Abstract
Multiple factors control the expression of the outer membrane porins OmpF and OmpC in Escherichia coli. In this work, we investigated the role of the mar-sox-rob regulon in regulating outer membrane porin expression in response to salicylate. We provide both genetic and physiological evidence that MarA and Rob can independently activate micF transcription in response to salicylate, leading to reduced OmpF expression. MarA was also found to repress OmpF expression through a MicF-independent pathway. In the case of OmpC, we found that its transcription was moderately increased in response to salicylate. However, this increase was independent of MarA and Rob. Finally, we found that the reduction in OmpF expression in a tolC mutant is due primarily to Rob. Collectively, this work further clarifies the coordinated role of MarA and Rob in regulating the expression of the outer membrane porins.
INTRODUCTION
Gram-negative bacteria can limit the uptake of membrane-impermeable antimicrobial compounds by modulating the composition of pores in their outer membranes (44). In Escherichia coli and closely related organisms, this alteration is accomplished in part by changing the relative expression of two porins, OmpF and OmpC (17, 44, 46). Both OmpF and OmpC form structurally similar outer membrane pores comprising trimers of 16-stranded β-barrels (2, 13). However, the two have different substrate specificities and diffusion rates (2, 13). The OmpF porin, which forms the larger pore, permits the diffusion of molecules at comparatively faster rates than OmpC (13).
The OmpF/OmpC ratio is primarily regulated at the transcriptional level by the EnvZ-OmpR two-component system in response to changes in the osmolarity of the growth medium (20, 26, 53, 55, 56). Numerous other factors also converge to transcriptionally regulate the differential expression of ompF and ompC in response to environmental conditions such as temperature and nutrient limitation (17, 46). Apart from transcriptional regulation, OmpF and OmpC are also translationally regulated through the action of two small regulatory RNAs (sRNA), MicF and MicC. These sRNA molecules, when expressed, are known to bind to the 5′ untranslated regions (5′-UTR) of ompF and ompC mRNAs and stop translation by preventing the ribosome from binding (7, 39). Although MicC is known to interact with the ompC mRNA, its expression is cryptic and has not been observed to substantially influence OmpC expression (7). MicF, on the other hand, is known to be a significant regulator of OmpF expression under certain environmental conditions, and the regulation of its expression has been extensively investigated (1, 14, 18, 49).
MicF expression is regulated by multiple factors. These include not only OmpR but also global transcription factors such as H-NS, Lrp, and IHF (16, 19, 27). In addition, MicF expression is regulated by three homologous AraC/XylS family transcription factors—MarA, SoxS, and Rob—when E. coli exhibits the porin-dependent antibiotic resistance phenotype (22, 29, 31, 34). These three transcription factors are the master regulators of the extensive mar-sox-rob regulon, involved in intrinsic multidrug resistance in enteric gammaproteobacteria (34). The regulation of MarA and SoxS expression is chiefly mediated at the level of transcription by the MarR repressor and SoxR redox sensor/activator, respectively (11, 42, 58). Rob, on the other hand, is expressed constitutively and regulated posttranslationally by a “sequestration-dispersion” mechanism (23, 29, 48, 50). Together, these three regulators control the expression of a number of downstream, overlapping target genes involved in intrinsic multidrug resistance.
In this work, we investigated the regulation of OmpF and OmpC expression by the mar-sox-rob regulon in response to the canonical inducer salicylate. We first provide both genetic and physiological evidence that the reductions in the levels of OmpF during salicylate exposure are through the parallel action of MarA and Rob in increasing micF transcription. We also demonstrate that MarA regulates ompF translation through a MicF-independent pathway. Finally, we found that the reduced levels of OmpF expression (and correlated increases in OmpC expression) in the absence of the TolC outer membrane efflux pore are primarily due to the action of Rob activating micF transcription.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All strains used in this work are listed in Table 1. Luria-Bertani liquid and solid media (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) were used for routine bacterial culture and genetic manipulation. Experiments were conducted in medium A (7 g/liter nutrient broth, 1 g/liter yeast extract, 2 g/liter glycerol, 3.7 g/liter K2HPO4, 1.3 g/liter KH2PO4) (28, 36). All bacterial cultures were grown at 37°C except for strains containing plasmids pKD46, pINT-ts, and pCP20, which were grown at 30°C. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 20 μg/ml; and chloramphenicol, 20 μg/ml. For some experiments, arabinose was supplied at 0.1% (wt/vol) and sodium salicylate (Sigma-Aldrich, St. Louis, MO) was introduced into growth medium at 5 mM.
