Abstract
Many Gram-negative enterobacteria produce surface-associated fimbriae that facilitate attachment and adherence to eucaryotic cells and tissues. These organelles are believed to play an important role during infection by enabling bacteria to colonize specific niches within their hosts. One class of these fimbriae is assembled using a periplasmic chaperone and membrane-associated scaffolding protein that has been referred to as an usher because of its function in fimbrial biogenesis. The presence of multiple types of fimbriae assembled by the chaperone/usher pathway can be found both within a single bacterial species and also among different genera. One way of controlling fimbrial assembly in these bacteria is at the genetic level by positively or negatively regulating fimbrial gene expression. This minireview considers the mechanisms that have been described to control fimbrial gene expression and uses specific examples to demonstrate both unique and shared properties of such regulatory mechanisms.
TEXT
Gram-negative enteric bacteria produce both fimbrial and nonfimbrial adherence factors that play a major role in facilitating attachment to biotic and abiotic surfaces (41, 42, 80). The presence of surface-associated fimbriae in these bacteria is due to one of three assembly pathways that have been described (8, 27, 64, 67, 71). Type IV pili, produced by a range of bacteria, are characterized by the presence of a modified amino acid at the N terminus of the major pilin subunit (25, 92). This subunit is produced within the bacterial cell as a prepilin polypeptide that undergoes proteolytic cleavage by a specific prepilin peptidase prior to assembly into a mature appendage (3). The assembly of type IV pili requires numerous additional pilus gene products, including an ATPase and inner and outer membrane proteins, and the assembly apparatus appears to be related to the type II secretion system described for enteric bacteria (4, 6, 19, 47). The second assembly pathway has been described as the nucleation pathway and is used by those bacteria that produce curli and curli-related appendages on their surface (8). Initiation of assembly is facilitated by a nucleation protein that allows the extracellular soluble subunit protein to be polymerized on the bacterium following secretion of the subunit protein. The secretion of the major subunit protein is mediated by a specific secretion pathway that also does not allow the subunits to be polymerized intracellularly (8, 32). The third pathway used by bacteria to assemble fimbriae is referred to as the chaperone/usher assembly pathway. This pathway is commonly used by a wide range of enterobacteria to produce functional adhesins that play a role in bacteria-host cell interactions. The pathway was first described in detail and characterized by Hultgren and coworkers (80), who studied the P pilus and type 1 fimbrial systems of uropathogenic Escherichia coli, and it has subsequently been shown to be present and utilized for the assembly of many different fimbrial types (67). The pathway is characterized by the presence of two genes within the relevant fimbrial gene cluster that encode a periplasmic chaperone and an outer membrane scaffolding protein. These two gene products are required for the ordered assembly of a functional adhesive organelle. In addition to the gene encoding the chaperone and the scaffolding protein, the fimbrial gene clusters of this family of fimbriae also possess determinants encoding the structural components of the fimbriae, including, in some cases, separate adaptor and adhesin gene products. Fimbriae assembled by the chaperone/usher pathway are frequently not constitutively produced but are assembled only under specific environmental conditions. Investigations into the production of these fimbriae have revealed that the regulation of the genes encoding the major fimbrial subunits is a key step in this process. However, studies by numerous groups on different fimbrial gene clusters have also demonstrated that the regulons, and their component parts, of fimbria production can be quite diverse. Consequently, over the last several years it has emerged that regulation of the genes encoding the major fimbrial subunits of those appendages assembled by the chaperone/usher pathway is comprised of regulatory pathways that in some cases are shared among different bacterial species but also are limited to specific fimbrial types. The currently identified regulatory mechanisms involved in controlling the expression of fimbriae assembled using the chaperone/usher pathway are the subject of this review. We describe below some of the common regulatory mechanisms that have been described and provide some specific examples. It is beyond the scope of this review to cite all fimbrial systems and their regulatory mechanisms, but we have aimed to use a few fimbrial types to illustrate both the diversity of regulation and also common features involved in fimbrial gene regulation. Table 1 provides a more comprehensive list of known regulatory proteins that have been implicated in affecting genes that comprise determinants encoding fimbriae assembled by the chaperone/usher pathway.
Table 1.
Known regulators of chaperone/usher fimbrial systems
| Mechanism of regulation | Fimbria type | Known regulator(s) | Reference(s) |
|---|---|---|---|
| Invertible DNA elements | E. coli CS18 | FotST | 37 |
| E. coli Fim | FimBE, IcsR, Lrp, IHF | 1, 2, 10, 26, 31, 93 | |
| H-NS | |||
| K. pneumoniae Fim | FimK | 74 | |
| K. pneumoniae Kpc | KpcI | 91 | |
| P. mirabilis Mrp | MrpI | 51, 54, 96 | |
| DNA methylation | E. coli Pap | PapI, Lrp | 34, 69, 70 |
| E. coli K88 | FaeA, Lrp | 38, 39 | |
| E. coli K99 | Mbf | 18 | |
| E. coli Sfa | SfaX | 77, 78, 88 | |
| S. Typhimurium Pef | PefI | 9, 66 | |
| S. Typhimurium Std | SeqA, HdfR, RosE | 7, 23, 45 | |
| Cyclic di-GMP | K. pneumoniae Mrk | MrkJ | 46 |
| P. aeruginosa CupA | PA1120, MorA, PvrR, MvaT | 60, 86 | |
| P. aeruginosa CupB | RocS1, RocA1, RocR, MvaT | 50, 86 | |
| P. aeruginosa CupC | RocS1, RocA1, RocR, MvaT | 50, 86 | |
| P. aeruginosa CupD | PvrR, RcsB, RcsC | 61, 65 | |
| Additional DNA binding regulatorsa | Bordetella pertussis Fha | BvgAS | 15–17 |
| E. coli 987P | FasH | 28, 37 | |
| E. coli CFA/I | Rns | 21 | |
| E. coli CS1 | Rns, H-NS | 20, 62, 63 | |
| E. coli CS2 | Rns | 20, 62 | |
| E. coli CS17 | Rns | 13 | |
| E. coli CS19 | Rns | 13 | |
| E. coli PCO71 | Rns | 13 | |
| E. coli F1C | FocB | 40, 57 | |
| E. coli Lpf | Ler, H-NS | 83 | |
| S. Typhimurium Fim | FimZYW | 75, 81, 95 |
Fimbrial gene regulators not associated with mediating DNA inversion, requiring DNA methylation for binding, or involved in cyclic di-GMP metabolism.
