by SsuR was identified as ssuDP, preceding the ssuDBC cluster that encodes aliphatic sulfonate transport and desulfonation functions (18).
Here we describe the identification of other genes whose expression is specifically dependent on the SsuR transcription factor in B. cenocepacia. An attempt to identify a recognition sequence for SsuR is also presented.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
B. cenocepacia strains used in this study are shown in Table 1. Growth conditions were essentially the same as those described previously (18). The modified sulfur-free M9 medium was supplemented with the alternative sole sulfur sources listed in Table 2, used at a concentration of 0.25 mM (sodium salts) each or 0.1 mM for l-cystine or l-methionine. Utilization of particular sulfur sources was tested in liquid cultures by inoculation of bacteria from a single colony in 2 ml of appropriate medium to a starting A600 of <0.02, followed by determining the A600 after overnight growth (16 to 18 h) at 37°C with aeration.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description (genotype or relevant features) | Reference(s) or source |
|---|---|---|
| B. cenocepacia strains | ||
| 715j | CF isolate, prototroph | 9,11,25 |
| 715j-ssuR::Tp | 715j with dfrB2 gene inserted in ssuR | 18 |
| 715j-cysB::Tp | 715j with dfrB2 gene inserted in cysB | 18 |
| Plasmids | ||
| pGEM-T Easy | Vector for direct cloning of PCR fragments; Apr | Promega |
| pKAGd4 | Broad-host-range lacZ transcription fusion vector; Apr Cmr | 1,27 |
| pMH611 | Contains whole intergenic region preceding the ssuD ORF (ssuDP, amplified with primers SSUDfw1 and SSUDrev2) in pKAGd4 | 18 |
| pMH639 | Contains ORF cysBBc with His6 codons at the 3′ terminus in the ptrc99A vector under promoter trcP | This study |
| pMH654 | Contains ORF ssuRBc with His6 codons at the 3′ terminus in the ptrc99A vector under promoter trcP | This study |
| pMH689 | tauD1 promoter region (amplified with primers TAUD1fw1 and TAUD1rev2) in the pKAGd4 BamHI site | This study |
| pMH802 | tauD2b promoter region (amplified with primers TAUD2Bfw1 and TAUDBrev2) in the pKAGd4 HindIII-BamHI sites | This study |
| pMH803 | bcal1736 promoter region amplified with primers BCAL1736fw1 and BCAL1736rev2 in the pKAGd4 HindIII-BamHI sites | This study |
| pMH804 | ssuDP2 fragment (amplified with primers SSUDfw21 and SSUDrev22) inserted in the pKAGd4 HindIII-BamHI sites | This study |
| pMH805 | ssuDP1 fragment (amplified with primers SSUDfw25 and SSUDrev2) inserted in the pKAGd4 HindIII-BamHI sites | This study |
| pRI32 | tauD2a promoter region (amplified with primers TAUD2Afw1 and TAUDArev2) in the pKAGd4 BamHI site | This study |
| pRI33 | tauD3 promoter region (amplified with primers TAUD3fw1 and TAUD3rev2) in the pKAGd4 BamHI site | This study |
| pRI34 | tauA promoter region (amplified with primers TAUAfw1 and TAUArev2) in the pKAGd4 BamHI site | This study |
| pRI35 | atsA promoter region (amplified with primers ATSAfw1 and ATSArev2) in the pKAGd4 BamHI site | This study |
| pRI45 | atsAP1 fragment (amplified with primers ATSAfw16 and ATSArev17) in the pKAGd4 HindIII-BamHI sites | This study |
Table 2.
Oligonucleotides used in this studya
| Oligonucleotide | Sequence | Region (segment) [ORF] |
|---|---|---|
| ATSAfw1 | 5′-GGGATCCACCGCGTTCTTCCT-3′ | atsAP |
| ATSArev2 | 5′-GGGATCCACGATGCGAGCGAC-3′ | atsA [BCAM0569] |
| ATSAfw14 | 5′-GCATATTCCCCCACCC-3′ | atsAP (1) |
| ATSArev15 | 5′-CCCGCGCAGCGCCCAA-3′ | atsAP (1) |
| ATSAfw16 | 5′-GTAAGCTTGCATATTCCCCCACCCC-3′ | atsAP (1) |
| ATSArev17 | 5′-TTGGATCCGAGCGGCGATCTTAGC-3′ | atsAP (1) |
| BCAL1736fw1 | 5′-GTAAGCTTTCGTCTTCAGGACC-3′ | bcal1736P |
| BCAL1736rev2 | 5′-GTGGATCCTTCCACTTTCGCAC-3′ | BCAL1736 |
| SSUDfw1 | 5′-GGAATTCGGATAGGTGCGC-3′ | ssuDP |
| SSUDrev2 | 5′-AAGGATCCAGAACACATTC-3′ | ssuD [BCAL1619] |
| SSUDfw11 | 5′-CCATCCCGATTCGAAT-3′ | ssuDP (1) |
| SSUDrev12 | 5′-TCGCGTCGCGGAGGAG-3′ | ssuDP (1) |
| SSUDfw13 | 5′-CGCCATTATAGGGC-3′ | ssuDP (4) |
| SSUDrev14 | 5′-TGATTAGCACGACGCT-3′ | ssuDP (4) |
| SSUDfw15 | 5′-CAGAAACAGCCGTGC-3′ | ssuDP (2) |
| SSUDrev16 | 5′-CTCGCCGGTCAGGCC-3′ | ssuDP (2) |
| SSUDfw17 | 5′-CGAAAGCGGAAAACC-3′ | ssuDP (3) |
| SSUDrev18 | 5′-AGGAATCAGTCTAGG-3′ | ssuDP (3) |
| SSUDfw21 | 5′-AGCTTCCTCAGAAACAGCCGTGCGATCGCGTCCTCGCCCGCTGCACGGCCTGACCGGCGAGCCATCCCGATTCGAATGCAG-3′ | ssuDP (1-2) |
| SSUDrev22 | 5′-GATCCTGCATTCGAATCGGGATGGCTCGCCGGTCAGGCCGTGCAGCGGGCGAGGACGCGATCGCACGGCTGTTTCTGAGGA-3′ | ssuDP (1-2) |
| SSUDfw25 | 5′-AAAGCTTCCATCCCGATTCGAAT-3′ | ssuDP (1) |
| TAUAfw1 | 5′-GGATCCGAGCATCGCACAC-3′ | tauAP |
| TAUArev2 | 5′-GGATCCGGATGAAGCTGAAA-3′ | tauA [BCAL0711] |
| TAUAfw10 | 5′-CAATCCGGTCGAGCGT-3′ | tauAP (1) |
| TAUArev11 | 5′-TGCGAAGCTATCGATG-3′ | tauAP (1) |
| TAUD1fw1 | 5′-GGATCCCAAGGACACCTTCGAAGGCT-3′ | tauD1P |
| TAUD1rev2 | 5′-GGATCCTCCAGCAGTTCCGCACCGAT-3′ | tauD1P [BCAL1738] |
