Abstract
Myxococcus xanthus belongs to the delta class of the proteobacteria and is notable for its complex life-style with social behaviors and relatively large genome. Although previous observations have suggested the existence of horizontal gene transfer in M. xanthus, its ability to take up exogenous DNA via natural transformation has not been experimentally demonstrated. In this study, we achieved natural transformation in M. xanthus using the autonomously replicating myxobacterial plasmid pZJY41 as donor DNA. M. xanthus exopolysaccharide (EPS) was shown to be an extracellular barrier for transformation. Cells deficient in EPS production, e.g., mutant strains carrying ΔdifA or ΔepsA, became naturally transformable. Among the inner barriers to transformation were restriction-modification systems in M. xanthus, which could be partially overcome by methylating DNA in vitro using cell extracts of M. xanthus prior to transformation. In addition, the incubation time of DNA with cells and the presence of divalent magnesium ion affected transformation frequency of M. xanthus. Furthermore, we also observed a potential involvement of the type IV pilus system in the DNA uptake machinery of M. xanthus. The natural transformation was totally eliminated in the ΔpilQ/epsA and Δtgl/epsA mutants, and null mutation of pilB or pilC in an ΔepsA background diminished the transformation rate. Our study, to the best of our knowledge, provides the first example of a naturally transformable species among deltaproteobacteria.
INTRODUCTION
Myxococcus xanthus is a motile Gram-negative soil bacterium with a fascinating life cycle, including many social behaviors such as swarming, predation, and development (30, 31). M. xanthus cells move on a solid surface by gliding (17, 43), which is regulated independently by the A (adventurous) and the S (social) motility systems (25). Under vegetative conditions, cells secrete antibiotics, bioactive compounds, or degradative enzymes in amounts sufficient to immobilize and digest other bacteria (53, 55). When deprived of nutrients, most cells aggregate to form multicellular fruiting bodies, within which the metabolically dormant spherical myxospores are developed (13). Reflective of its complex life-style, M. xanthus has an unusually large genome (9.14 Mb) within the sequenced deltaproteobacteria, which (90% of the genome) consists of 7,388 predicted protein coding sequences (CDSs) (21). In addition to frequent duplication events, at least 1.4 Mb of genomic expansion in M. xanthus may arise from extensive horizontal (lateral) gene transfer (HGT) (21). Further examination of the evolutionary history of 78 genes essential for its fruiting body development revealed that 22% of the genes, mostly encoding metabolic enzymes, exhibit an altered codon bias and/or phylogeny suggestive of HGT (20).
While predation behavior has been suggested to enhance the HGT process of M. xanthus (20), direct experimental evidence is still lacking. Under laboratory conditions, M. xanthus has been shown to obtain genes from Escherichia coli by transduction (58) and conjugation (5). However, the natural gene transfer of M. xanthus has not been experimentally demonstrated (20). In addition to conjugation and transduction, uptake and incorporation of exogenous DNA by cells via transformation are one of the major mechanisms for bacterial genetic exchange in nature (22), which is widely distributed in both Gram-positive and Gram-negative bacteria (16, 35). Many transformable bacteria become transiently competent for genetic transformation under specific conditions (10), except for neisseriae, which are competent during all phases of growth (22). The natural transformation process normally comprises several discrete steps, including DNA binding, fragmentation, uptake, and integration (2). In most competent bacteria, the DNA uptake machinery is related to type IV pili (TFP) or type II secretion systems (T2SSs) (9, 16). At the same time, active barriers against such DNA transfer are present at different phases of the transformation process (47, 59).
Although there was no previously known evidence to show that M. xanthus regulates competence and uptake of DNA in certain phases of growth, some of the new findings motivated our investigations of such a possibility, including the discovery of an autonomously replicating plasmid, pMF1, in Myxococcus (69) and the extracellular DNA forming reticulated structures with exopolysaccharide (EPS) in M. xanthus extracellular matrix (W. Hu and W. Shi, unpublished data). In this study, we sought to determine whether M. xanthus can acquire new genes by natural transformation and to investigate the factors affecting transformation efficiency.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The M. xanthus strains used in this study are listed in Table 1. Plasmid pJW01 carrying an in-frame deletion of epsA was constructed using SW810 (ΔepsA) genomic DNA as the PCR template and EpsA-IFD1 and -2 as primers (Table 1). The amplified region was digested with EcoRI and BamHI, ligated into pBJ113, and confirmed by sequencing. The plasmid was electroporated (32) into ΔpilA, ΔpilB, ΔpilC, ΔpilQ, ΔpilT, and Δtgl mutants, which were all null mutants in the DK1622 background, to construct SW2051 (ΔepsA/ΔpilA), SW2052 (ΔepsA/ΔpilB), SW2053 (ΔepsA/ΔpilC), SW2054 (ΔepsA/ΔpilQ), SW2055 (ΔepsA/ΔpilT), and SW2056 (ΔepsA/Δtgl), respectively. Deletion mutants were selected by their galactose-resistant and kanamycin (Km)-sensitive phenotypes as described previously (61). The insertion mutant SW2057 was constructed by electroporation (32) of plasmid pJW02 into SW810 (ΔepsA) and selected by kanamycin resistance phenotype. All the mutants were confirmed with PCR and sequence analyses.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant feature(s) or sequence | Reference or source |
|---|---|---|
| Strains | ||
| M. xanthus | ||
| DK1622 | Wild type | |
| DK10405 | DK1622, Δtgl, missing PilQ and surface pili | 64 |
| DK10409 | DK1622, ΔpilT, overpiliated | 67 |
| DK10410 | DK1622, ΔpilA, missing PilA | 66 |
| DK10416 | DK1622, ΔpilB, missing surface pili | 67 |
| DK10417 | DK1622, ΔpilC, missing surface pili | 67 |
| DK3088 | DK1622, stk, overproducing EPS | 11 |
| DK8615 | DK1622, ΔpilQ, missing surface pili | 63 |
| SW504 | DK1622, ΔdifA, missing EPS | 68 |
| SW505 | DK1622, difA::Tn5, Kmr | 68 |
| SW810 | DK1622, ΔepsA, missing EPS | 39 |
| SW810-41 | DK1622, transformant of SW810 (ΔepsA) by plasmid pZJY41, Kmr | This study |
| SW2051 | DK1622, ΔepsA/ΔpilA | This study |
| SW2052 | DK1622, ΔepsA/ΔpilB | This study |
| SW2053 | DK1622, ΔepsA/ΔpilC | This study |
| SW2054 | DK1622, ΔepsA/ΔpilQ | This study |
| SW2055 | DK1622, ΔepsA/ΔpilT | This study |
| SW2056 | DK1622, ΔepsA/Δtgl | This study |
| SW2057 | DK1622, SW810 (ΔepsA)::pJW02, constructed by electroporation, Kmr | This study |
| E. coli DH5α | Host for cloning and plasmid preparation | 23 |
| Plasmids | ||
| pBJ113 | Gene replacement vector with KG cassette, Kmr | 29 |
