Abstract
Deletion of Mycobacterium marinum MMAR2333 resulted in the loss of three of four subclasses of lipooligosaccharides (LOSs). The mutant was unable to extend an intermediate (LOS-II*) by addition of caryophyllose. These data and the predicted domain structure suggest that MMAR2333 is a glycosyltransferase involved in the generation of a lipid-linked caryophyllose donor.
TEXT
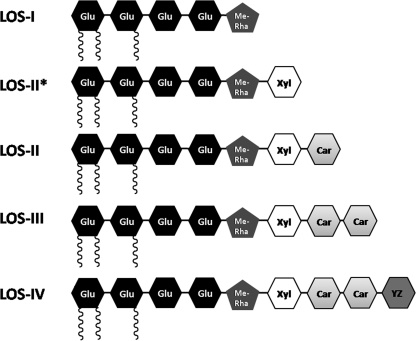
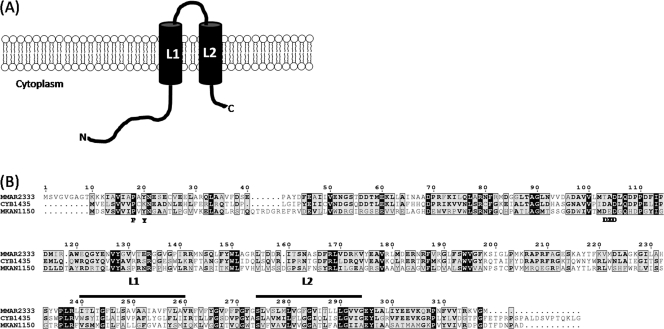
The cell envelope of members of the genus Mycobacterium, including the tubercle bacillus Mycobacterium tuberculosis, consists of a characteristic lipid-rich cell wall that constitutes an effective permeability barrier, imparting resistance to many therapeutic agents (3), and contributes to virulence (10). This distinct cell envelope consists of a covalently linked mycolyl-arabinogalactan-peptidoglycan (mAGP) complex, which in turn intercalates with various noncovalently bound complex lipids that vary between different mycobacterial species and within strains of the same species. These include the highly polar lipooligosaccharides (LOSs), which are produced by a number of mycobacteria, including the opportunistic pathogen Mycobacterium kansasii, the poikilotherm pathogen Mycobacterium marinum, and the M. tuberculosis complex strain Mycobacterium canetti (4, 11, 16). Studies on LOS-deficient M. kansasii strains, M. marinum LOS mutants, and purified LOSs also suggest a role in virulence (6, 19, 20). The vast majority of biochemical and genetic information on LOS biosynthesis comes from studies on M. marinum, which produces four types of LOSs (4), LOS-I, LOS-II, LOS-III, and LOS-IV, each containing a common acylated glycan core consisting of four glucose residues and one methylated rhamnose (Fig. 1). A d-Xylp (xylopyranase) residue is present in LOS-II, LOS-III, and LOS-IV in addition to the glycan core. LOS-II contains a further caryophyllose sugar [3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose; previously referred to as sugar X] (20), while LOS-III and LOS-IV contain two caryophyllose residues, with LOS-IV containing a novel N-acetylated dideoxy galactose (4,6-dideoxy-Galp replaced by a 3-hydroxy-3-methylated-pyrrolidone cycle; previously referred to as the “YZ” component) (21). Whether LOSs from other mycobacteria such as M. canetti also contain caryophyllose, the YZ sugar, or related components remains to be determined. Additionally, Rombouts et al. (20) identified further subclasses of LOS-II, LOS-III, and LOS-IV in M. marinum which contained a hydroxylated equivalent of caryophyllose. Transposon mutagenesis screens and studies on a natural strain with altered LOS patterns have revealed a genetic locus associated with LOS biosynthesis (4, 19, 20). Given the sugar-rich composition of LOSs, numerous glycosyltransferases potentially play a role in LOS biosynthesis pathways. Until now, losA (MMAR2313) was the only glycosyltransferase-encoding gene shown to be involved in LOS biosynthesis in M. marinum; a losA transposon mutant failed to synthesize LOS-IV, suggesting that LosA was likely involved in the transfer of the terminal N-acetylated dideoxy galactose residue to LOS-III, resulting in the formation of LOS-IV (4). Also present in this gene cluster is MMAR2333, encoding a putative glycosyltransferase. Initially annotated as wcaA (a glycosyltransferase gene involved in the biosynthesis of the exopolysaccharide colanic acid in enteric bacteria [23]), the encoded protein is predicted to contain two transmembrane domains located near the C terminus (Fig. 2), suggesting that the enzyme is membrane anchored, an attribute of many mycobacterial glycosyltransferases involved in the biosynthesis of cell wall-associated glycolipids and carbohydrate polymers. Additionally, the MMAR2333 sequence revealed domains characteristic of eukaryotic dolichol phosphate mannose (DPM) synthases (Fig. 2), members of the GT-2 family of glycosyltransferases that catalyze the transfer of sugars to dolichol phosphate by using nucleotide sugars as