INVERTIBLE DNA ELEMENTS
E. coli type 1 fimbriae.
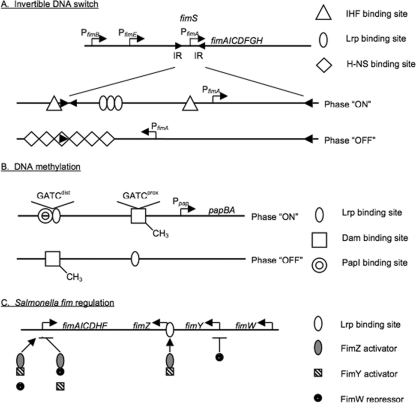
The fim gene cluster of Escherichia coli encodes the production of type 1 (or mannose-sensitive) fimbriae, which have been shown to play a major role in bacterium-host cell interactions (11, 58). The gene cluster is illustrated in Fig. 1A, and it possesses determinants that encode the characteristic chaperone (fimC) and scaffolding proteins (fimD) as well as structural components of the fimbriae, including the major fimbrial subunit (fimA). Transcription of fimA is dependent upon the orientation of an invertible DNA segment (fimS), referred to as the fim switch, which carries the fimA promoter and is flanked by two 9-bp inverted repeats (26, 35). In one orientation transcription can occur, whereas in the opposite orientation no transcription of fimA is possible. The inversion of fimS is mediated by two fimbrial site-specific recombinases (FimB and FimE) that act independently and bind to regions close to and overlapping the 9-bp repeats flanking the 314-bp fimS. FimE primarily mediates the inversion of the switch from the on-to-off orientation, while FimB is believed to play a role in mediating the inversion of fimS in both directions (1, 2). In addition to these two recombinases, the inversion is also influenced by the bacterial nucleoproteins leucine-responsive regulatory protein (Lrp), integration host factor (IHF), and histone-like nucleoid-structuring (H-NS) protein. The interesting question of whether the inversion, with resultant fimbrial phase variation, is stochastic or subject to programming was recently evaluated by Corcoran and Dorman (26). It has been suggested that the degree of supercoiling of the DNA switch, mediated by DNA gyrase activity, plays a role in the involvement of Lrp, IHF, and H-NS binding to this region. Since the negative supercoiling activity of DNA gyrase has been described to be a property of stationary-phase growth, the inversion to the phase-on orientation might be favored during this growth phase. Also, levels of Lrp and IHF increase as bacteria transition to the stationary phase. These conditions may therefore favor fimbrial expression and could explain the production of richly fimbriate bacteria at these times, validating the early observations by Duguid and colleagues (68) of outgrowth of type 1 fimbriate E. coli cells in broth cultures incubated over prolonged periods of time. A proposed model for the switch structure and switch bias has been developed. Both the IHF and Lrp proteins are proposed to facilitate the establishment of the phase-on bias, whereas the H-NS protein plays a role in maintaining the orientation of fimS in the phase-off position (10, 26, 31, 35).
Fig. 1.
Examples of fimbrial gene regulation involving invertible DNA elements, DNA methylation, and DNA-protein as well as protein-protein interactions. (A) The expression of the E. coli type 1 fimbrial gene cluster (fim) is dependent on the orientation of the fimS invertible DNA fragment flanked by two inverted repeat sequences (IR). Recombination at these sites is mediated by the fimbria-specific recombinases FimB and FimE. In addition, the global regulators IHF and Lrp facilitate fim gene transcription, whereas H-NS binding inhibits gene expression, depending on the orientation of the fragment. (B) E. coli pap gene expression is dependent upon the methylation status of two GATC sites upstream of the pap operon. Lrp and Dam compete for binding at these sites, and the pap-encoded protein PapI is a positive regulator of gene expression. (C) Expression of the Salmonella type 1 fimbrial gene cluster (fim) is controlled by two fim-encoded positive activators (FimZ and FimY) and one negative regulator (FimW). FimZ is a DNA binding protein and also interacts with FimW. The Fim regulators also control expression of the three regulatory genes via feedback loops. In addition, Lrp has been shown to bind to the promoter region of fimZ to affect its expression.
Since FimB and FimE play critical roles in mediating the orientation of the fim switch, it would be expected that regulators controlling the expression of these recombinases influence the type 1 fimbrial phenotype of E. coli. IcsR is a regulatory protein that has been shown to influence E. coli biofilm formation by affecting type 1 fimbria production (93). In the presence of limiting iron conditions this metalloprotein is proposed to bind to the fimE promoter region and activate expression of fimE, leading to an increased inversion rate of the fim switch to the Fim-off orientation. This would favor decreased type 1 fimbrial production under iron-limiting conditions, whereas bacterial growth under iron-replete conditions would result in reduced FimE production and a general influence of the fimS region to the phase-on position. Such a model could explain the observed decrease in biofilm and fimbria production by E. coli under iron-limiting conditions (93). Although FimB and FimE were originally described as the recombinases mediating the fimbrial switch inversion, it has been shown that additional recombinases also influence the orientation of the invertible element (51). Consequently, type 1 fimbria production in E. coli could be the result of the relative concentrations of several recombinases that are produced, depending upon environmental factors.
