Abstract
Actin-based motility of the melioidosis pathogen Burkholderia pseudomallei requires BimA (Burkholderia intracellular motility A). The mechanism by which BimA mediates actin assembly at the bacterial pole is ill-defined. Toward an understanding of the regions of B. pseudomallei BimA required for intracellular motility and the binding and polymerization of actin, we constructed plasmid-borne bimA variants and glutathione-S-transferase fusion proteins with in-frame deletions of specific motifs. A 13-amino-acid direct repeat and IP7 proline-rich motif were dispensable for actin binding and assembly in vitro, and expression of the mutated proteins in a B. pseudomallei bimA mutant restored actin-based motility in J774.2 murine macrophage-like cells. However, two WASP homology 2 (WH2) domains were found to be required for actin binding, actin assembly, and plaque formation. A tract of five PDASX direct repeats influenced the polymerization of pyrene-actin monomers in vitro and was required for actin-based motility and intercellular spread, but not actin binding. None of the mutations impaired surface expression or polar targeting of BimA. The number of PDASX repeats varied in natural isolates from two to seven. Such repeats acted additively to promote pyrene-actin polymerization in vitro, with stepwise increases in the rate of polymerization as the number of repeats was increased. No differences in the efficiency of actin tail formation could be discerned between strains expressing BimA variants with two, five, or seven PDASX repeats. The data provide valuable new insights into the role of conserved and variable motifs of BimA in actin-based motility and intercellular spread of B. pseudomallei.
INTRODUCTION
Burkholderia pseudomallei is a Gram-negative facultative intracellular pathogen that causes melioidosis, a severe invasive disease of animals and humans that is endemic in Southeast Asia and northern Australia (reviewed in reference 21). In common with selected species of Listeria, Shigella, Rickettsia, and Mycobacterium, some members of the genus Burkholderia are capable of intracellular actin-based motility (reviewed in reference 22). Such motility promotes cell-to-cell spread in the absence of immune surveillance and, in some cases, escape from autophagy. Although B. pseudomallei was the first of the Burkholderia species to be described to form actin-rich bacteria-containing membrane protrusions (11), actin-based motility is also a feature of infection by the glanders pathogen Burkholderia mallei and the avirulent saprophyte Burkholderia thailandensis (17, 23). Actin binding and assembly in these species is mediated by BimA (Burkholderia intracellular motility A), a putative type V secreted protein exhibiting carboxyl-terminal homology to the Yersinia autosecreted adhesin YadA (17, 20, 23, 24).
Mechanisms of bacterial actin-based motility often converge on activation of the cellular actin-related protein 2/3 (Arp2/3) complex. Activation of the Arp2/3 complex requires cellular nucleation-promoting factors (NPFs), such as Wiskott-Aldrich syndrome protein (WASP) family members (reviewed in reference 3). Pathogens capable of actin-based motility may mimic the activity of NPFs (e.g., Listeria ActA and RickA in selected Rickettsia species) or recruit and activate them at the bacterial pole (e.g., Shigella IcsA) (reviewed in reference 22). Recent studies have indicated that Listeria ActA not only mimics the activity of the cellular NPF N-WASP but is also regulated in a similar fashion by casein kinase 2 (CK2)-mediated phosphorylation, which enhances its affinity for the Arp2/3 complex (4). By using green fluorescent protein fusions and immunocytochemistry, Arp2/3 components were found to be localized throughout B. pseudomallei-induced actin tails (2). However, the role of the Arp2/3 complex in intracellular motility of B. pseudomallei is unclear, as overexpression of a dominant negative domain of the cellular NPF Scar1, which inhibits actin tail formation by Shigella flexneri, Listeria monocytogenes, and Rickettsia conorii, does not interfere with actin-based motility of B. pseudomallei (2). Further, in contrast to Shigella, N-WASP is not recruited to the surface of intracellular B. pseudomallei, and normal actin-based motility occurs in N-WASP−/− fibroblasts (2). Fibroblasts lacking vasodilator-stimulated phospho-protein (Ena/VASP) also support actin-based motility of B. pseudomallei, and Ena/VASP is not recruited to the tails, in contrast to those formed by Listeria and Shigella (2). Taken together, these observations imply that actin-based motility of B. pseudomallei occurs by a mechanism that may be distinct to that used by other intracellular pathogens.
The molecular basis of actin-based motility of B. pseudomallei may differ not only from that of other bacterial genera but also between closely related Burkholderia species. BimA from B. mallei (BimAma) is markedly shorter and more proline rich, whereas an amphipathic central and acidic (CA) domain is uniquely found in B. thailandensis BimA (BimAth) (20, 23). Although both variants can restore actin-based motility to a B. pseudomallei bimA mutant (23), only BimAth sequestered Arp3 from murine splenic extracts (20, 23), consistent with the fact that some cellular and pathogen-associated NPFs rely on a CA domain and one or more WASP homology 2 (WH2) domains to recruit and activate the Arp2/3 complex (3, 15). We recently confirmed that B. thailandensis BimA stimulates the polymerization of pyrene-actin monomers in a manner dependent on both the Arp2/3 complex and the CA domain in vitro (20). The CA domain was required for actin-based motility, though not for BimAth to interact with actin (20). A truncated version of B. pseudomallei BimA (BimAps) fused to glutathione-S-transferase (GST) binds monomeric actin and stimulates its assembly in vitro independently of the Arp2/3 complex (20, 24). Other pathogen-associated WH2-containing Arp2/3-independent actin nucleators have been described, including the type III secreted effector proteins TARP (translocated actin-recruiting phosphoprotein), which mediates Chlamydia invasion via actin remodeling (10), and Vibrio cholerae VopF and Vibrio parahaemolyticus VopL, which disrupt actin homeostasis (13, 25). Some eukaryotic NPFs, such as formins, Spire, cordon-bleu, and leiomodin, stimulate actin polymerization in an Arp2/3-independent fashion (reviewed in reference 3). Recent studies indicated that the putative autosecreted surface antigen Sca2 of R. rickettsii is required for actin-based motility (12) and mimics the function of eukaryotic formins by nucleating unbranched actin filaments and protecting them from capping (7). These data imply that in some bacteria, Arp2/3-independent modes of actin-based motility have evolved.