substrates. In bacteria, homologues of DPM synthases use polyprenol phosphate rather than dolichol phosphate, and one such enzyme from M. tuberculosis is Ppm1, which catalyzes the generation of polyprenol monophosphomannose from GDP-mannose and polyprenolphosphate for subsequent use as substrates for biosynthesis of lipomannan and lipoarabinomannan (15). Surprisingly, the best matches obtained from a BLAST search using the MMAR2333 amino acid sequence as the query were putative glycosyltransferases from cyanobacteria, which showed a higher homology to MMAR2333 than glycosyltransferases from other LOS-producing mycobacteria (the best match is shown in Fig. 2). Given the proximity of MMAR2333 to genes involved in LOS biosynthesis and its similarity to DPM synthases, it was likely that the MMAR2333-encoded glycosyltransferase was involved in the generation of a lipid-bound sugar moiety, utilized subsequently as a donor for the addition of one or more sugar residues found in M. marinum LOSs.
Fig. 1.
Schematic representation of four subclasses of LOSs and one LOS intermediate (LOS-II*) produced by M. marinum. Glu, glucose; Me-Rha, methyl rhamnose; Xyl, xylose; Car, caryophyllose; YZ, pyrrolidone-substituted dideoxy galactose. Acyl chains are shown as black coils.
Fig. 2.
(A) Predicted topology of MMAR2333. The predicted transmembrane domains are denoted by L1 and L2. N, N terminus; C, C terminus. (B) Alignment of the MMAR2333 amino acid sequence with that from the putative Synechococcus sp. glycosyltransferase (CYB1435) and the Mycobacterium kansasii homologue MKAN1150. Characteristic sugar binding residues are indicated below the alignment. The sequences spanning the transmembrane domains of MMAR2333, depicted as L1 and L2 in panel A, are indicated by bars above the sequences. Numbers are amino acid coordinates of MMAR2333. Black boxes show identity for all three proteins. Grey boxes indicate similar or identical residues for two of the three proteins; bold type indicates identical or similar residues.
Generation of an M. marinum MMAR2333 null mutant.
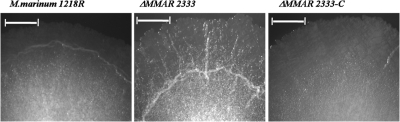
To determine whether MMAR2333 played a role in LOS biosynthesis in M. marinum, we first generated an allelic exchange plasmid, pΔMMAR2333, which consisted of PCR-amplified flanks upstream and downstream of MMAR2333 cloned on either side of a hygromycin resistance cassette (hyg) in the vector p004S (17). pΔMMAR2333 was then packaged in the temperature-sensitive mycobacteriophage phAE159 to generate a recombinant phage, phΔMMAR2333, designed for replacement of MMAR2333 with hyg by using specialized transduction, a highly efficient phage delivery-based knockout method for mycobacteria (2). Hygromycin-resistant (75-μg/ml) colonies obtained after transduction of M. marinum 1218R (ATCC 927; referred to as the wild type) using protocols described by Larsen et al. (17) were confirmed by Southern blot analysis, and one such strain, the ΔMMAR2333 mutant, was used for further analysis. The mutant strain exhibited an altered colony morphology with a slightly “rough” appearance (Fig. 3), suggesting potential alterations in the cell wall. The colony morphology was restored to that of the parental type upon introduction of a plasmid-borne copy of MMAR2333 into ΔMMAR2333, indicating that the phenotype observed in the mutant was due solely to the loss of MMAR2333 function (Fig. 3; ΔMMAR2333-C contains a cloned copy of MMAR2333 in the mycobacterial replicative plasmid pMV261 [24] delivered by electroporation). This study also demonstrated the utility of specialized transduction to generate null mutants of M. marinum. While phage delivery methods have been used to generate transposon mutants in M. marinum (1, 19), this is the first report of the use of phages for delivering allelic exchange substrates for targeted gene knockouts in this species. This tool should facilitate the generation of targeted mutants in M. marinum, which has relied so far on laborious two-step selection methods based on sacB counterselection (8, 14, 18).