Klebsiella pneumoniae type 1 fimbriae.
The Klebsiella pneumoniae type 1 fimbrial gene cluster has recently been identified as playing a role in the uropathogenesis of the organism (74). K. pneumoniae Top52 was found to form biofilm-like intracellular bacterial communities (IBC) in a murine cystitis model in a type 1 fimbria-dependent manner (74). Several elements of the E. coli type 1 fimbrial system are also found in K. pneumoniae: the recombinases FimBE, an invertable DNA switch (fimS), and similarly arranged structural genes. Two differences which were found in K. pneumoniae were the absence of a homolog to a third E. coli recombinase (FimX) and the presence of a putative regulatory gene immediately downstream of the K. pneumoniae fim cluster, fimK. Overexpression of E. coli fimX or the deletion of fimK led to increased surface expression of type 1 fimbriae (74). This was found to be the result of increased inversion of the fimS switch to the on orientation. Strains which were type 1 hyperfimbriate more readily colonized the bladders and kidneys of mice, probably through increased IBC formation (74). Interestingly, fimK exhibits relatedness to EAL phosphodiesterases, which are involved in the metabolism of the second messenger, c-di-GMP (56, 72, 76, 89). This may indicate that FimK is involved in a c-di-GMP-mediated inversion of the fimS switch in K. pneumoniae.
Proteus mirabilis Mrp fimbriae.
In addition to type 1 fimbrial production in E. coli and K. pneumoniae, the presence of an invertible DNA fragment used to control fimbrial gene expression has been reported in other bacteria. The Mrp fimbriae of uropathogenic Proteus mirabilis are also subject to control by the effect of a specific recombinase that mediates inversion of a DNA fragment (51, 54, 96). Construction of MrpI mutants demonstrated that these mutants were either locked in the on phase or in the off phase for fimbrial production and were unable to invert the region of DNA possessing the promoter of the mrp operon. In this case MrpI appears to be the single recombinase involved in this activity and mediates inversion of a 251-bp DNA fragment immediately upstream of the gene encoding the fimbrial subunit MrpA. The level of MrpI produced is controlled at the transcriptional level by the oxygen concentration, and growth of bacteria under aerobic conditions results in increased MrpI transcript production. Since these conditions also favor growth of non-fimbriate-phase bacteria, it has been suggested that MrpI may favor inversion of the DNA fragment from the phase-on to the phase-off orientation (51).
K. pneumoniae Kpc fimbriae.
A recently identified fimbrial type from Klebsiella pneumoniae NTUH-K2044, termed Kpc fimbriae, has also been shown to be regulated by an invertible DNA element (91). Genes encoding these fimbriae were shown to be predominantly found in K1 serotypes which, interestingly, are significantly associated with the formation of liver abscesses in diabetic patients in Southeast Asia. Inversion of a 302-bp promoter-containing region between two 11-bp inverted repeats upstream of the major subunit gene is mediated by the recombinase KpcI (91). The inversion of the switch (kpcS) by recombinant KpcI in an E. coli background could be mediated from either off-to-on or on-to-off orientations (91). One of the inverted repeats lies within the KpcI reading frame, so when kpcS is in the on orientation, a portion of the predicted amino acid sequence changes (91). It must be noted that this inversion and the overexpression of KpcI leads to the detectable production of Kpc fimbriae, but this could not be shown in the K. pneumoniae NTUH-K2044 background.
DNA METHYLATION
E. coli Pap fimbriae.
E. coli P fimbriae (pili) are produced by the majority of uropathogenic isolates implicated in upper urinary tract infections and are believed to mediate adherence to uroepithelial cells in vitro. The pap operon possesses all of the genes necessary for the synthesis and assembly of these appendages (Fig. 1B). Control of expression of the major subunit gene (papA) is mediated by the methylation status of two GATC sites upstream of the papBA promoter. In addition, the transcription of the pap gene cluster is influenced by the concentrations of the global Lrp protein and the fimbria-specific regulator PapI. At high intracellular concentrations of Lrp and relatively low concentrations of the Pap-specific regulator PapI, Lrp has been proposed to bind efficiently to the region of the GATC site closest to the pap promoter, resulting in inhibition of pap transcription (69, 70). This binding can only occur if the site is not methylated, and therefore Lrp binding competes with the DNA adenine methylase (Dam) enzyme for binding at this site. Following a round of DNA replication and cell division, at low concentrations of Lrp in the presence of PapI, Lrp binds to the hemimethylated GATC site distal to the Pap promoter, preventing methylation of this site by Dam but allowing full methylation of the second proximal site. In this state (phase on), transcription of the pap operon can occur. Consequently, phase variation of P pili is predicted to be significantly influenced by the concentrations of Lrp, which in turn have been shown to be dependent upon growth conditions. For example, as bacteria reach stationary phase during growth in rich medium, the concentrations of Lrp increase, favoring the transition from phase on to off as the bacteria enter stationary phase. In addition, it has been shown that the off- to on-phase orientation occurs with a greater frequency following growth in urine with a high amino acid content than during bacterial growth in minimal medium (36). Since P pili are produced by many uropathogenic isolates, the higher transition rates to the phase-on phenotype are likely to facilitate more efficient colonization of the urinary tract. However, it has yet to be determined what effect growth in the urinary tract has on Lrp production by bacteria.