| TAUD2Afw1 | 5′-GTGGATCCACGCTGCAATAACG-3′ | tauD2aP |
| TAUD2Arev2 | 5′-GTGGATCCATCACGTCGACCG-3′ | tauD2a [BCAM1121] |
| TAUD2Bfw1 | 5′-GTAAGCTTATTCATTGGAATTCGC-3′ | tauD2bP |
| TAUD2Brev2 | 5′-TTGGATCCAAAACGACGGAGCGG-3′ | tauD2b [BCAM1122] |
| TAUD3fw1 | 5′-GTGGATCCATAGTCGGCTCGCG-3′ | tauD3P |
| TAUD3rev2 | 5′-GTGGATCCGGCACGATGTCG-3′ | tauD3 [BCAS0426] |
| TAUD3fw10 | 5′-TCTTTATCACCGCAGC-3′ | tauD3P (1) |
| TAUD3rev11 | 5′-GGCGTGCGTCAACGAAG-3′ | tauD3P (1) |
DNA manipulations.
Standard procedures (2) were used for restriction enzyme digestions, ligation, 5′-end labeling of DNA fragments, and transformation of E. coli. Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, and DNase were obtained from MBI Fermentas or Invitrogen. Taq polymerase was purchased from MBI Fermentas, and High Fidelity PCR master mix was from Roche. [γ-32P]ATP used for DNA 5′-end labeling was from Amersham Pharmacia Biotech. All other chemicals (of the highest purity grade available) were from Sigma-Aldrich, Fluka, Promega, or Merck. Oligonucleotide synthesis and DNA sequencing (using the dideoxy chain termination method and an ABI Prism 3730 DNA sequencer [Applied Biosystems]) were performed at the Institute of Biochemistry and Biophysics, Warsaw, Poland.
Plasmids used in this study are listed in Table 1. All the B. cenocepacia 715j sequences used for plasmid constructions were amplified by PCR from SacII-digested total genomic DNA as a template and the appropriate oligonucleotide primers listed in Table 2. Routinely, the PCR products were ligated with pGEM-T Easy vector and independent isolates of each construct were sequenced to ensure that no undesired mutations had been introduced during PCR analysis. Inserts were recovered from pGEM-T Easy derivatives by restriction enzyme digestions, and they were subsequently cloned into appropriate vectors. In some cases, DNA fragments obtained by annealing two complementary oligonucleotides were cloned directly into pKAGd4 vector. The resultant pKAGd4 derivatives were introduced to the B. cenocepacia 715j strain and to the isogenic ssuR and cysB mutants via conjugation according to the procedure described previously (18).
β-Galactosidase assays.
The inocula for performing β-galactosidase assays on B. cenocepacia strains harboring plasmid-encoded promoter-lacZ fusions were prepared from three independent transformants containing each pKAGd4 derivative and grown in M9 with glucose (0.5%), l-djenkolic acid (S,S′-methylene-bis-cysteine) (1 mM) as a sulfur source, 18 amino acids (10 μg/ml each, except cysteine and methionine), and chloramphenicol (50 μg/ml). Following overnight growth, the cultures were diluted into fresh medium to a starting A600 of <0.02, grown to early log phase (A600, 0.3 to 0.4), chilled, and used directly for the enzyme assay. β-Galactosidase activities were determined according to the Miller method (26) with o-nitrophenyl-β-galactoside (ONPG) as a substrate. All assays were repeated at least twice.
Protein preparations.
Plasmids encoding B. cenocepacia CysB and SsuR that were C-terminally tagged with 6 histidine residues (pMH639 and pMH654, respectively) were used for overproduction of recombinant proteins in E. coli according to a procedure described previously (18). Protein extracts enriched with CysB(His6) and SsuR(His6) (protein fractions precipitated with 229 mg/ml of ammonium sulfate) were applied to the 2-ml columns with nickel-nitrilotriacetic acid agarose (Ni-NTA; Qiagen) and fractionated by imidazole elution according to the manufacturer's recommendation. The purity of obtained CysB(His6) and SsuR(His6) preparations was estimated as >90% by SDS-PAGE and Coomassie blue staining. Proteins were stored in aliquots at −20°C in 50 mM Tris-Cl (pH 7.5), 1 mM Na2EDTA, and 30% glycerol at a protein concentration of 1 to 2 mg/ml.
Electrophoretic mobility shift assays (EMSAs).
Fragments of DNA containing promoter regions of interest were labeled at the 5′ ends by [γ-32P]ATP and polynucleotide kinase. Reaction mixtures (20 μl) contained approximately 10 ng of labeled DNA fragment and 2 μg of sonicated calf thymus DNA per ml (as a nonspecific competitor) in a buffer consisting of 40 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol, and 100 μg bovine serum albumin per ml, plus CysB(His6) or SsuR(His6) at final concentrations of 1 to 3 μg/ml. Reaction mixtures were incubated at 37°C for 5 min and then separated in a 5% acrylamide-bisacrylamide (82:1) nondenaturing gel in 0.05 M Tris-borate-EDTA buffer (pH 8.3) for 1.5 h at 10 V/cm. Radiolabeled bands were visualized by autoradiography. In some assays utilizing short (∼50-bp) DNA fragments, nonradioactive DNA probes were used for incubation with SsuR protein and the reaction mixture products were separated on 1.5% agarose gels (High Resolution BioShop) and visualized by SYBRGold (Invitrogen) staining.
