Abstract
Inteins are the protein equivalent of introns. Their protein splicing activity is essential for the host protein's maturation and function. Inteins are grouped into three classes based on sequence signature and splicing mechanism. The sequence signature of the recently characterized class 3 inteins is a noncontiguous Trp-Cys-Thr (WCT) motif and the absence of the standard class 1 Cys1 or Ser1 N-terminal nucleophile. The intein N-terminal Cys1 or Ser1 residue is essential for splicing in class 1 inteins. The mycobacteriophage Catera Gp206, Nocardioides sp. strain JS614 TOPRIM, and Thermobifida fusca YX Tfu2914 inteins have a mixture of class 1 and class 3 motifs. They carry the class 3 Trp-Cys-Thr motif and have the standard class 1 N-terminal Ser1 or Cys1. This study determined which class the mycobacteriophage Catera Gp206 and Nocardioides sp. JS614 TOPRIM inteins belong to based on catalytic mechanism. The mycobacteriophage Catera Gp206 intein (starting with Ser1) is a class 3 intein, and its Ser1 residue is not required for splicing. Based on phylogenetic analysis, we propose that class 3 inteins arose from a single mutated intein that was spread by phage into predominantly helicase genes in various phages and their hosts.
INTRODUCTION
Intein-mediated protein splicing is a posttranslational reaction that excises an intervening sequence (the intein) from a protein precursor and simultaneously joins the flanking host protein fragments (the exteins). The ligated exteins are connected by a standard peptide bond in the absence of any intein scar. Inteins are frequently inserted into conserved motifs in essential genes, which means that splicing must occur efficiently for extein function and viability of the host organism. Over 550 inteins have been identified in archaea, eubacteria, eukaryotes, phages, and viruses; a listing of inteins and their properties is available in the InBase database (http://www.neb.com/neb/inteins.html) (14). A numbering system for residues in intein precursors was established to ease comparisons between inteins. It consists of separate numbering schemes for each region of the precursor, with the N terminus of the intein starting at 1 and continuing sequentially to the C terminus of the intein, and with all C-extein residues having a plus sign, starting with +1 at the N terminus of the C-extein. There are four conserved motifs present in all inteins, called either blocks A, B, F, and G (14) or N1, N3, C2, and C1 (15). Conserved intein residues can also be referred to by use of the block letter and the block position separated by a colon.
The intein and the first C-extein residue are required for splicing; they act as a single-turnover enzyme requiring no external cofactors or energy sources. The mechanism of protein splicing (Fig. 1) has been reviewed extensively (10, 12). Inteins were recently divided into 3 classes based on sequence and splicing mechanism (20). Most inteins are members of class 1 and are sometimes called standard inteins; inteins from the other classes are often referred to as atypical inteins (14). The class 1 protein splicing mechanism consists of four steps involving three conserved nucleophiles: (i) Ser1 or Cys1 at the intein N terminus; (ii) normally, Asn at the intein C terminus, but occasionally Gln or Asp; and (iii) Ser+1, Thr+1, or Cys+1 at the beginning of the C-extein (14). Although a natural example of a Thr1 intein has not been observed (14), Thr can substitute for Ser1 to yield a functional intein (6).
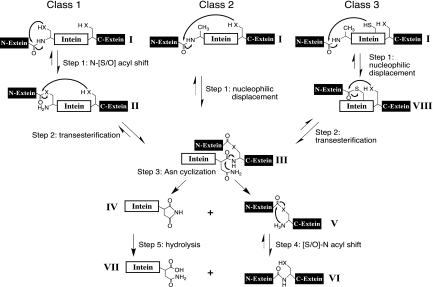
Fig. 1.
Splicing mechanisms for the three classes of inteins. The majority of inteins follow the class 1 intein-mediated protein splicing mechanism, which consists of four coordinated nucleophilic displacement reactions. Class 2 and class 3 inteins form the block G BI (III) by different pathways because they generally start with amino acids incapable of forming the linear (thio)ester intermediate (II). Class 3 inteins form the class-specific block F BI (VIII) prior to the block G BI. Once the block G BI is formed, the remainder of the splicing reaction is the same in all inteins. Residues within the intein assist in these enzymatic reactions. However, in all classes, there is variability in the residues and positions within the intein that facilitate each reaction. Tetrahedral intermediates are not shown. “X” represents the sulfur or oxygen atom in the side chain of Ser, Thr, or Cys, and solid arrows represent steps that have been verified experimentally.
The class 1 splicing mechanism (Fig. 1) is initiated by an acyl rearrangement of the intein N-terminal Cys1 or Ser1 residue to form a linear (thio)ester intermediate (II). A transesterification reaction shifts the N-extein onto the side chain of Cys+1, Ser+1, or Thr+1, joining the exteins with a (thio)ester bond. The resultant branched intermediate (BI) (III; block G BI) is resolved by Asn cyclization to yield a free intein (IV) plus the ligated exteins (V). A peptide bond is formed between the exteins after another acyl shift (VI).