E. coli K88 (or F4) fimbriae.
The K88 fimbriae in enterotoxigenic E. coli (ETEC) facilitate adherence to specific receptors present on the brush borders of villous enterocytes, allowing for colonization of the small intestine (87). It is this colonization that leads to infections in young pigs that result in diarrhea and relatively high mortality. Much like the Pap fimbriae, the regulation of the K88 fimbriae has been shown to involve Lrp and the operon-specific PapI homolog FaeA (38). The expression of the fae operon is controlled by the cooperative binding of the Lrp/FaeA complex to the faeB promoter region (39). There are distinct differences between the FaeA-mediated regulation of the K88 fimbriae and the Pap pili. FaeA negatively controls the expression of the K88 fimbriae by binding within the promoter region of faeB, which is in contrast to the positive effects of PapI on Pap expression. This repression is thought to be the result of Lrp/FaeA binding to a site between the −35 to −10 consensus region within the faeB promoter (39). Methylation of this site results in destabilization of the complex, leading to decreased promoter binding of Lrp/FaeA. Similar to the Pap system, faeA transcription is promoted by the PapB homolog FaeB (39).
E. coli K99 fimbriae.
As for the K88 fimbriae, some enterotoxigenic E. coli strains express a fimbrial type termed the K99 fimbriae. These appendages, similar to the K88 fimbriae, are implicated in diarrheal diseases via the colonization of the small intestines in neonatal calves, lambs, and piglets (30). While little is known about the molecular biology of K99 gene expression, previous genetic studies indicated that the K99 operon (fan) is positively regulated by the Lrp homolog Mbf (methylation blocking factor) (18). Mutants of Mbf were found to be significantly downregulated for K99 expression, and introduction of Lrp restored K99 expression. These results were found for both chromosomal fan transcription and plasmid-borne K99 surface expression (18).
E. coli Sfa fimbriae.
S fimbriae encoded by the sfa operon are frequently associated with E. coli strains implicated in causing neonatal meningitis (49). Dam-dependent methylation influences the methylation of two GATC sites located in the promoter region of the sfa operon (88). Binding of Lrp is required for transcription of the sfa operon and is modulated by the methylation state of these sites by Dam. In phase-on cells the GATC I site is unmethylated, whereas the GATC II site is unmethylated in phase-off cells. Interestingly, SfaX is encoded by the terminal gene of the operon and belongs to a family of Mar-like regulatory proteins (77, 78). However, SfaX mutants are not impaired in S fimbria production, and so the role of this protein in sfa gene expression is not clear. However, it has been shown that SfaX does affect transcription of the fimA gene of the E. coli type 1 fimbrial gene cluster, possibly mediating its effect through the activity of the FimBE recombinases. SfaX is one of the few proteins encoded by one fimbrial gene cluster that has been shown to control expression of fimbrial genes from a heterologous cluster (77).
Salmonella enterica serovar Typhimurium Std fimbriae.
The Std fimbriae of S. enterica serovar Typhimurium are necessary for persistence of the bacterium in mouse intestines (90). In a dam mutant the std genes are significantly derepressed, and these genes exhibit one of the highest levels of expression among Dam-regulated genes (7). Examination of the predicted promoter region of stdA revealed three GATC sites within a 25-bp region. Analysis of transposon insertions showed that SeqA, a protein that binds GATC sites, repressed stdA expression (45). In contrast HdfR, a member of the LysR-like regulators, was shown to activate stdA transcription (45). Whether this activation occurs by the direct binding of HdfR to the regulatory region of the sfa operon or is due to an indirect effect is unknown.
Interestingly, a second transposon screen for regulators of expression of Std fimbriae found genes that are not involved in DNA methylation, in addition to the expected dam mutant (23). Insertion mutations in csgC, setB, lrhA, and rosE were found. The determinant csgC is a gene of unknown function within the cluster encoding proteins for the production of thin curled fimbriae (curli-like), and setB encodes a sugar transporter; mutations in this gene have been shown to have pleiotropic effects (29, 73). The function of the S. Typhimurium lrhA gene product is unknown, but in E. coli LrhA represses expression of type 1 fimbriae following binding to the promoter regions of the fimBE genes and the fimS region itself (12). RosE exhibits relatedness to transcriptional regulators and is able to bind DNA located at bp −225 to 375 upstream of the stdA start codon, indicating that it may be involved in regulation of expression of stdA (23). Since RosE repressed a std::lacZ reporter fusion plasmid and RosE mutants assemble Std fimbriae, it is possible that this protein is involved in the negative regulation of Std fimbriae.
S. Typhimurium Pef fimbriae.
The plasmid-encoded fimbriae (Pef) of S. Typhimurium are involved in attachment to mouse intestinal cells and are expressed in a phase-variable manner (9). Like the E. coli pap gene cluster, pef has two GATC sites (I and II) separated by 102 bp and is located in the upstream regulatory region of the gene cluster. One of the pef genes encodes PefI, which is a homolog of the Pap regulatory protein PapI (66). The pef gene regulatory region also has an additional GATC site (X) which is located 12 bp from GATC II. Single mutations of GATC I or X to GCTC had little effect on Pef production. Mutating both GATC I and X allowed Pef production under neutral pH growth conditions, unlike the wild-type strain, which required an acidic pH for Pef production. No Pef was produced in the GCTC II mutant, indicating that methylation of this site is required for fimbria production. It has been proposed that PefI is required for Lrp binding to pef GATC sites, which subsequently prevents methylation of the DNA (66).