To identify conserved and variable portions of bimA, we previously surveyed the prevalence and sequence diversity of the gene in clinical and environmental isolates from areas of endemicity (19). Intraspecies conservation of BimA was found to be high in natural populations of B. pseudomallei, B. mallei, and B. thailandensis (19, 20), with the exception that a B. mallei-like BimA variant was found in B. pseudomallei isolates restricted to the Northern Territory of Australia, but not specific multilocus sequence types (19). ClustalW alignment of predicted BimA protein sequences indicated that tandem WH2 domains predicted to mediate the binding of actin monomers were conserved in sequenced B. pseudomallei strains (19). A proline-rich motif (IP7; also defined as a PRM1 motif [reviewed in reference 9]) was also conserved. PRM1 motifs interact with profilin, a G-actin-binding protein that funnels actin monomers to the tip of actin filaments and that aids the formin-like activity of R. rickettsii Sca2 (7). A tandem repeat of 13 amino acids adjacent to the IP7 motif on the amino-terminal side (NIPVPPPMPGGGA) was conserved in most B. pseudomallei strains, but a single copy was found in one strain (19). Variation was also detected in the number of PDASX repeats adjacent to the WH2 domains on the carboxyl-terminal side. Five PDASX repeats exist in the BimA of the sequenced prototype B. pseudomallei strain K96243 and the strain in which BimA was first characterized (24), but two copies were detected in strain BCC215 and seven were found in strain MSHR305 (19). When juxtaposed the PDASX repeats form predicted casein kinase 2 target sites (SxxD/E), the number of which therefore varies between one and six in the predicted BimAps proteins described to date.
Toward an understanding of the role played by conserved and variable portions of B. pseudomallei BimA, in-frame deletions of the motifs described above were generated and their impacts on actin-based motility, intercellular spread, and the binding and polymerization of actin in vitro were evaluated.
MATERIALS AND METHODS
Bacterial strains, plasmids, cell lines, and media.
Burkholderia pseudomallei strain 10276 was isolated from a human melioidosis patient and obtained from Ty Pitt, Health Protection Agency, Colindale, United Kingdom. A null mutant of this strain with an insertion of the suicide replicon pDM4 in bimA (bpss1492) has been described (24). Full-length bimA genes harboring in-frame deletions were placed under the control of a tac promoter in plasmid pME6032, which contains p15A and pVS1 origins of replication and stably replicates in B. pseudomallei (8). Plasmid pME6032-bimA contains the intact bimA gene from B. pseudomallei 10276 and fully restores actin-based motility to 10276 bimA::pDM4, as described previously (20, 23, 24). Plasmid pCR2.1-TOPO and chemically competent Escherichia coli K-12 TOP10 cells were used for topoisomerase-mediated cloning of internal portions of bimA genes harboring in-frame deletions of specific motifs (Invitrogen Life Technologies, Paisley, United Kingdom). Such regions were subcloned into pGEX-4T-1 to create fusions to the carboxy terminus of GST (Amersham Biosciences, Buckinghamshire, United Kingdom). A plasmid construct for expression of residues 54 to 455 of B. pseudomallei strain 10276 BimA as a GST fusion was recently described (20). E. coli K-12 Rosetta2(DE3)pLysS cells expressing rare amino-acyl tRNAs were used for protein expression (Merck Biosciences, Nottingham, United Kingdom). Bacterial strains were amplified in Luria-Bertani broth (LB) containing, as appropriate, ampicillin at 100 μg/ml, chloramphenicol at 50 μg/ml, kanamycin at 50 μg/ml, and/or tetracycline at 50 μg/ml. All strains and plasmids are listed in Table 1. J774.2 murine macrophage-like cells (ECACC 85011428) and A549 human lung epithelial carcinoma cells (ECACC 86012804) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum and 2 mM l-glutamine at 37°C in a humidified 5% CO2 atmosphere.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Bacterial strains | ||
| 10276 bimA::pDM4 | bimA insertion mutant in B. pseudomallei strain 10276 | Stevens et al. (23) |
| 10276 bimA::pDM4 [pME6032-bimA] | bimA mutant complemented with full-length bimA | Stevens et al. (23) |
| Rosetta2(DE3)pLysS | E. coli BL21 derivative containing tRNAs for rare codons, Cmr | Novagen |
| Plasmids | ||
| pME6032 | IPTG-inducible expression plasmid, Tetr | Heeb et al. (8) |
| pME6032-bimA | pME6032 expressing bimA | Stevens et al. (23) |
| pME6032-bimAΔ13AA | pME6032 expressing bimAΔ13AA | This study |
| pME6032-bimAΔPRM1 | pME6032 expressing bimAΔPRM1 | This study |
| pME6032-bimAΔWH2-1 | pME6032 expressing bimAΔWH2-1 | This study |
| pME6032-bimAΔWH2-2 | pME6032 expressing bimAΔWH2-2 | This study |
| pME6032-bimAΔWH2-1+2 | pME6032 expressing bimAΔWH2-1+2 | This study |
| pME6032-bimA 2PDASX | pME6032 expressing bimA 2PDASX | This study |
| pME6032-bimA 7PDASX | pME6032 expressing bimA 7PDASX | This study |
| pCR2.1-TOPO | Topoisomerase-mediated cloning vector, Ampr Kanr | Invitrogen Life Technologies |
| pGEX-4T-1 | IPTG-inducible vector for expression of GST fusion proteins, Ampr | GE Healthcare |
| pGEX-bimA54–455 | pGEX expressing GST-BimA54–455 protein | Sitthidet et al. (19) |
| pGEX-bimAΔ13AA54–455 | pGEX expressing GST-BimAΔ13AA54–455 protein | This study |
| pGEX-bimAΔPRM154–455 | pGEX expressing GST-BimAΔPRM154–455 protein | This study |
| pGEX-bimAΔWH2-154–455 | pGEX expressing GST-BimAΔWH2-154–455 protein | This study |
| pGEX-bimAΔWH2-254–455 | pGEX expressing GST-BimAΔWH2-254–455 protein | This study |
| pGEX-bimAΔWH2-1+254–455 | pGEX expressing GST-BimAΔWH2-1+254–455 protein | This study |
| pGEX-bimAΔPDASX54–455 | pGEX expressing GST-BimAΔPDASX54–455 protein | This study |
| pGEX-bimA 2PDASX54–455 | pGEX expressing GST-BimA 2PDASX54–455 protein | This study |
| pGEX-bimA 7PDASX54–455 | pGEX expressing GST-BimA 7PDASX54–455 protein | This study |
bimA is the B. pseudomallei bimA gene. Tetr, tetracycline resistance; CMr, chloramphenicol resistance; Ampr, ampicillin resistance; pGex, pGex-4T-1.
In-frame deletion of BimA motifs.