The N-terminal Cys1, Ser1, or Thr1 is absent in class 2 and class 3 inteins. Additionally, class 3 inteins have a noncontiguous WCT motif consisting of TrpB:12, CysF:4, and ThrG:5 (Table 1), and all but one have Ser+1 or Thr+1 instead of the more common Cys+1 residue (2, 20). Class 2 inteins do not have the WCT motif (19). More importantly, the 3 classes follow different reaction pathways (Fig. 1). Splicing of the class 2 prototype (Methanococcus jannaschii KlbA intein) is initiated by a direct attack on the N-terminal splice junction peptide bond by Cys+1 instead of the initial acyl shift that initiates splicing of class 1 inteins; the remaining reaction is identical to the class 1 mechanism (19). The splicing mechanism of class 3 inteins was defined using the mycobacteriophage Bethlehem DnaB intein (20) and the Deinococcus radiodurans Snf2 intein (2). Their splicing reaction is initiated by a nucleophilic attack on the N-terminal splice junction peptide bond by the WCT motif CysF:4, which results in formation of the class-specific branched intermediate (block F BI; VIII) (Fig. 1). The block G BI (III) is then formed by a transesterification reaction in which the N-extein is transferred to the side chain of the +1 residue. The remainder of the pathway is the same as in class 1 inteins. The splicing mechanisms for the 3 classes of inteins differ only by the reaction pathway leading to the block G BI.
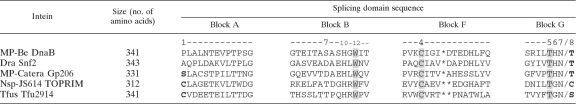
Table 1.
Comparison of conserved motifs in known WCT inteinsa
The four protein splicing domain motifs are depicted for selected WCT inteins. The WCT motif (positions B:12, F:4, and G:5) is shaded. Standard class 1 nucleophiles at the intein N terminus and the C-extein N terminus are shown in bold. The C-terminal splice junction is indicated by a slash (/). The position number in each block for a residue known to be important for catalysis is indicated above the position. By consensus in the intein field, residues within a block can be referred to by using the block letter and the block position separated by a colon.
The mycobacteriophage Catera (MP-Catera) Gp206 intein, the Nocardioides sp. JS614 TOPRIM (Nsp-JS614 TOPRIM) intein, and the Thermobifida fusca YX hypothetical protein 2914 (Tfus Tfu2914) intein have a WCT motif and begin with Ser1 or Cys1, combining signature sequences of both class 1 and class 3 inteins (Table 1). Biochemical characterization indicated that the Tfus Tfu2914 intein is a class 1 intein (16). The present study examined the splicing of the MP-Catera Gp206 and Nsp-JS614 TOPRIM inteins to determine whether they fall into class 1 or class 3 and to establish the phylogenetic relationship between class 1 and class 3 inteins.
MATERIALS AND METHODS
Cloning and mutagenesis.
All clones were sequenced by the New England BioLabs core facility, and all enzymes were obtained from New England BioLabs (Ipswich, MA) and used as described by the manufacturer. The genes encoding the MP-Catera Gp206 intein from the precursor with the locus tag Catera_gp206, the Nsp-JS614 TOPRIM intein from the precursor DNA primase with the locus tag Noca_1947, and the Tfus Tfu2914 intein from the precursor with the locus tag Tfu_2914, plus five native extein residues flanking each side of the element, were synthesized by GeneArt, Inc. (Burlingame, CA). The MP-Catera Gp206 intein native N-extein sequence included was Pro-Val-Glu-Leu-Lys, and the native C-extein sequence included was Thr-Gln-Asn-Ser-Arg. The Nsp-JS614 TOPRIM intein native N-extein sequence included was Arg-Gly-Phe-Phe-His, and the native C-extein sequence included was Cys-Phe-Gly-Cys-Ser. The Tfus Tfu2914 intein native N-extein sequence included was Ala-Asp-Ile-Gly-Tyr, and the native C-extein sequence included was Ser-Phe-Gly-Ala-Cys. Mutations were made in the homing endonuclease domain active site to block endonuclease activity (for the MP-Catera Gp206 intein, Asp125Ala and Asp209Ala; for the Nsp-JS614 TOPRIM intein, Asp121Ala and Asp190Ala; and for the Tfus Tfu2914 intein, Asp135Ala) (5, 14). The synthesized DNAs were digested with XhoI and SpeI, purified from agarose gels with a Wizard SV gel and PCR cleanup system (Promega, Madison, WI), and ligated into pMP1 (18), which was also digested with the same enzymes. This resulted in pMCP, with the MP-Catera Gp206 intein; pMNP, with the Nsp-JS614 TOPRIM intein; and pMTP, with the Tfus Tfu2914 intein. These clones all had their respective inteins flanked by the Escherichia coli maltose-binding protein (M) and the ΔSal fragment of Dirofilaria immitis paramyosin (P).
All mutations were constructed using a Phusion site-directed mutagenesis kit (New England BioLabs), with pMCP, pMNP, or pMTP as the template and with appropriate primers to introduce the desired mutation. Primers were obtained from Integrated DNA Technologies (San Diego, CA).
Expression, purification, and protein characterization.
All fusions were expressed in the E. coli NEB Turbo strain (New England BioLabs) by induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.4 to 0.6 in 10 ml LB medium containing 100 μg/ml ampicillin for 2 h at 37°C or 15°C overnight. Cell pellets were disrupted by sonication in buffer A (20 mM Na2HPO4, pH 8.0, and 500 mM NaCl). Soluble lysates were prepared by centrifugation and directly electrophoresed or purified over amylose resin (New England BioLabs) equilibrated in buffer A at pH 6, with elution with buffer A containing 10 mM maltose.
