Abstract
The human pathogen Burkholderia pseudomallei possesses multiple type III secretion system (T3SS) gene clusters. One of these, the B. pseudomallei T3SS2 (T3SS2bp) gene cluster, which apparently plays no role in animal virulence, is also found in six additional Burkholderia spp. and is very similar to T3SSs found in phytopathogenic Xanthomonas spp. and Ralstonia solanacearum. The T3SS2bp gene cluster also encodes an AraC-type regulatory protein (HrpBbp) that is an ortholog of HrpB, the master regulator of the R. solanacearum T3SS (T3SSrso) and its secreted effectors. Transcriptome analysis showed that HrpBbp activates the expression of T3SS2bp genes, as well as their orthologs in R. solanacearum. In addition to activating T3SS2bp, HrpBbp also upregulates the expression of ∼30 additional B. pseudomallei genes, including some that may confer production of adhesive pili, a polyketide toxin, several putative T3SS2bp-secreted effectors, and components of a regulatory cascade. T3SS2bp promoter regions were found to contain a conserved DNA motif (p2bp box) identical in sequence and position to the hrpII box required for HrpB-dependent T3SSrso transcription activation. The p2bp box is also present in the promoter regions of the essentially identical T3SS found in the very closely related species Burkholderia thailandensis (T3SS2bt). Analysis of p2bp box mutants showed that it is essential for HrpBbp-mediated transcription activation in both species. Although it has been suggested that T3SS2bp and T3SS2bt may function in phytopathogenicity, we were unable to demonstrate a phytopathogenic phenotype for B. thailandensis in three different plant hosts.
INTRODUCTION
Burkholderia pseudomallei is a pathogenic betaproteobacterium primarily found in wet soil and water in southeast Asia and northern Australia (32, 75). Infection with B. pseudomallei by inhalation or through open wounds results in melioidosis, a potentially deadly human disease predominately affecting individuals with preexisting conditions, such as diabetes or renal disease (14). Melioidosis is characterized by septicemia, pneumonia, and pulmonary, splenic, and/or hepatic abscesses (14, 75). No vaccine for this disease is available, and the disease is also difficult to diagnose and treat (75). The CDC lists B. pseudomallei as a category B select agent restricted to biosafety level 3 (BSL3) containment (54), underscoring the threat posed by this pathogen.
Type III secretion systems (T3SSs) are specialized macromolecular machines essential for virulence of many Gram-negative animal- and plant-pathogenic bacteria. T3SSs are comprised of 20 to 25 proteins, 9 of which are conserved among all known T3SSs (19). T3SSs deliver proteins, termed effectors, into the cytoplasm of eukaryotic cells via a structure variously termed a needle, filament, or pilus (19). Effector functions vary widely and include altering signal transduction, transcriptional activities, and protein turnover in host cells (2, 6).
The large (∼8-Mb) genome of B. pseudomallei encodes three T3SSs, compared to only 1 encoded by most other pathogenic bacteria (29). One of these, T3SS3, is orthologous to the Inv/Mxi-Spa T3SSs in Salmonella and Shigella spp. and is required for virulence in mice and hamsters (64, 71). The other two (T3SS1 and B. pseudomallei T3SS2 [T3SS2bp]) show similarity to the T3SS found in the plant pathogen Ralstonia solanacearum (T3SSrso), and neither is involved in animal virulence (52, 71). T3SS1 is unique to B. pseudomallei, while T3SS2bp, the focus of this study, is also found in Burkholderia thailandensis, a very closely related soil-dwelling bacterium from Thailand which was initially classified as B. pseudomallei. B. thailandensis is capable of causing disease in animal models, although its 50% lethal dose (LD50) is at least 105-fold higher than that of B. pseudomallei (7, 8). Since B. thailandensis apparently does not cause disease in humans, it is used as a surrogate for B. pseudomallei infection (28, 74).
T3SSrso in R. solanacearum, like T3SS2bp, has an AraC-type transcriptional regulator (HrpB) as part of the T3SS cluster. HrpB is the master regulator of transcription of T3SSrso and its secreted effectors. It binds to a conserved DNA motif called the hrpII box that is located 46 bp upstream of the transcriptional start of the T3SSrso operons or effector genes it regulates (21). Xanthomonas spp., which are also plant pathogens, have a very similar AraC-type transcriptional regulator, HrpX, which regulates T3SS transcription via a nearly identical DNA motif called the PIP box (73). Expression of HrpB in R. solanacearum is induced by either growth in minimal medium (3) or plant-cell contact (41). Induction via plant-cell contact is mediated by a regulatory cascade in which an unknown signal is sensed by the outer membrane receptor protein PrhA and transduced through the transmembrane protein PrhR, which in turn activates the extracytoplasmic sigma factor PrhI (1, 9). PrhI is required for expression of prhJ (9). PrhJ is a LuxR/UhpA-type regulator that activates transcription of the response regulator hrpG, the product of which activates expression of hrpB (9).
To investigate the significance and role of T3SS2bp in B. pseudomallei, we explored its regulation by the HrpB ortholog HrpBbp. We found that HrpBbp activates T3SS2bp gene expression in a manner analogous to HrpB activation of T3SS expression in R. solanacearum, utilizing a DNA motif identical in sequence and position to the hrpII box utilized by HrpB in R. solanacearum. We also defined the HrpBbp regulon in B. pseudomallei by identifying ∼30 additional genes whose expression is HrpBbp activated.
MATERIALS AND METHODS
Growth conditions.
Bacterial strains and plasmids used are listed in Table 1. Escherichia coli, B. pseudomallei, and B. thailandensis were grown at 37°C in Luria-Bertani medium (LB) (42), LB supplemented with 2% glycerol (LBG), or M9 medium (57) with 20 mM glutamate instead of glucose. R. solanacearum and Pseudomonas putida were grown at 30°C in 1% Bacto peptone, 0.1% Casamino Acids, 0.1% yeast extract, and 0.5% glucose (BG) (36). When necessary, antibiotics were used at the following concentrations: kanamycin at 50 μg/ml (Km50) for Burkholderia spp. and 25 μg/ml (Km25) for R. solanacearum; 20 μg/ml polymyxin B (Pm20), 20 μg/ml gentamicin (Gm20), and chloramphenicol at 20 μg/ml (Cm20) for E. coli and 50 μg/ml (Cm50) for B. thailandensis.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference and/or source |
|---|---|---|
| Strains | ||
| B. pseudomallei DD503 | Δ(amrR-oprA) rpsL (Smr) Kms | 43 |
| B. thailandensis DW503 | Δ(amrR-oprA) rpsL (Smr) Kms Cms Gms | 11 |
| B. thailandensis LL102 | Derivative of DW503, hrpBbt::pGHrpBm Gmr | This study |
| B. thailandensis E264 | Wild type | 7 |
| R. solanacearum GMI1000 | Wild type | 56 |
| P. putida ATCC 12633 | Wild type | 63 |
| E. coli NEB5-alpha | Cloning strain | Invitrogen |
| E. coli S17-1 | Tra+ Pms | 62 |
| E. coli HB101(pRK2013) | Tra+ mobilization strain, Pms Kmr | 26 |
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector, Kmr Ampr | Invitrogen |
| pCR-XL-TOPO | Cloning vector, Kmr Zeor | Invitrogen |
| pSCRhaB2K | Broad-host-range expression vector derived from pSCRhaB2, Kmr | W. Nierman, reference 12 |
| pPR9TT | Vector with promoterless lacZ, Cmr Ampr | 58 |
| pGSV3 | Suicide vector, Gmr | 24 |
| pGHrpBm | 406-bp internal fragment of hrpB in pGSV3, Gmr | This study |
| pSCR1610a | −160 bp relative to ATG start codon to +31 bp relative to TGA stop codon of BPSS1610 in pSCRhaB2K, Kmr | This study |
| pPR1606 | −356 to +62 bp relative to ATG start codon of BPSS1606 in pPR9TT, Cmr | This study |
| pPR0764 | −279 to +62 bp relative to ATG start codon of BTH_II0764 in pPR9TT, Cmr | This study |
| pPR0764Md | −160 to +62 bp relative to ATG of BTH_II0764 in pPR9TT, Cmr | This study |
| pPR0764Tr | −114 to +62 bp relative to ATG of BTH_II0764 in pPR9TT, Cmr | This study |
| pPR0764A2b | pPR0764 with T to A at −48 and −49, Cmr | This study |
| pPR0764B2b | pPR0764 with T to A at −68 and −69, Cmr | This study |
| pPR0764C2b | pPR0764 with T to A at −48, −49, −68, and −69, Cmr | This study |
| pPR07641ab | pPR0764 with C to A at −67, Cmr | This study |
| pPR07648ab | pPR0764 with G to T at −65, G to A at −62, and G to T at −51, Cmr | This study |
Km, kanamycin; Gm, gentamicin; Pm, polymyxin B; Cm, chloramphenicol; Zeo, Zeocin; Amp, ampicillin; Sm, streptomycin; S, sensitive; R, resistant.