In order to create in-frame deletions of specific residues, the strategy of PCR-ligation-PCR was adopted to fuse amplicons spanning the deletion site (1). In each case, the regions 5′ and 3′ of the residues to be deleted were separately amplified using Pfu proofreading DNA polymerase using pME6032-bimA plasmid DNA as template and the oligonucleotides listed in Table 2. For amplicons on the 5′ side of the deletion site, pCompE1.2 was used as the forward primer together with a reverse primer ending immediately 5′ of the deletion site. Similarly, a forward primer starting immediately 3′ of the deletion site was used to amplify the portion of bimA downstream of the deletion, using pCompXho.2 as the reverse primer. The blunt-ended first-round PCR products were resolved by agarose gel electrophoresis, excised, and purified. They were then phosphorylated with T4 polynucleotide kinase in the presence of ATP and ligated with T4 DNA ligase (Promega, Madison, WI), according to the manufacturer's instructions. Ligated molecules were then amplified by PCR with Pfu using pCompE1.2 and pCompXho.2. Juxtaposition of the amplicons on either side of the deletion site removed the following amino acid residues in frame: Δ13aa repeat, residues 63 to 88; ΔPRM1, residues 90 to 97; ΔWH21, residues 168 to 201; ΔWH22, residues 202 to 231; ΔWH21+2, residues 168 to 231; ΔPDASX, repeats 257 to 281. Products harboring these deletions were then subcloned under the Ptac promoter in pME6032 via EcoRI and XhoI sites introduced at the ends of bimA via primers pCompE1.2 and pCompXho.2. Faithful amplification and deletion of specific residues were confirmed by sequencing of both strands of the mutated bimA genes using vector- and bimA-specific primers (data not shown).
Table 2.
Oligonucleotides used in this study
| Oligonucleotide name | Sequence (5′-3′) | Orientation to bimA | Purpose |
|---|---|---|---|
| pCompE1.2 | CTCGAATTCCATGCGTGCAATAGCTG | Forward | Cloning |
| pCompxho.2 | CTTCTCGAGTGCTTACCATTGCCAGCTCAT | Reverse | Cloning |
| Bim117 | GGTGCCGCCCGGCGGTT | Reverse | Δ13AA |
| Bim118 | AACATCCCGCCCCCGC | Forward | Δ13AA |
| Bim119 | GTTCGCGCCGCCGCCCGGCA | Reverse | ΔPRM1 |
| Bim120 | GGCGGCATTGGCGGCGCAA | Forward | ΔPRM1 |
| Bim123 | GTTCGCATCGGGCGCCAGATT | Reverse | ΔWH21 |
| Bim124 | GCATACCTGCCTGCCGAGCG | Forward | ΔWH21 |
| Bim125 | CGGAGCGGCATGGCCGGCG | Reverse | ΔWH22 |
| Bim126 | CAGCCGCGGTCGACACCAC | Forward | ΔWH22 |
| Bim123 | GTTCGCATCGGGCGCCAGATT | Reverse | ΔWH21+2 |
| Bim126 | CAGCCGCGGTCGACACCAC | Forward | ΔWH21+2 |
| Bim121 | TACGTTCGGGTTGGGCGCGG | Reverse | ΔPDASX |
| Bim122 | CCCAGCCGACCTGCCCCT | Forward | ΔPDASX |
| 121+2PDAST | CGTCGATGCATCAGGCGTCGATGCGTCGGGTACGTTCGGGTTGGGCGCGG | Reverse | 2× PDASX |
| 122+3PDAST | CCCGACGCATCGACGCCTGATGCATCGACGCCTGACGCTTCGACGCCCAGCGACCTGCCCCT | Reverse | 7× PDASX |
| 4PDAST | GGCGCGGGTTGGGCTTGCATGGGCTGCGTAGCTGCGGGCTGCGTAGCTGCGGGCTGCGTAGCTGCGGGCTGCGTAGCTGC | Forward | 7× PDASX |
| BimA pGEX start | GCGCGCGGATCCATGAATCCCCCCGAACCGCCGGGC | Forward | Cloning |
| BimA pGEX stop | GCGCGCGAATTCTTAGCGCGCGGTGTCGGTG | Reverse | Cloning |
Variations in the number of PDASX repeats.
In natural populations of B. pseudomallei, the number of direct repeats of PDASX has been observed to vary from two to seven; however, the cognate bimA genes harbor other polymorphisms that preclude analysis of the impact of the number of repeats in isolation. Variants of B. pseudomallei strain 10276 bimA differing only in the number of PDASX repeats were therefore created by PCR-ligation-PCR, essentially as described above. To produce a construct with two PDASX repeats, primer pairs pCompE1.2 with 121+2PDAST and Bim122 with pCompXho.2 were used to amplify the portions upstream and downstream of the PDASX repeats, respectively, deleting three PDASX repeats upon ligation. To produce a construct with seven PDASX repeats, primer pairs pCompE1.2 with 122+3PDAST and 4PDAST with pCompXho.2 were used to generate the first-round products, adding an additional two repeats upon ligation. The joined amplicons were amplified by a second round of PCR with pCompE1.2 and pCompXho.2, subcloned as an EcoRI-XhoI fragment into similarly restricted pME6032, and verified by sequencing of both strands (data not shown).
Construction of protein expression vectors.
We previously demonstrated that a GST fusion to residues 54 to 455 of B. pseudomallei BimA sequesters actin from murine splenic extracts, binds monomeric actin, and stimulates polymerization of pyrene-labeled actin monomers in vitro (20). We therefore amplified the regions corresponding to these residues from the plasmid constructs harboring the in-frame deletions of motifs or two to seven PDASX repeats by using primers BimApGEX-start and BimApGEX-stop and Advantage-GC2 DNA polymerase (BD Biosciences Clontech, Oxford, United Kingdom). In the first instance, products were cloned by a topoisomerase-mediated method into pCR2.1-TOPO. The truncated bimA genes harboring deletions or insertions were then subcloned into pGEX-4T-1 via BamHI and EcoRI sites incorporated in the primers. Plasmid constructs were verified by sequencing on both strands (data not shown).
Expression and purification of GST-BimA fusion proteins.
Plasmid constructs for expression of BimA54–455 variants harboring in-frame deletions or insertions were separately transferred to E. coli K-12 Rosetta2(DE3)pLysS cells. Use of this strain improved both the yield and stability of the GST fusion proteins relative to BL21 derivatives that do not express rare amino-acyl tRNAs (data not shown). LB cultures were induced to express the proteins at late logarithmic phase for 3 h at ambient temperature, which proved optimal for recovery of intact proteins using glutathione-Sepharose 4B beads (Amersham Pharmacia Biotechnology, St. Albans, United Kingdom), as per the manufacturer's instructions. After washing, protein-coated beads were used directly for actin-binding assays. To evaluate the ability of the proteins to polymerize actin, each fusion was eluted from glutathione-Sepharose beads with 10 mM reduced glutathione as directed by the manufacturer and concentrated using Novagen U-tube concentrator units. The protein concentration was determined using the Bradford reagent (Sigma, Poole, United Kingdom) by analysis of absorbance at 595 nm relative to bovine serum albumin (BSA) standards of known concentrations. GST expressed from pGEX-4T-1 was used as a control throughout.
Cell infection studies and fluorescence microscopy.