DNase I protection assay.
The ssuD “promoter fragment” (from position −280 to +12, relative to the first base of the ATG translation start codon) was amplified by PCR with pMH611 as a template and primers SSUDfw1 and SSUDrev2, of which either the former or the latter was 5′ labeled with [γ-32P]ATP and polynucleotide kinase. Incubation mixtures (20 μl) contained approximately 50 ng (1.5 × 105 dpm) of the ssuD fragment and 2 μg sonicated calf thymus DNA in a buffer consisting of 40 mM Tris-HCl (pH 8.0), 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, and 100 μg/ml bovine serum albumin. DNase I (Promega) was used at a final concentration of 3.3 × 10−6 U/μl, and digestion was performed as described previously (5) in the absence or presence of the SsuR protein, either purified SsuR(His6) or a cellular extract enriched with native SsuR (as described previously [18]). The products of DNase digestion were analyzed by electrophoresis in 6% polyacrylamide-7 M urea gels alongside a sequencing reaction standard generated with pMH611 template and 32P-labeled primer SSUDfw1 or SSUDrev2.
RESULTS AND DISCUSSION
Utilization of sulfur sources by wild-type (wt) B. cenocepacia 715j and the ssuR-null mutant.
A growth screen performed with the wild-type B. cenocepacia strain 715j revealed that this strain is able to utilize a number of inorganic and organic substrates to satisfy its sulfur requirement (Table 3). An analogous screen of the ssuR-null mutant showed that the function of the SsuR regulator is negligible for the sulfate/thiosulfate assimilatory pathway but is required for utilization of several organosulfur compounds. In this respect, SsuR seems to be a functional counterpart of the E. coli transcription factor Cbl, whose targets (in E. coli) include the tauABCD and ssuEADCB operons involved in utilization of taurine and aliphatic sulfonates. The growth test also suggested that SsuR of B. cenocepacia controls a number of genes required for acquisition of sulfur from both environmental sources (aliphatic and aromatic sulfonates, aromatic sulfate esters, dimethyl sulfoxide [DMSO]) and sources of human origin (taurine, glycosamine sulfate esters, and sulfated glycosaminoglycans).
Table 3.
Utilization of sulfur sources by B. cenocepacia 715j (wild type) and ssuR-null mutant
| Sulfur source | Growth for indicated B. cenocepacia straina |
|
|---|---|---|
| 715j | ssuR-null mutant | |
| Sulfate | +++ | +++ |
| Sulfite | +++ | +++ |
| Thiosulfate | +++ | +++ |
| Sulfide | +++ | +++ |
| l-Cystine | +++ | +++ |
| l-Djenkolic acid | +++ | +++ |
| l-Methionine | +++ | + |
| Ethanesulfonate | +++ | − |
| Taurine | +++ | − |
| SDS | +++ | − |
| DMSO | +++ | − |
| Benzenesulfonate | +++ | − |
| p-Nitrophenyl sulfate | +++ | − |
| N-Acetylglucosamine-6-sulfate | +++ | − |
| N-Acetylgalactosamine-6-sulfate | +++ | − |
| Heparan sulfate | + | − |
| Chondroitin-6-sulfate | +++ | − |
Bacterial growth was determined spectrophotometrically following overnight growth at 37°C. +++, full growth (A600 > 1.5); +, partial growth (A600, 0.3 to 0.4); −, no growth (A600 < 0.05).
Candidate target genes for SsuR-mediated regulation.
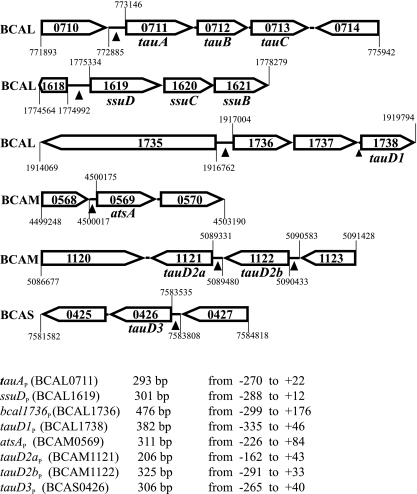
In our search for potential target genes for SsuR-mediated control we focused here on (i) the functions predicted to be involved in taurine utilization as a sulfur source and (ii) the function(s) that could account for utilization of aromatic and/or carbohydrate sulfate esters. The inspection of the genomic sequence of B. cenocepacia strain J2315 (available at http://www.sanger.ac.uk/Projects/B_cenocepacia/) (14) allowed us to recognize the presence and organization of genes potentially involved in these functions (Fig. 1).
Fig. 1.
Organization of B. cenocepacia genes investigated in this study. The gene locus tags, direction of transcription, and sequence coordinates are shown according to the published genomic sequence of B. cenocepacia J2315, available at the Wellcome Trust Sanger Institute website. The gene designations are proposed here on the basis of homology of the predicted products to their counterparts in E. coli (or other bacteria; see the text). Intergenic regions used for construction of fusions to the lacZ reporter gene and for binding of CysB and SsuR proteins are indicated by arrowheads. Sizes of these regions (bp) and sequence coordinates relative to the first base of the translation initiation codon of the following ORFs are shown below. Sequences of four of these regions (indicating segments that were analyzed further) are shown in Fig. 4.