Soluble lysates and purified proteins were boiled for 5 min in SDS sample buffer plus dithiothreitol (DTT) (New England BioLabs), loaded onto 10 to 20% Tris-glycine polyacrylamide gels (Invitrogen, Carlsbad, CA), and either stained with Simply Blue Safe stain (Invitrogen) or transferred to nitrocellulose membranes for Western blot analysis with antiserum against maltose-binding protein or paramyosin, as described previously (19).
Analysis of branched intermediates.
The pH of the purified BI samples was adjusted to 9.0 by adding 0.5 M Na2HPO4, and the samples were incubated at 4°C for 14 h with 1 mM DTT and 100 μM CGC649 peptide (New England BioLabs) in the presence or absence of 50 mM 2-mercaptoethanesulfonic acid (MESNA). The CGC649 peptide is commercially available (New England BioLabs) and has the sequence NH2-Cys-Gly-Cys(649)-CONH2. Cys(649) is a cysteine with a Dyomics DY-649-03 fluorophore covalently attached to its sulfhydryl group by use of maleimide chemistry. After the reaction, samples were electrophoresed in a 10 to 20% Tris-glycine polyacrylamide gel. The gel was scanned using an Odyssey infrared imaging system (Li-Cor Biotechnology, Lincoln, NE) at 700 nm and then stained with Simply Blue Safe stain.
Phylogenetic analysis.
Bayesian inference analysis was performed using the Geneious Pro 5.1 suite of programs (Geneious, Auckland, New Zealand). Intein motif sequences present in InBase (14; http://www.neb.com/neb/inteins.html) as of 8 August 2010 and intein insertion sites listed in InBase were used. Intein splicing domain motifs A, B, F, and G were concatenated to produce a single 49-amino-acid sequence for each intein. Due to the variable size of block F, which contains a loop between two beta strands, only the first and last 7 positions of block F were included. One hundred forty-eight intein sequences were compared, including all class 3 inteins, the 3 inteins in this study, all phage inteins, selected inteins with a Cys residue at F:4, and selected helicase inteins (see Fig. S1 in the supplemental material). MrBayes (7) was used with default parameters to create trees, using the Tfus RecA2 intein as the outgroup, with a final standard deviation of split frequencies of 0.01 or less.
RESULTS
Splicing of wild-type inteins.
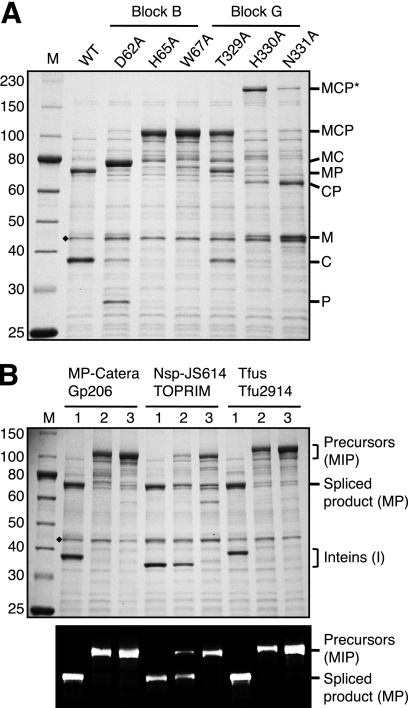
The MP-Catera Gp206 intein (331 amino acids) begins with Ser1, while the Nsp-JS614 TOPRIM (312 amino acids) and Tfus Tfu2914 (341 amino acids) inteins begin with Cys1. These three intein genes plus five natural extein residues flanking both sides were each cloned between the maltose-binding protein (M) and a fragment of paramyosin (P) to yield model precursors MCP (MP-Catera Gp206 intein), MNP (Nsp-JS614 TOPRIM intein), and MTP (Tfus Tfu2914 intein). Expression of each precursor was induced with IPTG in E. coli incubated at 37°C for 2 h or at 15°C overnight. Unless indicated, splicing was similar at both temperatures. Two temperatures were tested because splicing is occasionally temperature dependent in model precursors. Heterologous exteins can also result in decreased splicing activity and increased off-pathway cleavage reactions (Fig. 2), as can mutations in the intein or the +1 extein residue.
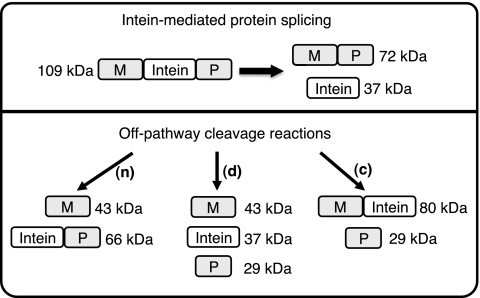
Fig. 2.
Schematic representation of splicing and off-pathway cleavage reactions for the MP-Catera Gp206 intein in a model precursor consisting of the maltose-binding protein as the N-extein (M) and a fragment of paramyosin (P) as the C-extein. Protein splicing yields ligated MP and free intein, while off-pathway N-terminal splice junction cleavage (n) yields M plus CP and off-pathway C-terminal splice junction cleavage (c) yields MC plus P. Cleavage of the (thio)ester linkage in branched intermediates also yields M plus CP. Some mutations result in double cleavage (d) at both splice junctions to yield M plus C plus P. Molecular masses of the MP-Catera Gp206 intein MCP precursor and potential products are listed.