To determine the impact of in-frame deletions or insertions in bimA on actin-based motility, pME6032-based constructs were separately introduced into 10276 bimA::pDM4 by electroporation with selection for tetracycline and chloramphenicol resistance. Strains were grown to stationary phase in Luria-Bertani broth, washed in phosphate-buffered saline (PBS), adjusted to the same optical density, and then used to infect J774.2 murine macrophage-like cells at a multiplicity of approximately 10 per cell in DMEM containing 10% (vol/vol) fetal calf serum at 37°C in a humidified 5% CO2 atmosphere. The actual number of bacteria added was confirmed by retrospective plating of serial dilutions of each culture to LB agar. After 30 min, infected cells were washed and overlaid with medium containing 250 μg/ml kanamycin to kill extracellular bacteria. During bacterial culture and cell infection, expression of BimA proteins was induced with 0.25 mM isopropyl-β-d-thiogalactoside (IPTG). Eight hours postinfection cells were washed, fixed with 4% (vol/vol) paraformaldehyde in PBS, and permeabilized with 0.5% (vol/vol) Triton X-100 in PBS for 15 min, and nonspecific binding sites were blocked with 0.5% (wt/vol) BSA in PBS. Bacteria were stained using mouse monoclonal anti-B. pseudomallei lipopolysaccharide antibody (Camlab, Cambridge, United Kingdom), which was detected with anti-mouse Ig-Alexa Fluor 488 or anti-mouse Ig-Alexa Fluor 568 (Molecular Probes, Leiden, Netherlands). F-actin was stained using phalloidin conjugated to Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes). To detect BimA, a panel of three monoclonal antibodies raised against a GST fusion to residues 48 to 384 of B. pseudomallei strain 10276 BimA was used (24). Bound anti-BimA was detected with anti-mouse Ig-Alexa Fluor 488, and bacteria stained with rabbit polyclonal anti-B. pseudomallei lipopolysaccharide antibody were detected with anti-rabbit Ig-Alexa Fluor 568. Images were captured using a Leica confocal laser scanning microscope with LAS AF v.2.0 software. B. pseudomallei 10276 and 10276 bimA::pDM4 control strains were included throughout the experiment.
To analyze the ability of the bimA mutant strain to spread from cell to cell upon trans-complementation with full-length bimA or variants harboring in-frame deletions or insertions, confluent A549 monolayers were infected at a multiplicity of infection of 100 with the same strains as above. Bacteria were gently driven into contact with the cells by centrifugation at 300 × g. IPTG was included during bacterial and cell culture to induce expression of the BimA proteins, and extracellular bacteria were killed by the addition of 250 μg/ml kanamycin from 30 min postinfection. Monolayers were incubated statically for 4 days and then stained with 1% (wt/vol) crystal violet in 20% (vol/vol) methanol for at least an hour.
Pull-down assays and immunoblotting.
A lysate of the spleens of C57BL/6 mice was prepared by homogenization of tissue in polymerization buffer (10 mM Tris [pH 7.5], 50 mM KCl, 2 mM MgCl2) and clearing of debris by high-speed centrifugation. Glutathione-Sepharose beads coated with GST, GST-BimA54–455, or variants of GST-BimA54–455 harboring in-frame deletions or insertions were mixed with murine splenic cell lysates supplemented with 100 μM CaCl2, 2 mM MgCl2, and 500 μM ATP for 30 min at room temperature. Beads were washed five times in ice-cold Tris-buffered saline. For the detection of actin sequestered by the beads, proteins were released by heating in Laemmli buffer, then resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis, and transferred to a nitrocellulose membrane by using a Bio-Rad semidry Trans-Blot Apparatus. After blocking, membranes were incubated with goat polyclonal antiactin antibody at 0.5 μg/ml (Autogen Bioclear, Wiltshire, United Kingdom). Bound antibody was detected using a 1:3,000 dilution of rabbit anti-goat Ig-horseradish peroxidase conjugate in Tris-buffered saline containing 0.1% (vol/vol) Tween 20. Bound secondary antibody was detected using an enhanced chemiluminescence method by exposure to Hyperfilm (GE Healthcare, Amersham, United Kingdom). Direct binding of the proteins to highly purified actin was confirmed by mixing coated beads with 1 μM rhodamine-labeled actin (cytoskeleton; Universal Biologicals, Cambridge, United Kingdom) in Tris-buffered saline supplemented with 100 μM CaCl2, 2 mM MgCl2, and 500 μM ATP. Five minutes after mixing, 10-μl samples were mounted onto slides and visualized by confocal microscopy as described above.
Assay of actin polymerization in vitro.
Polymerization of pyrene-actin monomers leads to emission of fluorescence that can be sensitively recorded over time. Lyophilized pyrene-actin, Arp2/3, and the verprolin-like central and acidic (VCA) domain of WASP were obtained from Cytoskeleton and prepared as per the manufacturer's instructions. Assay conditions were essentially as described previously (26). Briefly, 90-μl reaction mixtures were assembled in black opaque 96-well plates containing 100 nM GST, GST fusion protein or the VCA domain of WASP, 2 μM pyrene-actin, and 13 nM Arp2/3 as required. Reactions were initiated by the addition of 10 μl of 10× polymerization buffer (100 mM Tris [pH 7.5], 500 mM KCl, 20 mM MgCl2, 10 mM ATP), and the emission of fluorescence at 407 nm, after excitation at 365 nm, was followed every 30 s for 90 min using a Tecan Infinite M200 fluorescent plate reader with i-control software. Replicates were performed with independently purified sets of protein as specified below in Results. Rates of polymerization were calculated as the rise in fluorescence units per second during the linear phase of polymerization, and the means ± standard errors of the means are recorded. Results were analyzed by pair-wise Student t tests using R software (version 2.11), and P values of ≤0.05 were considered significant.
RESULTS
Roles of conserved and variable motifs of BimA in actin-based motility of B. pseudomallei.