| pSWU19 | 3-kb SmaI-SmaI attP fragment in pBGS18 at DraI sites, Kmr | 65 |
| pZJY41 | Shuttle plasmid for M. xanthus, Kmr, constructed from Myxococcus autonomously replicating plasmid pMF1 | 69 |
| pJW01 | epsA IFD PCR product cloned into pBJ113 using EcoRI and BamHI sites | This study |
| pJW02 | 940-bp PCR product homologous to DK1622 genome (part of MXAN_0606 and MXAN_0607) cloned into pBJ113 using EcoRI and BamHI sites | This study |
| Primers | ||
| EpsA-IFD | ||
| Primer 1 (sense, 5′–3′) | CCGGAATTCACAGACGAACGTCGGTGTCG | |
| Primer 2 (antisense, 5′–3′) | CGCGGATCCCCGCAGCGTAGAAGAGTGCC | |
| PMF-rep | ||
| Primer 1 (sense, 5′–3′) | ATGTGGAGCCGAGTGCC | |
| Primer 2 (antisense, 5′–3′) | CAGCGTTGGTCAGGTAAGTC | |
| Int | ||
| Primer 1 (sense, 5′–3′) | CCGGAATTCGAGCAGCGGCGTTTCATC | |
| Primer 2 (antisense, 5′–3′) | CGCGGATCCAAGAGGGTGGTGCCAATGAG | |
| DifA-tn | ||
| Primer 1 (sense, 5′–3′) | CCGGCTTGAAGTGGGAGGAC | |
| Primer 2 (antisense, 5′–3′) | CCGAGGACTATGAGTTGCTG |
Three plasmids were used as donor DNA in the transformation experiments. (i) Plasmid pSWU19 was constructed by ligating the 3-kb SmaI fragment into DraI-digested pBGS18 (65), which contains Mx8 attP and allows expression of Kmr by site-specific recombination and insertion of the plasmid into the M. xanthus genome. (ii) A DNA fragment (940 bp) randomly selected on the DK1622 (wild-type [WT]) genome, which contained part of MXAN_0606 (610 bp) and MXAN_0607 (230 bp), was amplified by PCR using DK1622 genomic DNA as the template and Int1 and -2 as primers (Table 1). The PCR product was cloned into pBJ113 using EcoRI and BamHI sites to construct the integration plasmid pJW02, which allowed expression of Kmr by homologous recombination. (iii) The myxobacterial shuttle plasmid pZJY41, which is derived from plasmid pMF1, an autonomously replicating plasmid isolated from Myxococcus fulvus 124B02 (69), was also used.
Media and growth conditions.
M. xanthus cells were grown in CYE medium (7) at 32°C on a rotary shaker at 300 rpm and harvested by centrifugation at 12,000 × g for 5 min. For transformation experiments, the cells were incubated on MMC agar plates (10 mM MOPS [morpholinepropanesulfonic acid], 4 mM MgSO4, 2 mM CaCl2, 1.5% agar, pH 7.6) (36), and transformants were screened on CYE agar containing 100 μg/ml kanamycin as described below.
Natural transformation of M. xanthus.
M. xanthus cells were harvested from 48-h CYE culture and concentrated to 5 × 109 cells/ml in MMC buffer (10 mM MOPS, 4 mM MgSO4, 2 mM CaCl2, pH 7.6) (36). The donor DNA was added to the cell suspension at a final concentration of 100 ng/μl (for most of the donor DNA) or 10 ng/μl (for plasmid pZJY41 isolated from SW810-41). The mixture was spread onto MMC agar and kept in a humidity chamber at 32°C for 7 days. The cells were scraped off agar plates and transferred onto CYE agar containing 100 μg/ml kanamycin. Serial dilutions plated onto nonselective solid medium in parallel were used to determine the total number of cells placed on the selective plates. Kanamycin-resistant colonies that arose on the selective plates were considered putative transformants. Transformation of cell extract (CFE)-modified pSWU19 (see below) was confirmed by colony PCR with the primer pairs described previously (12), which specifically target the Kmr gene (930 bp). Transformation of pZJY41 was further confirmed by colony PCR with the pair of primers PMF-rep1 and PMF-rep2 (Table 1), which specifically target the rep region on the plasmid (670 bp). At the same time, we performed plasmid extractions from all the putative pZJY41 transformants as previously described (69). PCR products and extracted plasmids were visualized by gel electrophoresis in 1% Tris-acetate-EDTA (TAE)–agarose with ethidium bromide. The transformation frequency was calculated as the proportion of confirmed transformants arising from the total cell population.
Plasmid DNA and EPS precipitation assay.
Crude EPS was prepared from wild-type DK1622 as described previously (8). To generate nucleic acid-free EPS, 200-μg/ml amounts of DNase I and RNase (Worthington) were added to the EPS, and samples were incubated at 37°C for 24 h followed by pronase treatment (38). Different amounts of pZJY41 plasmid DNA isolated from E. coli DH5α (10, 1, and 0.1 μg, respectively) were mixed with 100 μg of purified EPS each in 100 μl MMC buffer. The samples were vortexed for 30 s, incubated at 32°C for 120 min, and centrifuged at 12,000 × g for 30 min at 4°C. Thirty microliters of the supernatant was transferred into fresh tubes, and the plasmid DNA concentration was determined with a Quant-iT double-stranded DNA (dsDNA) high-sensitivity DNA assay kit (Invitrogen). Solution containing EPS (1 μg/μl) or plasmid DNA (0.1 μg/μl) alone was used as a negative control.
The EPS pellet precipitated from the buffer containing 100 μg/ml pZJY41 DNA was further treated with 50 μg/ml DNase I at 37°C for 6 h. The EPS pellets (precipitated from 100-μg/ml DNA buffer) with or without DNase I treatment were then subjected to staining, and EPS pellet (precipitated from buffer without DNA) was used as a control. A 5 μM concentration of Sytox green and 5 μg/ml of Alexa 633-conjugated derivatives of the wheat germ agglutinin (WGA) lectin (Molecular Probes) were used to label DNA and carbohydrates (40) present in EPS, respectively. Confocal laser scanning microscopy (CLSM) was employed to image the stained EPS pellets as previously described (40). The samples were viewed through a 40× oil-immersion objective (Plan-Neofluar/numerical aperture [NA], 1.3). Excitation at 488 nm with an argon laser in combination with a 505- to 530-nm band-pass emission filter was used for Sytox green fluorescence imaging. Excitation at 633 nm with a helium-neon laser and a 650-nm long-pass emission filter was used to observe Alexa 633 signals.
Trypan blue binding assay.
EPS production by different strains was measured using a previously described trypan blue binding assay (4). The cell pellets were vortexed, and total protein of the cells was used to determine the biomass of each sample. The relative amounts of EPS were calculated according to trypan blue bound to the wild-type cells.
Preparation of cell extracts of M. xanthus SW810 (ΔepsA).
Cell extracts (CFE) were prepared from M. xanthus cultures using a previously described assay (14) with minor modifications. SW810 cells were harvested from 100 ml 12-h CYE culture (32°C; 300 rpm; optical density at 600 nm [OD600], 0.6 to 0.8), and 0.5-g cell pellets were resuspended in 2.5 ml extraction buffer (14). Bacterial cell suspensions were sonicated 6 times for 10 s at a setting of 3 using the Sonic Dismembrator-60 (Fisher) on ice and then centrifuged at 12,000 × g at 4°C for 5 min. Supernatants were used immediately for in vitro plasmid DNA digestion or modification reactions (see below). Protein concentrations of the CFE were determined with the bicinchoninic acid (BCA) assay (Pierce) using bovine serum albumin as a standard.