Fig. 3.
Effect of deletion of MMAR2333 on colony morphology. Colonies of M. marinum wild-type (1218R), mutant (ΔMMAR2333), and complemented (ΔMMAR2333-C) strains on 7H10 agar plates. Bar, 1 mm.
Altered LOS profiles in the ΔMMAR2333 mutant.
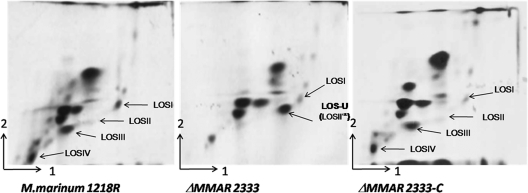
To assess the effects of the loss of MMAR2333 function on cell wall lipid composition, cultures of wild-type, mutant, and complemented strains grown at 30°C in 7H10 broth (Middlebrook 7H10 minus agar) were first pulsed with 1 μCi/ml [14C]acetate (57 mCi/mmol) to label lipids. Labeled polar and apolar lipids were extracted and analyzed by two-dimensional thin-layer chromatography (2D-TLC) using five solvent systems (A to E) as described by Dobson et al. (12). Differences in the lipid profiles of the wild-type and ΔMMAR2333 strains were visible only in TLCs run in solvent system E, which is designed to separate phospholipids and LOSs (Fig. 4). While LOS-I was present in the ΔMMAR2333 mutant, LOS-II, LOS-III, and LOS-IV were missing and the strain accumulated instead a 14C-labeled species that migrated to a position between that of LOS-I and LOS-II (Fig. 4). Complete LOS biosynthesis was restored in the complemented strain, indicating that the changes observed in the mutant were due solely to the loss of MMAR2333. Staining of solvent system E TLC plates of lipids extracted from the mutant strain with α-naphthol revealed that the new accumulating lipid species was a glycolipid (data not shown). As the appearance of this glycolipid was paralleled by the disappearance of LOS-II, -III, and -IV, it was quite likely that the accumulated glycolipid was a LOS intermediate, and we thus termed this unidentified lipid LOS-U (LOS-unknown).
Fig. 4.
2D-TLC analysis of 14C-labeled polar lipids from M. marinum strains. Polar lipids were extracted from wild-type (1218R), mutant (ΔMMAR2333), and complemented (ΔMMAR2333-C) strains, separated using solvent system E (direction 1, CHCl3:CH3OH:H2O [60:30:6]; direction 2, CHCl3:CH3COOH:CH3OH:H2O [40:25:3:6]), and visualized on X-ray films by autoradiography. LOSI, LOS-I.
Characterization of LOS-U.