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype or relevant characteristicsa | Source and/or referenceb |
|---|---|---|
| Strains | ||
| MG1655 | F− λ−ilvG rph-1 | CGSC 7740 |
| BW25141 | F− λ− Δ(araB-araD)567 ΔlacZ4787(::rrnB-3) Δ(phoB-phoR)580 galU95 ΔuidA3::pir+recA1 ΔendA9::FRTcrph-1 Δ(rhaB-rhaD)568 hsdR514 | CGSC 7635 |
| MDG147 | MG1655 Φ(ompF+-yfp+)30 Φ(ompC+-cfp+)31 | M. D. Goulian, 4 |
| CR701 | ΔtolC::cat | 10 |
| CR702 | ΔtolC::FRT | 10 |
| CR713 | attλ::[kan ompF′-yfp oriR6K] | |
| CR714 | attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR715 | attλ::[kan micF′-yfp oriR6K] | |
| CR716 | ΔmarRAB::kan (1617077–1618231) | |
| CR717 | ΔsoxRS::kan (4275083–4275950) | |
| CR718 | Δrob::cat (4632554–4633393) | |
| CR719 | ΔmicF::kan (2311106–2311203) | |
| CR720 | ΔompR::cat (3533885–3534603) | |
| CR721 | ΔmarRAB::FRT | |
| CR722 | ΔsoxRS::FRT | |
| CR723 | Δrob::FRT | |
| CR724 | ΔmicF::FRT | |
| CR725 | ΔmarRAB::FRT ΔsoxRS::FRT | |
| CR726 | ΔmarRAB::FRT Δrob::FRT | |
| CR727 | ΔsoxRS::FRT Δrob::FRT | |
| CR728 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT | |
| CR729 | ΔmicF::FRT ΔmarRAB::FRT | |
| CR730 | ΔmicF::FRT Δrob::FRT | |
| CR731 | ΔmicF::FRT ΔmarRAB::FRT Δrob::FRT | |
| CR732 | ΔtolC::FRT ΔmarRAB::FRT | |
| CR733 | ΔtolC::FRT ΔsoxRS::FRT | |
| CR734 | ΔtolC::FRT Δrob::FRT | |
| CR735 | ΔtolC::FRT ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT | |
| CR736 | ΔtolC::FRT ΔmicF::FRT | |
| CR737 | ΔmarRAB::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR738 | ΔsoxRS::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR739 | Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR740 | ΔmicF::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR741 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR742 | ΔmarRAB::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR743 | ΔsoxRS::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR744 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR745 | ΔmicF::FRT ΔmarRAB::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR746 | ΔmicF::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR747 | ΔmicF::FRT ΔmarRAB::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR748 | ΔtolC::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR749 | ΔtolC::FRT ΔmarRAB::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR750 | ΔtolC::FRT ΔsoxRS::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR751 | ΔtolC::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR752 | ΔtolC::FRT ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR753 | ΔtolC::FRT ΔmicF::FRT attλ::[kan ompF′-yfp oriR6K] | |
| CR754 | ΔmarRAB::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR755 | ΔsoxRS::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR756 | Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR757 | ΔmicF::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR758 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR759 | ΔmarRAB::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR760 | ΔsoxRS::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR761 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR762 | ΔmicF::FRT ΔmarRAB::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR763 | ΔmicF::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR764 | ΔmicF::FRT ΔmarRAB::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR765 | ΔtolC::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR766 | ΔtolC::FRT ΔmarRAB::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR767 | ΔtolC::FRT ΔsoxRS::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR768 | ΔtolC::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR769 | ΔtolC::FRT ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR770 | ΔtolC::FRT ΔmicF::FRT attλ::[kan ompF′-′yfp(hyb) oriR6K] | |
| CR771 | ΔmarRAB::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR772 | ΔsoxRS::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR773 | Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR774 | ΔmicF::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR775 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR776 | ΔmarRAB::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR777 | ΔsoxRS::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR778 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR779 | ΔmicF::FRT ΔmarRAB::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR780 | ΔmicF::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR781 | ΔmicF::FRT ΔmarRAB::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR782 | ΔtolC::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR783 | ΔtolC::FRT ΔmarRAB::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR784 | ΔtolC::FRT ΔsoxRS::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR785 | ΔtolC::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR786 | ΔtolC::FRT ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR787 | ΔtolC::FRT ΔmicF::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR788 | MDG147 ΔmarRAB::kan | |
| CR789 | MDG147 Δrob::cat | |
| CR790 | MDG147 ΔmarRAB::kan Δrob::cat | |
| Plasmids | ||
| pKD46 | bla PBADgam bet exo pSC101 ori(ts) | 15 |
| pCP20 | bla cat cI857 λPR′-flp pSC101 ori(ts) | 8 |
| pKD3 | bla rgnB FRT cat FRT oriR6K | 15 |
| pKD4 | bla rgnB FRT aph FRT oriR6K | 15 |
| pKD13 | bla rgnB FRT aph FRT oriR6K | 15 |
| pBAD30 | bla araC PBAD p15A ori | 24 |
| pMarA | pBAD30::RBS-marA | |
| pRob | pBAD30::RBS-rob | |
| pVenus | kan MCS yfp(venus) t0 attλ oriR6K | 52 |
| pVenus-ompF | kan MCS ompF′-yfp(venus) t0 attλ oriR6K | |
| pVenus-FY | kan MCS ompF′-′yfp(venus)(hyb) t0 attλ oriR6K | |
| pVenus-micF | kan MCS micF′-yfp(venus) t0 attλ oriR6K |
All strains are isogenic derivatives of E. coli K-12 strain MG1655. Numbers in parentheses indicate deletion endpoints as determined using the MG1655 genome sequence.
All strains and plasmids are from this work unless otherwise noted.
FRT, FLP recombination target.
Strain and plasmid construction.
All strains used in this work are isogenic derivatives of the sequenced E. coli K-12 strain MG1655. The generalized transducing phage P1vir was used in all genetic crosses according to standard methods (57). Targeted gene deletions and subsequent marker removal were made using the λRed recombinase method of Datsenko and Wanner (15). Site-specific integrations were made using the λInt/CRIM method of Haldimann and Wanner (25).