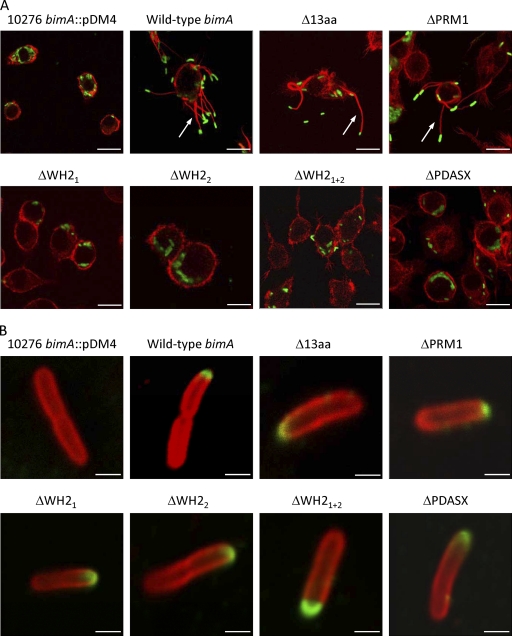
Previously we reported the plasmid-mediated trans-complementation of the actin-based motility defect of a B. pseudomallei strain 10276 bimA::pDM4 null mutant (20, 23, 24). Using this system, PCR-ligation-PCR was used to splice together amplicons flanking the 13-amino-acid direct repeat, an adjacent PRM1 motif, WH2 domains singly or in combination, or the PDASX repeats, producing in-frame deletions as described in Materials and Methods. Amplicons comprising bimA genes harboring such in-frame deletions were cloned in pME6032, sequenced on both strands, and confirmed to encode proteins of the expected size by immunoblotting of IPTG-induced cultures with BimA-specific monoclonal antibodies (data not shown). The constructs were separately introduced into B. pseudomallei 10276 bimA::pDM4 and J774.2 murine macrophage-like cells separately infected with each strain. F-actin tails were detected for the bimA null mutant trans-complemented with pME6032-based constructs for expression of intact full-length bimA, and those lacking the 13-amino-acid repeat and PRM1 motifs, implying that such motifs are not essential for actin-based motility of B. pseudomallei (Fig. 1A). Actin tails were not associated with intracellular bacteria harboring in-frame deletions of the WH2 domains (singly or in combination) or the PDASX repeats (Fig. 1A). Normal surface expression and polar targeting of BimA were detected in each of the strains expressing proteins with in-frame motif deletions (Fig. 1B), indicating that the loss of actin-based motility of the ΔWH2 and ΔPDASX mutants is unlikely to be a consequence of interference in these processes. We cannot exclude the possibility that the deletions affect the tertiary structure of BimA at the bacterial pole.
Fig. 1.
(A) Representative confocal laser scanning micrographs of J774.2 cells infected with the B. pseudomallei strain 10276 bimA::pDM4 mutant or 10276 bimA::pDM4 trans-complemented with wild-type bimA carried on pME6032, or variants thereof harboring in-frame deletions of conserved or variable motifs. IPTG was included during bacterial and cell culturing to induce expression of the proteins. Bacteria (green) were stained using mouse monoclonal anti-B. pseudomallei lipopolysaccharide antibody that was detected with anti-mouse Ig-Alexa Fluor 488. F-actin (red) was stained with Alexa Fluor 568-conjugated phalloidin. Arrows denote bacteria-tipped actin-rich membrane protrusions. Bar, 40 μm. (B) Polar targeting of BimA proteins harboring in-frame deletions of conserved or variable motifs. BimA (green) was detected with a panel of three GST-BimA48-384-specific monoclonal antibodies and anti-mouse Ig-Alexa Fluor 488. Bacteria (red) were stained using rabbit polyclonal anti-B. pseudomallei lipopolysaccharide antibody that was detected with anti-rabbit Ig-Alexa Fluor 568. Bar, 2 μm.
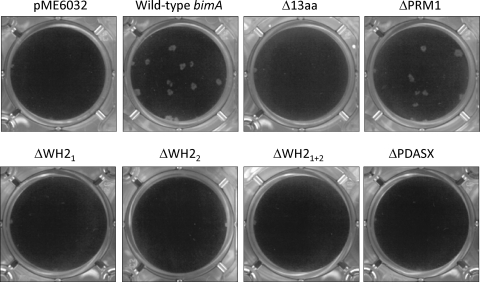
Consistent with the proposed role of actin-based motility in intercellular spread, we observed that the null bimA mutant of strain 10276 did not form plaques in A549 monolayers, whereas plaques could be detected upon introduction of pME6032-bimA (Fig. 2). trans-complementation of 10276 bimA::pDM4 with the bimA derivative lacking the PRM1 motif restored plaque formation; however, bimA genes containing all other mutations failed to restore intercellular spread under the assay conditions (Fig. 2).
Fig. 2.
Impact of in-frame deletions of conserved and variable motifs of BimA on intercellular spread of B. pseudomallei. A549 monolayers were infected with 10276 bimA::pDM4 trans-complemented with empty pME6032, pME6032 harboring intact bimA, or constructs identical to pME6032-bimA except for deletion of the 13-amino-acid repeat, PRM1 motif, WH2 domains (singly or in combination), or PDASX repeats. IPTG was included during bacterial and cell culturing to induce expression of the proteins. Plaques were visualized by crystal violet staining of the monolayers 4 days postinfection.
Role of conserved and variable motifs of B. pseudomallei BimA in actin binding.
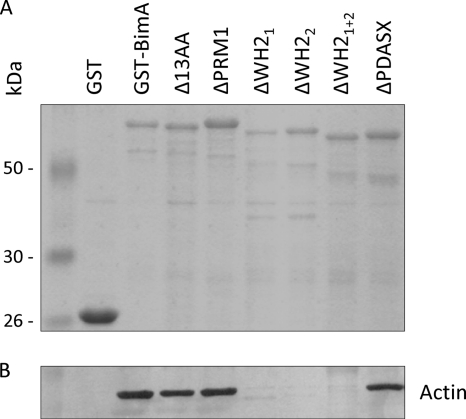
To evaluate the impact of the in-frame deletions on the ability of BimA to bind actin, the region corresponding to residues 54 to 455 was subcloned from each of the plasmids used above into pGEX-4T-1 as carboxyl-terminal fusions to GST. The proteins were affinity purified from E. coli expressing rare amino-acyl tRNAs by using glutathione-Sepharose beads as described in Material and Methods. SDS-PAGE analysis confirmed that proteins of the expected size were produced in all cases (Fig. 3A). Minor protein species migrating at a lower molecular weight were shown to be derived from the fusion protein by Western blotting with GST- or BimA-specific antiserum (data not shown). We analyzed the ability of beads coated with GST-BimA54–455, variants of GST-BimA54–455 harboring the deletions, or GST alone to sequester actin from a murine splenic extract. Immunoblotting of proteins associated with beads after mixing with the extract confirmed an earlier report that GST-BimA54–455 specifically sequesters actin (Fig. 3B) (20). GST-BimA54–455 fusions with deletions of the 13-amino-acid repeat, PRM1 motif, or PDASX repeats also sequestered actin; however, mutation of one or both WH2 domains abolished actin binding (Fig. 3B). This provides a potential explanation for the loss of actin-based motility upon mutation of the WH2 domains. The ability of the fusion proteins to interact with highly purified rhodamine-labeled actin was also evaluated, and these experiments confirmed that GST-BimA54–455 directly binds actin in the absence of other host proteins in a manner that requires the WH2 domains (data not shown).
Fig. 3.
(A) SDS-PAGE analysis of B. pseudomallei BimA54–455 and the corresponding regions of variants harboring in-frame motif deletions, fused to the carboxy terminus of GST. (B) Glutathione-Sepharose beads coated with these proteins, or GST alone, were examined for their ability to sequester actin from a murine splenic extract by Western blotting of bead-associated proteins with an actin-specific antibody.