For pPR0764 derivatives, numbers refer to nucleotide positions relative to the BTH_II0764 transcription start.
Plasmid construction. (i) pSCR1610a.
Primers 1610F and 1610R (see Table S1 in the supplemental material) were used to PCR amplify the BPSS1610 coding sequence (CDS) (hrpBbp), which was then cloned into pCR2.1-TOPO. This plasmid was digested with XbaI and HindIII, and the resultant fragment was ligated into similarly digested pSCRhaB2K. The accuracy of the cloned BPSS1610 DNA CDS was confirmed by DNA sequencing.
(ii) pGHrpBm.
A 406-bp internal fragment of the BTH_II0762 CDS (B. thailandensis hrpB [hrpBbt]) was PCR amplified with HrpBmF and HrpBmR primers and cloned into pCRXL-TOPO. A SpeI-NotI fragment from this plasmid was ligated into the similarly digested suicide vector pGSV3.
(iii) pPR1606, pPR0764, pPR0764Md, and pPR0764Tr.
The promoter regions of BTH_II0764 and BPSS1606 were PCR amplified with PromFLF/PromR and 1606PF/1606PR primers, respectively, and individually cloned into pCR2.1-TOPO. PstI-XhoI fragments from each plasmid were cloned into the similarly digested promoterless lacZ fusion vector pPR9TT, yielding pPR0764 and pPR1606. Similarly, two shorter promoter region fragments of BTH_II0764 were PCR amplified using primers PromMDF/PromR and PromTrF/PromR and individually cloned into pCR2.1-TOPO. PstI-XhoI fragments from each plasmid were cloned into similarly digested pPR9TT, to yield pPR0764Md and pPR0764Tr, respectively.
(iv) pPR0764A2, pPR0764B2, and pPR0764C2.
Splice overlap extension PCR (30) with primers incorporating specific nucleotide changes was used to introduce site-specific mutations into the promoter region of BTH_II0764. Briefly, PromFLF/MutA2 and MutA3/PromR primers were used to generate two overlapping amplicons that were gel purified and used in a second PCR with only the PromFL/PromR primers. This amplicon was cloned into pCR2.1-TOPO. The resultant plasmid was digested with PstI and XhoI and ligated into similarly digested pPR9TT, yielding pPR0764A2. This procedure was repeated using the PromFLF/MutB2 and MutB3/PromR primers for the construction of pPR0764B2 and the PromFLF/MutC2 and MutC3/PromR primers for pPR0764C2.
(v) pPR07641a and pPR07648a.
Error-prone PCR with PromFLF/PromR primers was used to enhance misincorporation of nucleotides in the promoter fragment of BTH_II0764. To do this, 0.5 mM MnCl2 was added to the PCR and the dCTP and dTTP concentrations were changed to 0.55 mM each. The resultant amplicon was gel purified, digested with PstI and XhoI, ligated into similarly digested pPR9TT, and transformed into E. coli NEB5-alpha cells. All pPR9TT plasmid inserts were subjected to DNA sequencing.
All DNA sequences were obtained from Integrated Microbial Genomes (IMG; http://img.jgi.doe.gov). PCR conditions were 98°C for 4 min, 30 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 1 min (except for the case of BPSS1610, for which it was 2 min), and a final extension of 72°C for 10 min.
Strain construction.
Chemically competent E. coli NEB5-alpha cells and E. coli S17-1 cells (62) were transformed as previously described (61). The pSCRhaB2K- and pPR9TT-based plasmids were transferred from E. coli NEB5-alpha into B. pseudomallei, B. thailandensis, or R. solanacearum recipients by triparental mating. Briefly, donor, recipient, and E. coli HB101(pRK2013) helper were grown overnight on antibiotic selection plates, resuspended in LBG or BG to an optical density at 600 nm (OD600) of 0.2, and shaken until the OD600 reached 0.4. Equal volumes of the cultures were mixed, and 30 μl was spotted onto LBG or BG plates. After 18 h, cells were resuspended in sterile water and spread onto selection plates containing the appropriate antibiotic to select for the desired plasmid and Pm20 to counterselect against the E. coli donor and helper strains.
To inactivate hrpBbt, the suicide plasmid pGHrpBm (Table 1; see above) was transferred into B. thailandensis from E. coli S17-1, as described above, and integrants were selected on plates with Gm20 and Pm20. Proper insertion of pGHrpBm into the genome of B. thailandensis was confirmed by PCR.
β-Galactosidase assays.
Cells from overnight plate cultures were suspended in LB or M9–20 mM glutamate to an OD600 of 0.2 and shaken until the OD600 reached 0.4. If induction of HrpBbp overexpression was needed, rhamnose was added to 0.2% and the cells were harvested 3 h later; otherwise, the cells were harvested at an OD600 of 1.0. β-Galactosidase assays were performed as previously described (15) and repeated at least three times.
Quantitative reverse transcription PCR (qRT-PCR).
Cells were grown overnight on plates, suspended in LBG or BG to an OD600 of 0.2, and shaken until the OD600 reached 0.6. Rhamnose was added to 0.2% to induce HrpBbp expression, and incubation continued until the OD600 was 1.2. After an equal volume of RNAprotect (Qiagen) was added to the culture, RNA was purified from pelleted cells by using the RNeasy kit (Qiagen) and treated with DNase I (Qiagen) according to manufacturer's instructions, except that the amount of DNase I was increased 3-fold. cDNA was synthesized using 1 μg of RNA and the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions. Quantitative PCR was performed on 22 target genes with an Applied Biosystems StepOne real-time PCR system using Sybr green under the following conditions: 10 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min. The amplification data were analyzed by the 2−ΔΔCT method to determine relative log2 changes in gene expression (38) in reference to rpoA for B. pseudomallei and B. thailandensis and the cytochrome oxidase gene (RSc0369) for R. solanacearum. We chose rpoA as a reference because of its stable expression under a variety of conditions (53). Means and standard deviations were calculated from at least three independent experiments.
Whole-genome expression profiling.
RNA from B. pseudomallei was prepared as described above. For microarray analysis, cDNA was synthesized, labeled with Cy3/Cy5, and hybridized to a whole-genome PCR amplicon microarray for B. pseudomallei, and raw data were analyzed as described previously (59). For massively parallel RNA sequencing (RNA-Seq) (76), the rRNA content was reduced by 2 rounds of treatment with MicrobExpress (Ambion). This mRNA-enriched fraction was used to synthesize cDNA by using SuperScript II (Invitrogen) with random hexamer primers and sequenced with a Genome Analyzer (Illumina Technologies). Data were analyzed by using the CLC Genomics workbench (CLC bio) allowing for 2 mismatches per read.
Determination of transcription start sites.
A 5′ rapid amplification of cDNA ends (RACE) system (Invitrogen) was used according to the manufacturer's instructions. Briefly, the antisense primers for BTH_II0764 and BPSS1622 mRNA (RACEFL1 and S1622race1, respectively; see Table S1 in the supplemental material) were used to prime synthesis of cDNA to the 5′ end of the transcript. A poly(C) tail was then added to the 3′ end of the cDNA by using terminal deoxynucleotidyl transferase. Primer RACEFL2 or S1622raceb2 (for BTH_II0764 or BPSS1622, respectively) and a primer specific for the poly(C) tail (provided with the kit) were used to amplify the cDNA. The amplicons were then cloned into pCR2.1-TOPO and subjected to DNA sequencing.
Plant pathogenicity assays.