Preparation of different donor DNAs for transformation.
Plasmid DNA from E. coli DH5α was prepared with the SV Minipreps DNA purification system (Promega). Plasmid pZJY41 was isolated from SW810-41 according to the published protocol (69), and genomic DNA of SW505 (difA::Tn5) was prepared as described previously (3). The SW505 genome (1 μg/μl) was partially digested by DNase I (2 × 10−5 U/μl) at 37°C for 5 min, 10 min, and 15 min to obtain fragments of genomic DNA at >20 kb, 5 to 20 kb, and <5 kb in size, respectively, and fragment size was confirmed by 1% agarose gel electrophoresis. SW505 genomic DNA was used as a template to amplify an ∼5-kb region containing the difA::Tn5 insertion with the DifA-Tn1 and -2 primers (Table 1). The PCR fragments were purified with the Wizard SV gel and PCR cleanup system (Promega) by following the manufacturer's instructions. Double-stranded DNA (dsDNA) was denatured by alkaline treatment according to previously published procedures (49) to obtain single-stranded DNA (ssDNA).
Plasmid DNA was modified with M. xanthus SW810 CFE as previously described (14). In brief, 10 μg plasmid DNA, prepared from E. coli DH5α, was mixed with SW810 CFE (see above; 290 μg of protein) in a 50-μl methylation reaction solution containing 5 mM EDTA (14). Samples were incubated at 32°C for 5 h and then extracted with phenol-chloroform. Plasmid DNA was precipitated with ethanol and further purified using 1% TAE-agarose gel electrophoresis and a Wizard SV gel and PCR cleanup system (Promega).
Digestion of the M. xanthus genome and plasmid.
A modified digestion assay (42) was employed to test endonuclease activity of the CFE from SW810. Plasmid pZJY41 isolated from E. coli was used as substrate. Two different buffers were used as reaction solutions: digestion I (10 mM Tris, 10 mM MgCl2, pH 8.0) and digestion II (10 mM Tris, 5 mM EDTA, pH 8.0). SW810 CFE (150 μg of protein) was adjusted to 50 μl digestion I (without EDTA) or digestion II (with EDTA) containing 500 ng DNA and incubated at 32°C for 2 h. The products were directly analyzed by gel electrophoresis in 1% TAE-agarose with ethidium bromide.
To test the resistance of genomic and plasmid DNA to certain restriction enzymes, the DNA samples were digested with HindIII, SalI, PstI, and XbaI (Fermentas) at 37°C for 2 h, respectively. In 50 μl digestion buffer (Fermentas), 20 units of enzyme was added together with 500 ng plasmid DNA or 4 μg genomic DNA. All products were analyzed by gel electrophoresis in 1% TAE-agarose with ethidium bromide.
Plasmid transformation of M. xanthus SW810 (ΔepsA) under different conditions.
To investigate the effect of precultivation time for recipient cells on transformation efficiency, SW810 cells were harvested from 12-, 24-, 48-, and 72-h CYE liquid cultures, respectively, and concentrated to 5 × 109 cells/ml in MMC buffer. Their transformation frequencies with 10 ng/μl pZJY41 (isolated from SW810-41) were determined on MMC medium after 7 days of incubation (see above). To investigate the effect of incubation time of cells with DNA on MMC medium, SW810 cells harvested from 48-h CYE cultures were mixed with 10 ng/μl pZJY41 (isolated from SW810-41) and incubated on MMC agar at 32°C for 3 days, 7 days, 11 days, and 15 days, respectively. Then, the transformation frequency was determined (see above). To test the effect of divalent cations, different concentrations of MgSO4 and CaCl2 were added in resuspension buffer and incubation medium used in the transformation assay described above. Heat (45°C for 5 or 15 min) or UV (30- or 90-s) treatment was applied to SW810 cells before they were mixed with DNA, and the effect of stress conditions on the transformation efficiency was determined.
Western blot assay.
To collect whole-cell protein, 1 × 107 M. xanthus cells were lysed by being boiled in 1% SDS for 10 min. Cell surface proteins from 1 × 109 cells were purified as previously described (66). All samples were analyzed for pilin protein with Western blot assays using anti-PilA antibody (37) according to the published protocol (66).
RESULTS
EPS is an extracellular barrier to plasmid natural transformation of M. xanthus.
The stable E. coli-M. xanthus shuttle plasmid pZJY41, which was originally derived from the Myxococcus autonomously replicating plasmid pMF1 (69), was chosen as donor DNA for studying possible natural transformation of M. xanthus. Our initial attempts to transform wild-type DK1622 cells using various general transformation protocols for other bacteria (1, 28, 44, 50) (data not shown) as well as the transformation assay that we designed as described in Materials and Methods (Fig. 1A) failed. During the course of our experiments, we discovered that EPS of M. xanthus, which is abundant in wild-type strains, has the ability to interact and colocalize with extracellular DNA (Hu and Shi, unpublished). This discovery led to our hypothesis that EPS could affect transformation of M. xanthus.
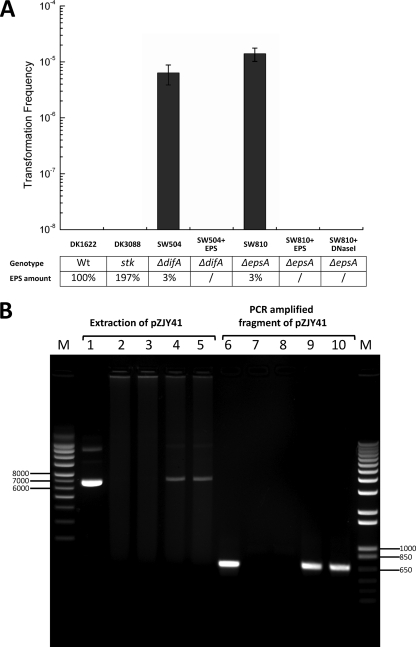
Fig. 1.
The EPS-deficient M. xanthus mutants are transformable by plasmid pZJY41 (isolated from E. coli DH5α) at 100 ng/μl. (A) pZJY41 transformation frequencies of different strains were determined as described in Materials and Methods. Column 1, DK1622 (WT, normal EPS); column 2, DK3088 (stk, EPS overproduction); column 3, SW504 (ΔdifA, EPS deficient); column 4, SW504 cells were premixed with 1 μg/μl EPS before transformation; column 5, SW810 (ΔepsA, EPS deficient); column 6, SW810 cells were premixed with 1 μg/μl EPS; column 7, 5 ng DNase I was added in the transformation system of SW810. The relative EPS amount was determined with a trypan blue binding assay. Results are from three independent experiments, and averages ± standard deviations are plotted. (B) Putative transformants of SW504 (ΔdifA) and SW810 (ΔepsA) transformed by pZJY41 were confirmed by plasmid extraction (lanes 1 to 5) and colony PCR (lanes 6 to 10). Lane 1, pZJY41 plasmid isolated from E. coli DH5α; lanes 2 and 3, plasmid extraction from SW504 and SW810, respectively; lanes 4 and 5, plasmid extraction from transformants of SW504 and SW810, respectively; lane 6, PCR product using PMF-rep as primer and pZJY41 as template; lanes 7 to 10, colony PCR using the same set of primers was performed on SW504 (lane 7), SW810 (lane 8), SW504 putative transformant (lane 9), and SW810 putative transformant (lane 10). Only representative results were shown. Lanes M, supercoiled DNA ladder (left) and 1 Kb Plus DNA ladder (right), both from Invitrogen.