Deletion cassettes were generated with the plasmid templates pKD3, pKD4, and pKD13 using standardized priming sites (15). The ΔmarRAB, ΔsoxRS, Δrob, ΔmicF, and ΔompR deletion cassettes were generated by PCR using the primer pairs 5′-CTT GAA CCG ATT TAG CAA AAC GTG GCA TCG GTC AAT TCA TTG TAG GCT GGA GCT GCT TCG-3′ and 5′-GGG AAG TTA ATA AGC CCC GAG ATG TCG GGG CCA GAA CAA ACA TAT GAA TAT CCT CCT TAG-3′, 5′-AGC AAT TAC CCG CGC GGG AGT TAA CGC GCG GGC AAT AAA ATG TAG GCT GGA GCT GCT TCG-3′ and 5′-ACC GGA AAA CAA ACT AAA GCG CCC TTG TGG CGC TTT AGT TCA TAT GAA TAT CCT CCT TAG-3′, 5′-CTC CCG CTT TGG CAT CTT CTG CCG GGT AGT ATC GCT CAA TTG TAG GCT GGA GCT GCT TCG-3′ and 5′-CTC TAC TAA GAA AAA AAC ACT GAA TGC TAA AAC AGC AAA ACA TAT GAA TAT CCT CCT TAG-3′, 5′-TGT CAA AAC AAA ACC TTC ACT CGC AAC TAG AAT AAC TCC CAT TCC GGG GAT CCG TCG ACC-3′ and 5′-AGT TTT TCT GTG GTA GCA CAG AAT AAT GAA AAG TGT GTA ATG TAG GCT GGA GCT GCT TCG-3′, and 5′-GCT TAC AAA TTG TTG CGA ACC TTT GGG AGT ACA AAC AAT GTG TAG GCT GGA GCT GCT TCG-3′ and 5′-TAC GGG CAA ATG AAC TTC GTG GCG AGA AGC GCA ATC GCC TCA TAT GAA TAT CCT CCT TAG-3′, respectively. All cassettes were transformed into MG1655 cells expressing λRed recombinase from the pKD46 helper plasmid. Deletions were verified by PCR using primers in the antibiotic resistance marker and sites adjacent on the host chromosome. All deletions were subsequently transduced into a clean MG1655 background prior to antibiotic cassette removal using the FLP recombinase expressing pCP20 helper plasmid.
Single-copy transcriptional and translational fusions were constructed in trans using the pVenus integration vector (52). Transcriptional fusions to the PompF and PmicF promoters were made by PCR amplifying the promoter regions of the ompF and micF genes using primers 5′-ATA GGT ACC ACG TGC TGG ACG AGC GTA TG-3′ and 5′-ATA GAA TTC AGC AGG GAC GAT CAC TGC-3′ and 5′-ATA GGT ACC ACC TGA GTT TCA CCT TTG AA-3′ and 5′-ATA GAA TTC TGC GAG GCA TCC GGT TGA AA-3′, respectively. Following amplification, the PCR products were digested with KpnI and EcoRI (sequences underlined) and ligated into the corresponding restriction sites of pVenus to produce pVenus-ompF and pVenus-micF. The translational fusion of ompF to yfp [ompF′-′yfp(hyb)] was produced in two steps. First, a fragment containing the promoter region and the first 39 bases of ompF was generated by PCR using primers 5′-ATA GGT ACC ACG TGC TGG ACG AGC GTA TG-3′ and 5′-CAG TGA AAA GTT CTT CTC CTT TAC TAG CAG GGA CGA TCA CTG C-3′. The resulting product also contained an overhang complementary to 25 base pairs after the first 6 bases of yfp(venus). Second, the ompF-yfp′ fragment was used to amplify the entire yfp(venus) and t0 terminator region from pVenus using the reverse primer 5′-CTC GCA ATC CAG TGC AAA-3′. The ompF′-′yfp(hyb) fragment was then cloned into the KpnI (sequence underlined) and NheI sites of pVenus to produce pVenus-FY. The pVenus derivatives described above were then integrated into the phage λ attachment site in MG1655 cells expressing λInt from the pINT-ts helper plasmid. Single-copy integrations were verified by PCR using primers described by Haldimann and Wanner (25). Resulting single-copy fusions were transduced back into a clean MG1655 background.
Complementation vectors for expression of MarA and Rob were constructed using the medium-copy-number, arabinose-inducible pBAD30 plasmid (24). The marA and rob genes were PCR amplified using forward and reverse primers 5′-ATA GAA TTC TTT ATA AGG AGG AAA AAC ATA TGA CGA TGT CCA GAC GC-3′ and 5′-ATA TCT AGA CTA GCT GTT GTA ATG ATT TAA TGG A-3′ and 5′-ATA GAG CTC TTT ATA AGG AGG AAA AAC ATA TGG ATC AGG CCG GCA TTA T-3′ and 5′-ATA GGT ACC TTA ACG ACG GAT CGG AAT CA-3′, respectively. The marA and rob products both contain strong, synthetic ribosome binding sites (RBS) to ensure high-level translation. Resulting PCR products were treated with EcoRI and XbaI (sequences underlined) for marA and SacI and KpnI (sequences underlined) for rob. The digested products were then ligated into the corresponding restriction sites downstream of the PBAD promoter in pBAD30 to produce the plasmids pMarA and pRob.
Fluorescence-based promoter activity assays.
Cells were grown overnight in medium A to saturation and subcultured 1:200 in fresh medium with or without 5 mM salicylate. For experiments, 0.5 ml was dispensed to individual wells of microtiter plates with 96 deep, square wells (VWR; 82006-448). Plates were sealed with Breath-Easy membranes (Diversified Biotech) to reduce evaporation, placed on a high-speed, microplate shaker (VWR), and shaken at 1,000 rpm and 37°C.
To measure fluorescence and optical density (OD), 250 μl of culture was transferred from the deep-well plates to black, clear-bottomed Costar 96-well microtiter plates. Fluorescence (excitation/emission λ, 515/530 nm) and OD (600 nm) were measured using a Tecan Safire2 microplate reader. Fluorescence measurements are reported as the relative fluorescence normalized to the optical density of the sample to correct for differences in cell density. All experimental data presented are the averages and standard deviations of four replicate samples.
Small-scale envelope preparation and SDS-PAGE.