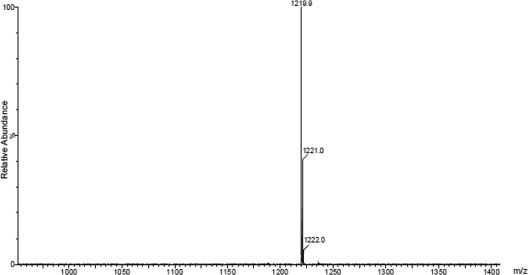
Another LOS biosynthesis intermediate, LOS-II*, was also reported to migrate to an intermediate position between LOS-I and LOS-II on 2D-TLC plates (19). Isolated from M. marinum MRS1178, a transposon mutant of MMAR2332, LOS-II* is a precursor of LOS-II and contains d-Xylp attached to the glycan core but lacks the caryophyllose found in LOS-II. When mixed samples of 14C-labeled lipids from the MRS1178 and ΔMMAR2333 strains were separated on the same 2D-TLC plate, LOS-II* and LOS-U migrated to the same position, appearing as one spot (data not shown), suggesting that LOS-U and LOS-II* were likely the same glycolipid species. In order to ascertain this by determining the chemical nature of LOS-U, we first purified LOS-U using a combination of column chromatography and preparative TLC, and per-O-methylated LOS-U was analyzed by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) as described earlier for other LOS subclasses (4). A prominent signal was obtained at m/z ∼1,219.9 [M + Na]+ (Fig. 5), which was within the same range (m/z 1,219 ± 1) as that obtained for LOS-II* (m/z 1,219.4) (19). These data indicated that LOS-U, the intermediate we isolated from the ΔMMAR2333 strain, was the LOS-II precursor LOS-II*, and like the MMAR2332 transposon mutant, the MMAR2333 deletion mutant also accumulated LOS-II*. In other words, the addition of the first caryophyllose residue to the d-Xylp glycan core did not occur in the ΔMMAR2333 mutant. Given the similarity of MMAR2333 to DPM-like glycosyltransferases, these results suggest that MMAR2333 was likely involved in the cytoplasmic transfer of a nucleotide-bound caryophyllose residue (or its precursor) to a polyprenol phosphate or other lipid-based unit for subsequent use as a sugar donor by another glycosyltransferase to extend LOS-II* to LOS-II. Additionally, the identical LOS patterns of the MMAR2332 transposon mutant and the ΔMMAR2333 strain on 2D-TLC plates made it likely that MMAR2332, which encodes a putative protein homologous to thiamine pyrophosphate (TPP)-requiring carboxylases, was involved in the biosynthesis of the unique caryophyllose sugar.
Fig. 5.
Estimation of the molecular size of LOS-U by MALDI-MS analysis of per-O-methylated LOS-U. The sample was prepared and processed as described by Ren et al. (19).
Effects of MMAR2333 deletion on virulence.
Transposon-mediated disruption of MMAR2332, which also led to the accumulation of LOS-II*, did not alter the ability to survive inside cultured macrophages (19). Similarly, we did not observe any differences in the ability of the ΔMMAR2333 strain to survive in murine bone marrow-derived macrophages (data not shown). Additionally, we did not observe any differences in bacterial loads between zebrafish embryos infected with wild-type and mutant strains (see the supplemental material), suggesting that the loss of LOS-II, LOS-III, and LOS-IV and the parallel accumulation of LOS-II* in the mutant strain did not affect the survival of the mutant strain in the above-described models of infection.
Later stages of LOS biosynthesis may occur in an extracytoplasmic environment.
The similarity of MMAR2333 to bacterial DPM-like synthases suggested that the glycosyltransferase was not directly involved in the transfer of caryophyllose to LOS-II* but instead was likely to catalyze the transfer of nucleotide-bound caryophyllose to a lipid (polyprenol) carrier. Alternately, MMAR2333 could catalyze the formation of a polyprenol-bound precursor of caryophyllose, which is subsequently modified to caryophyllose. Efforts to confirm this by in vitro enzyme assays are currently limited by the unavailability of nucleotide-bound caryophyllose substrates. In addition to MMAR2333, two other genes, MMAR2311 and MMAR2313 (losA), also encode DPM-like glycosyltransferases. The presence of this class of glycosyltransferases in the LOS biosynthesis cluster suggests that some glycosyltransferases may be involved in the generation of lipid-bound sugar substrates which are “flipped” to the extracytoplasmic side of the membrane to be subsequently used by extracytoplasmic glycosyltransferases to extend the oligosaccharide moiety of LOSs. Indeed, the losA mutant is devoid of LOS-IV, suggesting that LosA could likely be involved in the generation of a lipid-bound YZ sugar which is subsequently transferred by an extracellular glycosyltransferase to LOS-III. This affords a model for LOS biosynthesis wherein acylated hexasaccharide comprising LOS-II* is synthesized intracellularly and transported across the membrane. This process could be initiated by MMAR2342, which encodes a transmembrane protein belonging to a group of larger mycobacterial proteins, termed MmpL proteins, which are involved in the transport of mycobacterial glycolipids or their intermediates (5, 7, 9, 13, 22). On the extracytoplasmic side, LOS-II* would then be extended by specific glycosyltransferases that use lipid-bound sugars (caryophyllose or the YZ sugar) as sugar donors to yield LOS-II, LOS-III, and LOS-IV. In an alternative model, LOS-I could be transported by the MmpL protein, with xylose being the first sugar added on the extracytoplasmic side, a process that would require the generation of a lipid-bound xylose substrate. MMAR2311, the third putative DPM-like glycosyltransferase in the LOS cluster, is a potential candidate for this function. In summary, later stages of LOS biosynthesis may involve a distinct set of glycosyltransferases that catalyzes the formation of lipid-bound sugar donors and another set that extends the LOSs. The generation of mutants of MMAR2311 and other putative glycosyltransferase genes in the LOS cluster will shed more light on the biosynthesis of these carbohydrate-rich mycobacterial glycolipids.