To test our hypothesis, we investigated the interaction between pZJY41 DNA and M. xanthus EPS in vitro. As shown in Table 2, significant reductions of the pZJY41 amount in the supernatant were detected for different EPS-to-DNA ratios after the precipitation of EPS. The following control experiments were performed to confirm the presence of DNA in the EPS pellet precipitated from the buffer containing 100 μg/ml pZJY41. DNA (labeled with Sytox green) was observed in the EPS pellet (labeled with WGA) incubated with the pZJY41 buffer while the EPS-only pellet displayed very little staining by Sytox green (see Fig. S1 in the supplemental material). The DNA signals in the EPS pellet could be drastically reduced by additional DNase I treatment (Fig. S1). Furthermore, several M. xanthus strains with different levels of EPS production were tested for their abilities to take up pZJY41 DNA, which was determined by whether a plasmid-encoded kanamycin resistance marker could be transformed into the cells. Results showed that M. xanthus cells with a normal (100% in DK1622, WT) or an excessive (197% in DK3088, stk) EPS amount did not yield detectable transformants when incubated in the presence of 100 ng/μl pZJY41 DNA isolated from E. coli DH5α (Fig. 1A). However, EPS− strains (SW504-ΔdifA and SW810-ΔepsA) produced putative transformants with an average efficiency of ∼10−5, a level that is more than 1,000 times the limit of detection (Fig. 1A). When the EPS− cells (both ΔdifA and ΔepsA) were premixed with EPS before plasmid DNA was added, no transformants were detected (Fig. 1A). Since DNase terminates or reduces the bacterial transformation through hydrolysis of exogenous DNA (10, 19), we explored the sensitivity of observed M. xanthus transformation to additive DNase I. After 5 ng DNase I was added in the mixture of cells and pZJY41, SW810 (ΔepsA) failed to produce any transformants (Fig. 1A). At the same time, the frequency of spontaneous kanamycin resistance was below the limit of detection (<10−8), as estimated on the same MMC medium in the absence of added pZJY41 DNA. Putative transformants were further analyzed by a colony PCR screen with primers (PMF-rep1 and -2) specific to the rep-pMF1 region of pZJY41 (Table 1). The appearance of PCR products of the predicted size indicated that transformation had occurred in all putative transformants (representative results are shown in lanes 9 and 10 of Fig. 1B). Since pZJY41 is an autonomously replicating plasmid, the natural transformation was further confirmed in the transformants by isolation of the plasmid (representative results are shown in lanes 4 and 5 of Fig. 1B). Our results suggest that EPS may act as an extracellular barrier to natural transformation of M. xanthus by binding exogenous DNA, while M. xanthus cells deficient in EPS production are naturally transformable by the plasmid pZJY41.
Table 2.
Precipitation of plasmid DNA by purified M. xanthus EPS
| EPS (μg/100 μl) | pZJY41 DNAa (μg/100 μl) | Residual DNA (%, ±SD)b |
|---|---|---|
| 0 | 10 | 99.4 ± 1.0 |
| 100 | 10 | 53.2 ± 5.4 |
| 100 | 1 | 32.0 ± 4.3 |
| 100 | 0.1 | 7.7 ± 7.5 |
Isolated from E. coli DH5α.
The amount of residual DNA was calculated as the percentage of DNA remaining in the supernatant compared to the total amount of DNA that was initially added to the sample. Triplicate experiments were conducted.
Transformation of EPS− strain SW810 (ΔepsA) depends on the type of donor DNA.
In bacterial natural transformation, different DNA molecules can act as donors, including plasmids, genome, and PCR products (52). We wanted to determine what type of DNA would be taken up and incorporated most efficiently by SW810 (ΔepsA) cells. Genomic DNA was isolated from M. xanthus strain SW505 (containing a Tn5 insertion with Kmr on difA) (68), and fragments of genomic DNA in different sizes were prepared by partial digestion of the SW505 genome with DNase I. With use of SW505 genomic DNA as the template, PCR DNA (about 5 kb in size) was produced by amplification of the region surrounding the difA gene carrying a Tn5 insertion. In addition, three plasmids were tested: the site-specific integration plasmid pSWU19 isolated from E. coli DH5α (65), the integration plasmid pJW02 (Table 1) isolated from E. coli DH5α, and the autonomously replicating plasmid pZJY41 isolated from E. coli DH5α or SW810-41 (pZJY41 transformant of SW810-ΔepsA).
As shown in Table 3, transformation of SW810 (ΔepsA) with SW505 (difA::Tn5) genomic DNA was not detected, which could be due to the low number of Kmr gene copies within the genomic fragments. In order to increase the number of Kmr gene copies in the donor DNA, a PCR product of difA::Tn5 was used. However, no transformants were detected using either ds or alkaline-denatured ss PCR product as donor, despite the fact that ssDNA has been described as being able to increase the efficiency of homologous recombination in certain bacteria (49). Since the transformation of linear DNA (genomic fragments or PCR product) depended on double crossover events, an integration vector (pJW02) in ds or ss form was used as the donor; still, no transformants were detected. To rule out the chance that the lack of transformants could be due to adverse effects caused by integration of pJW02 into the M. xanthus chromosome, the plasmid was introduced into SW810 (ΔepsA) via electroporation (strain SW2057, Table 1). Typical transformation efficiencies of 5.5 × 103 colonies/μg pJW02 plasmid were obtained. Although plasmid pZJY41 DNA isolated from E. coli gave a relatively high frequency, transformation with ds or ss plasmid pSWU19 DNA (a site-specifically integrative plasmid) was not observed. More interestingly, when used as donor DNA, pZJY41 isolated from M. xanthus SW810-41 was about 600-fold more efficient at transforming the EPS− strain than was pZJY41 isolated from E. coil (Table 3).
Table 3.
Efficiencies of natural transformation of SW810 (ΔepsA) by different donor DNAsa
| DNA formb | Hostc | Pretreatment and description | Mean transformation frequencyd | Range |
|---|---|---|---|---|
| Genomic DNA | ||||
| Genome | SW505 (difA::Tn5) | Untreated | —e | — |
| DNase I digested | >20 kb in size | — | — | |
| 5–20 kb in size | — | — | ||
| <5 kb in size | — | — | ||
| PCR product | 5 kb of difA::Tn5 | Untreated, ds | — | — |
| Alkaline denatured, ss | — | — | ||
| Plasmid DNA | ||||
| pJW02 | DH5α | Untreated, ds | — | — |
| Alkaline denatured, ss | — | — | ||
| Modified by SW810 CFE | — | — | ||
| pSWU19 | DH5α | Untreated, ds | — | — |
| Alkaline denatured, ss | — | — | ||
| Modified by SW810 CFE | 0.9 | 0–3 | ||
| pZJY41 | DH5α | Untreated | 140 | 100–190 |
| SW810-41 | Untreated | 85,000 | 37,000–130,000 | |
| DH5α | Modified by SW810 CFE | 1,100 | 120–2,300 |
In this experiment, the observed transformation frequency could vary by as much as several orders of magnitude, and so both the means and ranges of frequencies were reported.