Envelope fractions were prepared as described by Slauch and Silhavy with minor modifications (56). Briefly, cells were grown overnight in medium A and subcultured 1:200 in 10 ml of fresh medium with or without 5 mM salicylate. Cultures were grown to mid-log phase (OD = 0.4 to 0.5). Sample volumes were normalized to the lowest optical density to allow for comparison of outer membrane protein quantities across strain backgrounds. Normalized cultures were then pelleted at 3,800 × g. The pellet was washed once in 30 mM Tris-HCl (pH 8.1) and repelleted. Cell pellets were then resuspended in 30 mM Tris-HCl–20% sucrose buffer, followed by the addition of 10 μl of 20-mg/ml lysozyme–0.1 mM EDTA (pH 7.3) and incubated on ice for 30 min. Following lysozyme treatment, 3 ml of 3 mM ETDA (pH 7.3) was added and the resulting extract was disrupted with a single 20-s pulse using a microtip sonicator (Fisher Scientific). A 1.5-ml fraction of the extract was then centrifuged at 16,000 × g for 60 min. Envelope fractions were collected as centrifuged precipitate and resuspended in 40 μl of Laemmli SDS sample buffer (30). Samples were boiled at 100°C for 5 min prior to SDS-PAGE display. Finally, 10-μl aliquots were displayed on 10% acrylamide–6 M urea-1% SDS gels at 150 V for 80 min.
RESULTS
Salicylate decreases the expression of OmpF and increases the expression of OmpC.
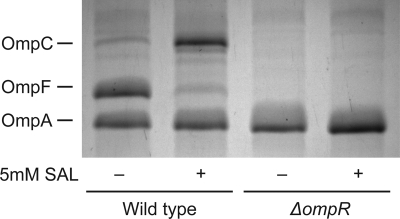
Previous reports have shown that exposure to salicylate decreases the amount of OmpF in the outer membrane (12, 47, 49, 54). Although these reports all observe a reduction in OmpF, discrepancies exist regarding changes in the expression of OmpC. To determine the effects of salicylate on OmpF/OmpC expression, we harvested insoluble membrane fractions from cells grown in a rich, low-osmolarity medium (28) in the presence or absence of 5 mM salicylate (Fig. 1). Under these conditions, we observed a decrease in the levels of OmpF and an increase in the levels of OmpC in the outer membrane.
Fig. 1.
The levels of OmpC, OmpF, and OmpA in the outer membrane in the presence or absence of salicylate. Cells were grown overnight in medium A and subcultured 1:200 in fresh medium A in the presence or absence of 5 mM salicylate (SAL). OmpC, OmpF, and OmpA protein bands are indicated. Strains used in this experiment were MG1655 and CR720.
Most likely, these discrepancies between our work and those in the literature regarding OmpC are due to differences in the strains employed. In our experiments, we used strain MG1655. In those involving strain MC4100, salicylate was found to decrease OmpC despite increases in ompC transcription (49). However, in derivatives of strain AG100, no changes in OmpC expression were observed (12).
We additionally explored the effects of OmpR on regulating OmpF/OmpC expression under salicylate exposure (Fig. 1). Consistent with the observations of Rosner and coworkers (49), both OmpF and OmpC are not expressed in the absence of OmpR, irrespective of whether salicylate is present or not.
MarA and Rob are functionally redundant regulators of MicF and OmpF expression.
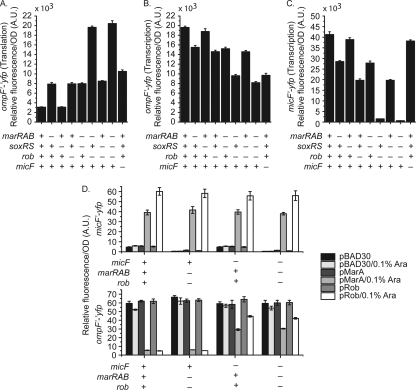
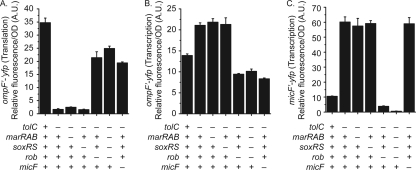
The MicF sRNA is known to repress the expression of OmpF. The transcription of micF, in turn, is activated by MarA, SoxS, and Rob. Of the three, only MarA expression is known to be directly responsive to salicylate (12, 33, 35). However, Cohen and coworkers found that the reduction of OmpF expression in response to salicylate was not solely dependent on increased expression of MarA (12). Based on this observation, we hypothesized that Rob may also be involved. Specifically, we have observed that Rob is indirectly activated by salicylate, independent of either MarA or MarR (9). To test this hypothesis, we measured the expression from single-copy transcriptional fusions of ompF and micF and a translational fusion of OmpF to the fast-folding yellow fluorescing protein (YFP) variant Venus (41). We performed these experiments in a series of genetic backgrounds where the marRAB, soxRS, and rob regulatory components of the mar-sox-rob network were systematically deleted. In addition, we tested the expression of these fusions in a strain lacking micF. Consistent with our hypothesis, we found that both MarA and Rob work in parallel to decrease OmpF expression in the presence of 5 mM salicylate (Fig. 2A and 3). We also found that MarA and Rob both increase ompF transcription (Fig. 2B). No change, however, was observed upon loss of SoxS under identical conditions. Specifically, we observed a 2.5-fold increase in OmpF expression, as determined using translational fusions to Venus upon loss of either MarA (ΔmarRAB) or Rob (Δrob) (Fig. 2A). Likewise, we observed a 20% reduction in ompF transcription upon loss of either transcription factor (Fig. 2B). Moreover, their contributions were additive with respect to both OmpF expression and ompF transcription. The increases in OmpF expression upon loss of either factor were also reflected by the changes in micF transcription, where we found that the loss of MarA or Rob decreased transcription 1.4- or 2.1-fold, respectively (Fig. 2C). In mutants lacking both MarA and Rob, we found that OmpF expression was increased greater than 6-fold, with correlated decreases in micF transcription of greater than 26-fold.
Fig. 2.