Tomato seeds (Solanum lycopersicum var. cerasiforme) were surface sterilized with 15% bleach for 15 min, germinated, and grown for 4 weeks at 25°C on Murashige and Skoog medium (MS) solidified with 0.8% agar (46). Resultant plantlets were inoculated with bacteria as previously described (34). Briefly, plantlets with 2 or 3 leaves were placed in 50-ml Falcon tubes containing 5 ml of MS with 106 cells/ml of each test bacterium, and symptom development was monitored at 30°C for 7 days. Commercial tomato plants (“Better Boy”) with 7 or 8 leaves were inoculated via the stub of a cut petiole (55) with a 2-μl cell suspension, and symptoms were monitored at 30°C for 7 days. Arabidopsis thaliana seeds (Col-0; Lehle Seeds) were incubated in moist Arabidopsis growing medium (Lehle Seeds) at 4°C for 2 days and then germinated at 25°C. Leaves of 3-week-old plants were infiltrated with each test bacterium, and disease was monitored as described previously (27).
To evaluate bacterial multiplication in plants, 0.3-cm2 leaf discs from A. thaliana or stem sections from tomato were removed, flame sterilized, macerated or cut into ∼1-mm sections, and agitated for 30 min at 25°C in sterile water. Serial dilutions of the supernatant were plated and colonies counted. Hypersensitive-response assays were performed on fully expanded leaves of Nicotiana tabacum (tobacco) by infiltration of each test bacterium into the leaves and monitored as previously described (31).
Microarray data accession number.
The microarray data were deposited into the ArrayExpress database (49) under the accession number E-MTAB-484.
RESULTS
T3SS2bp is remarkably similar to the T3SSs in the phytopathogens R. solanacearum and Xanthomonas species.
Nearly identical clusters of genes orthologous to the T3SS2bp cluster are found not only in B. thailandensis but also in Burkholderia mallei, Burkholderia ubonensis, Burkholderia oklahomensis, Burkholderia ambifaria, and Burkholderia dolosa. None of these are reported to be plant pathogens, and all except B. mallei and B. dolosa were isolated from soil. Burkholderia spp. that do not have a T3SS orthologous to T3SS2bp include Burkholderia cenocepacia, Burkholderia multivorans, Burkholderia xenovorans, Burkholderia graminis, Burkholderia glumae, Burkholderia phytofirmans, Burkholderia phymatum, Burkholderia vietnamensis, and Burkholderia rhizoxinica. Of the characterized plant pathogens, R. solanacearum and Xanthomonas spp., but not Pseudomonas syringae or Erwinia spp., have T3SSs that are also orthologous to T3SS2bp.
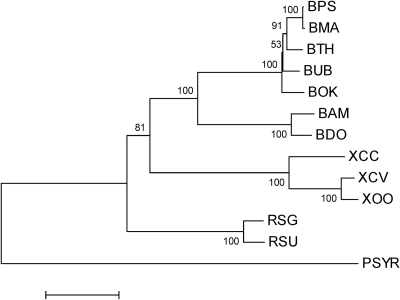
Given the relationship of T3SS2bp to plant pathogen T3SSs, we examined the evolutionary relationships between the Burkholderia T3SS2bp orthologs and those in well-characterized plant pathogen T3SSs. A neighbor-joining phylogenetic tree was constructed using 3 of the 9 core T3SS genes that are conserved among all bacterial T3SSs (Fig. 1). Pairwise distances suggest that the orthologous T3SSs of B. mallei, B. thailandensis, B. oklahomensis, and B. ubonensis are the most closely related to T3SS2bp in B. pseudomallei, followed by the T3SSs of B. ambifaria and B. dolosa. The DNA sequence identities for hrcV orthologs are 99% between B. pseudomallei and B. mallei, ∼94% between B. pseudomallei, B. thailandensis, B. oklahomensis, and B. ubonensis, and ∼75% between B. pseudomallei, B. ambifaria, and B. dolosa, indicating a high degree of conservation. This example is illustrative of the DNA sequence identities between all the core T3SS genes among these Burkholderia spp. and largely reflects phylogenetic distance based on 16S rRNA sequence analysis (67). Of the well-characterized T3SSs in plant pathogens, the one in R. solanacearum (T3SSrso) is the most closely related to T3SS2bp.
Fig. 1.
Phylogenetic tree of 3 conserved T3SS genes from plant pathogens and Burkholderia species. Multiple sequence alignments of hrcV, hrcN, and hrcC orthologs from B. pseudomallei K96243 (BPS), B. mallei ATCC 23344 (BMA), B. thailandensis E264 (BTH), B. ubonensis Bu (BUB), B. oklahomensis C6786 (BOK), B. ambifaria AMMD (BAM), B. dolosa AU0158 (BDO), R. solanacearum UW551 (RSU), R. solanacearum GMI1000 (RSG), X. campestris pv. campestris ATCC 33913 (XCC), X. oryzae pv. oryzae MAFF311018 (XOO), X. campestris pv. vesicatoria 85-10 (XCV), and Pseudomonas syringae pv. tomato DC3000 (PSYR) were trimmed and concatenated, and a neighbor-joining phylogenetic tree was built using MEGA4 (65). The numbers at nodes are the percent bootstrap support values (1,000 replicates). Scale bar, 10% substitution. All sequences were obtained from IMG.
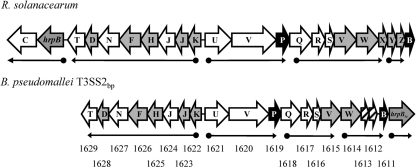
Nineteen of the 20 genes in the T3SS2bp gene cluster have orthologs in the above-mentioned Burkholderia spp., and 18 of the 20 have orthologs in the phytopathogen R. solanacearum. Overall, the order and orientation of these genes are conserved between R. solanacearum and the Burkholderia spp. described above (Fig. 2). The only differences between the T3SS2bp gene cluster, the T3SSrso gene cluster, and the orthologous clusters in B. mallei, B. thailandensis, B. oklahomensis, and B. ubonensis are the translocation of hrpBbp to the opposite end of the T3SS cluster and the translocation of the hrcC secretin gene to a site 19 kbp upstream. In the remaining Burkholderia spp., the location of hrpB and the secretin gene are inverted relative to what is found in B. pseudomallei, B. mallei, B. thailandensis, B. oklahomensis, and B. ubonensis.
Fig. 2.
Organization of the orthologous T3SS gene clusters of R. solanacearum and B. pseudomallei. The R. solanacearum “hrc” genes (i.e., the nine core genes conserved in all bacterial T3SS clusters) are denoted by white arrows, the “hrp” genes by gray arrows, and the “hpa” genes by black arrows. Their orthologs in the B. pseudomallei T3SS2bp cluster are similarly colored. The striped arrows indicate genes whose products do not have >25% amino acid sequence identity with any T3SSrso proteins. The determined transcription units for R. solanacearum, predicted transcription units for B. pseudomallei, and the direction of transcription are denoted by thin black arrows; the filled circles indicate transcription starts. Numbers are the BPSS locus tags for B. pseudomallei genes from IMG. The B. pseudomallei ortholog of hrcC is located 19 kbp upstream from the T3SS2bp cluster (not pictured). The T3SS2bt cluster in B. thailandensis (not pictured) is identical to the T3SS2bp cluster.
The nine core proteins of the T3SS of R. solanacearum are encoded by the hrc genes. Over half of these proteins average ∼60% amino acid identity with their B. pseudomallei orthologs (Table 2). The remaining core T3SS2bp proteins have amino acid identities of ∼40% with their orthologs in T3SSrso, and all are assigned to the same clusters of orthologous groups (COGs) families. A majority of the 10 remaining T3SS2bp proteins have amino acid identities ranging from 33 to 37% with their R. solanacearum counterparts. Finally, the predicted transcription units of T3SS2bp (51) are also nearly identical in composition and direction to the experimentally determined ones of T3SSrso.
Table 2.