All the donor DNA was used at 100 ng/μl in the transformation assay.
Represents the host cells from which donor DNA was isolated or the target of the PCR product.
Average of three experiments (×107).
—, not detected.
The R/M system is an inner barrier to natural plasmid transformation of M. xanthus.
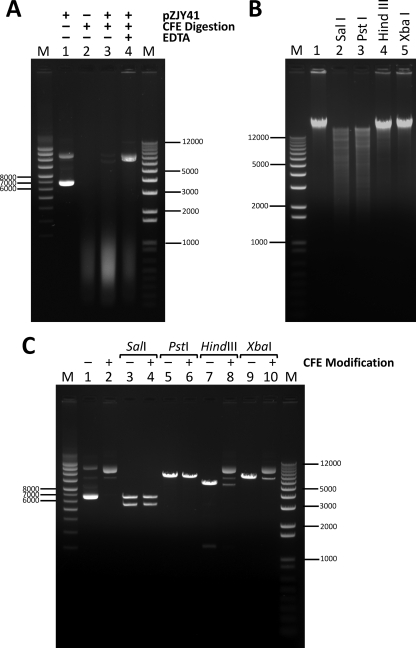
The observation that M. xanthus was more transformable by pZJY41 isolated from M. xanthus SW810-41 than by the same plasmid isolated from E. coli DH5α suggested that the restriction-modification (R/M) system might interfere with the transformation of SW810 (ΔepsA) cells, a phenomenon that has been documented in different bacterial species (47, 59). Considerable restriction endonuclease activities were found in myxobacterial strains (42), and several restriction enzymes and DNA methyltransferases were identified or predicted in M. xanthus DK1622 (WT) (54). In our experiments, the treatment of plasmid pZJY41 isolated from E. coli with SW810 cell extracts (CFE) in digestion buffer I (without EDTA) resulted in rapid degradation (lane 3, Fig. 2A), which was prevented by adding EDTA to the buffer (digestion buffer II, lane 4, Fig. 2A). This was consistent with the finding in Helicobacter pylori that adding EDTA to chelate divalent cations inhibited the nuclease activity of CFE (14). In the presence of EDTA, plasmid pZJY41 was mostly converted from the supercoiled monomer form (covalently closed circular DNA, lane 1, Fig. 2A) to the open circular form (lane 4, Fig. 2A). This change might be due to the activity of nicking enzymes in M. xanthus DK1622 (parent strain of SW810) (54).
Fig. 2.
Restriction-modification systems in M. xanthus SW810 (ΔepsA). (A) Agarose gel electrophoresis of pZJY41 DNA (isolated from E. coli DH5α) incubated with cell extracts (CFE) from SW810 in the digestion buffer without (lane 3) or with (lane 4) 5 mM EDTA. Lane 1, undigested pZJY41; lane 2, SW810 CFE. The absence or presence of plasmid, CFE, and EDTA is indicated above each lane by a minus or plus sign, respectively. (B) Restriction endonuclease digestions of SW810 genome by SalI (lane 2), PstI (lane 3), HindIII (lane 4), and XbaI (lane 5). Lane 1, undigested SW810 genome; lane M, 1 Kb Plus DNA ladder. (C) In vitro methylation of pZJY41 (isolated from E. coli DH5α) by SW810 CFE results in partial resistance to HindIII and XbaI digestion. The absence or presence of CFE treatment in methylation buffer with 5 mM EDTA is indicated above each lane by a minus or plus sign, respectively. Lanes 1 and 2, pZJY41 without or with CFE modification, respectively. Samples to which SalI, PstI, HindIII, or XbaI was added are indicated above the appropriate lanes. Lanes M in panels A and C, supercoiled DNA ladder (left) and 1 Kb Plus DNA ladder (right).
Based on the prototypes of M. xanthus restriction enzymes indicated in the REBASE database (54), four restriction enzymes were chosen to digest genomic DNA of SW810 (ΔepsA). As shown in Fig. 2B, SW810 genomic DNA was sensitive to SalI and PstI (lanes 2 and 3, respectively) and resistant to HindIII and XbaI (lanes 4 and 5, respectively). Since EDTA did not interfere with DNA methyltransferase activity, pZJY41 from E. coli was treated with SW810 CFE in a methylation reaction solution containing 5 mM EDTA (14), and the CFE-modified pZJY41 was extracted (lane 2, Fig. 2C). As shown in Fig. 2C, the untreated pZJY41 was completely digested by all four enzymes (lanes 3, 5, 7, and 9), while the modified pZJY41 was sensitive to digestion by SalI (lane 4) and PstI (lane 6) and partially resistant to digestion by HindIII (lane 8) and XbaI (lane 10). These results suggested that pZJY41 from E. coli modified in vitro by methyltransferases in SW810 CFE would acquire the same pattern of sensitivity/resistance to selected restriction enzymes as that characteristic of SW810 genomic DNA.
Furthermore, transformation frequencies with the SW810 CFE-methylated pZJY41, pJW02, and pSWU19 (all isolated from E. coli DH5α) were determined. As expected, the modified pZJY41 was more effective than untreated plasmid in transforming EPS null strains (about 8-fold increase in frequency [Table 3]). While no transformation was detected with untreated pSWU19, transformants transformed by CFE-modified pSWU19 were observed on the selective plate, albeit with a low frequency (Table 3). These results suggested that the R/M system was an inner barrier to natural plasmid transformation of M. xanthus, which could be overcome by direct isolation of plasmids from M. xanthus strains or modification of plasmids from E. coli with M. xanthus CFE. This finding is consistent with the previous suggestion that variation in plasmid transformation frequency reflects the involvement of DNA R/M mechanisms (24).
Plasmid transformation of M. xanthus SW810 (ΔepsA) under different conditions.
Using the plasmid pZJY41 isolated from M. xanthus as efficient donor DNA, we were able to investigate basic physiological characteristics of plasmid transformation of SW810 (ΔepsA). As shown in Table 4, when provided with the same amounts of donor DNA, the cells collected from liquid CYE medium after cultivation for more than 24 h produced transformants and 48-h-cultivated cells gave the highest transformation frequency. After the cells were mixed with pZJY41, a relatively long incubation on starvation medium seemed to be necessary, since the highest transformation frequency was obtained only after 7 days of incubation on MMC agar (Table 4) and no transformants were detected on nutritious medium, such as CYE agar (data not shown). SW810 (ΔepsA) cells failed to form fruiting bodies due to their EPS deficiency (39), making it unlikely that the induction of competence in M. xanthus cells depended on developmental processes.
Table 4.