The putative taurine transport system is encoded by chromosome 1 of J2315 by the cluster of ORFs (BCAL0711 to -0713) encoding proteins sharing a significant degree of identity/similarity with constituents of the ABC-type taurine transporter of E. coli, TauA (38%/68%), TauB (50%/80%), and TauC (49%/74%), respectively. It seems likely that the corresponding genes of B. cenocepacia (designated here as tauA, tauB, and tauC) (Fig. 1) form an operon whose expression might be regulated by elements within the intergenic region (260 bp) that precedes BCAL0711 and is designated here as the tauA promoter (tauAP). Unlike the E. coli situation, where the single tauABCD operon also encodes the taurine dioxygenase TauD (an enzyme involved in desulfonation of taurine), four tauD-like ORFs can be found in several locations in the B. cenocepacia genome (Fig. 1). The deduced products of these ORFs display comparable degrees of identity/similarity with E. coli TauD, as follows: BCAL1738 (proposed gene designation tauD1), 38%/61%; BCAM1121 (tauD2a), 35%/59%; BCAM1122 (tauD2b), 36%/54%; and BCAS0426 (tauD3), 38%/67%. Among them, tauD1 is likely to be expressed as a part of an operon containing three ORFs (BCAL1736 to -1738), since the distances between BCAL1736 and BCAL1737 as well as BCAL1737 and BCAL1738 are very short (5 bp and 12 bp, respectively). The candidate promoter of this operon is located between BCAL1735 and BCAL1736 (bcal1736P).
Three genes were identified in the genome of B. cenocepacia J2315 that encode putative sulfatases (BCAM0569, BCAM1158, BCAM2313). Among these, we focused our investigation on expression from the promoter region preceding BCAM0569. Sequences homologous to BCAM0569 are highly conserved (over 90% identity at the level of the encoded products) among all Burkholderia genomes sequenced so far (38). The protein encoded by BCAM0569 shares 38%/52% identity/similarity with arylsulfatase AtsA of Pseudomonas aeruginosa and with proteins annotated as “arylsulfatases” in other microorganisms, e.g., Acinetobacter baumannii (40%/52%), Mycobacterium avium (38%/51%), and some Bacteroides species (33%/48%). For this reason, we propose the gene designation atsA for BCAM0569.
Expression of SsuR-regulated genes in vivo.
To isolate putative regulatory regions preceding the ORFs of interest, the appropriate pairs of forward and reverse oligonucleotide primers (Table 2) were designed on the basis of the genomic sequence of B. cenocepacia strain J2315. These primers were used for PCR-mediated amplification of homologous gene fragments from genomic DNA of B. cenocepacia strain 715j. Nucleotide sequence analysis of the obtained promoter fragments confirmed their strict homology to the corresponding intergenic fragments of strain J2315 in accordance with the known close relatedness of these two strains. Altogether, seven long promoter fragments (including at least 160 bp of DNA sequence positioned upstream of the respective ORFs) (Fig. 1) were individually inserted upstream of the promoterless lacZ gene in the pKAGd4 transcriptional reporter plasmid. The resultant pKAGd4 derivatives were introduced into B. cenocepacia 715j and into the isogenic ssuR-null and cysB-null mutants, and the expression of lacZ driven by the individual promoter regions was monitored by measurements of β-galactosidase activities in strains carrying the respective plasmids. As shown in Table 4, the empty vector (control) used to construct the lacZ fusions allows for expression of some background β-galactosidase activity that might be due to transcription initiating from the multiple-cloning site (MCS) of pKAGd4. A relatively high level of reporter gene expression (above the control) directed from intergenic regions preceding tauA, tauD3, atsA, and BCAL1736 was observed in the wild-type (wt) strain as well as in the cysB mutant, whereas this expression was entirely abolished in the ssuR mutant. Expression from the other three promoter regions tested, tauD1P, tauD2aP, and tauD2bP, was rather low in comparison to that from tauAP, and a negative effect of ssuR mutation (a further 2-fold decrease in expression) was observed only in the case of tauD2bP. It may be noticed that among the four putative tauD homologs found in the B. cenocepacia genome (Fig. 1), the tauD2a gene is not upregulated by SsuR when expressed from its own promoter and there is no promoter activity in the region preceding the tauD1 gene (BCAL1738) (Table 4). However, an SsuR-activated promoter is located upstream of BCAL1736, and it is therefore likely that tauD1 is regulated by SsuR as part of an operon (BCAL1736 to -1738) under the control of bcal1736P (Fig. 1). Thus, the newly identified targets of SsuR include an operon potentially involved in taurine transport (tauABC), a gene encoding a putative arylsulfatase (atsA), three tauD-type genes potentially involved in taurine desulfonation (i.e., tauD1, tauD2b, and tauD3), and BCAL1736 and BCAL1737 genes encoding, respectively, a protein of unknown function and a putative quinone oxidoreductase. Among the members of the SsuR regulon identified so far, the ssuDBC operon expressed from the ssuD promoter appears to be most strongly activated by SsuR. We also found that lacZ expression directed by the ssuDP, tauAP, tauD3P, bcal1736P, and atsAP promoters in the wt strain was not affected by the presence of inorganic sulfate in the growth medium (Table 4). Consequently, the genes identified here as SsuR-responsive targets cannot be categorized among the sulfate starvation-induced (ssi) genes in contrast to the tauABCD and ssuEADCB operons of E. coli (35, 36, 37) and atsA of P. aeruginosa (15, 32). The molecular basis of sulfate starvation-induced expression of the ssu and tau genes in E. coli has been explained by the fact that the activity of the Cbl protein (acting as an essential transcriptional activator of the tau and ssu promoters) is abolished in the presence of adenosine 5′ phosphosulfate (APS), the first intermediate in the sulfate assimilatory pathway (5, 31). If the SsuR regulator of B. cenocepacia can be considered a functional counterpart of Cbl, it is noteworthy that SsuR seems to act in a constitutive manner, independent of the presence of sulfate and/or APS.
Table 4.