Supplementary Material
Acknowledgments
A.B. and G.S.B. acknowledge support from the Medical Research Council (United Kingdom) and the Wellcome Trust. G.S.B. also acknowledges support in the form of a Personal Research Chair from James Bardrick and a Royal Society Wolfson Research Merit Award. D.S. and J.C. are funded by a Ph.D. studentship from the Darwin Trust of Edinburgh.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Alexander D. C., Jones J. R., Tan T., Chen J. M., Liu J. 2004. PimF, a mannosyltransferase of mycobacteria, is involved in the biosynthesis of phosphatidylinositol mannosides and lipoarabinomannan. J. Biol. Chem. 279:18824–18833 [DOI] [PubMed] [Google Scholar]
- 2. Bardarov S., et al. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 3. Brennan P. J., Nikaido H. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29–63 [DOI] [PubMed] [Google Scholar]
- 4. Burguiere A., et al. 2005. LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 280:42124–42133 [DOI] [PubMed] [Google Scholar]
- 5. Camacho L. R., et al. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845–19854 [DOI] [PubMed] [Google Scholar]
- 6. Collins F. M., Cunningham D. S. 1981. Systemic Mycobacterium kansasii infection and regulation of the alloantigenic response. Infect. Immun. 32:614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Converse S. E., et al. 2003. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. U. S. A. 100:6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cosma C. L., Klein K., Kim R., Beery D., Ramakrishnan L. 2006. Mycobacterium marinum Erp is a virulence determinant required for cell wall integrity and intracellular survival. Infect. Immun. 74:3125–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83 [DOI] [PubMed] [Google Scholar]
- 10. Daffe M., Draper P. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131–203 [DOI] [PubMed] [Google Scholar]
- 11. Daffe M., McNeil M., Brennan P. J. 1991. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30:378–388 [DOI] [PubMed] [Google Scholar]
- 12. Dobson G., et al. 1985. Systematic analysis of complex mycobacterial lipids, p. 237–265 In Goodfellow M., Minnikin D. E. (ed.), Chemical methods in bacterial systematics. Academic Press, London, United Kingdom [Google Scholar]
- 13. Domenech P., et al. 2004. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279:21257–21265 [DOI] [PubMed] [Google Scholar]
- 14. Gao L. Y., et al. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677–1693 [DOI] [PubMed] [Google Scholar]
- 15. Gurcha S. S., et al. 2002. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem. J. 365:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunter S. W., Murphy R. C., Clay K., Goren M. B., Brennan P. J. 1983. Trehalose-containing lipooligosaccharides. A new class of species-specific antigens from Mycobacterium. J. Biol. Chem. 258:10481–10487 [PubMed] [Google Scholar]
- 17. Larsen M. H., Biermann K., Tandberg S., Hsu T., Jacobs W. R., Jr 2007. Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. 10:A.2.1–A.2.21 [DOI] [PubMed] [Google Scholar]
- 18. Ramakrishnan L., Federspiel N. A., Falkow S. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436–1439 [DOI] [PubMed] [Google Scholar]
- 19. Ren H., et al. 2007. Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 63:1345–1359 [DOI] [PubMed] [Google Scholar]
- 20. Rombouts Y., et al. 2009. Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-alpha secretion in macrophages. J. Biol. Chem. 284:20975–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rombouts Y., et al. 2010. Structural analysis of an unusual bioactive N-acylated lipo-oligosaccharide LOS-IV in Mycobacterium marinum. J. Am. Chem. Soc. 132:16073–16084 [DOI] [PubMed] [Google Scholar]
- 22. Sonden B., et al. 2005. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 58:426–440 [DOI] [PubMed] [Google Scholar]
- 23. Stevenson G., Andrianopoulos K., Hobbs M., Reeves P. R. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stover C. K., et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.