Effect of incubation time on efficiency of natural transformation of SW810 (ΔepsA) by plasmid pZJY41a
| Cultured time (h) for the cells as recipientb | Incubation time (days) in transformation assayc | Mean transformation frequencyd | Range |
|---|---|---|---|
| 12 | 7 | —e | — |
| 24 | 7 | 210 | 170–530 |
| 48 | 7 | 7,600 | 3,300–12,000 |
| 72 | 7 | 1,400 | 350–2,700 |
| 48 | 3 | 870 | 170–1,800 |
| 11 | 5,700 | 2,000–9,800 | |
| 15 | 1,200 | 150–3,000 |
Plasmid was isolated from SW810-41 and used at 10 ng/μl in the transformation assay.
Cells were grown in CYE medium at 32°C on a rotary shaker at 300 rpm.
Cells with plasmid pZJY41 were coincubated on MMC medium.
Average of three experiments (×107).
—, not detected.
In some bacteria, transformation is dependent on the presence of divalent cations (50). We investigated the effect of Mg2+ and Ca2+ on the efficiency of transformation of SW810 (ΔepsA) by plasmid pZJY41. As shown in Table 5, SW810 cells were not transformable in the absence of Mg2+ and Ca2+ in buffer and medium. Addition of Ca2+ at a 2 to 6 mM concentration yielded only a very low transformation frequency, which was about 0.5% of the frequency on MMC medium (containing 4 mM Mg2+ and 2 mM Ca2+; Table 5). Addition of 2 mM Mg2+ partially restored transformation up to 80% of the frequency on MMC medium, and 4 or 6 mM Mg2+ gave a transformation with a frequency similar to that on MMC medium, indicating that 4 mM was likely the saturating concentration for Mg2+.
Table 5.
Effect of divalent cations on efficiency of natural transformation of SW810 (ΔepsA) by plasmid pZJY41a
| Transformation medium and bufferb |
Mean transformation frequencyc | Range | |
|---|---|---|---|
| MgSO4 (mM) | CaCl2 (mM) | ||
| 0 | 0 | —d | — |
| 2 | 0 | 5,900 | 1,700–9,300 |
| 4 | 0 | 7,300 | 2,700–11,000 |
| 6 | 0 | 7,100 | 3,600–9,900 |
| 0 | 2 | 36 | 25–53 |
| 0 | 4 | 31 | 16–46 |
| 0 | 6 | 39 | 24–56 |
| 4 | 2 | 7,600 | 3,300–12,000 |
Plasmid was isolated from SW810-41 and used at 10 ng/μl in the transformation assay.
The buffer was used to suspend cells, and the solid medium was used to incubate cells with DNA in the transformation assay (see Materials and Methods). Both contained 10 mM MOPS at pH 7.6.
Average of three experiments (×107).
—, not detected.
There are several reports suggesting that natural transformation in bacteria has evolved as a DNA repair system (15, 45). We investigated the transformability of SW810 (ΔepsA) cells subjected to UV irradiation, a treatment that could induce DNA damage. Heat shock at 45°C was used as a control. As shown in Table 6, both treatments decreased the survival of SW810 cells at different levels after incubation for 7 days on MMC medium. Heat treatment of cells at 45°C for 5 min gave a transformation frequency similar to that of untreated cells. A slight increase in frequency was observed in the 30-s-UV-irradiated cells, while a lower transformation frequency was detected for SW810 cells subjected to 15 min of heat shock at 45°C or 90 s of UV irradiation treatment. Although natural transformation of M. xanthus likely occurs at late stationary phase (Table 4), when the DNA damage and cell death could be detected (46, 56), an obvious correlation between stress conditions and competence induction was not observed.
Table 6.
Effect of heat and UV treatments on efficiency of natural transformation of SW810 (ΔepsA) by plasmid pZJY41a
| Pretreatment of cells | Total no. of cells/ml after incubation for 7 daysc | Mean transformation frequencyd | Range |
|---|---|---|---|
| Heat | |||
| 45°C, 5 min | 2.1 × 107 | 7,400 | 2,700–14,000 |
| 45°C, 15 min | 9.5 × 105 | 2,300 | 680–4,900 |
| UVb | |||
| 30 s | 5.6 × 106 | 8,200 | 1,900–16,000 |
| 90 s | 1.2 × 105 | 5,400 | 1,100–9,200 |
| None | 7.0 × 107 | 7,600 | 3,300–12,000 |
Plasmid was isolated from SW810 transformant and used at 10 ng/μl in the transformation assay.
UV treatment was performed in an XL-1000 UV cross-linker (Fisher).
Total cell number was determined by CFU counting on CYE plates without kanamycin.
Average of three experiments (×107).
Natural transformation of M. xanthus is correlated with certain components of the type IV pilus system.
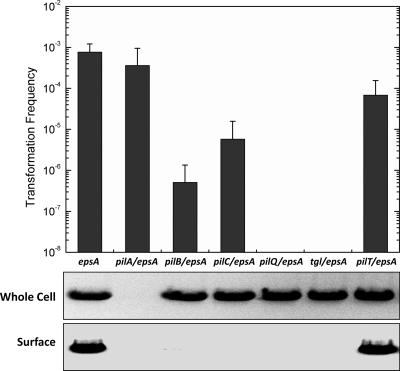
After establishing an efficient natural transformation assay for M. xanthus, we sought to explore the components involved in the DNA uptake apparatus. In most Gram-negative bacteria, transport of exogenous DNA through the outer membrane involves TFP or a TFP-like apparatus (9, 16). In M. xanthus, TFP is the engine to power S motility (65), while its possible involvement in DNA uptake has not been investigated. In order to evaluate the transformability of a group of defined TFP-related null mutants, the epsA gene was inactivated in all the mutants to generate EPS-deficient cells. The TFP biogenesis of the new generated mutants was examined by immunoblotting assays using anti-PilA antibody. As shown in Fig. 3 (panel designated “Whole Cell”), in an immunoblot prepared with whole-cell protein from strains, the 25-kDa band corresponding to PilA was absent from strain SW2051 carrying null mutations of the pilA and epsA genes. PilA was expressed at a level similar to that from the ΔepsA mutant in all the other five double mutants. Immunoblot analysis was also used to detect PilA in surface pili prepared by vortexing (panel designated “Surface” in Fig. 3). Similar levels of expressed surface pili were detected in preparations from ΔepsA and ΔepsA/ΔpilT mutants, but no surface PilA was detected in preparations from all the other five double mutants. These results of double mutants were identical to the PilA expression patterns in epsA, pilA, pilB, pilC, pilQ, pilT, and tgl null single mutants (39, 63, 66), respectively. This indicated that the deletion of the epsA gene did not change the PilA expression and surface pilus biogenesis (assembly of PilA) in this group of TFP-related null mutants.
Fig. 3.
Plasmid pZJY41 (isolated from SW810-41, 10 ng/μl) transformation frequencies of different mutants. Full names of genes represent in-frame deletions of corresponding genes in DK1622 (WT). Results are from three independent experiments, and averages + standard deviations are plotted. The bottom panels show immunoblots of M. xanthus whole-cell protein (top) and sheared-off surface protein (bottom) probed with anti-PilA polyclonal antibody. Lanes are identical to the labels in the x axis of the upper plot.