Full repression of ompF translation during salicylate exposure requires both MarA and Rob. (A) Levels of ompF′-′yfp translation. (B) Transcriptional activity of the PompF promoter. (C) Transcriptional activity of the PmicF promoter. Cells were grown overnight in medium A and subcultured 1:200 in medium A containing 5 mM salicylate for 4 h prior to fluorescence and optical density measurements. Presence or absence of genes is denoted by + or −, respectively. Strains used in this experiment were CR713 to CR715, CR737 to CR744, CR754 to CR761, and CR771 to CR778. (D) Transcriptional activity of PmicF and levels of ompF′-′yfp translation during ectopic complementation of MarA and Rob in the presence and absence of MicF. Cells were grown in medium A overnight and subcultured 1:200 in fresh medium A with and without 0.1% arabinose. Strains used in this experiment are CR715, CR774, CR776, CR781, CR714, CR757, CR759, and CR764.
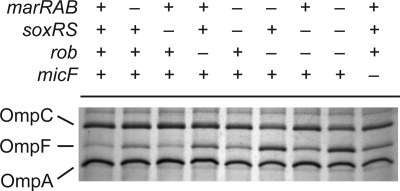
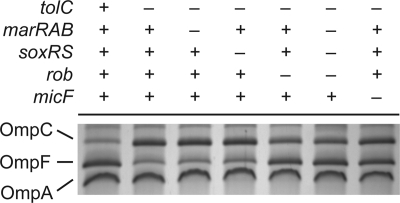
Fig. 3.
Both MarA and Rob are required to fully repress OmpF expression in the outer membrane during salicylate exposure. Presence or absence of genes is indicated by + and −, respectively. Cells were grown overnight in medium A and subcultured 1:200 in fresh medium A containing 5 mM salicylate. Cultures were grown to mid-logarithmic phase prior to envelope extraction. Envelope fractions were displayed on 10% acrylamide–6 M urea-1% SDS gels and stained with Coomassie R250. Strains used were MG1655 and CR721 to CR728.
We also measured OmpF expression in mutants lacking micF in the presence of 5 mM salicylate. Given the current regulatory model, disruptions in micF should result in levels of OmpF translation comparable to those observed in a marRAB rob double mutant or marRAB soxRS rob triple mutant. Surprisingly, we found that the double and triple mutants exhibited 2-fold-higher levels of OmpF expression than the micF mutant (Fig. 2A). These results demonstrate that MarA and Rob do not regulate ompF translation in response to salicylate solely through a MicF-dependent pathway. This conclusion is further supported by the phenotypic observation that OmpF levels in the presence of 5 mM salicylate are higher in mutants lacking both MarA and Rob than in mutants lacking MicF alone (Fig. 3).
Interestingly, we found that ompF transcription was reduced in a micF mutant in the presence of salicylate (Fig. 2B), opposite to what we see at the levels of translation and protein expression. Consistent with these results, the changes in ompF transcription observed in a marRAB rob double mutant and a marRAB soxRS rob triple mutant were nearly identical to those in a micF mutant. These results are surprising, as Cohen and coworkers previously observed that increased micF transcription decreases ompF transcription (12). One possible explanation is that MicF stabilizes the mRNA of our OmpF transcriptional fusion. Regardless, we suspect that the effect we observe is not physiologically significant.
The experiments described above were performed in the presence of 5 mM salicylate. As a control, we also performed identical experiments in the absence of salicylate. In this case, we found that both OmpF expression and ompF transcription were mostly unchanged in the different mutant backgrounds (see Fig. S1 in the supplemental material). The difference was no greater than 10% in the case of the translational fusions and 20% in the case of the transcriptional fusions. Likely, micF expression is too weak and OmpF expression too high for there to be any change in the absence of salicylate. Indeed, micF expression is significantly reduced in the absence of salicylate. One interesting observation, though, is that OmpF expression is almost 2-fold higher in the absence of salicylate in wild-type cells than in mutants in the presence of salicylate. These results suggest that salicylate also represses OmpF expression through an alternate mechanism.
We also tested whether MarA and Rob could independently repress OmpF expression when ectopically expressed from an arabinose-inducible promoter on a plasmid in the absence of salicylate (Fig. 2D). To account for different background levels of OmpF expression, we also performed these experiments in the presence of 200 mM NaCl (see Tables S1 and S2 in the supplemental material). In genetic backgrounds containing micF, we found that the ectopic expression of MarA or Rob led to a 10-fold increase in micF expression irrespective of whether NaCl was present or not. This 10-fold increase correlates well with the corresponding 10-fold decrease in OmpF expression that we also observed. In strains lacking micF, the ectopic expression of MarA led to an approximately 50% reduction in OmpF expression whereas the ectopic expression of Rob led to an approximately 30% reduction in OmpF expression. While the level of repression is significantly reduced, these results suggest, as discussed below, that MarA and possibly Rob can repress OmpF expression independent of MicF.
MarA regulates OmpF expression through a MicF-independent pathway.
We observed that the ectopic expression of MarA or Rob could reduce OmpF expression in a micF mutant (Fig. 2D), suggesting that the two may function through a MicF-independent pathway. To further explore this putative mechanism, we constructed marRAB and rob mutants in otherwise micF null genetic backgrounds and monitored OmpF expression and ompF and micF transcription in the presence of salicylate.
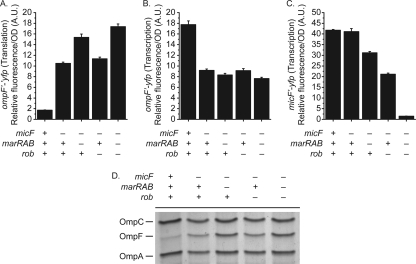
We found that loss of Rob had no effect on OmpF expression in the absence of MicF (Fig. 4A and D), indicating that it functions upstream of MicF. In the case of MarA, however, we found that deleting it could affect OmpF translation in the absence of MicF. Specifically, we observed a 50% increase in OmpF expression upon loss of MarA in an otherwise micF background (Fig. 4A and D), indicating that MarA represses OmpF translation independent of MicF. Lastly, we found that the decrease in micF transcription upon loss of MarA or Rob is independent of MicF (Fig. 4C).
Fig. 4.