Roles of conserved and variable motifs of B. pseudomallei BimA in actin polymerization.
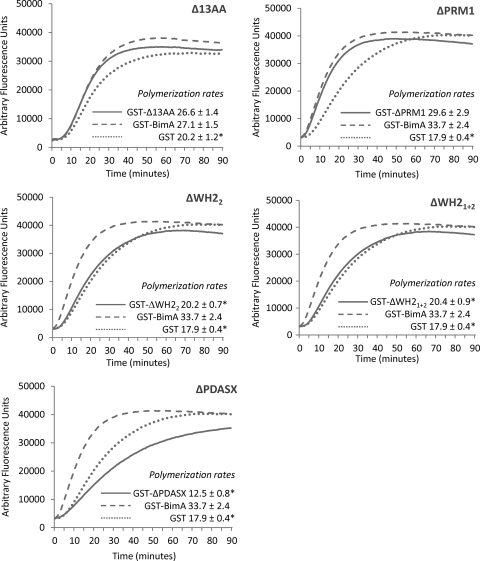
We previously demonstrated that GST-BimA54–455 and GST-BimA48-384 fusion proteins stimulate the Arp2/3-independent polymerization of pyrene-labeled actin monomers, which can be detected by the emission of fluorescence over time (20, 24). Variants of GST-BimA54–455 with in-frame deletions were eluted from glutathione-Sepharose beads and adjusted to the same molar concentration. The fusion protein harboring a deletion of WH2 domain 1 could not be purified in adequate quantities in an intact form and was omitted from this analysis, as the mutated proteins lacking both domains or WH2 domain 2 were available. The ability of each protein to stimulate pyrene-actin polymerization was then tested relative to control proteins of GST alone or GST-BimA54–455. The mean rates of polymerization from at least three independent protein preparations tested in triplicate are recorded under the curves in Fig. 4. A positive control comprising the VCA domain of WASP stimulated rapid actin polymerization on all occasions (data not shown), as expected (20). Pyrene-actin spontaneously polymerizes under the assay conditions; however, a statistically significant increase in the rate of polymerization was detected in the presence of GST-BimA54–455 relative to GST (Fig. 4) (P ≤ 0.05), as expected given previous observations with GST-BimA54–455 (20) and GST-BimA48-384 (24). For the fusion proteins harboring each deletion, the emission of fluorescence is shown relative to GST or GST-BimA54–455 tested at the same time. Deletion of the 13-amino-acid repeat or PRM1 motif did not significantly alter the rate of pyrene-actin polymerization relative to GST-BimA54–455 (Fig. 4). However, deletion of WH2 domains or the PDASX repeats significantly reduced the rate of pyrene-actin polymerization compared to GST-BimA54–455 (Fig. 4) (P ≤ 0.05). In the case of the WH2 domain mutants, the rate of polymerization was comparable to the GST control. The protein harboring the deletion of PDASX repeats stimulated polymerization at a rate lower than for the GST control.
Fig. 4.
Emission of fluorescence due to polymerization of pyrene-labeled actin monomers by GST-BimA54–455 proteins harboring motif deletions (solid lines), in comparison with GST-BimA54–455 (dashed lines) and GST (dotted lines). Three independent preparations of fusion protein were each tested in triplicate, and the graphs show the mean emission levels of fluorescence over time. Rates of polymerization were calculated as the rise in fluorescence units per second during the linear phase of polymerization. The mean rates of fluorescence units/s (± the standard errors of the means) from at least nine replicates per protein are recorded under the curves. Asterisks denote a significant difference in the rate of polymerization compared to GST-BimA54–455 (P ≤ 0.05).
PDASX repeats act additively to promote actin polymerization.
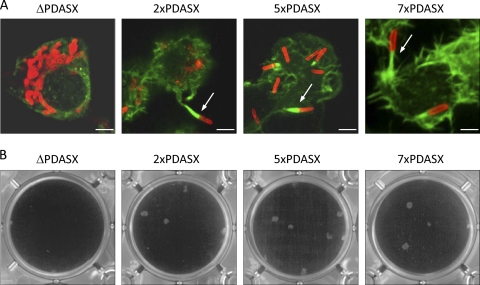
To evaluate the impact of variation in the number of PDASX repeats in the absence of other polymorphisms, PCR-ligation-PCR was used to create variants of B. pseudomallei strain 10276 bimA (which encodes five PDASX repeats in its native form), containing either two or seven PDASX repeats at the same location (as found in strains BCC215 and MSHR305, respectively) (19). trans-complementation of the 10276 null bimA mutant with pME6032-based constructs encoding BimA with two, five, or seven PDASX repeats restored actin-based motility (Fig. 5A) and plaque formation in A549 monolayers (Fig. 5B). No differences in the morphology of tails or plaques could be discerned.
Fig. 5.
(A) Representative confocal laser scanning micrographs of J774.2 cells infected with the B. pseudomallei 10276 bimA::pDM4 mutant strains trans-complemented with pME6032-bimA plasmids encoding zero, two, five, or seven PDASX repeats. IPTG was included during bacterial and cell culturing to induce expression of the proteins. Bacteria (red) were stained using mouse monoclonal anti-B. pseudomallei lipopolysaccharide antibody, which was detected with anti-mouse Ig-Alexa Fluor 568. F-actin (green) was stained with Alexa Fluor 488-conjugated phalloidin. Arrows denote bacteria-tipped actin-rich membrane protrusions. Bar, 10 μm. (B) Plaque formation in A549 monolayers infected with the same strains.
The region corresponding to residues 54 to 455 was subcloned from the constructs harboring two or seven repeats into pGEX-4T-1 and GST fusion proteins expressed and purified in E. coli. Proteins of the expected size were detected relative to proteins harboring the in-frame deletion of PDASX repeats or native GST-BimA54–455 (Fig. 6A). All four proteins sequestered actin from a murine splenic extract (Fig. 6B) and directly bound rhodamine-labeled actin (data not shown). Two independent preparations of the four fusion proteins containing zero, two, five, or seven PDASX repeats were then tested in duplicate for their ability to stimulate polymerization of pyrene-actin monomers in vitro. As observed above, the rate of polymerization was significantly lower than with GST-BimA54–455 when all five PDASX were deleted (Fig. 6C). The rate of polymerization in the presence of proteins containing two or seven PDASX repeats was significantly different from GST-BimA54–455 (Fig. 6C) (P ≤ 0.05), with stepwise increases in the rate of polymerization being detected as the number of PDASX repeats was increased from zero to two to five to seven, implying that they act in an additive manner to promote actin assembly.
Fig. 6.