As shown in Fig. 3 (upper plot), pilA/epsA (SW2051) and epsA (SW810) mutants showed similar transformation frequencies, and the frequency of the pilT/epsA (SW2055) mutant was slightly lower than that of the epsA mutant. A decrease of transformation frequency was observed in both pilB/epsA (SW2052) and pilC/epsA (SW2053) mutants. PilB is an ATPase responsible for PilA assembling to pilus (41), and PilC is a inner membrane protein for the pilus filament's extension in M. xanthus (66). Interestingly, plasmid transformation was abolished in both pilQ/epsA (SW2054) and tgl/epsA (SW2056) mutants. In M. xanthus, PilQ forms a large oligomeric channel through which TFP filaments span the outer membrane (63). The Tgl is required for PilQ assembly, and no PilQ complexes are present in a tgl mutant (48). These results suggest that part of the TFP system may be involved in the DNA uptake machinery of M. xanthus.
DISCUSSION
In the proteobacteria, natural transformation has been discovered in the species of the alpha, beta, gamma, and epsilon classes (27). We demonstrate that M. xanthus cells are able to take up exogenous DNA via natural transformation under specific conditions, a finding which, to our knowledge, provides the first example of a naturally transformable species in the deltaproteobacteria. The prevalence of natural transformation ability in proteobacteria, with the only exception being the zeta lineage (represented by many sequences in environmental small-subunit rRNA genes and infrequent cultured isolates [18]), supports the hypothesis that the ancestor of this monophyletic group may possess this property (27).
Consistent with the notion that barriers against natural transformation are present (47, 59), our study demonstrated that natural transformation of M. xanthus is also affected by some barriers. EPS, as an extracellular component, blocks the natural plasmid transformation of M. xanthus, possibly by interacting with exogenous DNA and lowering its local effective concentration. Restriction, which is an important barrier to bacterial natural transformation (9), has been speculated to eliminate the possibility of HGT in M. xanthus (21). Under our transformation conditions, only the autonomously replicating plasmid pZJY41 could be efficiently transformed into EPS− M. xanthus cells (Table 3). Many plasmids carry R/M systems (34) that potentially affect transfer to new hosts (9), but proteins related to the R/M system were not identified among the products encoded by all 23 predicted open reading frames (ORFs) on pMF1 (parent plasmid of pZJY41) (69). At the same time, a remarkable difference was observed in the transformation frequencies with pZJY41 isolated from E. coli and M. xanthus (Table 3). This was likely due to the modification of plasmid by M. xanthus cells (Fig. 2), which is consistent with the observation that DNA from the same or closely related bacterial species is more efficient in transformation (59). In addition, the incorporation efficiency of incoming DNA in recipient cells also affected transformation frequency in M. xanthus. The donor DNA that integrated into the genome by homologous or site-specific recombination did not generate any transformants under the conditions tested, and only the CFE-modified site-specific recombinant plasmid transformed at a very low frequency. In contrast, plasmid pZJY41 generated a relatively high frequency of transformation, which could be due to its ability to replicate in M. xanthus (69) and therefore avoid recombination into a replicon like the chromosome (59).
In M. xanthus, PilQ belongs to the superfamily of general secretion pathway secretins (51, 60) and forms a membrane-associated channel involved in the extrusion of TFP (63). Tgl is a lipoprotein necessary for PilQ assembly (48). The natural transformation of M. xanthus was totally eliminated in the pilQ and tgl mutants, which is consistent with the observation in several microorganisms that secretins are involved in the DNA uptake process (9). The traffic nucleoside triphosphatase(s) (NTPase[s]) plays important roles in both TFP functions (62) and DNA uptake (9). Two traffic ATPases, PilB and PilT, are required in M. xanthus to assemble the pilin (PilA) subunits into a filament and depolymerize the TFP filament (6, 26), respectively. It was shown that only a mutation in pilB diminished the transformation rate of M. xanthus. In some bacterial species, like Neisseria gonorrhoeae, PilF (equivalent to PilB in M. xanthus) and PilT in the TFP apparatus are both required for DNA uptake (9). However, in other bacteria, like Klebsiella oxytoca (57), the second traffic NTPase (equivalent to PilT in TFP) is absent in the T2SS responsible for DNA uptake (9). PilC is designated a polytopic membrane protein in M. xanthus and is important for the pilus filament's extension outwards from the cytoplasmic membrane (66). The M. xanthus pilC mutant produced natural transformants with a decreased frequency. In N. gonorrhoeae, mutation in pilG (equivalent to pilC in M. xanthus) abolishes the cell's transformation ability (22). Unexpectedly, the deletion of the pilA (pilin) gene did not remarkably affect the transformation frequency of M. xanthus, while the expression of pilin protein is normally essential for DNA uptake in many other bacterial species (9).
Natural transformation has been suggested to be one of the major ways for bacteria to acquire genetic material and to be the driving force in evolution (27). However, there are still many questions that need to be answered before we can understand its real function in the expansion of the genome and ecological diversification of M. xanthus. Since EPS has an inhibiting effect on transformation, in natural habitats, the wild-type (EPS+) M. xanthus may use the EPS layer as protection to prevent the uptake of foreign DNA fragments. Another possibility would be that M. xanthus cells regulate and reduce their EPS amount to be transformable. However, while some studies have indicated that EPS production is regulated under different growth conditions (33), nothing is known about similar processes in Myxococcus bacteria in their natural environments. The existence of a complicated R/M system in M. xanthus limits the source of donor DNA to similar bacterial species, which is consistent with the finding that much of the genome expansion of M. xanthus is specific to the lineage of myxobacteria (21). In this study, only the plasmid pZJY41 has yielded efficient transformation, and it is important to note that the existence of an autonomously replicating plasmid in Myxococcus is very rare (69). The donor DNA that can partake in M. xanthus transformation in natural environments is still unknown.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mitch Singer, Lawrence Shimkets, Daniel Wall, and Dale Kaiser for providing bacterial strains and Hongwei Pan and Zhe Yang for helpful discussion, as well as Muhaiminu Hossain for careful review of the manuscript. We also thank three anonymous reviewers for their constructive comments.