CYCLIC DI-GMP
K. pneumoniae type 3 fimbriae.
The type 3 fimbrial appendages of K. pneumoniae have been shown to facilitate adherence and subsequent biofilm formation on human extracellular matrix-coated surfaces (44, 52). Additionally, these fimbriae have also been shown to be an important determinant in biofilm formation on abiotic surfaces (46). Recently, it was shown that the K. pneumoniae strain IApc35 encodes a functional cyclic diguanylate phosphodiesterase (MrkJ) which, when deleted, leads to intracellular accumulation of c-di-GMP (46). The increased intracellular concentrations of c-di-GMP resulted in the increased transcription of mrkA (the gene encoding the major fimbrial subunit) and surface expression of type 3 fimbriae. Also, analysis of K. pneumoniae genomes indicates the presence of two additional genes (mrkH and mrkI) contiguous with the mrk gene cluster which are predicted to encode proteins that contain domains involved in cyclic di-GMP metabolism and sensing. This includes the PilZ domain-containing protein MrkH, which we have recently found to be involved in both c-di-GMP binding and transcription of the type 3 fimbrial genes (unpublished results).
Sfa fimbriae.
As mentioned above, the sfaX determinant is part of the sfa gene cluster encoding the S fimbriae associated with strains of pathogenic E. coli causing neonatal meningitis. Located between sfaX and the genes responsible for assembly and production of S fimbriae is sfaY, which has recently been suggested to encode a putative phosphodiesterase (78). Expression of this gene in a Vibrio cholerae host resulted in a reduced ability of the bacterium to form a biofilm. These observations led to the speculation that SfaY is involved in the turnover of intracellular c-di-GMP in E. coli cells possessing the sfa fimbrial gene cluster (78).
ADDITIONAL DNA BINDING FIMBRIAL REGULATORS
S. Typhimurium type 1 fimbriae.
Pathogenic S. Typhimurium is able to establish infections in humans and animals by first binding to mucosal surfaces in the intestines, followed by colonization and, in some cases, invasion of epithelial cells. Analysis of the S. Typhimurium genome has led to the identification of 12 putative fimbrial operons encoding appendages assembled by the chaperone/usher pathway, namely, fim, pef, lpf, bcf, saf, stb, stc, std, stf, sth, sti, and stj (43, 90). Analysis of S. Typhimurium mutants has shown that these operons are responsible for functions that establish an infection through persistence in host animals (5, 22, 53, 90).
S. Typhimurium type 1 fimbriae, produced by the fim operon, are mannose-sensitive hemagglutinins that decorate the surface of the cell. The regulatory genes fimW, fimY, and fimZ encode proteins that affect the expression of the major subunit gene fimA (75, 81, 82, 94). Additionally, upstream of fimW is fimU, which encodes a tRNA that is required for adequate expression of the regulatory proteins (79). FimZ possesses a signal domain and a DNA binding domain associated with response regulators of two-component regulatory systems, but identification of its cognate histidine kinase has yet to be achieved. FimY has no recognizable domains and exhibits little homology to any proteins in the available databases, whereas FimW does possess a putative DNA binding domain (Fig. 1C).
Both fimZ and fimY mutants exhibit a nonfimbriate phenotype, indicating that they are both positive regulators for type 1 fimbriae (81, 94). A fimW mutant is more strongly fimbriate than the parent strain, suggesting that FimW is a negative regulator of type 1 fimbriae (81). FimZ binds directly to the promoter region of fimA to regulate expression, but neither FimW nor FimY has been shown to bind in vitro to this region (95). However, FimW and FimZ were shown to interact in a bacterial two-hybrid assay, whereas FimY and FimZ did not show any demonstrable interaction (81). Possibly, FimW inhibits FimZ activation of fimA by interaction with FimZ and prevention of binding to the fimA promoter. FimY may prevent this interaction and facilitate fimA expression.
Recent studies have shown that FimW, FimY, and FimZ also play a role in affecting the expression of the regulatory genes themselves. Specifically, FimZ and FimY are able to induce each other's expression (75). FimY is also able to induce expression of fimW, which results in repression of PfimY promoter activity. Rao and colleagues (75) suggested that type 1 fimbrial expression is controlled by a positive feedback loop involving FimY and FimZ as well as a negative feedback loop between FimW and FimY. In addition, the expression of fimZ itself has been shown to be controlled at the level of transcription by Lrp, and Lrp-negative mutants show no fim expression (59). Lrp binds to a region upstream (between bp 65 and 170) of the fimZ transcription start site and is proposed to facilitate fimZ expression, leading to increased type 1 fimbrial production due to activation by FimZ.
ETEC fimbriae.
Strains of ETEC are characterized by their abilities to produce enterotoxins and a range of fimbrial colonization antigens that are assembled by the chaperone/usher pathway (14, 33). The regulation of expression of some, but not all, of the genes encoding these fimbriae has been examined and has been shown to involve a DNA binding protein that is a transcriptional regulator. For example, the production of CS1 fimbriae by ETEC is controlled by the production of the DNA binding protein Rns (13). Promoters of fimbrial genes regulated by Rns are characterized by the presence of specific nucleotide sequences at the site of binding that enable Rns to interact with RNA polymerase in order to activate transcription. This binding site has been found in a range of colonization factors produced by ETEC strains (13), and a second separate Rns binding site has also been localized in some of these strains. Many of the genes encoding colonization antigens associated with ETEC isolates have now been shown to be regulated by Rns. Some of these fimbriae include not only CS1 but also all so-called class 5 fimbriae of ETEC strains, e.g., CS17, CS19, CFA/I, and PCO71 fimbriae.