Percent amino acid sequence identities of B. pseudomallei T3SS2bp proteins with their orthologs in R. solanacearum and X. campestris pv. vesicatoria
| B. pseudomallei protein type and corresponding locus tag |
R. solanacearum GMI1000 |
X. campestris pv. vesicatoria |
||
|---|---|---|---|---|
| Protein | % amino acid identity | Protein | % amino acid identity | |
| Hrc protein orthologs | ||||
| BPSS1629 | HrcT | 41 | HrcT | 40 |
| BPSS1627 | HrcN | 67 | HrcN | 72 |
| BPSS1624 | HrcJ | 60 | HrcJ | 50 |
| BPSS1621 | HrcU | 47 | HrcU | 45 |
| BPSS1620 | HrcV | 60 | HrcV | 63 |
| BPSS1618 | HrcQ | 30 | HrcQ | 50 |
| BPSS1617 | HrcR | 62 | HrcR | 61 |
| BPSS1616 | HrcS | 57 | HrcS | 53 |
| BPSS1592 | HrcC | 44 | HrcC | 46 |
| Hrp protein orthologs | ||||
| BPSS1628 | HrpD | 33 | HrpB7 | <10 |
| BPSS1626 | HrpF | 37 | HrcL | 32 |
| BPSS1625 | HrpH | 33 | HrpB4 | 28 |
| BPSS1623 | HrpJ | 30 | HrpB2 | 29 |
| BPSS1622 | HrpK | 30 | HrpB1 | 31 |
| BPSS1615 | HrpV | 26 | HpaA | 27 |
| BPSS1614 | HrpW | 34 | HrcD | 33 |
| BPSS1610 | HrpB | 38 | HrpX | 38 |
| Hpa protein orthologs | ||||
| BPSS1619 | HpaP | 36 | HpaC | 27 |
| BPSS1611 | HpaB | 53 | HpaB | 54 |
BPSS1612 and BPSS1613 are in a genome location analogous to that of hrpY and hrpX of T3SSrso, which encode the pilin subunit and a pilus assembly protein, respectively (Fig. 2) (68, 69). However, the corresponding amino acid identities are only 18%. Although pilin proteins of plant pathogen T3SSs lack amino acid sequence similarity, predicted amphiphilic helices and coiled-coil secondary structures are usually predicted in most of these proteins, including HrpY (69). BPSS1613 contains a predicted amphiphilic helix and coiled-coil domain (39, 60), indicating that it could be the pilin subunit.
HrpB of R. solanacearum (HrpBrso), the AraC-type activator that is the master regulator of T3SSrso expression, has 38% amino acid identity with its B. pseudomallei ortholog, encoded by BPSS1610 (HrpBbp). In addition to the amino acid sequence similarities described above, there are also DNA sequence similarities in the T3SS promoter regions of B. pseudomallei and R. solanacearum (see below). Taken together, these data suggest that the T3SS2bp genes encode all the proteins needed for the assembly and regulation of a functionally complete plant pathogen-like type III secretion system analogous to the one in R. solanacearum.
Transcriptome analysis of B. pseudomallei overexpressing HrpBbp reveals its regulon.
Because HrpB positively regulates the transcription of the T3SSrso genes and many of its secreted effectors (21), we determined whether HrpBbp activates the expression of T3SS2bp genes in B. pseudomallei. Total mRNA from B. pseudomallei cells overexpressing HrpBbp was compared to total mRNA from cells with the empty vector by microarray analysis. Overall, 48 genes were upregulated >4-fold when HrpBbp was overexpressed ∼10-fold from plasmid pSCR1610a (Table 3). Bulk sequence analysis of cDNA populations derived from one set of the same RNA preparations used as described above with an Illumina Genome Analyzer (RNA-Seq) (76) gave very similar results, except that the magnitude of upregulation in general was >10-fold higher (Table 3). Sixteen of the 20 T3SS2bp genes in all five transcription units were upregulated an average of ∼14-fold. Only one of the ∼40 genes in the other two T3SSs (BPSS1409 from T3SS1bp) appeared to be upregulated. Thus, HrpBbp activates the transcription of the genes encoding T3SS2bp but not the other T3SSs.
Table 3.
B. pseudomallei and select B. thailandensis genes showing increased transcription in response to HrpBbp overexpression
| Locus tag(s) | p2bp boxa | Description/product(s)b | Log2 relative expressionc |
||
|---|---|---|---|---|---|
| Microarray | qRT-PCR | RNA-Seq | |||
| BPSS1611-BPSS1629 | + | T3SS2bp cluster | 3.5 | NA | 9.4 |
| BPSS1604-BPSS1609 | + | Hypothetical proteins and a two-component regulatory system | 3.3 | NA | 9.3 |
| BPSS1593-BPSS1601 | − | Type IV pilus-encoding cluster | 2.9 | NA | 5.5 |
| BPSS0998-BPSS1004 | + | Polyketide biosynthesis cluster | 1.7 | NA | 4.8 |
| BPSS1614 (BTH_II0758d) | + | Ortholog of R. solanacearum hrpW | Spot absent | 10.6 ± 0.7 (11.5 ± 0.3) | 10.6 |
| BPSS1621 (BTH_II0750d) | + | Ortholog of R. solanacearum hrcU | 3.4 ± 0.05 | 10.5 ± 0.1 (10.6 ± 0.6) | 9.8 |
| BPSS1623 (BTH_II0748d) | + | Ortholog of R. solanacearum hrpJ | 4.4 ± 0.01 | 12.2 ± 0.1 (12.0 ± 0.5) | 11.8 |
| BPSS1592 | + | T3SS2bp secretin | 4.5 ± 0.1 | 8.1 ± 0.8 | 8.2 |
| BPSS1606 (BTH_II0764d) | + | Hypothetical protein | 4.2 ± 0.1 | 13.2 ± 0.2 (12.8 ± 0.5) | 13.1 |
| BPSS1003 | + | Hypothetical protein | 1.3 ± 0.03 | 4.7 ± 1.8 | 4.2 |
| BPSS1004 | + | Malonyl CoA-acyl carrier protein transacylase | 1.5 ± 0.08 | 6.3 ± 1.0 | 8.3 |
| BPSS1323 | + | Hypothetical protein | 3.9 ± 0.2 | 13.4 ± 0.4 | 13.3 |
| BPSS0740 | + | Hypothetical protein | 3.0 ± 0.3 | 9.6 ± 0.2 | 10.5 |
| BPSS0310 | + | Putative SET domain protein | 3.2 ± 0.2 | ND | 2.3 |
| BPSS0312 | + | Autoinducer-binding transcriptional regulator | 2.4 ± 0.4 | 3.2 ± 0.4 | 4.3 |
| BPSS1389 | + | Putative hemagglutinin/hemolysin | 1.9 ± 0.6 | 6.2 ± 0.5 | 6.8 |
| BPSS1384 | − | Putative membrane protein | 4.0 ± 0.1 | 7.6 ± 1.3 | 7.8 |
| BPSS0804 | − | Hypothetical protein | 4.3 ± 0.03 | ND | 10.0 |
| BPSS1322 | − | Transcriptional regulator, AraC family | 4.3 ± 0.01 | ND | 8.6 |
| BPSS0803 | − | Hemolysin III-like integral membrane protein | 3.3 ± 0.04 | ND | 6.8 |
| BPSL0006 | − | Putative soluble lytic transglycosylase | 2.1 ± 0.1 | ND | 2.8 |
| BPSS1409 | − | Hypothetical protein | 2.0 ± 0.2 | ND | 6.4 |
| BPSS0995 | − | AraC family transcriptional regulator | 1.9 ± 0.1 | ND | 5.1 |
| BPSS0996 | − | Rieske family iron-sulfur cluster-binding protein | 2.0 ± 0.3 | ND | 4.4 |
+, present; −, absent.
The first 4 genes were manually annotated; the descriptions of the remaining genes are based upon data from IMG (http://img.jgi.doe.gov).
Log2 ratios (± standard deviations) of gene transcript levels in B. pseudomallei or B. thailandensis strains overexpressing HrpBbp relative to that of the wild type. The first four rows of data represent the average upregulation of all genes in the indicated cluster. NA, not applicable; ND, not done.
Locus tag of the orthologous gene in B. thailandensis.
Interestingly, 14 of the 17 genes in the 19-kbp region upstream of the T3SS2bp operons (BPSS1593-BPSS1609) were also highly upregulated in response to overexpression of HrpBbp (Table 3). BPSS1593-BPSS1601, which is predicted to be an operon encoding a type IV pilus, was upregulated ∼8-fold; this surface structure could be involved in adhesion (50). BPSS1604 and BPSS1605 encode a two-component regulatory system that was upregulated ∼8-fold, suggesting that they might comprise downstream components in an HrpBbp regulatory cascade. The type IV pilus operon and two-component regulatory system are also present in the same genome position in all the Burkholderia species that have orthologous T3SS2bp gene clusters. Three additional transcriptional regulators were also upregulated in response to HrpBbp overexpression: BPSS0312 (LuxR family) and BPSS1322 and BPSS0995 (both AraC family). These genes were upregulated 9-, 19-, and 4-fold, respectively, and also may be components of an HrpBbp regulatory cascade.