Measurements of reporter gene expression from promoter regions preceding the sulfur metabolism-related genes in B. cenocepacia
| Fusion carried by pKAGd4 | β-Galactosidase activity (Miller units) in indicated B. cenocepacia 715j strainc |
||||
|---|---|---|---|---|---|
| Wild type |
ssuR-null mutant |
cysB-null mutant −sulfate | |||
| −sulfate | +sulfate | −sulfate | +sulfate | ||
| None (empty vector) | 95 ± 5 | ND | 97 ± 6 | ND | 90 ± 10 |
| tauAP-lacZa | 1,273 ± 169 | 1,238 ± 125 | 84 ± 5 | 90 ± 5 | 944 ± 71 |
| tauD1P-lacZa | 105 ± 8 | ND | 113 ± 10 | ND | 111 ± 12 |
| bcal1736P-lacZa | 479 ± 12 | 506 ± 18 | 112 ± 10 | ND | 501 ± 16 |
| tauD2aP-lacZa | 227 ± 15 | ND | 199 ± 9 | ND | ND |
| tauD2bP-lacZa | 172 ± 19 | ND | 88 ± 5 | ND | 188 ± 10 |
| tauD3P-lacZa | 1,396 ± 75 | 1,435 ± 82 | 93 ± 8 | 119 ± 10 | 1,425 ± 120 |
| atsAP-lacZa | 1,788 ± 250 | 1,436 ± 20 | 90 ± 8 | 124 ± 22 | 1,790 ± 190 |
| ssuDP-lacZa | 4,684 ± 120 | 4,639 ± 150 | 31 ± 5 | ND | 4,690 ± 199 |
| atsAP1-lacZb | 650 ± 80 | ND | 56 ± 6 | ND | 626 ± 70 |
| ssuDP1-lacZb | 778 ± 90 | ND | 91 ± 5 | ND | 681 ± 50 |
| ssuDP2-lacZb | 341 ± 25 | ND | 92 ± 8 | ND | 351 ± 10 |
Fusions containing full-length promoter regions preceding particular ORFs (lengths of fragments inserted in pKAGd4 as indicated in Fig. 1).
Fusions containing promoter region segments (lengths of fragments designated atsAP1, ssuDP1, and ssuDP2 that were inserted in pKAGd4 as indicated in Fig. 4).
ND, not determined; −sulfate, without sulfate; +sulfate, with 0.25 mM sulfate in the culture medium.
Binding of SsuR to DNA at target promoter regions.
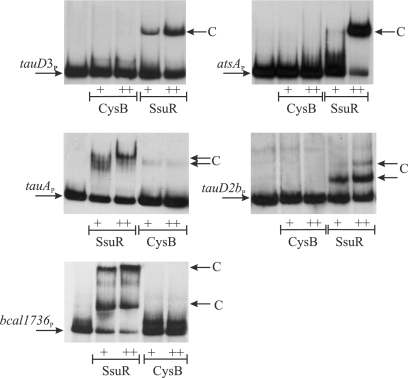
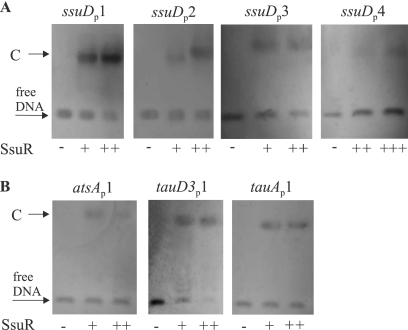
Since B. cenocepacia CysB and SsuR were previously demonstrated to act jointly at some target promoters (18), we decided to test the binding abilities of both regulators at tauAP, tauD2bP, tauD3P, atsAP, and bcal1736P. CysB and SsuR were purified as C-terminally His6-tagged recombinant proteins and they were used in an electrophoretic mobility shift assay (EMSA) with 32P-labeled promoter probes. The probes encompassed the same DNA regions as those used to construct the respective promoter-lacZ fusions. As shown in Fig. 2, all five promoter fragments tested were able to produce complexes (visualized as shifted bands) with SsuR protein, and none of them formed a complex with CysB. The relative rates of migration of complexes detected by the EMSAs were similar for SsuR bound to tauD3P and atsAP regions, and they were also similar to the EMSA result for the SsuR-ssuDP complex obtained previously (18). In the case of tauAP, at least two complexes of different mobilities were observed that resemble the EMSA result of the SsuR-sbpP interaction obtained previously (18). Two or three discrete shifted bands were produced by SsuR with the tauD2bP and bcal1736P probes, respectively, suggesting the formation of several DNA-protein complexes at these promoter regions. Since the sizes of all promoter probes used in the EMSA were comparable (around 300 bp), it seems evident that complexes formed by SsuR with different targets employ a different stoichiometry of promoter-regulator interactions. Assuming that the fast-migrating band observed in the EMSA with tauD2bP and bcal1736P may represent a primary complex with SsuR (one protein molecule bound to a single binding site), then the slower-migrating shifted bands could be considered higher-order complexes involving additional SsuR molecules. It seems, therefore, possible that the discrete “slow” complexes produced by SsuR with all five promoter regions tested (Fig. 2) represent SsuR interaction with more than one recognition element (binding site).
Fig. 2.
Binding of SsuR and CysB to DNA at target promoter regions. The electrophoretic mobility shift assays (EMSAs) were performed with radiolabeled promoter probes corresponding to the DNA fragments listed in Fig. 1. Purified SsuR(His6) or CysB(His6) proteins were used at a final concentration of 1 μg/ml (+) or 3 μg/ml (++). Arrows indicate complexes (C) observed as shifted bands above the band representing unbound probe.
Identification of SsuR-binding sites in the ssuD regulatory region.