DISCUSSION
The chaperone/usher assembly pathway of bacterial fimbriae is one of three pathways that have been described to account for the production of surface-associated fimbriae (67, 80). This pathway is used by a large number of enterobacterial species to assemble organelles that are primarily used to adhere to receptor molecules on host cells and tissues. The production of these fimbriae requires the presence of gene clusters on the bacterial genome, and the regulation of the genes comprising these clusters can utilize quite diverse regulatory mechanisms. It is becoming apparent that the regulation of fimbrial gene expression involves complex regulatory circuits with multiple components required for optimal activity. In those cases that have been investigated in detail, the presence of numerous proteins that affect fimbrial gene expression has been described (Table 1). In many instances, presumably either directly or indirectly, these proteins interact with each other to form integrated circuits. Whole-genome sequencing of many strains of enterobacteria has also indicated that the presence of multiple fimbrial gene clusters is the norm in these bacteria, and yet very little is known about interfimbrial gene regulation. The ability of bacteria to produce one fimbrial type and inhibit production of another type may occur at both the level of gene expression and appendage assembly. Consequently, regulation of a single fimbrial gene cluster can use multiple regulatory proteins and, in addition to this, a level of more global regulation between clusters is likely to occur. For example, it has been shown that H-NS can repress multiple fimbrial gene clusters in E. coli, and a related protein, MvaT in P. aeruginosa, also controls expression of fimbrial genes (48, 85, 86). Recent investigations of the regulatory interactions between multiple fimbrial gene clusters encoding distinct adhesins of uropathogenic E. coli have begun to examine interfimbrial gene regulators (57, 84). Interestingly, it has been suggested that one of the pap operon regulators, PapI, is subject to positive selection and that allelic variants of papI exhibit different abilities to activate Pap-related systems in E. coli. In addition, observations indicate that regulators from two distinct gene clusters (pap and foc) can form heterodimers to repress one of the gene clusters. These investigations are beginning to address the question of how the enterobacteria ensure that an array of adhesins is not produced at one time by enterobacteria and cross talk between fimbrial regulators is achieved.
Besides regulation within and between fimbrial gene clusters, there is also evidence that bacteria can coordinate the control of fimbrial and nonfimbrial phenotypes. There are now several reported examples of gene products that were originally characterized as fimbrial regulatory proteins that also affect the production of other surface factors. For example, the fimbrial regulatory proteins MrpJ, FimZ, and SfaX, originally described as affecting fimbria production in Proteus mirabilis, S. Typhimurium, and pathogenic E. coli, respectively, have also been implicated in controlling flagellum production at the level of gene expression (24, 55, 78). Besides both fimbria and flagellum gene expression, FimZ has also been shown to control invasion gene expression by acting at the level of another regulator, HilE (75).
Above, we have cited some specific examples of the regulatory mechanisms utilized to control fimbrial gene expression. It is important to point out that many of these mechanisms exhibit a large degree of overlap. For example, many DNA binding regulatory proteins are only active at sites that are methylated, or vice versa. The orientation of invertible DNA fragments can also have a profound effect on whether protein complexes recognize their target operator sites. In summary, the regulation of fimbrial gene expression is dependent upon a complex regulatory cascade that is likely to involve both fimbria-specific regulators (both at the level of a single gene cluster and multiple gene clusters) and non-fimbria-specific regulators. In addition, each system appears to also have evolved to employ unique circuit designs to achieve gene regulation. The elucidation of these circuits at the molecular level may provide one way to develop novel strategies to inhibit the colonization and growth of enterobacterial pathogens.
ACKNOWLEDGMENTS
We apologize to any researchers whose work was not cited in this minireview and thank the many laboratories whose research has contributed to our understanding of fimbrial regulation and assembly.
The research in the laboratory of S.C. is supported by NIH grants GM084318, AI1050011, and AI074693. J.J. is supported by NIH training grant T32 AI007511.
Footnotes
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Aberg A., Shingler V., Balsalobre C. 2008. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67:1223–1241 [DOI] [PubMed] [Google Scholar]
- 2. Adiciptaningrum A. M., Blomfield I. C., Tans S. J. 2009. Direct observation of type 1 fimbrial switching. EMBO Rep. 10:527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akahane K., Sakai D., Furuya N., Komano T. 2005. Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Mol. Genet. Genomics 273:350–359 [DOI] [PubMed] [Google Scholar]
- 4. Alphonse S., et al. 2010. Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J. Struct. Biol. 169:75–80 [DOI] [PubMed] [Google Scholar]
- 5. Althouse C., Patterson S., Fedorka-Cray P., Isaacson R. E. 2003. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71:6446–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ayers M., Howell P. L., Burrows L. L. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5:1203–1218 [DOI] [PubMed] [Google Scholar]
- 7. Balbontin R., et al. 2006. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:8160–8168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barnhart M. M., Chapman M. R. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baumler A. J., et al. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blomfield I. C., Calie P. J., Eberhardt K. J., McClain M. S., Eisenstein B. I. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blomfield I. C., McClain M. S., Eisenstein B. I. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439–1445 [DOI] [PubMed] [Google Scholar]
- 12. Blumer C., et al. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287–3298 [DOI] [PubMed] [Google Scholar]