MarA functions through MicF-dependent and MicF-independent pathways to reduce the levels of OmpF during salicylate exposure. (A) Levels of ompF′-′yfp translation (strains CR757 and CR762 to CR764). (B) Transcriptional activity of the PompF promoter (strains CR740 and CR745 to CR747). (C) Transcriptional activity of the PmicF promoter (strains CR774 and CR779 to CR781). (D) Levels of OmpC, OmpF, and OmpA in the envelope fraction displayed on a 10% acrylamide–6 M urea-1% SDS gel (strains MG1655, CR724, and CR729 to CR731). Cells were grown in medium A overnight and subcultured 1:200 in fresh medium A with 5 mM salicylate. Cultures were grown for 4 h prior to fluorescence and optical density measurements or to mid-log phase prior to envelope extraction.
We also found that the decrease in ompF transcription upon loss of MarA or Rob is due to MicF (Fig. 4B). In the absence of MicF, MarA or Rob had no effect on ompF transcription. This epistasis indicates that both MarA and Rob function upstream of MicF with regard to ompF transcription. It also suggests that MicF activates ompF transcription, though this is likely an artifact of our transcriptional fusions as discussed above.
Increases in ompC transcription are independent of MarA or Rob.
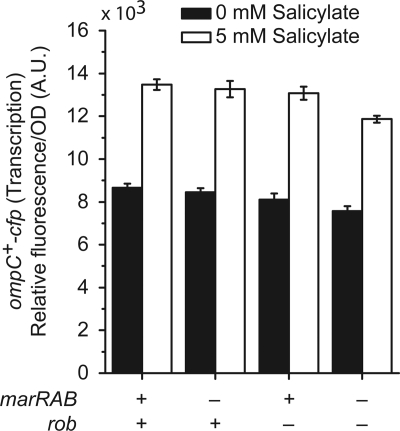
Previous results have shown increases in ompC transcription during salicylate exposure (49). Despite the close proximity of the MarA/SoxS/Rob binding site in the divergently arranged PmicF promoter, the role of MarA and Rob in mediating this increase in ompC transcription has not been previously explored. To determine whether the salicylate-induced increase in ompC transcription is MarA or Rob dependent, we monitored ompC transcription in mutants lacking marRAB or rob using a cyan fluorescing protein (CFP) gene fused downstream of the ompC coding region. This fusion has previously been shown to have minimal effects on OmpC expression and to provide an accurate measure of ompC transcription (3, 4). Consistent with the previous findings, we observed modest increases in ompC transcription in the presence of 5 mM salicylate (Fig. 5). However, we found that this increase is independent of MarA and Rob. These results indicate that MarA and Rob do not regulate ompC transcription in response to salicylate.
Fig. 5.
Increases in ompC transcription are independent of MarA and Rob. Cells were grown overnight in medium A and subcultured 1:200 in fresh medium A with or without 5 mM salicylate. Cultures were grown for 4 h prior to fluorescence and optical density measurements. Strains used in this experiment were MDG147 and CR788 to CR790.
The reduction in OmpF expression in tolC mutants is due to Rob.
E. coli mutants lacking TolC are known to have altered outer membrane porin compositions. Specifically, the expression of OmpF in the outer membrane is significantly reduced regardless of medium osmolarity (40). Misra and Reeves previously showed that the reduction in OmpF expression in a tolC mutant was due to MicF (38). However, they did not determine what caused micF transcription to increase upon loss of TolC. Recent data from Rosner and Martin suggest that the increase in micF transcription in tolC mutants is due to the upregulation of MarA, SoxS, and Rob (51). Based on these findings, we wished to determine which of the mar-sox-rob systems contribute to the decreased expression of OmpF observed in a tolC mutant. Specifically, we studied the effects of MarA, SoxS, Rob, and MicF on the expression of transcriptional and translational fusions described previously.
In the absence of tolC, we observed that OmpF expression was decreased and micF transcription was increased, consistent with previous findings (Fig. 6A and C). Introducing marRAB, soxRS, and rob deletions into the tolC mutant background indicated that Rob is the primary, though not sole, factor increasing micF transcription and, as a consequence, decreasing OmpF expression (Fig. 6A and C). Examining OmpF expression in the outer membranes of these mutants also supports this conclusion (Fig. 7). Collectively, these data indicate that Rob is the primary regulator involved in increased MicF expression in tolC mutants. The role of MarA and SoxS in this instance appears to be minor.
Fig. 6.
Reduction in ompF translation by MicF in tolC mutants is a result of Rob-dependent activation of micF gene expression. (A) Levels of ompF′-′yfp translation. (B) Transcriptional activity of the PompF promoter. (C) Transcriptional activity of the PmicF promoter. Cells were grown overnight in medium A and subcultured 1:200 in medium A containing 5 mM salicylate for 4 h prior to fluorescence and optical density measurements. Strains used in this experiment were CR713 to CR715, CR748 to CR753, CR765 to CR769, and CR782 to CR787.
Fig. 7.
MicF-dependent reduction of OmpF expression in tolC mutants is a result of Rob activation of micF gene expression. Cells were grown overnight in medium A and subcultured 1:200 in fresh medium A containing 5 mM salicylate. Cultures were grown to mid-logarithmic phase prior to envelope extraction. Envelope fractions were displayed on 10% acrylamide–6 M urea-1% SDS gels and stained with Coomassie R250. Strains used were MG1655, CR702, and CR732 to CR736.
We also found that ompF transcription was increased in tolC mutants, though this increase was rob and micF dependent (Fig. 6B). Specifically, we can attribute the increase in ompF transcription to increased MicF expression through Rob. As we have mentioned, increased MicF expression leads to increased ompF transcription, though this effect is likely an artifact of our transcriptional reporter.
Though Rob is the key factor regulating MicF expression in a tolC mutant, these findings do not directly indicate the source of Rob activation. Whether this is caused by increased intracellular metabolites or perturbation of other elements of cellular physiology is still unknown (10). Interestingly, MarA is upregulated in a tolC mutant but does not affect MicF expression (51).