MCP, MTP, and MNP precursors all yielded spliced products (MP plus C, T, or N) in vivo. MCP and MTP precursors were spliced to completion at both 37°C and 15°C (Fig. 3A and data not shown), while some MNP precursor remained at 37°C but MNP was spliced to completion at 15°C (Fig. 3B and data not shown). These results were confirmed by Western blotting, using antisera against both maltose-binding protein and paramyosin (Fig. 3C and data not shown).
Fig. 3.
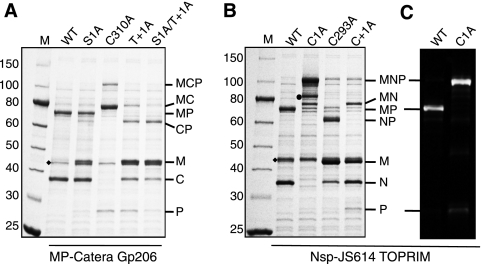
Splicing of wild-type (WT) and mutated MP-Catera Gp206 (A) and Nsp-JS614 TOPRIM (B and C) inteins in model MCP and MNP precursors, respectively. Each precursor was expressed at 15°C overnight. Soluble proteins were electrophoresed by SDS-PAGE and stained with Simply Blue Safe stain. (C) The Nsp-JS614 TOPRIM sample was probed in a Western blot with anti-paramyosin serum. A black dot marks a product of in vivo proteolytic cleavage of MNP. A diamond marks the endogenous E. coli maltose-binding protein, which migrates slightly faster than M from MCP and slightly slower than M from MNP. Mutations are listed above each lane. Multiple mutations in the same protein are separated by a slash. Abbreviations are the same as in Fig. 2.
Mutations used to classify each intein.
To determine whether these inteins are class 3 inteins, alanine substitutions were made at position 1, position +1, or the conserved block F Cys (F:4) residue. If these inteins are class 1 inteins, mutation of Ser1 or Cys1 should block splicing and N-terminal cleavage, mutation of the +1 residue should block BI formation (although it may allow N-terminal cleavage), and mutation of the block F Cys could affect several steps. In contrast, if they are class 3 inteins, mutation of Ser1 or Cys1 should allow splicing, mutation of the +1 residue should allow BI formation and N-terminal cleavage, and mutation of CysF:4 should block splicing, N-terminal cleavage, and BI formation.
The first test was splicing in the absence of Ser1 or Cys1. The MP-Catera Gp206 intein still spliced efficiently with the Ser1Ala mutation, but the Nsp-JS614 TOPRIM intein did not splice or yield N-terminal cleavage products when Cys1 was mutated to Ala (Fig. 3). These results indicate that like the Tfus Tfu2914 intein (16), the Nsp-JS614 TOPRIM intein is a class 1 intein. The MP-Catera Gp206 intein could be a class 2 or 3 intein because the Ser1Ala mutation did not block splicing.
The second test was to mutate the +1 position to Ala. This yielded N-terminal (M plus CP or NP), C-terminal (MC or MN plus P), or double-cleavage (M plus C or N plus P) products (Fig. 3). This result is not diagnostic, because an N-terminal Ser1 can potentially form the linear ester intermediate (intermediate II) even in atypical inteins, which can lead to N-terminal cleavage (19). The Ser1Ala/Thr+1Ala double mutation in the MP-Catera Gp206 intein yielded N-terminal cleavage products and double-cleavage products, indicating that neither Ser1 nor Thr+1 is required for N-terminal cleavage (Fig. 3A). This result clearly indicates that the MP-Catera Gp206 intein is a class 3 intein, not a class 2 intein, because class 2 inteins require the +1 amino acid for N-terminal cleavage (19) and class 3 inteins do not (2, 20).
Further proof of class status was obtained by mutating the block F CysF:4 (from the WCT motif) to Ala (Fig. 3). In class 1 and 2 inteins, mutations at F:4 inhibit splicing because this residue assists reactions at one or both splice junctions, depending on the individual intein (2, 8, 13, 19–21). In class 3 inteins, Ala substitution for this residue blocks all N-terminal cleavage reactions, including BI formation. N-terminal cleavage occurred in the mutated Cys293Ala Nsp-JS614 TOPRIM intein. However, in the MP-Catera Gp206 intein, mutation of the F:4 Cys310 to Ala blocked all N-terminal cleavage and resulted in mainly C-terminal cleavage products. Taken together, these results indicate that the MP-Catera Gp206 intein is a class 3 intein and the Nsp-JS614 TOPRIM intein is a class 1 intein.
Mutagenesis of conserved residues in block B.
Residues in intein block B (Table 1) activate the N-terminal splice junction for both on- and off-pathway cleavage (2, 9, 10, 12, 17, 19, 20). These residues include the conserved HisB:10 residue in all types of inteins (9, 17) and the WCT motif TrpB:12 residue in class 3 inteins (2, 20). The B:7 residue is not highly conserved but assists in N-terminal reactions, to various extents, in all classes of inteins (2, 14, 15, 20).
To examine the role of block B residues in splicing of the MP-Catera Gp206 intein, especially the importance of the class 3 TrpB:12 residue, either Asp62 (B:7), His65 (B:10), or Trp67 (B:12) was mutated to Ala (Table 1 and Fig. 4A). All of these mutations inhibited splicing and N-terminal cleavage, to various degrees. The Asp62Ala mutant yielded mainly off-pathway C-terminal cleavage products (MC plus P), with small amounts of precursor (MCP) and spliced products (MP plus C). The spliced MP band was detected by Western blotting only (data not shown) and was not abundant enough for detection in gels stained with Simply Blue Safe stain. Both His65Ala and Trp67Ala mutations resulted in mainly precursor (MCP), with a small amount of off-pathway C-terminal cleavage products (MC plus P).