- 13. Bodero M. D., Harden E. A., Munson G. P. 2008. Transcriptional regulation of subclass 5b fimbriae. BMC Microbiol. 8:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boedeker E. C. 2005. Vaccines for enterotoxigenic Escherichia coli: current status. Curr. Opin. Gastroenterol. 21:15–19 [PubMed] [Google Scholar]
- 15. Boucher P. E., Maris A. E., Yang M. S., Stibitz S. 2003. The response regulator BvgA and RNA polymerase alpha subunit C-terminal domain bind simultaneously to different faces of the same segment of promoter DNA. Mol. Cell 11:163–173 [DOI] [PubMed] [Google Scholar]
- 16. Boucher P. E., Yang M. S., Schmidt D. M., Stibitz S. 2001. Genetic and biochemical analyses of BvgA interaction with the secondary binding region of the fha promoter of Bordetella pertussis. J. Bacteriol. 183:536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boucher P. E., Yang M. S., Stibitz S. 2001. Mutational analysis of the high-affinity BvgA binding site in the fha promoter of Bordetella pertussis. Mol. Microbiol. 40:991–999 [DOI] [PubMed] [Google Scholar]
- 18. Braaten B. A., et al. 1992. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:4250–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Campos M., Nilges M., Cisneros D. A., Francetic O. 2010. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc. Natl. Acad. Sci. U. S. A. 107:13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caron J., Coffield L. M., Scott J. R. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:963–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caron J., Scott J. R. 1990. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect. Immun. 58:874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chessa D., Winter M. G., Jakomin M., Baumler A. J. 2009. Salmonella enterica serotype Typhimurium Std fimbriae bind terminal α(1,2)fucose residues in the cecal mucosa. Mol. Microbiol. 71:864–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chessa D., Winter M. G., Nuccio S. P., Tukel C., Baumler A. J. 2008. RosE represses Std fimbrial expression in Salmonella enterica serotype Typhimurium. Mol. Microbiol. 68:573–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clegg S., Hughes K. T. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen N. D., et al. 2008. Association of soil concentrations of Rhodococcus equi and incidence of pneumonia attributable to Rhodococcus equi in foals on farms in central Kentucky. Am. J. Vet. Res. 69:385–395 [DOI] [PubMed] [Google Scholar]
- 26. Corcoran C. P., Dorman C. J. 2009. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol. Microbiol. 74:1071–1082 [DOI] [PubMed] [Google Scholar]
- 27. Craig L., Li J. 2008. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edwards R. A., Schifferli D. M. 1997. Differential regulation of fasA and fasH expression of Escherichia coli 987P fimbriae by environmental cues. Mol. Microbiol. 25:797–809 [DOI] [PubMed] [Google Scholar]
- 29. Espeli O., Nurse P., Levine C., Lee C., Marians K. J. 2003. SetB: an integral membrane protein that affects chromosome segregation in Escherichia coli. Mol. Microbiol. 50:495–509 [DOI] [PubMed] [Google Scholar]
- 30. Gaastra W., de Graaf F. K. 1982. Host-specific fimbrial adhesins of noninvasive enterotoxigenic Escherichia coli strains. Microbiol. Rev. 46:129–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gally D. L., Rucker T. J., Blomfield I. C. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol. 176:5665–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gibson D. L., White A. P., Rajotte C. M., Kay W. W. 2007. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology 153:1131–1140 [DOI] [PubMed] [Google Scholar]
- 33. Hacker J. 1992. Role of fimbrial adhesins in the pathogenesis of Escherichia coli infections. Can. J. Microbiol. 38:720–727 [DOI] [PubMed] [Google Scholar]
- 34. Hernday A. D., Braaten B. A., Low D. A. 2003. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol. Cell 12:947–957 [DOI] [PubMed] [Google Scholar]
- 35. Holden N., et al. 2007. Comparative analysis of FimB and FimE recombinase activity. Microbiology 153:4138–4149 [DOI] [PubMed] [Google Scholar]
- 36. Holden N., Totsika M., Dixon L., Catherwood K., Gally D. L. 2007. Regulation of P-fimbrial phase variation frequencies in Escherichia coli CFT073. Infect. Immun. 75:3325–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Honarvar S., Choi B. K., Schifferli D. M. 2003. Phase variation of the 987P-like CS18 fimbriae of human enterotoxigenic Escherichia coli is regulated by site-specific recombinases. Mol. Microbiol. 48:157–171 [DOI] [PubMed] [Google Scholar]
- 38. Huisman T. T., Bakker D., Klaasen P., de Graaf F. K. 1994. Leucine-responsive regulatory protein, IS1 insertions, and the negative regulator FaeA control the expression of the fae (K88) operon in Escherichia coli. Mol. Microbiol. 11:525–536 [DOI] [PubMed] [Google Scholar]
- 39. Huisman T. T., de Graaf F. K. 1995. Negative control of fae (K88) expression by the ‘global’ regulator Lrp is modulated by the ‘local’ regulator FaeA and affected by DNA methylation. Mol. Microbiol. 16:943–953 [DOI] [PubMed] [Google Scholar]
- 40. Hultdin U. W., et al. 2010. Structure of FocB: a member of a family of transcription factors regulating fimbrial adhesin expression in uropathogenic Escherichia coli. FEBS J. 277:3368–3381 [DOI] [PubMed] [Google Scholar]
- 41. Hultgren S. J., Normark S. 1991. Biogenesis of the bacterial pilus. Curr. Opin. Genet. Dev. 1:313–318 [DOI] [PubMed] [Google Scholar]
- 42. Hultgren S. J., Normark S., Abraham S. N. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383–415 [DOI] [PubMed] [Google Scholar]
- 43. Humphries A. D., et al. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357–1376 [DOI] [PubMed] [Google Scholar]
- 44. Jagnow J., Clegg S. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149:2397–2405 [DOI] [PubMed] [Google Scholar]
- 45. Jakomin M., Chessa D., Baumler A. J., Casadesus J. 2008. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190:7406–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnson J. G., Clegg S. 2010. Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J. Bacteriol. 192:3944–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kagami Y., Ratliff M., Surber M., Martinez A., Nunn D. N. 1998. Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol. Microbiol. 27:221–233 [DOI] [PubMed] [Google Scholar]
- 48. Korea C. G., Badouraly R., Prevost M. C., Ghigo J. M., Beloin C. 2010. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 12:1957–1977 [DOI] [PubMed] [Google Scholar]