DISCUSSION
In this work, we investigated the role of the mar-sox-rob regulon in regulating outer porin expression, where the focus was on the MicF-dependent regulation of OmpF expression upon salicylate exposure. We found that MarA and Rob can independently activate micF transcription in response to salicylate, leading to reduced OmpF expression. MarA was also found to repress OmpF expression though a MicF-independent pathway. In the case of OmpC, salicylate increased its transcription though this effect was independent of MarA and Rob. Finally, we were able to show that the reduction in OmpF expression in a tolC mutant is due primarily to Rob.
A key finding of this study was that MarA is not the sole factor regulating MicF expression in response to salicylate. Rob is also capable of activating MicF expression in response to salicylate. Both function in parallel regulatory pathways, where their effects on OmpF expression are additive. Previous studies have, in fact, shown that the salicylate-induced reduction in OmpF expression is not solely due to MarA and that some other factor is involved (12). The surprising finding here was that Rob is one of the factors. While salicylate is known to induce MarA expression through the derepression of MarR and is often taken as the canonical inducer for MarA, salicylate has not previously been shown to directly activate Rob to the best our knowledge. The known activating ligands for Rob are bile salts, fatty acids, and 2,2′-dipyridyl (48, 50). Whether salicylate directly binds and activates Rob, however, is unknown.
We have found that MarA also activates a MicF-independent pathway to reduce OmpF expression. Rob can too but only when ectopically expressed from a plasmid, a result not surprising given the common binding targets for both regulators (5, 29, 34). These observations suggest that MarA utilizes both MicF and the MicF-independent pathway simultaneously to achieve levels of OmpF reduction similar to that which Rob accomplishes through MicF alone. Through the combined action of these factors, the parallel MarA and Rob-dependent pathways may serve to ensure OmpF reduction in the presence of a variety of toxic chemicals.
How MarA is able to work through a MicF-independent pathway to inhibit translation of ompF mRNA is unknown. As this regulation occurs at the level of ompF translation, MarA likely regulates an additional sRNA not detected by previous microarray analyses. Currently, the only well-characterized sRNA regulator of ompF mRNA translation is MicF. Although our data suggest the possibility of an additional sRNA regulator, they do not discount the possibility of MarA-regulated factors that may work to destabilize the ompF mRNA. Future implementation of sRNA detection strategies during salicylate exposure will help to differentiate between these possibilities.
OmpF expression is decreased in the absence of the outer membrane efflux pore TolC (40). Misra and Reeves previously demonstrated that this reduction in OmpF expression is due to MicF (37, 38). In the present study, we showed that the reduction in OmpF expression in the absence of TolC is due primarily to Rob. These results are consistent with recent observations made by Rosner and Martin, who have also shown that MarA and SoxS expression and Rob activation are elevated approximately 2-fold in tolC null mutants (51). The increase in mar-sox-rob regulon activation has been attributed in part to the elevated intracellular levels of intermediary metabolites that serve as inducers for these three systems (10, 51). What was surprising was that the tolC effect could be almost solely ascribed to Rob even though the MarA expression is also increased under these conditions.
Although this work has more clearly defined roles for MarA and Rob in regulating OmpF expression in response to salicylate, there are still a number of unresolved issues. For one, we found that ompC transcription is increased during salicylate exposure in a MarA/Rob-independent manner. As the mar-sox-rob regulon is extensive, it may be possible that additional downstream elements cause indirect changes in the expression of ompC. Alternatively, parallel regulators responsive to salicylate, such as EmrR (MprA), may instead regulate ompC transcription (32). Another point of interest may be the convergence of additional two-component systems at the ompF promoter that may be stimulated by salicylate and other extracellular toxins. A number of systems, such as the CpxAR and RstAB two-component systems, have been shown to directly and indirectly change the activity of the ompF promoter (4, 21, 43). Additionally, salicylate may stimulate other extracytoplasmic stress systems. However, minimal overlap exists between extracytoplasmic stress and salicylate transcriptional responses based on genome-wide microarray data (6, 45). Whether MarA and Rob serve an auxiliary role in changing the activity of these systems or EnvZ-OmpR activation remains to be seen.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. M. Slauch for helpful discussions and advice regarding this work. Also, we thank M. D. Goulian for providing strain MDG147.