This work was supported by U.S. National Institutes of Health grant GM54666 (to W.S.) and Chinese National Natural Science Foundation grant 30870020 (to W.H.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Aas F. E., Lovold C., Koomey M. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46:1441–1450 [DOI] [PubMed] [Google Scholar]
- 2. Averhoff B., Friedrich A. 2003. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 180:385–393 [DOI] [PubMed] [Google Scholar]
- 3. Avery L., Kaiser D. 1983. Construction of tandem genetic duplications with defined endpoints in Myxococcus xanthus. Mol. Gen. Genet. 191:110–117 [DOI] [PubMed] [Google Scholar]
- 4. Black W. P., Yang Z. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 186:1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breton A. M., Jaoua S., Guespin-Michel J. 1985. Transfer of plasmid RP4 to Myxococcus xanthus and evidence for its integration into the chromosome. J. Bacteriol. 161:523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bulyha I., et al. 2009. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol. Microbiol. 74:691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campos J. M., Geisselsoder J., Zusman D. R. 1978. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J. Mol. Biol. 119:167–178 [DOI] [PubMed] [Google Scholar]
- 8. Chang B. Y., Dworkin M. 1994. Isolated fibrils rescue cohesion and development in the Dsp mutant of Myxococcus xanthus. J. Bacteriol. 176:7190–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen I., Dubnau D. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241–249 [DOI] [PubMed] [Google Scholar]
- 10. Claverys J. P., Martin B., Polard P. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33:643–656 [DOI] [PubMed] [Google Scholar]
- 11. Dana J. R., Shimkets L. J. 1993. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J. Bacteriol. 175:3636–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. den Hartigh A. B., et al. 2004. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect. Immun. 72:5143–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diodati M. E., Gill R. E., Plamann L., Singer M. 2008. Initiation and early developmental events, p. 43–76 In Whitworth D. E. (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 14. Donahue J. P., Israel D. A., Peek R. M., Blaser M. J., Miller G. G. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37:1066–1074 [DOI] [PubMed] [Google Scholar]
- 15. Dorer M. S., Fero J., Salama N. R. 2010. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 6:e1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dubnau D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217–244 [DOI] [PubMed] [Google Scholar]
- 17. Dworkin M. 1973. Cell-cell interactions in myxobacteria. Symp. Soc. Gen. Microbiol. 23:125–147 [Google Scholar]
- 18. Emerson D., et al. 2007. A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2:e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaasbeek E. J., et al. 2010. Nucleases encoded by the integrated elements CJIE2 and CJIE4 inhibit natural transformation of Campylobacter jejuni. J. Bacteriol. 192:936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldman B., Bhat S., Shimkets L. J. 2007. Genome evolution and the emergence of fruiting body development in Myxococcus xanthus. PLoS One 2:e1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldman B. S., et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamilton H. L., Dillard J. P. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376–385 [DOI] [PubMed] [Google Scholar]
- 23. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 24. Heuermann D., Haas R. 1995. Genetic organization of a small cryptic plasmid of Helicobacter pylori. Gene 165:17–24 [DOI] [PubMed] [Google Scholar]
- 25. Hodgkin J., Kaiser D. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 171:177–191 [Google Scholar]
- 26. Jakovljevic V., Leonardy S., Hoppert M., Sogaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnsborg O., Eldholm V., Havarstein L. S. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767–778 [DOI] [PubMed] [Google Scholar]
- 28. Johnsborg O., Havarstein L. S. 2009. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 33:627–642 [DOI] [PubMed] [Google Scholar]
- 29. Julien B., Kaiser A. D., Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaiser D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45–54 [DOI] [PubMed] [Google Scholar]
- 31. Kaplan H. B. 2003. Multicellular development and gliding motility in Myxococcus xanthus. Curr. Opin. Microbiol. 6:572–577 [DOI] [PubMed] [Google Scholar]
- 32. Kashefi K., Hartzell P. L. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF- defect. Mol. Microbiol. 15:483–494 [DOI] [PubMed] [Google Scholar]
- 33. Kim S. H., Ramaswamy S., Downard J. 1999. Regulated exopolysaccharide production in Myxococcus xanthus. J. Bacteriol. 181:1496–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kovacs A. T., Smits W. K., Mironczuk A. M., Kuipers O. P. 2009. Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries. Environ. Microbiol. 11:1911–1922 [DOI] [PubMed] [Google Scholar]
- 36. Kuner J. M., Kaiser D. 1982. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J. Bacteriol. 151:458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Lux R., Pelling A. E., Gimzewski J. K., Shi W. 2005. Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili. Microbiology 151:353–360 [DOI] [PubMed] [Google Scholar]
- 38. Li Y., et al. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu A., et al. 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 55:206–220 [DOI] [PubMed] [Google Scholar]
- 40. Lux R., Li Y., Lu A., Shi W. 2004. Detailed three-dimensional analysis of structural features of Myxococcus xanthus fruiting bodies using confocal laser scanning microscopy. Biofilms 1:293–303 [Google Scholar]
- 41. Mauriello E. M., Zusman D. R. 2007. Polarity of motility systems in Myxococcus xanthus. Curr. Opin. Microbiol. 10:624–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mayer H., Reichenbach H. 1978. Restriction endonucleases: general survey procedure and survey of gliding bacteria. J. Bacteriol. 136:708–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McBride M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49–75 [DOI] [PubMed] [Google Scholar]
- 44. Meibom K. L., Blokesch M., Dolganov N. A., Wu C. Y., Schoolnik G. K. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 45. Michod R. E., Wojciechowski M. F., Hoelzer M. A. 1988. DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics 118:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nariya H., Inouye M. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55–66 [DOI] [PubMed] [Google Scholar]
- 47. Nielsen K. M. 1998. Barriers to horizontal gene transfer by natural transformation in soil bacteria. APMIS Suppl. 84:77–84 [DOI] [PubMed] [Google Scholar]
- 48. Nudleman E., Wall D., Kaiser D. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60:16–29 [DOI] [PubMed] [Google Scholar]
- 49. Oh S. H., Chater K. F. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palmen R., Vosman B., Buijsman P., Breek C. K., Hellingwerf K. J. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295–305 [DOI] [PubMed] [Google Scholar]
- 51. Peabody C. R., et al. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072 [DOI] [PubMed] [Google Scholar]
- 52. Pollack-Berti A., Wollenberg M. S., Ruby E. G. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol. 12:2302–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reichenbach H., Dworkin M. 1992. The myxobacteria, p. 3416–3487 In Balows A., Truper H. G., Dworkin M., Harder W., Schleifer K. H. (ed.), The prokaryotes. Springer-Verlag, New York, NY [Google Scholar]
- 54. Roberts R. J., Vincze T., Posfai J., Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234–D236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosenberg E., Baron M. 1984. Antibiotics and lytic enzymes, p. 109–125 In Rosenberg E. (ed.), Myxobacteria. Springer-Verlag, New York, NY [Google Scholar]
- 56. Rosenbluh A., Rosenberg E. 1993. Developmental lysis and autocides, p. 213–234 In Dworkin M., Kaiser D. (ed.), Myxobacteria II. American Society for Microbiology, Washington, DC [Google Scholar]
- 57. Sandkvist M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271–283 [DOI] [PubMed] [Google Scholar]
- 58. Shimkets L. J., Gill R. E., Kaiser D. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. U. S. A. 80:1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thomas C. M., Nielsen K. M. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
- 60. Trindade M. B., Job V., Contreras-Martel C., Pelicic V., Dessen A. 2008. Structure of a widely conserved type IV pilus biogenesis factor that affects the stability of secretin multimers. J. Mol. Biol. 378:1031–1039 [DOI] [PubMed] [Google Scholar]
- 61. Ueki T., Inouye S., Inouye M. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153–157 [DOI] [PubMed] [Google Scholar]
- 62. Wall D., Kaiser D. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1–10 [DOI] [PubMed] [Google Scholar]
- 63. Wall D., Kolenbrander P. E., Kaiser D. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wall D., Wu S. S., Kaiser D. 1998. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J. Bacteriol. 180:759–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu S. S., Kaiser D. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547–558 [DOI] [PubMed] [Google Scholar]
- 66. Wu S. S., Kaiser D. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu S. S., Wu J., Kaiser D. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109–121 [DOI] [PubMed] [Google Scholar]
- 68. Yang Z., Geng Y., Xu D., Kaplan H. B., Shi W. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123–1130 [DOI] [PubMed] [Google Scholar]
- 69. Zhao J. Y., et al. 2008. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl. Environ. Microbiol. 74:1980–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.