Another notable HrpBbp-regulated gene cluster (BPSS0998-BPSS1004) is predicted to include polyketide biosynthesis genes. This cluster was upregulated an average of ∼3-fold and is found only in B. pseudomallei and B. mallei. This gene cluster is similar in organization and composition to a predicted polyketide biosynthesis gene cluster in the distantly related algicidal flavobacterium Kordia algicida. BPSS1003 and BPSS1004 from this cluster were upregulated 19- and 313-fold, respectively, in the RNA-Seq analysis.
Several monocistronic genes were also upregulated in response to overexpression of HrpBbp. BPSL0006, which is found in all Burkholderia spp. harboring T3SS2bp, was upregulated 4-fold and encodes a putative soluble lytic transglycosylase whose ortholog in Xanthomonas oryzae is predicted to remodel peptidoglycan to allow for assembly of the T3SS apparatus in the cell wall (78). Another gene, BPSS0310, was upregulated 9-fold and encodes a putative SET domain protein, and these are best known for altering eukaryotic gene expression by methylating lysine residues on histones (25). Proteins similar to those encoded by BPSL0006 and BPSS0310 are found in R. solanacearum (with >30% amino acid identity), but neither is regulated by HrpBrso (48). Finally, some of the genes most strongly activated by HrpBbp (>8-fold), BPSS1323, BPSS1609, BPSS0740, and BPSS1384, encode hypothetical proteins without PFAM domains or COG assignments. Interestingly, all of these encoded hypothetical proteins, as well as that encoded by BPSS0310, are predicted to be T3SS-secreted effectors by the Effective T3 prediction tool (http://www.effectors.org/index.jsp), based upon their 50 N-terminal amino acids (4).
To verify and more accurately quantify upregulation measurements from the microarray analysis, mRNA levels of select genes were measured by qRT-PCR. Four T3SS2bp genes from separate predicted transcription units (BPSS1623, BPSS1621, BPSS1614, and BPSS1592) each showed an expression increase of >250-fold when HrpBbp was overexpressed (Table 3). As a control, the expression levels of BPSS1180, a non-T3SS2bp gene which was not upregulated in the microarray, was measured by qRT-PCR and showed no increase in expression. We also verified the HrpBbp-dependent transcription activation of 8 non-T3SS2bp genes. Four showed expression increases of >190-fold, two others were upregulated ∼75-fold, and the remainder were upregulated ∼15-fold. These values are >10-fold higher than the values obtained from the microarray analysis. However, this is not unexpected, as microarray analysis has been shown to underestimate the extent of mRNA expression change (77).
HrpBbp regulates transcription of T3SS2bt and T3SSrso.
Given the high (>90%) nucleotide sequence identity and synteny between the T3SS2bp cluster in B. pseudomallei and the T3SS cluster in B. thailandensis (T3SS2bt), we overexpressed HrpBbp in B. thailandensis and used qRT-PCR to measure HrpBbp-dependent upregulation of the orthologs of the T3SS2bp genes from B. pseudomallei that were analyzed (BTH_II0748, BTH_II0750, and BTH_II0758). We also measured the expression change of a gene adjacent to the T3SS2bt cluster (BTH_II0764), whose ortholog in B. pseudomallei (BPSS1606) was the most highly upregulated gene. All of these genes were upregulated >1,000-fold (Table 3), suggesting that HrpBbp also strongly regulates T3SS2bt in B. thailandensis.
These results and the similarities between T3SS2bp regulation and T3SSrso regulation prompted the analysis of the ability of HrpBbp to activate transcription of T3SS genes in the heterologous host R. solanacearum. The mRNA levels of three T3SSrso genes in R. solanacearum overexpressing HrpBbp were measured by qRT-PCR. These genes, hrpK, hrpW, and hrcV, are in the same transcription units as the T3SS2bp genes tested in B. pseudomallei and B. thailandensis. Transcription of these genes increased ∼4.5-fold when HrpBbp was overexpressed. Although HrpBbp shares only 38% amino acid identity with HrpB from R. solanacearum, it can still activate the expression of T3SSrso genes, albeit much less than in B. pseudomallei.
Several HrpBbp-regulated promoters have a conserved DNA motif.
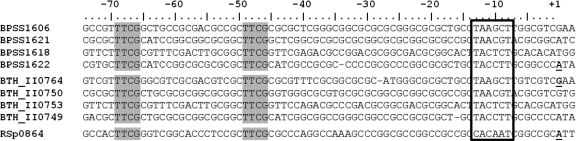
In R. solanacearum, HrpB positively regulates transcription of the genes encoding the T3SS and its secreted effectors via a conserved DNA sequence called the hrpII box located 46 bp upstream from the transcription start site (21). Manual alignment of the promoter region sequences of HrpBbp-regulated genes in B. pseudomallei and B. thailandensis revealed the presence of a motif identical to the hrpII box (Fig. 3). This motif (designated the p2bp box) consists of two 4-bp direct repeats separated by 16 bp (TTCG-N16-TTCG) and is located ∼32 bp upstream from a putative −10 region.
Fig. 3.
p2bp boxes in the promoter regions of HrpBbp-regulated genes. Predicted promoter regions of HrpBbp-regulated genes were aligned manually by using the ATG start codon as an anchor. BPSS promoter regions shown are from genes upregulated >10 fold according to the microarray analysis. B. thailandensis (BTH_II) promoter regions shown are from genes orthologous to the B. pseudomallei (BPSS) genes. RSp0864 is the promoter region from the HrpB-regulated gene hrcU in R. solanacearum. Shading indicates the conserved direct repeats, and the open box indicates the predicted −10 region. Transcription start sites are indicated by +1, and those which were experimentally determined are underlined and in bold.
We employed 5′ RACE to determine the transcription start sites of two highly HrpBbp-regulated genes identified by both microarray and qRT-PCR analysis, BPSS1622, a B. pseudomallei T3SS2bp gene, and BTH_II0764, a B. thailandensis gene encoding a hypothetical protein adjacent to the two-component regulatory system that is also highly regulated. For both genes, the transcription start site was mapped to a position 45 bp downstream from the 3′ end of the p2bp box (Fig. 3). This confirmed the location of the predicted −10 region, which is nearly identical to that found in hrpII box promoters in R. solanacearum (21).
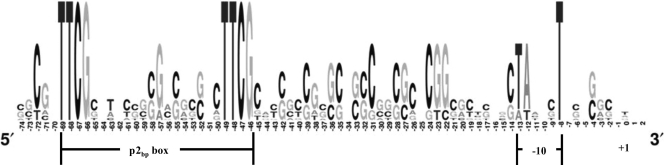
Several HrpBbp-regulated genes not associated with T3SS2bp also possess a p2bp box in their predicted promoter regions. Using Weblogo (20), we analyzed the nucleotide sequence of promoter regions of the seven most highly HrpBbp-regulated genes determined by qRT-PCR (Fig. 4). This analysis identified most, if not all, of the conserved nucleotides in these HrpBbp-regulated promoters. In addition to the direct repeats and the −10 region, other conserved positions around position −72, between positions −58 and −55, and between positions −22 and −39, especially positions −22 to −31, were identified. These additional regions of sequence conservation may contribute to promoter strength because they are not conserved in weakly upregulated genes with a p2bp box (data not shown). One feature that is notably absent from these promoters is a −35 consensus sequence. However, this feature is also absent in the hrpII box promoters of R. solanacearum (21).
Fig. 4.
Sequence conservation in the promoter regions of HrpBbp-regulated genes. The figure was generated by using a manual alignment of the promoter regions of seven B. pseudomallei genes upregulated >200-fold when HrpBbp is overexpressed and Weblogo (20). The locations of the p2bp box, −10 region, and transcription start site (+1) are indicated.
The p2bp box is essential for transcription activation by HrpBbp.