This work was supported in part by grants from the National Science Foundation and the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Andersen J., Forst S. A., Zhao K., Inouye M., Delihas N. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961–17970 [PubMed] [Google Scholar]
- 2. Basle A., Rummel G., Storici P., Rosenbusch J. P., Schirmer T. 2006. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J. Mol. Biol. 362:933–942 [DOI] [PubMed] [Google Scholar]
- 3. Batchelor E., Goulian M. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U. S. A. 100:691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batchelor E., Walthers D., Kenney L. J., Goulian M. 2005. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins OmpF and OmpC. J. Bacteriol. 187:5723–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennik M. H., Pomposiello P. J., Thorne D. F., Demple B. 2000. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J. Bacteriol. 182:3794–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bury-Mone S., et al. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5:e1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen S., Zhang A., Blyn L. B., Storz G. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186:6689–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 9. Chubiz L. M. 2010. The role of coordinated regulation and aromatic metabolites in activating the mar/sox/rob regulon of Escherichia coli. Ph.D. thesis University of Illinois at Urbana-Champaign, Urbana, IL [Google Scholar]
- 10. Chubiz L. M., Rao C. V. 2010. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J. Bacteriol. 192:4786–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen S. P., Hachler H., Levy S. B. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen S. P., Levy S. B., Foulds J., Rosner J. L. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cowan S. W., et al. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727–733 [DOI] [PubMed] [Google Scholar]
- 14. Coyer J., Andersen J., Forst S. A., Inouye M., Delihas N. 1990. micF RNA in ompB mutants of Escherichia coli: different pathways regulate micF RNA levels in response to osmolarity and temperature change. J. Bacteriol. 172:4143–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deighan P., Free A., Dorman C. J. 2000. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 38:126–139 [DOI] [PubMed] [Google Scholar]
- 17. De la Cruz M. A., Calva E. 2010. The complexities of porin genetic regulation. J. Mol. Microbiol. Biotechnol. 18:24–36 [DOI] [PubMed] [Google Scholar]
- 18. Delihas N., Forst S. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1–12 [DOI] [PubMed] [Google Scholar]
- 19. Ferrario M., et al. 1995. The leucine-responsive regulatory protein of Escherichia coli negatively regulates transcription of ompC and micF and positively regulates translation of ompF. J. Bacteriol. 177:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garrett S., Taylor R. K., Silhavy T. J. 1983. Isolation and characterization of chain-terminating nonsense mutations in a porin regulator gene, envZ. J. Bacteriol. 156:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerken H., Charlson E. S., Cicirelli E. M., Kenney L. J., Misra R. 2009. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol. Microbiol. 72:1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gillette W. K., Martin R. G., Rosner J. L. 2000. Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J. Mol. Biol. 299:1245–1255 [DOI] [PubMed] [Google Scholar]
- 23. Griffith K. L., Fitzpatrick M. M., Keen E. F., III, Wolf R. E., Jr 2009. Two functions of the C-terminal domain of Escherichia coli Rob: mediating “sequestration-dispersal” as a novel off-on switch for regulating Rob's activity as a transcription activator and preventing degradation of Rob by Lon protease. J. Mol. Biol. 388:415–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haldimann A., Wanner B. L. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall M. N., Silhavy T. J. 1981. Genetic analysis of the ompB locus in Escherichia coli K-12. J. Mol. Biol. 151:1–15 [DOI] [PubMed] [Google Scholar]
- 27. Huang L., Tsui P., Freundlich M. 1990. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J. Bacteriol. 172:5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawaji H., Mizuno T., Mizushima S. 1979. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J. Bacteriol. 140:843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwon H. J., Bennik M. H., Demple B., Ellenberger T. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424–430 [DOI] [PubMed] [Google Scholar]
- 30. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 31. Li Z., Demple B. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J. Biol. Chem. 269:18371–18377 [PubMed] [Google Scholar]
- 32. Lomovskaya O., Lewis K., Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin R. G., Rosner J. L. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. U. S. A. 92:5456–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin R. G., Rosner J. L. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611–1624 [DOI] [PubMed] [Google Scholar]
- 35. Martin R. G., Rosner J. L. 2004. Transcriptional and translational regulation of the marRAB multiple antibiotic resistance operon in Escherichia coli. Mol. Microbiol. 53:183–191 [DOI] [PubMed] [Google Scholar]
- 36. Miller J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related organisms. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 37. Misra R., Reeves P. 1985. Molecular characterisation of the Stc- mutation of Escherichia coli K-12. Gene 40:337–342 [DOI] [PubMed] [Google Scholar]
- 38. Misra R., Reeves P. R. 1987. Role of micF in the tolC-mediated regulation of OmpF, a major outer membrane protein of Escherichia coli K-12. J. Bacteriol. 169:4722–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizuno T., Chou M. Y., Inouye M. 1984. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U. S. A. 81:1966–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morona R., Reeves P. 1982. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J. Bacteriol. 150:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagai T., et al. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90 [DOI] [PubMed] [Google Scholar]
- 42. Nunoshiba T., Hidalgo E., Amabile Cuevas C. F., Demple B. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174:6054–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ogasawara H., et al. 2007. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol. 189:4791–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pages J. M., James C. E., Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 6:893–903 [DOI] [PubMed] [Google Scholar]
- 45. Pomposiello P. J., Bennik M. H., Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pratt L. A., Hsing W., Gibson K. E., Silhavy T. J. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911–917 [DOI] [PubMed] [Google Scholar]
- 47. Ramani N., Boakye K. 2001. Salicylate inhibits the translation and transcription of ompF in Escherichia coli. Can. J. Microbiol. 47:1053–1057 [PubMed] [Google Scholar]
- 48. Rosenberg E. Y., Bertenthal D., Nilles M. L., Bertrand K. P., Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609–1619 [DOI] [PubMed] [Google Scholar]
- 49. Rosner J. L., Chai T. J., Foulds J. 1991. Regulation of ompF porin expression by salicylate in Escherichia coli. J. Bacteriol. 173:5631–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosner J. L., Dangi B., Gronenborn A. M., Martin R. G. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosner J. L., Martin R. G. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J. Bacteriol. 191:5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saini S., Pearl J. A., Rao C. V. 2009. Role of FimW, FimY, and FimZ in regulating the expression of type i fimbriae in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sarma V., Reeves P. 1977. Genetic locus (ompB) affecting a major outer-membrane protein in Escherichia coli K-12. J. Bacteriol. 132:23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sawai T., Hirano S., Yamaguchi Y. 1987. Repression of porin synthesis by salicylate in Escherichia coli, Klebsiella pneumoniae, and Serratia marcescens. FEMS Microbiol. Lett. 40:233–237 [Google Scholar]
- 55. Slauch J. M., Garrett S., Jackson D. E., Silhavy T. J. 1988. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J. Bacteriol. 170:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Slauch J. M., Silhavy T. J. 1989. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 210:281–292 [DOI] [PubMed] [Google Scholar]
- 57. Thomason L. C., Costantino N., Court D. L. 2007. E. coli genome manipulation by P1 transduction, p. 1–9 In Ausubel F. M., et al. (ed.), Current protocols in molecular biology, chapter 1, unit 1.17. Greene Publishing Associates, New York, NY: [DOI] [PubMed] [Google Scholar]
- 58. Wu J., Weiss B. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.