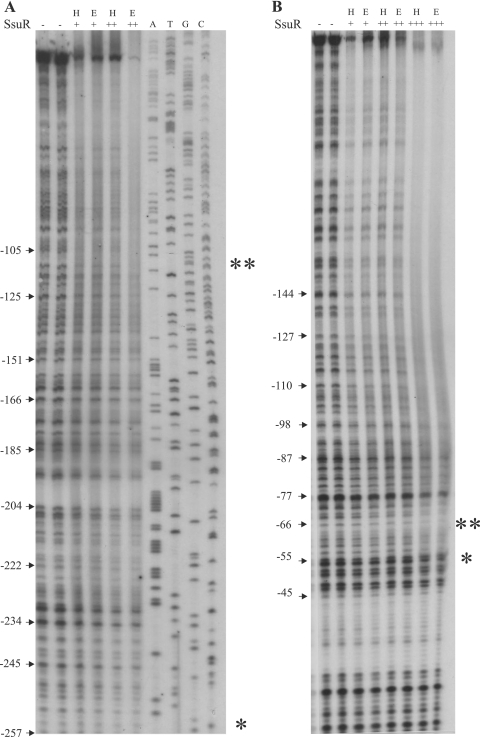
The first attempt to identify the recognition sequence for SsuR was undertaken through DNase I footprinting experiments using the ssuDP region as a target, as it appeared to be the most strongly SsuR-activated promoter among SsuR-responsive promoters identified so far (Table 4). As shown in Fig. 3, the protection area of ssuDP by SsuR extended over a large part of the examined fragment, suggesting several possible binding sites for this regulator. Protection patterns observed with SsuR(His6) and native SsuR were equivalent (Fig. 3), indicating that the His6 tag does not affect the DNA-regulator affinity. An examination of protection of nontranscribed and transcribed strands led us to notice that the boundaries of the SsuR footprint (at the highest SsuR concentration employed) can be detected as G (−254) and A (−55) with respect to A of the ATG initiation start codon of ssuD; these upstream and downstream boundaries of protection are indicated by single asterisks in Fig. 3 and also in the ssuDP nucleotide sequence shown in Fig. 4. However, a careful comparison of areas of weaker and stronger protection, detectable at the lowest SsuR concentration employed, suggested that a shorter region, downstream of C (−105) and upstream of A (−66) (Fig. 3A and B, positions indicated by a double asterisk), may represent a preferable binding site for SsuR; this region is also indicated between brackets in the ssuDP sequence shown in Fig. 4. Since the footprinting experiments using the long ssuDP fragment could not clearly define any discrete SsuR-binding sites upstream of A (−55), we decided to separately analyze the shorter segments of ssuDP. No clear σ70-dependent promoter has been identified in the sequence preceding the ssuD ORF (Fig. 4). However, the ssuDP sequence contains at least three pairs of hexamers (Fig. 4, shading), spaced by 16 to 17 bp and showing an imperfect match to −35 and −10 elements of σ70-dependent promoters. The sequence indicated between brackets that appeared to be relatively most strongly protected by SsuR (in the footprinting experiment) is located just upstream of the putative −35 element localized most proximally to the ssuD ORF (Fig. 4). In order to test whether this sequence, as well as other ssuDP segments, would be sufficient to produce a complex with SsuR, the DNA region preceding the ssuD ORF was dissected into four shorter fragments embracing sequences, numbered 1 to 4 in Fig. 4. Each of these ssuDP segments was amplified by PCR with the corresponding pair of forward and reverse primers, and the products were used in EMSAs with purified SsuR(His6) protein. As shown in Fig. 5A, a shifted band corresponding to a DNA-protein complex was observed with each of the ssuDP segments examined by EMSA, consistently with a large protection area observed in footprinting experiments with the whole ssuDP region. However, the SsuR concentration used to detect a shifted band with segment 4 of ssuDP was relatively high, which argued against a specific affinity of the regulator to this DNA fragment.
Fig. 3.
DNase I footprinting analysis of the SsuR-regulated promoter region ssuDP. The DNA fragment subjected to footprinting was amplified with primers SSUD1fw and SSUD2rev of which either the former or the latter was 5′ labeled with 32P to label the nontranscribed (A) or transcribed (B) strand, respectively. The nucleotide sequence of this fragment is shown in Fig. 4. Purified SsuR(His6) protein (designated H) was used at concentrations of 5 μg/ml (+), 10 μg/ml (++), or 20 μg/ml (+++). A protein extract (designated E) containing over 50% of native SsuR (18) was used at concentrations of 10 μg/ml (+), 20 μg/ml (++), or 40 μg/ml (+++). The boundaries of protection area detectable at the highest SsuR concentration are shown by single asterisks, and the boundaries of the most pronounced protection area detectable at the lowest SsuR concentration are shown by double asterisks on both strands. Sequence coordinates are indicated with respect to the first base of the ATG translation initiation codon of ssuD. The sequencing reaction (A, T, G, C) of the fragment with primer 32P-SSUD1fw was run on the same gel (shown at the top right of panel A). The sequencing reaction with primer 32P-SSUDrev2 (data not shown) was used to indicate sequence coordinates for the transcribed strand in panel B.
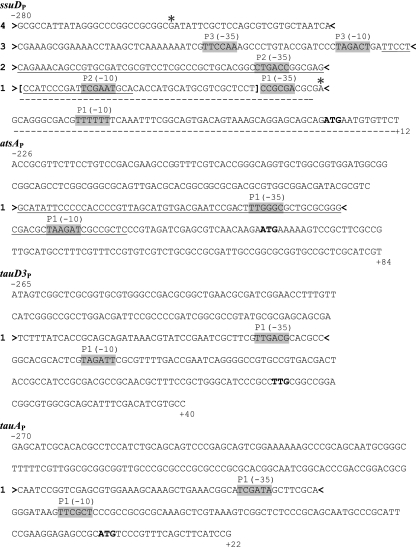
Fig. 4.
Sequences of the ssuD, atsA, tauD3, and tauA regulatory regions. Sequence coordinates are with respect to the first base of the respective translation initiation codon (shown in bold). The predicted elements of the σ70-dependent promoters are indicated as (−35) and (−10) and shaded. Four short segments of ssuDP region used in EMSA experiments with SsuR are indicated between > and < signs and denoted as 1, 2, 3, and 4 to the left of each sequence. Single asterisks indicate G (−254) and A (−55) as boundaries of the region protected from DNase at highest SsuR concentration employed. Square brackets indicate boundaries of region protected from DNase detectable at the lowest SsuR concentration (compare with Fig. 3). Short segments of the atsA, tauD3, and tauA regulatory regions used in the EMSA are indicated between > and < signs and denoted as 1 to the left of each corresponding sequence. Selected fragments of ssuDP and atsAP that were used to construct short promoter-reporter fusions are underlined by a solid line (ssuDP2 and atsAP1 fragments) or a dotted line (ssuDP1 fragment).