Fig. 4.
(A) Mutation of selected MP-Catera Gp206 intein block B and G residues to alanine. Only the 15°C samples are shown; there were no significant differences between the 37°C samples and 15°C samples. Soluble proteins were electrophoresed by SDS-PAGE and stained with Simply Blue Safe stain. (B) The effect of replacement of the WCT motif TrpB:12 by Ala was examined with each of the indicated inteins. Soluble proteins were electrophoresed by SDS-PAGE and stained with Simply Blue Safe stain (upper panel) or Western blotted with anti-paramyosin serum (lower panel). Lanes 1, 15°C samples of wild-type inteins; lanes 2, 15°C samples of inteins with Ala substitution at TrpB:12; lanes 3, 37°C samples of inteins with Ala substitution at TrpB:12. Abbreviations are the same as in Fig. 2, with the addition of MCP*, which is the branched intermediate. A diamond marks the endogenous E. coli maltose-binding protein.
A WCT motif B:12 Trp residue is present in the Nsp-JS614 TOPRIM and Tfus Tfu2914 inteins, although they are not class 3 inteins. Ala substitutions were made to see if this residue is important for splicing or N-terminal cleavage in these class 1 inteins (Fig. 4B). The Tfus Tfu2914 intein Trp72Ala mutant yielded only precursor at both 37 and 15°C, while the Nsp-JS614 TOPRIM intein Trp69Ala mutant yielded spliced products (MP plus N) only at 15°C.
Mutagenesis of conserved residues in block G.
The block G motif is composed of the C-terminal part of the intein and the N-terminal residue of the C-extein (Table 1). It includes three essential catalytic residues: the intein penultimate His (G:6) residue, the intein C-terminal Asn (G:7) residue, and the Cys, Ser, or Thr residue at the +1 position (G:8). The penultimate HisG:6 residue assists in C-terminal cleavage in most inteins (3, 10–12, 14). In class 3 inteins, the WCT motif Thr residue is present at G:5. Compared with mutations of the other WCT motif residues, mutating ThrG:5 had a much smaller effect on splicing (2, 20).
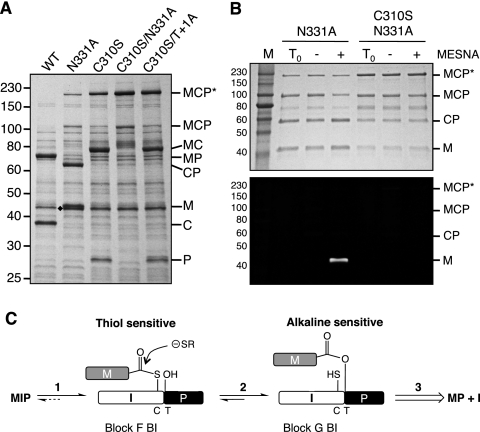
Ala scanning mutagenesis was performed on the last three residues of block G in the MP-Catera Gp206 intein (Fig. 4A). Mutation of Thr329 (G:5) impaired splicing, with more MCP precursor present than spliced products (MP plus C). Mutation of the penultimate His330 (G:6) residue dramatically decreased C-terminal cleavage, which resulted in accumulation of the branched intermediate (MCP*) as well as small amounts of N-terminal cleavage products and MCP precursor. MCP* migrates slowly in SDS-PAGE gels due to its unusual branched structure. The spliced product (MP) was detectable only in Western blots (data not shown). Ala mutation of the C-terminal Asn331 yielded mostly N-terminal cleavage products (M plus CP), with small amounts of branched intermediate (MCP*) and precursor (MCP). These results are consistent with the known properties of these block G residues.
Analysis of the MP-Catera Gp206 intein branched intermediate.
The block F BI and the block G BI have similar mobilities in SDS-PAGE gels. Therefore, the identity of any BI must be deciphered based on a mutation at either branch point or by differential reactivity. Since the native block F BI is always a thioester and the block G BI is usually an oxygen ester in class 3 inteins, these intermediates can be distinguished based upon the differential sensitivity to thiol reagents of esters versus thioesters. No block F BI was observed in the Thr+1Ala mutant (Fig. 3A), consistent with previous observations that BIs with Cys branch points break down in vivo to form N-terminal cleavage products, while BIs accumulate when the branch point is an ester (2, 4, 20). In previous studies, mutation of a Cys branch-point residue to Ser allowed accumulation of the BI (2, 4).
Identification of a block F BI would directly prove that an intein is a class 3 intein. To see if the block F BI could be trapped in the MP-Catera Gp206 intein, Cys310 (F:4) was mutated to Ser. The Cys310Ser mutation yielded a slow-migrating BI (MCP*) (Fig. 5A). Although the Cys310Ser mutation allowed BI accumulation, C-terminal cleavage predominated over splicing, suggesting that Ser310 inhibited the subsequent transesterification step. The BI was the major product when the Cys310Ser mutation was combined with the Asn331Ala mutation that blocks C-terminal cleavage (Fig. 5A). The Cys310Ser/Thr+1Ala double mutation specifically trapped the block F BI (Fig. 5A). Although the BI in the Asn331Ala, Cys310Ser, and Cys310Ser/Asn331Ala mutants could be either the block F BI, the block G BI, or both, the BI in the Cys310Ser/Thr+1Ala double mutant must be the block F BI, because Ala at the block G branch point (the +1 position) precludes the generation of a block G BI.