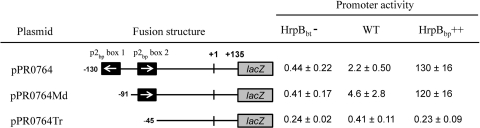
The data described above imply that the p2bp box is homologous to the hrpII box and likely functions with HrpBbp to activate transcription in a manner similar to HrpB-mediated transcription activation in R. solanacearum. To confirm this, several lacZ reporter fusions that either have or lack the p2bp box were constructed and transferred into wild-type, HrpBbt (HrpBbp ortholog) mutant, or HrpBbp-overexpressing HrpBbt mutant B. thailandensis strains and assayed for HrpBbp-dependent promoter activity by using β-galactosidase activity. B. thailandensis was used because we could not find or readily construct a fusion vector approved for use in B. pseudomallei and because B. pseudomallei and B. thailandensis are so closely related (>90% DNA sequence identity). Relative to the wild type, the HrpBbp-dependent promoter activities of pPR1606 and pPR0764 (Table 1), which contain promoter fragments from orthologous B. pseudomallei and B. thailandensis genes (BPSS1606 and BTH_II0764, respectively), decreased ∼5-fold when HrpBbt was inactivated but increased ∼83-fold in the HrpBbp-overexpressing strain, suggesting HrpBbp-mediated transcription activation. The promoter region of BTH_II0764 is predicted to have a divergent promoter with two p2bp boxes, oriented in opposite directions. p2bp box 1 is likely associated with the divergently transcribed upstream gene, BTH_II0763, and deleting it (pPR0764Md) produced no significant change in HrpBbp-dependent promoter activity (Fig. 5). However, deleting p2bp box 2 (pPR0764Tr) showed dramatically reduced promoter activity, demonstrating that p2bp box 2 is required for HrpBbp-dependent transcription activation of BTH_II0764. Furthermore, the reciprocal recognition of B. pseudomallei and B. thailandensis p2bp box promoter fragments by HrpBbp and HrpBbt indicates homology of these transcriptional regulators.
Fig. 5.
Analysis of p2bp box requirement for transcription activation by HrpBbp. Mutants with 5′ deletions of the promoter region of BTH_II0764 were cloned into a lacZ reporter plasmid and transferred into wild-type (WT), HrpBbt-deficient (HrpBbt−), and HrpBbt-deficient but HrpBbp-overexpressing (HrpBbp++) B. thailandensis strains. Promoter activity was monitored by β-galactosidase assay, and activities are reported in Miller units/100. Numbers indicate nucleotide positions relative to the transcription start; white arrows indicate the orientation of the p2bp box. +1, transcription start site; +135, first nucleotide of lacZ CDS.
For R. solanacearum, growth in minimal medium with glutamate increases the expression of HrpB-regulated genes relative to growth in rich media. To determine if this is also the case for B. thailandensis, we assayed the HrpBbt-dependent promoter activities of wild-type B. thailandensis (pPR0764) grown in LB and M9–20 mM glutamate. There was no difference in HrpBbt-regulated promoter activity in cells from these two types of media (data not shown), suggesting that HrpBbt expression, as well as subsequent induction of HrpBbt-regulated genes in B. thailandensis, is regulated by different environmental signals than the mechanism in R. solanacearum.
Determination of nucleotides involved in HrpBbp-dependent transcription activation.
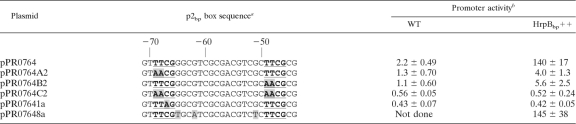
To assess the functional importance of specific nucleotides in the direct repeats of the p2bp box, we mutated the p2bp box in plasmid pPR0764 by using site-directed mutagenesis or error-prone PCR and measured promoter activities in various B. thailandensis strains (Table 4). In the HrpBbp-overexpressing strain, when TT in either the first or second repeat of the p2bp box (positions −68 and −69 or −48 and −49) was changed to AA, the promoter activity decreased ∼30-fold. When TT was changed to AA in both repeats, HrpBbp activation of the promoter was lost. Changing the third base in the first repeat, from C to A at position −67, also abolished the ability of the promoter to be activated by HrpBbp. A mutant harboring three mutations in the spacer between the direct repeats of the p2bp box retained HrpBbp-dependent activity. This suggests that specific bases at −49, −48, and −67 to −69 are crucial for HrpBbp-dependent transcription activation from this promoter but that the sequence of the spacer between the repeats is not critical for promoter activity.
Table 4.
Effect of nucleotide substitutions in p2bp box direct repeats on HrpBbp-dependent promoter activity
The bold, underlined nucleotides are the direct repeats of the p2bp box; shadowed nucleotides indicate substitutions. Nucleotide positions relative to transcription start are given above.
β-Galactosidase activity directed by each plasmid in wild-type (WT) and HrpBbp-overexpressing (HrpBbp++) B. thailandensis strains in Miller units/100.
B. thailandensis fails to demonstrate a phytopathogenic phenotype.
Given the remarkable similarity of T3SS2bp to the T3SSs required for virulence of several plant pathogens, it is plausible that T3SS2bp in Burkholderia spp. is also involved in plant pathogenesis. Lee et al. (34) found that soaking tomato plantlets in suspensions of B. thailandensis E264 or B. pseudomallei for 7 days produced disease-like symptoms (i.e., leaf vein blackening, wilting, and necrosis) and that development of these symptoms was delayed when using a T3SS2bp mutant. We treated tomato plantlets identically with the same B. thailandensis E264 strain but observed only a wilting symptom. However, soaking plantlets in suspensions of nonphytopathogenic P. putida ATCC 12633 or media alone also caused wilting. Moreover, we found that after 36 h, populations of all strains in the soaking solution exceeded 108 cells ml−1. When we petiole inoculated ∼106 cells of B. thailandensis E264, B. thailandensis DW503, or P. putida into larger, more mature tomato plants, they remained asymptomatic for 7 days. This is in stark contrast to the plants inoculated with ∼104 cells of R. solanacearum, which showed extensive wilting after only 3 days. These wilted plants harbored >109 cells g−1 of stem, while populations of B. thailandensis strains in asymptomatic plants were only 104 cells g−1. We tested a second plant host (Arabidopsis thaliana) by infiltrating leaves with 107 or 108 cells ml−1 of the B. thailandensis strains used as described above. After 21 days, these plants were asymptomatic. Consistent with previous reports (10), plants infiltrated with the same amounts of R. solanacearum were chlorotic and wilted by day 12. Symptomatic A. thaliana plants harbored 10-fold more cells than asymptomatic plants.
A hallmark of nearly all plant-pathogenic bacteria is the ability to produce a hypersensitive response in tobacco (13, 23, 79). Tobacco leaves infiltrated with 109 cells ml−1 of B. thailandensis or P. putida did not elicit a hypersensitive response after 7 days, while those infiltrated with the same amount of R. solanacearum developed a hypersensitive response after 2 days. Those leaves infiltrated with 107 or 108 cells ml−1 of R. solanacearum showed a hypersensitive response by day 4. Taken together, these results suggest that B. thailandensis is not an aggressive or typical plant pathogen.
DISCUSSION
We investigated the organization and regulation of the T3SS2bp gene cluster, one of the three T3SS gene clusters in B. pseudomallei. Orthologous gene clusters containing this T3SS cluster are found in B. mallei, B. thailandensis, B. oklahomensis, B. ubonensis, B. ambifaria, and B. dolosa but not in any of the 12 other sequenced Burkholderia species. Although our analyses clearly showed that this T3SS is very closely related in gene content, organization, and sequence to one required by the plant pathogens R. solanacearum (T3SSrso) and Xanthomonas spp. for causing disease, the role of T3SS2bp in plant pathogenesis or any other biological process involving any of the Burkholderia spp. harboring it remains unclear.
In addition to finding genetic similarities between the T3SS2bp and T3SSrso gene clusters, we also found remarkable regulation similarities. In R. solanacearum, transcription activation of >20 operons and genes associated with T3SSrso is directly mediated by the AraC-type regulator HrpB via interaction with the conserved hrpII box motif (21). We identified an identical motif in B. pseudomallei (p2bp box) that is similarly required for HrpBbp-dependent transcription activation of T3SS2bp genes, and we showed that, just as hrpII box mutations negatively affect transcription activation by HrpB in R. solanacearum (21), p2bp box mutations eliminate HrpBbp-dependent transcription activation. The PIP box (TTCGC-N15-TTCGC) in Xanthomonas spp. is nearly identical to the hrpII and p2bp boxes and is likewise required for HrpX (the ortholog of HrpB and HrpBbp)-mediated activation of T3SS transcription (66). HrpB from R. solanacearum can partially complement a HrpX mutant in X. campestris pv. vesicatoria (73), and we showed that HrpBbp from B. pseudomallei can activate T3SSrso gene expression. Thus, HrpB and the sequences it uses to regulate transcription are highly conserved across at least 3 genera. Additionally, Acidovorax avenae subsp. citrulli AAC00-1, another plant pathogen in the Betaproteobacteria, also has an HrpB ortholog and PIP boxes upstream from some of its T3SS genes. However, HrpB regulation in A. avenae subsp. citrulli has not been explored.