Fig. 5.
Binding of SsuR to short segments of four promoter regions. The EMSAs were performed on agarose gels with DNA probes (about 50 ng) containing portions of the ssuD, atsA, tauD3, and tauA promoter regions. Probes were preincubated with SsuR(His6) protein at a concentration of 5 μg/ml (+), 10 μg/ml (++), or 20 μg/ml (+++). Arrows indicate unbound probe (free DNA) and complexes (C) observed as shifted bands. (A) Four ssuDP probes containing sequences indicated as segments 1, 2, 3, and 4 in Fig. 4. (B) Probes designated atsAP1, tauD3P1, and tauAP1 contained segments indicated as sequence 1 (between > and <) in Fig. 4. No shifted band was produced by SsuR with a 50-bp fragment internal to ssuD ORF (data not shown).
In order to test if the detection of SsuR-binding sites within particular segments of ssuDP correlates with regulation of promoter activities in vivo, we have chosen for further analysis two partially overlapping fragments of the ssuD regulatory region (Fig. 4, sequences underlined by dotted and solid lines, respectively), which contained either the putative promoter P2 plus 46 bp upstream or the putative promoter P1 plus 40 bp upstream. The respective sequences were amplified by PCR and each of them was used to construct a promoter-reporter fusion in pKAGd4. The measurements of expression of ssuDP1-lacZ and ssuDP2-lacZ fusions (Table 4) confirmed that ssuD regulatory region contains at least two functional promoters, P1 and P2, and both of them are upregulated by SsuR. However, the combined activities of P1 and P2 measured separately by lacZ expression in B. cenocepacia strain 715j (in the presence of SsuR) are still not sufficient to account for the high-level activation of ssuD expression observed when the entire regulatory region is intact (compare the level of ssuDP-lacZ expression). Since in vivo activity of the putative P3 promoter was not investigated in this study, we can only speculate that either the presence of the upstream SsuR-binding site(s), i.e., the site detected in segment 3 of ssuDP (Fig. 4 and Fig. 5A), contributes significantly to maximum expression of ssuD or the effect of SsuR at P1 and P2 promoters is synergistic.
SsuR-binding sites in other gene regulatory regions.
In order to test whether SsuR is able to form complexes with isolated segments of atsAP, tauD3P, and tauAP (besides ssuDP), short fragments of these regions (Fig. 4, sequences indicated by 1) that were located upstream of, and overlapping, sequences resembling σ70-dependent promoters were amplified by PCR analysis and used as probes in the EMSA with SsuR. As shown in Fig. 5B, each of these probes produced a discrete shifted band corresponding to a DNA-SsuR complex. In addition, a longer segment of atsAP containing the sequence 1 plus the core promoter region (Fig. 4, underlined fragment of atsAP) was used to construct the respective promoter-reporter fusion in pKAGd4. As shown in Table 4, expression of this fusion (designated atsAP1-lacZ) revealed that the predicted atsAP1 promoter is functional and upregulated by SsuR. However, analogous to the situation observed with ssuDP1 and ssuDP2 (see “Identification of SsuR-binding sites in the ssuD regulatory region”), upregulation of atsAP1 was lower than that observed when the entire atsAP regulatory region was present. It is possible that some upstream SsuR-binding sites contribute to full upregulation of atsA expression, similar to that proposed for ssuDP.
The combined results obtained with selected “short” promoter probes tested by EMSA and by the expression pattern measured using lacZ reporter fusions lead us to conclude that sequences of approximately 44 bp preceding and (possibly) overlapping the promoter −35 elements are sufficient to bind SsuR. We note that both the position and length of the SsuR-binding sequences at SsuR-responsive promoter regions are analogous to those established for the single binding site of the E. coli Cbl regulator at the responsive promoter ssuEP in E. coli (5). Since SsuR of B. cenocepacia is closely related to Cbl, and the ssuE promoter of E. coli (ssuEPEc) was previously shown to be upregulated in vivo by SsuR (18), it seems likely that SsuR recognizes the target sequence for Cbl in ssuEPEc. It is noteworthy that the observed upregulation of some SsuR-responsive genes (i.e., ssuD) may involve an interaction of the regulator with multiple binding sites.
The identification of several target genes for SsuR in B. cenocepacia encouraged us to compare the regions assumed to represent the SsuR-binding sequences located upstream of the predicted −35 hexamer of eight SsuR-responsive promoters. The alignment (Fig. 6) includes six DNA segments identified experimentally by EMSA as those containing SsuR target sites (ssuDP1, ssuDP2, ssuDP3, atsAP1, tauD3P1, tauAP1) and two DNA segments containing putative SsuR-binding elements (tauD2bP and bcal1736P). The overall homology between aligned sequences is limited, although there does appear to be a high level of similarity between the corresponding regions at atsAP and tauD2bP. We believe that the most conserved central motif of the putative SsuR-recognition sequence (Fig. 6, asterisks) might be helpful for preliminary prediction of further SsuR target genes in the B. cenocepacia genome.
Fig. 6.
Alignment of identified and putative SsuR-binding sites at target promoter regions. Putative elements of σ70-dependent promoters in regions analyzed are indicated as (−35) and (−10). Coordinates relative to the translation start codons of downstream ORFs are indicated below the downstream limit of the −10 region in each case. The region of ssuDP protected by SsuR from DNase digestion (detectable at the lowest concentration of SsuR) is underlined. Nucleotides identical in at least five SsuR-regulated promoter regions are shown in bold and indicated by asterisks below the aligned sequences. The highest identity observed between atsAP and tauD2bP sequences is illustrated by vertical bars. For a comparison, the ssuE promoter region of E. coli (ssuEPEc) that is upregulated by CblEc (5) and SsuRBc (18) is also shown with the Cbl-binding site/putative SsuR-binding site underlined.