Fig. 5.
(A) Detection of block F BI in the MP-Catera Gp206 intein. Expression of each mutated MCP precursor was induced for 2 h at 37°C, and the soluble protein was electrophoresed by SDS-PAGE and stained with Simply Blue Safe stain. Mutations are indicated above each lane. Multiple mutations in the same protein are separated by a slash. A diamond marks the endogenous E. coli maltose-binding protein. Abbreviations are the same as in Fig. 4. (B) Analysis of MP-Catera Gp206 branched intermediates (MCP*). Asn331Ala and Cys320Ser/Asn331Ala samples were purified by affinity chromatography on amylose resin and then adjusted to pH 9. Purified samples were incubated overnight at 4°C to examine the breakdown of the BI (thio)ester linkage to yield M plus CP in response to 50 mM MESNA in the presence of 100 mM CGC649 peptide, which has an N-terminal Cys and a fluorescent tag. Gels were scanned using an Odyssey infrared imaging system at 700 nm to detect peptide ligation (lower panel) and then stained with Simply Blue Safe stain (upper panel). Lanes T0, time zero samples; lanes −, samples incubated with the CGC649 peptide and without MESNA; lanes +, samples incubated with MESNA and the CGC649 peptide. Abbreviations are the same as in Fig. 4. (C) Proposed pathway for thiol-induced cleavage of block G BI. Steps 1 and 2 are reversible, with the equilibrium in step 2 favoring the block G BI. Step 3, which includes Asn cyclization and the O-N acyl shift, is irreversible. In the Asn331Ala mutant, the block G BI can be isolated and unfolded with urea, making step 2 irreversible. The observed decay of the block G BI in response to thiols is due to elimination of the block F BI by thiols, which stimulates a reverse step 2 reaction in order to maintain the equilibrium between the two BIs. The Cys branch point in block F (Cys310 in the MP-Catera Gp206 intein) and the Thr+1 branch point in block G are indicated below the rectangles as C and T, respectively.
In the previously studied class 3 inteins, there was a reversible equilibrium (Fig. 5C) between the block F and block G BIs (2, 20). The oxygen ester in the block G BI should not be cleaved by thiols under mild conditions, whereas the thioester in the block F BI should be sensitive to cleavage by thiols under mild conditions. Although the equilibrium favored the block G ester-linked BI in previous studies, the BI decayed when it was incubated with thiols because thiol elimination of the block F BI drove the reverse reaction, which converts the block G BI into the block F BI (2, 20). To examine whether the block G BI in the MP-Catera Gp206 intein can also perform this reverse reaction to become sensitive to thiols, the Asn331Ala block G BI was purified over amylose resin. The purified protein sample was incubated with 50 mM MESNA and 100 mM CGC649 peptide (Fig. 5B). Under these conditions, expressed protein ligation should occur between the C terminus of M and the Cys at the N terminus of the CGC649 peptide if M is linked to CP by a thioester bond in the BI. Incubation of the Asn331Ala samples with MESNA and the peptide resulted in decay of the BI, as observed in gels stained with Simply Blue Safe stain and by fluorescence of the M band ligated to the fluorescent CGC649 peptide. As a control, the Cys310Ser/Asn331Ala sample was treated identically. However, because both BIs from this double mutation are oxygen esters, there was no decay of the Cys310Ser/Asn331Ala BI or peptide ligation after incubation with MESNA. These results indicate that the block G BI from the MP-Catera Gp206 intein can perform the reverse reaction to form the block F BI.
Phylogenetic analysis.
It was previously noted that class 3 inteins are present in proteins with putative helicase activity, including DnaB, RecG, Snf2, primases, and terminase large subunits of bacteriophages (20). A phylogenetic analysis was performed to examine the relationships between class 3 inteins, class 1 WCT motif inteins, inteins with a WCT motif CysF:4 instead of the more common AspF:4 in the absence of the other WCT motif residues, various types of DNA helicase inteins, and phage, prophage, and viral inteins. Block A, B, F, and G motif sequences listed in the InBase database (14) were concatenated to yield a single 49-amino-acid sequence for each of 148 inteins (see Table S1 in the supplemental material). This included all 39 class 3 inteins present in InBase as of 8 August 2010 as well as the 3 WCT motif inteins from this study, 29 class 1 inteins with Cys at F:4, 22 inteins from phages or prophages, 8 inteins in viral proteins, and 31 class 1 DnaB inteins, among other helicase and nonhelicase inteins. Trees were generated using a Bayesian inference analysis, and probability values at each node were expressed as consensus support percentages (see Fig. S1 in the supplemental material).
The most dramatic finding was that all class 3 inteins formed a single clade that excluded inteins from all other classes. The one exception was the uncharacterized Arthrobacter sp. FB24 DnaB intein, which begins with Gly1 but lacks a Cys at F:4. Class 3 DnaB inteins failed to show any phylogenetic association with class 1 DnaB inteins, even when these inteins were present at the same site in DnaB. Class 3 inteins also failed to cluster with other phage inteins, prophage-derived inteins, or class 1 inteins having Cys at F:4.