(A) SDS-PAGE analysis of B. pseudomallei BimA54–455 and the corresponding regions of variants harboring two, five, or seven PDASX repeats, fused to the carboxy terminus of GST. The fusion protein containing five repeats is derived from the native 10276 bimA. (B) Glutathione-Sepharose beads coated with these proteins, or GST alone, were examined for their ability to sequester actin from a murine splenic extract by Western blotting of bead-associated proteins with an actin-specific antibody. (C) Impact of variation of the number of PDASX repeats on the ability of GST-BimA54–455 to stimulate pyrene-actin polymerization. Two independent preparations of fusion protein were each tested in duplicate, and the graph shows the mean emission fluorescence over time. Rates of polymerization were calculated as the rise in fluorescence units per second during the linear phase of polymerization. The mean fluorescence units (± the standard errors of the means) of at least four replicates per protein are recorded under the curves. Asterisks denote a significant difference in the rate of polymerization compared to GST-BimA54–455 (P ≤ 0.05).
DISCUSSION
Actin-based motility influences the virulence of several intracellular bacterial pathogens and mediates intercellular spread and, in the case of Listeria, escape from autophagy (22, 27). We previously reported that B. pseudomallei relies on BimA for this process (24), and a null mutant exhibits attenuation in a murine model of melioidosis (G. Bancroft and E. Galyov, unpublished observations). In contrast, a B. mallei bimA mutant produced comparable lethality as the parent strain in Syrian hamsters after intraperitoneal infection (17); the reasons for this disparity are unclear. The formation of plaques in monolayers of A549 lung epithelial carcinoma cells was found here to require bimA, implying that the formation of bacteria-tipped actin-rich membrane protrusions aids bacterial spread to adjacent cells. We report the first structure-activity analysis of B. pseudomallei BimA, identifying conserved motifs that are vital and dispensable for the assembly of actin in vitro and in infected cells.
WASP homology 2 domains exist in many pathogen-associated and cellular nucleation-promoting factors. Consistent with the role they play in binding of monomeric actin in other proteins (reviewed in references 3 and 16), deletion of one or both WH2 domains from B. pseudomallei strain 10276 BimA abolished intracellular motility and intercellular spread without affecting the surface expression or polar targeting of the protein. By using fusion proteins based on a truncated form of BimA lacking the signal peptide and transmembrane domains (GST-BimA54–455), we also found that the WH2 domains were required for actin binding and the polymerization of pyrene-actin monomers in vitro. Interestingly, loss of either of the in silico-predicted WH2 domains (24) abolished actin binding (Fig. 3B). It is anticipated that the remaining WH2 domain in the ΔWH21 or ΔWH22 deletion constructs would be sufficient to mediate actin binding, as the actin-binding orthologues from B. mallei and B. thailandensis each possess a single predicted WH2 domain (23). This may be a consequence of cooperative interactions, whereby occupation of one site favors actin binding to the other domain. Alternatively, elements required for actin binding by each domain may overlap, meaning that in-frame deletions constructed herein resulted in a loss of function of both domains rather than each individual domain as intended.
In other NPFs, one or more WH2 domains act in concert with an amphipathatic CA domain to form a WCA [or verprolin (V)CA] domain that recruits and activates the Arp2/3 complex. BimAps lacks an invariant tryptophan residue typical of CA domains and is incapable of sequestering Arp2/3 components (20, 23), and further studies will be required to determine how B. pseudomallei BimA polymerizes the actin monomers it binds. Truncated BimAps fusion proteins can stimulate pyrene-actin polymerization in a concentration-dependent manner in the absence of the Arp2/3 complex in vitro (20, 24), indicating that BimAps may possess an intrinsic ability to assemble actin, perhaps in a manner akin to Chlamydia TARP, Vibrio VopF/L, and R. rickettsii Sca2, or cellular NPFs such as Spire, formins, cordon-bleu, and leiomodin. However, it is noteworthy that Arp2/3 components have been detected in B. pseudomallei actin tails (2), and we cannot exclude the possibility that the proteins used for in vitro assays lack the activities of BimA at the bacterial pole in infected cells. Indeed, the derivatives used in the in vitro assays are truncated and fused to GST (which may affect their conformation and predicted oligomerization) and expressed in E. coli with no scope for posttranslational modification by B. pseudomallei or cellular factors. These limitations arise from the challenge of producing adequate quantities of soluble protein for assays of actin binding and polymerization.
We previously reported that predicted BimA proteins from clinical and environmental isolates of B. pseudomallei vary in the number of PDASX repeats, from two to seven, with unknown consequences (19). Here, we have shown that the PDASX repeats are required for actin-based motility and intercellular spread, but not polar surface localization of BimAps. Significantly, we observed that the repeats are required for polymerization of pyrene-actin, but not for actin binding. Moreover, the repeats acted in an additive fashion, with stepwise increases in the rate of polymerization being detected as the number of repeats was increased. A similar observation has been made with respect to proline-rich repeats in the type III secreted effector TccP/EspFU, which mediates actin pedestal formation by selected attaching and effacing E. coli strains. The number of such repeats varies in natural isolates (6), and they act cooperatively to activate N-WASP and promote actin assembly (5). We could not discern any obvious differences in the frequency or morphology of actin tails in cells infected with B. pseudomallei strains expressing variants with two, five, and seven PDASX repeats or the efficiency of plaque formation in A549 monolayers. However, given the role of actin-based motility in virulence, it is possible that such variations may subtly influence pathogenesis.
It is noteworthy that deletion of PDASX repeats in GST-BimA54–455 produced a rate of pyrene-actin polymerization below that of the GST control. A similar phenotype was recently observed upon removal of the CA domain from B. thailandensis BimA, which also abolished actin polymerization but not actin binding (20). Pyrene-actin spontaneously polymerizes under the assay conditions used, and we hypothesize that GST-BimA54–455 lacking the PDASX repeats sequesters the monomers from the assay mixture in such a way that they can no longer spontaneously assemble. It is important to note that while PDASX repeats are predicted to form target sites for the host serine-threonine kinase casein kinase 2 (CK2) when juxtaposed, the pyrene-actin polymerization assays were performed using highly defined and purified reagents rather than cell extracts. No CK2 or other kinase is expected to have been present; therefore, the phenotype attributable to the PDASX repeats is not a consequence of their phosphorylation.