The expression of hrpB in R. solanacearum is induced either through a plant cell contact-dependent regulatory cascade or by an unidentified signal that can be mimicked by growth in minimal media. The B. pseudomallei and B. thailandensis genomes do not contain orthologs corresponding to any of the regulatory proteins involved in the plant cell contact-dependent pathway, and contrary to what was found for R. solanacearum (3), growth in minimal medium containing glutamate did not induce expression of hrpBbt in B. thailandensis. This suggests that the signal inducing hrpBbt expression in B. thailandensis, and by extension hrpBbp expression in B. pseudomallei, is different from the inducing signal in R. solanacearum.
In R. solanacearum, HrpB is the terminal regulatory protein for the activation of T3SS transcription. Transcriptome analysis of R. solanacearum overexpressing HrpB showed that it does not positively regulate any other regulators (48). However, HpaR, a transcriptional regulator that is positively regulated by HrpX in Xanthomonas campestris pv. campestris, is required for pathogenicity but does not regulate any T3SS genes (72). While there is no ortholog of HpaR in B. pseudomallei, our analysis did indicate that a two-component regulatory system, a LuxR family regulator, and two AraC-type regulators may be downstream components of a HrpBbp regulatory cascade. The multiple proteins that may be involved in the HrpBbp regulatory cascade underscore the greater complexity of the HrpBbp regulon and warrant further study.
HrpB in R. solanacearum positively regulates not only the expression of T3SSrso but also the expression of many effectors secreted through T3SSrso (22, 45). Transcriptome analyses, genetic screens, and bioinformatic searches for hrpII boxes has identified 72 HrpB-regulated effectors in R. solanacearum (44, 45, 48). In contrast, the transcriptome analysis we performed identified only 33 HrpBbp-upregulated genes in addition to the T3SS2bp cluster. Of these genes, BPSS0310, BPSS0740, BPSS1609, BPSS1323, and BPSS1384 all possess p2bp boxes in their predicted promoter regions, and the proteins they encode are predicted to be T3SS secreted. BPSS0310 is particularly interesting because it encodes a putative SET domain protein; some members of this family are histone lysine methyltransferases involved in modulating eukaryotic gene expression (25). It is tempting to speculate that this protein is a bona fide effector that could affect host gene expression after it is injected by T3SS2bp. None of the other predicted T3SS-secreted proteins show amino acid sequence identity to any known plant or animal pathogen T3SS effectors, or any other proteins, and thus may represent novel effectors.
Two notable gene clusters in B. pseudomallei that were also HrpBbp regulated are a type IV pilus (Tfp) gene cluster located 8 kbp away from T3SS2bp and a predicted polyketide biosynthesis cluster located ∼86 kbp away from T3SS2bp. This Tfp system is orthologous to the Tfp that contributes to virulence of Yersinia pseudotuberculosis in mice (18), where it was suggested to play a role in colonization of intestinal mucosa. The Tfp in B. pseudomallei could be involved in adhesion to host cells to facilitate injection of effectors by T3SS2bp.
The HrpBbp-regulated polyketide gene cluster may be involved in the synthesis of a type III polyketide, since it contains BPSS1002, encoding a predicted type III polyketide synthase with the Cys-His-Asn catalytic triad required for the condensation reactions (5). These proteins are in the chalcone synthase superfamily and generate aromatic polyketides by using various starter and extender substrates (i.e., long-chain fatty acyl coenzyme A [acyl-CoA] thioester starter and malonyl-CoA extenders) (5, 47). It is possible that BPSS1002 and its adjacent genes produce a toxin or antibiotic that may function independently or as an enhancer of T3SS2bp function.
Although B. thailandensis and B. pseudomallei were reported to cause disease-like symptoms on tomato plantlets (34), our analysis indicates that B. thailandensis does not produce symptoms characteristic of a plant pathogen on tomato, tobacco, or A. thaliana plants. Moreover, an attempt to demonstrate phytopathogenicity of B. thailandensis on Oryza sativa (rice) plants was similarly unsuccessful (34). These results, coupled with observation that, unlike those of R. solanacearum and Xanthomonas spp., the genomes of B. thailandensis and B. pseudomallei encode no plant cell wall-degrading enzymes (e.g., pectate lyases, polygalacturonases, or endoglucanases) (37, 70), call into question any suggestion that B. thailandensis and, by extension, B. pseudomallei are typical plant pathogens. In fact, of the >30 described Burkholderia spp., only 6 are reported to cause plant disease (17, 40), and none of these has a T3SS orthologous to T3SS2bp.
If B. pseudomallei is not a plant pathogen, then what could be the function of T3SS2bp? Shotgun proteomic analysis has shown that at least some HrpBbp-activated genes (encoding the secretins of Tfp and T3SS2bp) are not just transcribed pseudogenes or relics but are translated into protein and properly localized in the outer membrane only when HrpBbp is overexpressed in B. pseudomallei (unpublished data). A possible T3SS2bp function could be a role in pathogenesis or manipulation of other organisms, such as fungi, amoebae, or algae. Interestingly, treatment of pea seeds with B. ambifaria AMMD, which contains a T3SS orthologous to T3SS2bp, controls seed rot and damping-off caused by Pythium spp. and in some cases controls root rot caused by Aphanomyces euteiches (16, 33). Although it has been reported that B. pseudomallei can invade the spores and attach to the hyphae of the arbuscular mycorrhizal fungus Gigaspora decipiens (35) and that B. pseudomallei and B. thailandensis can survive inside the amoeba Dictyostelium discoideum (28a), the role of T3SS2bp in its interaction with these organisms remains to be investigated.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grant R21-AI069081 and USDA-NRI Plant Biosecurity award 07-21 55605-17843.