DISCUSSION
Class 3 inteins were previously identified based on a unique sequence signature (the absence of Ser1, Thr1, or Cys1 and the presence of the WCT motif) and a unique splicing mechanism that included the class-specific block F BI. The MP-Catera Gp206 intein, the Nsp-JS614 TOPRIM intein, and the Tfus Tfu2914 intein begin with Ser1 or Cys1 but also have the WCT motif, combining properties of both class 1 and class 3 inteins. A previous study showed that the Tfus Tfu2914 intein is a class 1 intein (16). The present study demonstrates that the Nsp-JS614 TOPRIM intein is also a class 1 intein, while the MP-Catera Gp206 intein is a class 3 intein. Classification of the Nsp-JS614 TOPRIM intein was based on the inability of Ala to substitute for Cys1 and, secondarily, on the presence of N-terminal cleavage in precursors with the Cys293Ala (F:4) mutation. Previous studies with class 2 and class 3 inteins all showed splicing activity when the intein N terminus was mutated to Ala or other residues (2, 19, 20), whereas class 1 inteins failed to splice without Ser1, Thr1, or Cys1 (2, 10, 12, 19, 20).
Proof that the MP-Catera Gp206 intein was a class 3 intein required testing of its mechanism of splicing. The essential characteristics of class 3 inteins are (i) cleavage at the N-terminal splice junction in the absence of all class 1 and class 2 splice junction nucleophiles at A:1, G:7, and G:8; (ii) activation of the N-terminal splice junction by a block B motif that includes the conserved HisB:10 residue and the WCT motif TrpB:12 residue; (iii) decay of the block G BI (intermediate III) in response to thiols, despite an ester linkage at the block G branch point; (iv) essentiality of the WCT motif CysF:4 for splicing, although Ser substitution allows block F BI formation; (v) the presence of both a block F BI (intermediate VIII) and a block G BI (intermediate III); and (vi) the fact that nonconservative mutation of CysF:4 prevents formation of both BIs, as well as off-pathway N-terminal cleavage. The MP-Catera Gp206 intein satisfied all of these criteria. Furthermore, the MP-Catera Gp206 intein block F BI was trapped and visualized directly in the Cys310Ser/Thr+1Ala precursor; this could only be the block F BI, because formation of the block G BI is not possible with Ala at the +1 position. Experiments with the Asn331Ala and Cys310Ser/Asn331Ala samples demonstrated that only the block F BI with a thioester linkage decayed in response to MESNA, not the block G BI or the block F BI with a Cys310Ser substitution. This was confirmed by expressed protein ligation (10, 12), as a fluorescently tagged N-terminal Cys peptide was ligated to the N-extein (M) in MESNA-treated Asn331Ala samples but not in Cys310Ser/Asn331Ala samples. These results also indicate that there was interconversion between the two BIs. The MP-Catera Gp206 intein is the first example, to our knowledge, of a class 3 intein that has a standard class 1 nucleophile at its N terminus. However, Ser1 was not required for activity. In class 3 inteins, TrpB:12 is involved in hydrophobic packing with surrounding residues (2, 20). Class 1 and 2 inteins also have a hydrophobic residue at the B:12 position (8, 11, 21), including Trp, which suggests that this position is involved in hydrophobic packing in all classes of inteins.
As of November 2010, all known class 3 inteins in the InBase database are from eubacteria or phages. Of the 40 class 3 inteins (including the MP-Catera DnaB intein), 15 are derived from phages or prophages and 37 are present in ATPase P-loop motifs. The observation that many class 3 inteins are of phage origin or are inserted in similar sites in host bacterial homologs suggested that class 3 inteins might have originated from a phage-derived intein that spread to host helicase genes and beyond. To verify this hypothesis, a phylogenetic tree based on conserved splicing motifs of 148 selected inteins was generated (see Fig. S1 and Table S1 in the supplemental material). Class 3 inteins clearly have a monophyletic distribution, while other phage or prophage-derived inteins and class 1 inteins having Cys at F:4 are polyphyletic. The failure of class 3 DnaB inteins to cluster with class 1 DnaB inteins was unexpected, because class 1 and class 2 inteins present at the same extein insertion site generally have the highest sequence similarities and have been shown to cluster together in phylogenetic analyses, even across domains of life (14, 15). This absence of association based on DnaB insertion site supports the hypothesis that class 3 DnaB inteins arose from a different progenitor than class 1 DnaB inteins or evolved so extensively in isolation from class 1 DnaB inteins that they no longer cluster. The one potential exception to the unity of the class 3 intein clade is the Arthrobacter sp. FB24 DnaB intein, which begins with Gly1 but does not have Cys at F:4. Further experimentation is required to determine whether it is a class 2 or class 3 intein.
These results support the hypothesis that class 3 inteins arose from a phage intein that lost its N-terminal nucleophile but contained or, more likely, accumulated secondary mutations that allowed splicing to occur efficiently. During this process, the lost extein activity could have been supplied by another phage or by the host cell. Many inteins are bifunctional proteins that have homing endonuclease activity, which leads to intein gene mobilization (homing) to similar insertion sites in homologs or paralogs. This would allow the ancestral class 3 intein to spread among various phages and their hosts through intein homing. As with other inteins, these class 3 inteins could have spread further to other sites as they picked up varied homing endonuclease specificities (1).