Proline-rich motifs are frequently found in cellular and pathogen-associated NPFs and influence the recruitment of actin monomers. We observed that one such motif, similar to that essential for actin-based motility mediated by Listeria ActA, is not required for actin-based motility by B. pseudomallei or for actin binding and polymerization in vitro. It should be noted, however, that BimAps contains numerous other proline-rich motifs (24), and functional redundancy may therefore exist. To determine if PRM-mediated recruitment of profilin occurs, it would be interesting to examine if profilin is localized to B. pseudomallei-induced actin tails, whether silencing of profilin expression impairs motility as described for R. parkeri (18), and to study the influence of purified profilin on GST-BimA54–455-mediated pyrene-actin polymerization in vitro. A 13-amino-acid direct repeat encoded immediately adjacent to the IP7 PRM1 motif on the amino-terminal side was not strictly necessary for actin-based motility. Intriguingly, plaque formation in A549 monolayers was impaired in the absence of the 13-amino-acid repeat, despite an ability of this mutant to form F-actin tails in A549 cells at 24 h postinfection (data not shown). It is possible that this may reflect subtle differences in the frequency or kinetics of tail assembly that cannot be detected by the methods used. Detailed measurements of the rate of actin-based motility by live cell imaging are hindered by the need for high containment and the absence of a cell-free system in which BimA-mediated motility of coated beads can be reconstituted. It was recently described that L. monocytogenes ActA is a target for autophagic recognition and that recruitment of the Arp2/3 complex and Ena/VASP promotes intracellular survival by masking ActA (27). Shigella IcsA is also a target for autophagic recognition, but it uses the type III secreted protein IcsB to mask IcsA (14). It is possible that deletion of the 13-amino-acid direct repeat may have no effect on actin-based motility early after infection but interferes with the recruitment of bacterial or cellular factors that may mask BimA from detection by the host autophagic pathway. In this context, it is noteworthy that the B. pseudomallei bimA null mutant was recovered from J774.2 and A549 cells in lower numbers than the parent strain at 24 h postinfection, and this defect can be partially restored by inducible expression of the native bimA allele in pME6032 (J. Stevens, unpublished observations), indicating that BimA may play a role in intracellular survival.
This study provides valuable new data on the motifs required for actin-based motility of B. pseudomallei and the possible consequences of natural variation in the number of repeats of domains. Importantly, motifs that are required for activity of BimAps were not absolutely conserved in the functional orthologues of BimA that exist in B. mallei and B. thailandensis (19, 23). In both these species, only one WH2 domain is present and PDASX direct repeats are absent, although other predicted target sites for CK2 and other cellular kinases exist. Taken together with our recent finding that a central acidic domain is conserved in, but not restricted to, B. thailandensis BimA, our data reinforce the notion that distinct molecular mechanisms may have evolved for actin-based motility among closely related Burkholderia species. Understanding such mechanisms will provide new insights into both the cellular control of actin assembly and how pathogens subvert actin dynamics during infection.
ACKNOWLEDGMENTS
We gratefully acknowledge the support of the National Science and Technology Development Agency (grant BT-B-01-MG-14-5123 to S.K.), the Royal Thai Golden Jubilee studentship scheme (grant PHD0188/2547 to C.S.), and the Biotechnology & Biological Sciences Research Council, United Kingdom (grant BB/E01212/1 to M.P.S. and J.M.S.).
We thank Pucharee Songprakhon at the Division of Medical Molecular Biology, Department of Research and Development, Faculty of Medicine, Siriraj Hospital, for confocal technical support and Veerachat Muangsombut for technical assistance.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Ali S. A., Steinkasserer A. 1995. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. Biotechniques 18:746–750 [PubMed] [Google Scholar]
- 2. Breitbach K., et al. 2003. Actin-based motility of Burkholderia pseudomallei involves the Arp2/3 complex, but not N-WASP and Ena/VASP proteins. Cell. Microbiol. 5:385–393 [DOI] [PubMed] [Google Scholar]
- 3. Campellone K. G., Welch M. D. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11:237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chong R., et al. 2009. Regulatory mimicry in Listeria monocytogenes actin-based motility. Cell Host Microbe 6:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garmendia J., Carlier M. F., Egile C., Didry D., Frankel G. 2006. Characterization of TccP-mediated N-WASP activation during enterohaemorrhagic Escherichia coli infection. Cell. Microbiol. 8:1444–1455 [DOI] [PubMed] [Google Scholar]
- 6. Garmendia J., et al. 2005. Distribution of tccP in clinical enterohemorrhagic and enteropathogenic Escherichia coli isolates. J. Clin. Microbiol. 43:5715–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haglund C. M., Choe J. E., Skau C. T., Kovar D. R., Welch M. D. 2010. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12:1057–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heeb S., Blumer C., Haas D. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holt M. R., Koffer A. 2001. Cell motility: proline-rich proteins promote protrusions. Trends Cell Biol. 11:38–46 [DOI] [PubMed] [Google Scholar]
- 10. Jewett T. J., Fischer E. R., Mead D. J., Hackstadt T. 2006. Chlamydial TARP is a bacterial nucleator of actin. Proc. Natl. Acad. Sci. U. S. A. 103:15599–15604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kespichayawattana W., Rattanachetkul S., Wanun T., Utaisincharoen P., Sirisinha S. 2000. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spread. Infect. Immun. 68:5377–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleba B., Clark T. R., Lutter E. I., Ellison D. W., Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 78:2240–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liverman A. D., et al. 2007. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. U. S. A. 104:17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogawa M., et al. 2005. Escape of intracellular Shigella from autophagy. Science 307:727–731 [DOI] [PubMed] [Google Scholar]
- 15. Panchal S. C., Kaiser D. A., Torres E., Pollard T. D., Rosen M. K. 2003. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol. 10:591–598 [DOI] [PubMed] [Google Scholar]
- 16. Paunola E., Mattila P. K., Lappalainen P. 2002. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 513:92–97 [DOI] [PubMed] [Google Scholar]
- 17. Schell M. A., et al. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64:1466–1485 [DOI] [PubMed] [Google Scholar]
- 18. Serio A. W., Jeng R. L., Haglund C. M., Reed S. C., Welch M. D. 2010. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe 7:388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sitthidet C., et al. 2008. Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J. Clin. Microbiol. 46:2418–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sitthidet C., et al. 2010. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J. Bacteriol. 192:5249–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stevens J. M., Stevens M. P. 2009. Burkholderia pseudomallei, p. 393–413 In Schaible U. E., Haas A. (ed.), Intracellular niches of microbes. Wiley VCH, Weinheim, Germany [Google Scholar]
- 22. Stevens J. M., Galyov E. E., Stevens M. P. 2006. Actin-dependent movement of bacterial pathogens. Nat. Rev. Microbiol. 4:91–101 [DOI] [PubMed] [Google Scholar]
- 23. Stevens J. M., et al. 2005. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 187:7857–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stevens M. P., et al. 2005. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56:40–53 [DOI] [PubMed] [Google Scholar]
- 25. Tam V. C., Serruto D., Dziejman M., Brieher W., Mekalanos J. J. 2007. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1:95–107 [DOI] [PubMed] [Google Scholar]
- 26. Yarar D., D'Alessio J. A., Jeng R. L., Welch M. D. 2002. Motility determinants in WASP family proteins. Mol. Biol. Cell 13:4045–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshikawa Y., et al. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11:1233–1240 [DOI] [PubMed] [Google Scholar]