ACKNOWLEDGMENT
This study was supported by a grant from the Polish Ministry of Science and Higher Education (project no. N303 074 32/2454).
Footnotes
Published ahead of print on 11 February 2011.
REFERENCES
- 1. Agnoli K., Lowe C. A., Farmer K. L., Husnain I., Thomas M. S. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an ECF σ factor which is a part of the Fur regulon. J. Bacteriol. 188:3631–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ausubel F. M., et al. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 3. Bhave D. P., Muse W. B., Carroll K. S. 2007. Drug targets in mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 7:140–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bogdan J. A., Nazario-Larrieu J., Sarwar J., Alexander P., Blake M. S. 2001. Bordetella pertussis autoregulates pertussis toxin production through the metabolism of cysteine. Infect. Immun. 69:6823–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bykowski T., van der Ploeg J., Iwanicka-Nowicka R., Hryniewicz M. M. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43:1347–1358 [DOI] [PubMed] [Google Scholar]
- 6. Chiarini L., Bevivino A., Dalmastri C., Tabacchioni S., Visca P. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14:277–286 [DOI] [PubMed] [Google Scholar]
- 7. Coenye T., Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719–729 [DOI] [PubMed] [Google Scholar]
- 8. Compant S., Nowak J., Coenye T., Clément C., Ait Barka E. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32:607–626 [DOI] [PubMed] [Google Scholar]
- 9. Darling P., Chan M., Cox A. D., Sokol P. A. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ejim L. J., et al. 2004. Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 72:3310–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farmer K. L., Thomas M. S. 2004. Isolation and characterization of Burkholderia cenocepacia mutants deficient in pyochelin production: pyochelin biosynthesis is sensitive to sulfur availability. J. Bacteriol. 186:270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gooder H., Gehring L. B. 1954. Inhibition by cysteine of lecithinase (alpha-toxin) production in Clostridium welchii (perfringens) BP6K. Nature 174:1054–1055 [DOI] [PubMed] [Google Scholar]
- 13. Grifantini R., et al. 2002. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 20:914–921 [DOI] [PubMed] [Google Scholar]
- 14. Holden M. T. G., et al. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191:261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hummerjohann J., Laudenbach S., Retey J., Leisinger T., Kertesz M. A. 2000. The sulfur-regulated arylsulfatase gene cluster of Pseudomonas aeruginosa, a new member of the cys regulon. J. Bacteriol. 182:2055–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imperi F., Tiburzi F., Visca P. 2010. Transcriptional control of the pvdS iron starvation sigma factor by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12:1630–1642 [DOI] [PubMed] [Google Scholar]
- 17. Iwanicka-Nowicka R., Hryniewicz M. M. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11–17 [DOI] [PubMed] [Google Scholar]
- 18. Iwanicka-Nowicka R., Zielak A., Cook A. M., Thomas M. S., Hryniewicz M. M. 2007. Regulation of sulfur assimilation pathways in Burkholderia cenocepacia: identification of transcription factors CysB and SsuR and their role in control of target genes. J. Bacteriol. 189:1675–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karlsson S., Lindberg A., Norin E., Burman L. G., Akerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect. Immun. 68:5881–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kredich N. M. 1996. Biosynthesis of cysteine, p. 514–527 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology Press, Washington, DC [Google Scholar]
- 21. Lestrate P., et al. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543–551 [DOI] [PubMed] [Google Scholar]
- 22. Mahenthiralingam E. T., Urban A., Goldberg J. B. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144–156 doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 23. Mahenthiralingam E. T., Vandamme P. 2005. Taxonomy and pathogenesis of the Burkholderia cepacia complex. Chron. Respir. Dis. 2:209–217 [DOI] [PubMed] [Google Scholar]
- 24. Mahenthiralingam E., Baldwin A., Dowson C. G. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539–1551 [DOI] [PubMed] [Google Scholar]
- 25. McKevitt A. I., Bajaksouzian S., Klinger J. D., Woods D. E. 1989. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect. Immun. 57:771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27. Santos P. M., Di Bartolo I., Błatny J. M., Zennaro E., Valla S. 2001. New broad-host-range promoter probe vectors based on plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91–96 [DOI] [PubMed] [Google Scholar]
- 28. Sokol P. A. 1986. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J. Clin. Microbiol. 23:560–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sokol P. A., Woods D. E. 1988. Effect of pyochelin on Pseudomonas cepacia respiratory infections. Microb. Pathog. 5:197–205 [DOI] [PubMed] [Google Scholar]
- 30. Soutourina O., et al. 2009. CymR, the master regulator of cysteine biosynthesis in Staphylococcus aureus, controls sulfur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73:194–211 [DOI] [PubMed] [Google Scholar]
- 31. Stec E., et al. 2006. Structural basis of the sulfate starvation response in E. coli: Crystal structure and mutational analysis of the cofactor-binding domain of the Cbl transcriptional regulator. J. Mol. Biol. 364:309–322 [DOI] [PubMed] [Google Scholar]
- 32. Tralau T., et al. 2007. Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J. Bacteriol. 189:6743–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valvano M. A., Keith K. E., Cardona S. T. 2005. Survival and persistence of opportunistic Burkholderia species in host cells. Curr. Opin. Microbiol. 8:99–105 [DOI] [PubMed] [Google Scholar]
- 34. Vandamme P., et al. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91–96 [DOI] [PubMed] [Google Scholar]
- 35. van der Ploeg J. R., et al. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Ploeg J. R., Iwanicka-Nowicka R., Kertesz M. A., Leisinger T., Hryniewicz M. M. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671–7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Ploeg J. R., Iwanicka-Nowicka R., Bykowski T., Hryniewicz M. M., Leisinger T. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358–29365 [DOI] [PubMed] [Google Scholar]
- 38. Winsor G. L., et al. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]