This paper expands the definition of class 3 inteins to include WCT motif inteins with an N-terminal Ser1. In the future, it is possible that other class 3 inteins will be identified that have Cys1 or Thr1, in addition to those that have Ser1. The presence of a WCT motif is insufficient to classify Cys1, Ser1, or Thr1 inteins as class 3 inteins, since the Tfus Tfu2914 Cys1-containing (16) and Nsp-JS614 TOPRIM WCT motif-containing inteins are class 1 inteins. Previous papers have suggested that biochemical analysis of reaction pathways is necessary to classify inteins (2, 20). Although this is the most stringent proof, the data presented here suggest that WCT inteins can be classified tentatively as class 3 inteins if they cluster with the other class 3 inteins after phylogenetic analysis, even if they have a standard class 1 N-terminal nucleophile. This is supported by the fact that only the MP-Catera Gp206 intein branched with the class 3 inteins, while the WCT class 1 Nsp-JS614 TOPRIM intein (Cys1) and the Tfus Tfu2914 intein (Cys1) did not.
Inteins have continued to provide surprises since their discovery more than 20 years ago. Variations in protein splicing mechanisms may be tolerated more easily because the enzyme and substrate are linked in a single precursor molecule, making the process of converting one enzyme active site to another easier in inteins than in standard enzyme systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank Manoj Cheriyan (New England BioLabs) for helpful discussions, Dan Distel (Ocean Genome Legacy Foundation), Nicole Wood (Ocean Genome Legacy Foundation), and Sanjay Kumar (New England BioLabs) for help with phylogenic analysis, and Don Comb (New England BioLabs) for support and encouragement.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 11 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Belfort M., Perlman P. S. 1995. Mechanisms of intron mobility. J. Biol. Chem. 270:30237–30240 [DOI] [PubMed] [Google Scholar]
- 2. Brace L. E., Southworth M. W., Tori K., Cushing M. L., Perler F. 2010. The Deinococcus radiodurans Snf2 intein caught in the act: detection of the class 3 intein signature block F branched intermediate. Protein Sci. 19:1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L., Benner J., Perler F. B. 2000. Protein splicing in the absence of an intein penultimate histidine. J. Biol. Chem. 275:20431–20435 [DOI] [PubMed] [Google Scholar]
- 4. Chong S., et al. 1996. Protein splicing involving the Saccharomyces cerevisiae VMA intein. The steps in the splicing pathway, side reactions leading to protein cleavage, and establishment of an in vitro splicing system. J. Biol. Chem. 271:22159–22168 [DOI] [PubMed] [Google Scholar]
- 5. Dalgaard J. Z., et al. 1997. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 25:4626–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hodges R. A., Perler F. B., Noren C. J., Jack W. E. 1992. Protein splicing removes intervening sequences in an archaea DNA polymerase. Nucleic Acids Res. 20:6153–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huelsenbeck J. P., Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 8. Johnson M. A., et al. 2007. NMR structure of a KlbA intein precursor from Methanococcus jannaschii. Protein Sci. 16:1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawasaki M., Nogami S., Satow Y., Ohya Y., Anraku Y. 1997. Identification of three core regions essential for protein splicing of the yeast Vma1 protozyme. A random mutagenesis study of the entire Vma1-derived endonuclease sequence. J. Biol. Chem. 272:15668–15674 [DOI] [PubMed] [Google Scholar]
- 10. Mills K. V., Perler F. B. 2005. The mechanism of intein-mediated protein splicing: variations on a theme. Protein Pept. Lett. 12:751–755 [DOI] [PubMed] [Google Scholar]
- 11. Mizutani R., et al. 2002. Protein-splicing reaction via a thiazolidine intermediate: crystal structure of the VMA1-derived endonuclease bearing the N- and C-terminal propeptides. J. Mol. Biol. 316:919–929 [DOI] [PubMed] [Google Scholar]
- 12. Paulus H. 2001. Inteins as enzymes. Bioorg. Chem. 29:119–129 [DOI] [PubMed] [Google Scholar]
- 13. Pearl E. J., Tyndall J. D., Poulter R. T., Wilbanks S. M. 2007. Sequence requirements for splicing by the Cne PRP8 intein. FEBS Lett. 581:3000–3004 [DOI] [PubMed] [Google Scholar]
- 14. Perler F. B. 2002. InBase: the intein database. Nucleic Acids Res. 30:383–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pietrokovski S. 1998. Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci. 7:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reitter J. N., Mills K. V. 2011. Canonical protein splicing of a class one intein that has a class three noncanonical sequence motif. J. Bacteriol. 193:994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Romanelli A., Shekhtman A., Cowburn D., Muir T. W. 2004. Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein-intein junction. Proc. Natl. Acad. Sci. U. S. A. 101:6397–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Southworth M. W., Amaya K., Evans T. C., Xu M. Q., Perler F. B. 1999. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques 27:110–114, 116,, 118–120 [DOI] [PubMed] [Google Scholar]
- 19. Southworth M. W., Benner J., Perler F. B. 2000. An alternative protein splicing mechanism for inteins lacking an N-terminal nucleophile. EMBO J. 19:5019–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tori K., et al. 2010. Splicing of the mycobacteriophage Bethlehem DnaB intein: identification of a new mechanistic class of inteins that contain an obligate block F nucleophile. J. Biol. Chem. 285:2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Roey P., et al. 2007. Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue. J. Mol. Biol. 367:162–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.