We thank W. Nierman and L. Losada for providing pSCRhaB2K and for performing transcriptome analysis. We also thank Jason Cloward and Dave Samuels for critically reading the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Aldon D., Brito B., Boucher C., Genin S. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alfano J. R., Collmer A. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385–414 [DOI] [PubMed] [Google Scholar]
- 3. Arlat M., et al. 1992. Transcriptional organization and expression of the large hrp gene cluster in Ralstonia solanacearum. Mol. Plant Microbe Interact. 2:187–193 [DOI] [PubMed] [Google Scholar]
- 4. Arnold R., et al. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 5:e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Austin M. B., Noel J. P. 2003. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 1:79–110 [DOI] [PubMed] [Google Scholar]
- 6. Block A., Li G., Fu Z. Q., Alfano J. R. 2008. Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11:396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brett P. J., DeShazer D., Woods D. E. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317–320 [DOI] [PubMed] [Google Scholar]
- 8. Brett P. J., Deshazer D., Woods D. E. 1997. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol. Infect. 118:137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brito B., Aldon D., Barberis P., Boucher C. A., Genin S. 2002. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant Microbe Interact. 15:109–119 [DOI] [PubMed] [Google Scholar]
- 10. Brito B., Marenda M., Barberis P., Boucher C., Genin S. 1999. prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31:237–251 [DOI] [PubMed] [Google Scholar]
- 11. Burtnick M. N., Bolton A. J., Brett P. J., Watanabe D., Woods D. E. 2001. Identification of the acid phosphatase (acpA) gene homologues in pathogenic and non-pathogenic Burkholderia spp. facilitates TnphoA mutagenesis. Microbiology 147:111–120 [DOI] [PubMed] [Google Scholar]
- 12. Cardona S. T., Valvano M. A. 2005. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54:219–228 [DOI] [PubMed] [Google Scholar]
- 13. Carney B. F., Denny T. P. 1990. A cloned avirulence gene from Pseudomonas solanacearum determines incompatibility on Nicotiana tabacum at the host species level. J. Bacteriol. 172:4836–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng A. C., Currie B. J. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clough S. J., Lee K. E., Schell M. A., Denny T. P. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coenye T., et al. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481–1490 [DOI] [PubMed] [Google Scholar]
- 17. Coenye T., Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719–729 [DOI] [PubMed] [Google Scholar]
- 18. Collyn F., et al. 2002. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 70:6196–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cornelis G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825 [DOI] [PubMed] [Google Scholar]
- 20. Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cunnac S., Boucher C., Genin S. 2004. Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186:2309–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cunnac S., Occhialini A., Barberis P., Boucher C., Genin S. 2004. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53:115–128 [DOI] [PubMed] [Google Scholar]
- 23. Deng W. L., Preston G., Collmer A., Chang C. J., Huang H. C. 1998. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 180:4523–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeShazer D., Waag D. M., Fritz D. L., Woods D. E. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253–269 [DOI] [PubMed] [Google Scholar]
- 25. Dillon S., Zhang X., Trievel R., Cheng X. 2005. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Figurski D. H., Helinski D. R. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forsyth A., et al. 2010. Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Mol. Plant Microbe Interact. 23:1545–1552 [DOI] [PubMed] [Google Scholar]
- 28. Haraga A., West T. E., Brittnacher M. J., Skerrett S. J., Miller S. I. 2008. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect. Immun. 76:5402–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a. Hasselbring B. M., Patel M. K., Schell M. A. Dictyostelium discoideum as a model system for identification of Burkholderia pseudomallei virulence factors. Infect. Immun., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holden M. T., et al. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horton R. M., et al. 1993. Gene-splicing by overlap extension. Methods Enzymol. 217:270–279 [DOI] [PubMed] [Google Scholar]
- 31. Huang H. C., et al. 1988. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 170:4748–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inglis T. J., Sagripanti J. 2006. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 72:6865–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. King E. B., Parke J. L. 1993. Biocontrol of aphanomyces root rot and pythium damping-off by Pseudomonas cepacia AMMD on four pea cultivars. Plant Dis. 77:1185–1188 [Google Scholar]
- 34. Lee Y. H., Chen Y., Ouyang X., Gan Y.-H. 2010. Identification of tomato plant as a novel host model for Burkholderia pseudomallei. BMC Microbiol. 10:28 doi:10.1186/1471-2180-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levy A., et al. 2003. Invasion of spores of the arbuscular mycorrhizal fungus Gigaspora decipiens by Burkholderia spp. Appl. Environ. Microbiol. 69:6250–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H., Kang Y., Genin S., Schell M. A., Denny T. P. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215–3229 [DOI] [PubMed] [Google Scholar]
- 37. Liu H., Zhang S., Schell M. A., Denny T. P. 2005. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol. Plant Microbe Interact. 18:1296–1305 [DOI] [PubMed] [Google Scholar]
- 38. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 39. Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 40. Maeda Y., et al. 2006. Phylogenetic study and multiplex PCR-based detection of Burkholderia plantarii, Burkholderia glumae and Burkholderia gladioli using gyrB and rpoD sequences. Int. J. Syst. Evol. Microbiol. 56:1031–1038 [DOI] [PubMed] [Google Scholar]
- 41. Marenda M., et al. 1998. PhrA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27:437–453 [DOI] [PubMed] [Google Scholar]
- 42. Miller J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Moore R. A., DeShazer D., Reckseidler S., Weissman A., Woods D. E. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mukaihara T., Tamura N., Iwabuchi M. 2010. Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23:251–262 [DOI] [PubMed] [Google Scholar]
- 45. Mukaihara T., Tamura N., Murata Y., Iwabuchi M. 2004. Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol. Microbiol. 54:863–875 [DOI] [PubMed] [Google Scholar]
- 46. Murashige T., Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15:473–497 [Google Scholar]
- 47. Nakano C., Ozawa H., Akanuma G., Funa N., Horinouchi S. 2009. Biosynthesis of aliphatic polyketides by type III polyketide synthase and methyltransferase in Bacillus subtilis. J. Bacteriol. 191:4916–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Occhialini A., Cunnac S., Reymond N., Genin S., Boucher C. 2005. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant Microbe Interact. 18:938–949 [DOI] [PubMed] [Google Scholar]
- 49. Parkinson H., et al. 2011. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 39:D1002–D1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 51. Price M. N., Huang K. H., Alm E. J., Arkin A. P. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rainbow L., Hart C. A., Winstanley C. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374–384 [DOI] [PubMed] [Google Scholar]
- 53. Ritz M., Garenaux A., Berge M., Federighi M. 2009. Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RT-qPCR and application to oxidative stress. J. Microbiol. Methods 76:196–200 [DOI] [PubMed] [Google Scholar]
- 54. Rotz L. D., Khan A. S., Lillibridge S. R., Ostroff S. M., Hughes J. M. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saile E., McGarvey J. A., Schell M. A., Denny T. P. 1997. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87:1264–1271 [DOI] [PubMed] [Google Scholar]
- 56. Salanoubat M., et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497–502 [DOI] [PubMed] [Google Scholar]
- 57. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold SpringHarbor, NY [Google Scholar]
- 58. Santos P. M., Di Bartolo I., Blatney J. M., Zennaro E., Valla S. 2001. New broad-host-range promoter probe vectors based the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91–96 [DOI] [PubMed] [Google Scholar]
- 59. Schell M. A., et al. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64:1466–1485 [DOI] [PubMed] [Google Scholar]
- 60. Schiffer M., Edmundson A. B. 1967. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys. J. 7:121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seidman C. E., Struhl K., Sheen J., Jessen T. 1997. Introduction of plasmid DNA into cells, unit 1.8. In Current protocols in molecular biology. John Wiley & Sons, Inc., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 62. Simon R., Priefer U., Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 63. Stanier R. Y., Palleroni N. J., Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271 [DOI] [PubMed] [Google Scholar]
- 64. Stevens M. P., et al. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649–659 [DOI] [PubMed] [Google Scholar]
- 65. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 66. Tsuge S., et al. 2005. Effects on promoter activity of base substitutions in the cis-acting regulatory element of HrpXo regulons in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 187:2308–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vandamme P., Govan J., LiPuma J. 2007. Diversity and role of Burkholderia spp. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 68. Van Gijsegem F., Vasse J., De Rycke R., Castello P., Boucher C. 2002. Genetic dissection of the Ralstonia solanacearum hrp gene cluster reveals that the HrpV and HrpX proteins are required for hrp pilus assembly. Mol. Microbiol. 44:935–946 [DOI] [PubMed] [Google Scholar]
- 69. Van Gijsegem F., Vasse J., Camus J. C., Marenda M., Boucher C. 2000. Ralstonia solanacearum produces Hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol. 36:249–260 [DOI] [PubMed] [Google Scholar]
- 70. Wang L., Rong W., He C. 2008. Two Xanthomonas extracellular polygalacturonases, PghAxc and PghBxc, are regulated by type III secretion regulators HrpX and HrpG and are required for virulence. Mol. Plant Microbe Interact. 21:555–563 [DOI] [PubMed] [Google Scholar]
- 71. Warawa J., Woods D. E. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101–108 [DOI] [PubMed] [Google Scholar]
- 72. Wei K., et al. 2007. hpaR, a putative marR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris. J. Bacteriol. 189:2055–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wengelnik K., Bonas U. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. West T. E., Frevert C. W., Liggitt H. D., Skerrett S. J. 2008. Inhalation of Burkholderia thailandensis results in lethal necrotizing pneumonia in mice: a surrogate model for pneumonic melioidosis. Trans. R. Soc. Trop. Med. Hyg. 102:S119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wiersinga W. J., van der Poll T., White N. J., Day N. P., Peacock S. J. 2006. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4:272–282 [DOI] [PubMed] [Google Scholar]
- 76. Wilhelm B. T., Landry J.-R. 2009. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48:249–257 [DOI] [PubMed] [Google Scholar]
- 77. Yuen T., Wurmbach E., Pfeffer R. L., Ebersole B. J., Sealfon S. C. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang J., Wang X., Zhang Y., Zhang G., Wang J. 2008. A conserved HpaII protein has lytic activity against the bacterial cell wall in phytopathogenic Xanthomonas oryzae. Appl. Microbiol. Biotechnol. 79:605–616 [DOI] [PubMed] [Google Scholar]
- 79. Zou L.-F., et al. 2006. Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72:6212–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.