- 49. Korhonen T. K., et al. 1986. Binding of Escherichia coli S fimbriae to human kidney epithelium. Infect. Immun. 54:322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kulasekara H. D., et al. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368–380 [DOI] [PubMed] [Google Scholar]
- 51. Lane M. C., Li X., Pearson M. M., Simms A. N., Mobley H. L. 2009. Oxygen-limiting conditions enrich for fimbriate cells of uropathogenic Proteus mirabilis and Escherichia coli. J. Bacteriol. 191:1382–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Langstraat J., Bohse M., Clegg S. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 69:5805–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ledeboer N. A., Frye J. G., McClelland M., Jones B. D. 2006. Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74:3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li X., Lockatell C. V., Johnson D. E., Mobley H. L. 2002. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol. Microbiol. 45:865–874 [DOI] [PubMed] [Google Scholar]
- 55. Li X., Rasko D. A., Lockatell C. V., Johnson D. E., Mobley H. L. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 20:4854–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lim B., Beyhan S., Meir J., Yildiz F. H. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331–348 [DOI] [PubMed] [Google Scholar]
- 57. Lindberg S., et al. 2008. Regulatory interactions among adhesin gene systems of uropathogenic Escherichia coli. Infect. Immun. 76:771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Madison B., Ofek I., Clegg S., Abraham S. N. 1994. Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect. Immun. 62:843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McFarland K. A., Lucchini S., Hinton J. C., Dorman C. J. 2008. The leucine-responsive regulatory protein, Lrp, activates transcription of the fim operon in Salmonella enterica serovar typhimurium via the fimZ regulatory gene. J. Bacteriol. 190:602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meissner A., et al. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol. 9:2475–2485 [DOI] [PubMed] [Google Scholar]
- 61. Mikkelsen H., Ball G., Giraud C., Filloux A. 2009. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One 4:e6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Munson G. P., Scott J. R. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391–1402 [DOI] [PubMed] [Google Scholar]
- 63. Murphree D., Froehlich B., Scott J. R. 1997. Transcriptional control of genes encoding CS1 pili: negative regulation by a silencer and positive regulation by Rns. J. Bacteriol. 179:5736–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nenninger A. A., Robinson L. S., Hultgren S. J. 2009. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U. S. A. 106:900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nicastro G. G., Boechat A. L., Abe C. M., Kaihami G. H., Baldini R. L. 2009. Pseudomonas aeruginosa PA14 cupD transcription is activated by the RcsB response regulator, but repressed by its putative cognate sensor RcsC. FEMS Microbiol. Lett. 301:115–123 [DOI] [PubMed] [Google Scholar]
- 66. Nicholson B., Low D. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728–742 [DOI] [PubMed] [Google Scholar]
- 67. Nuccio S. P., Baumler A. J. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71:551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Old D. C., Duguid J. P. 1970. Selective outgrowth of fimbriate bacteria in static liquid medium. J. Bacteriol. 103:447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Peterson S. N., Reich N. O. 2008. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J. Mol. Biol. 383:92–105 [DOI] [PubMed] [Google Scholar]
- 70. Peterson S. N., Reich N. O. 2006. GATC flanking sequences regulate Dam activity: evidence for how Dam specificity may influence pap expression. J. Mol. Biol. 355:459–472 [DOI] [PubMed] [Google Scholar]
- 71. Proft T., Baker E. N. 2009. Pili in Gram-negative and Gram-positive bacteria: structure, assembly and their role in disease. Cell. Mol. Life Sci. 66:613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rao F., et al. 2009. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 191:4722–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Romling U., Sierralta W. D., Eriksson K., Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 74. Rosen D. A., et al. 2008. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 76:3337–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saini S., Pearl J. A., Rao C. V. 2009. Role of FimW, FimY, and FimZ in regulating the expression of type i fimbriae in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Simm R., Lusch A., Kader A., Andersson M., Romling U. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sjostrom A. E., et al. 2009. The SfaXII protein from newborn meningitis E. coli is involved in regulation of motility and type 1 fimbriae expression. Microb. Pathog. 46:243–252 [DOI] [PubMed] [Google Scholar]
- 78. Sjostrom A. E., et al. 2009. Analysis of the sfaX(II) locus in the Escherichia coli meningitis isolate IHE3034 reveals two novel regulatory genes within the promoter-distal region of the main S fimbrial operon. Microb. Pathog. 46:150–158 [DOI] [PubMed] [Google Scholar]
- 79. Swenson D. L., Kim K. J., Six E. W., Clegg S. 1994. The gene fimU affects expression of Salmonella typhimurium type 1 fimbriae and is related to the Escherichia coli tRNA gene argU. Mol. Gen. Genet. 244:216–218 [DOI] [PubMed] [Google Scholar]
- 80. Thanassi D. G., Hultgren S. J. 2000. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods 20:111–126 [DOI] [PubMed] [Google Scholar]
- 81. Tinker J. K., Clegg S. 2000. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 68:3305–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tinker J. K., Clegg S. 2001. Control of FimY translation and type 1 fimbrial production by the arginine tRNA encoded by fimU in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 40:757–768 [DOI] [PubMed] [Google Scholar]
- 83. Torres A. G., et al. 2007. Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157:H7. J. Bacteriol. 189:5916–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Totsika M., Beatson S. A., Holden N., Gally D. L. 2008. Regulatory interplay between pap operons in uropathogenic Escherichia coli. Mol. Microbiol. 67:996–1011 [DOI] [PubMed] [Google Scholar]
- 85. Vallet-Gely I., Donovan K. E., Fang R., Joung J. K., Dove S. L. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:11082–11087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vallet I., et al. 2004. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186:2880–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Van den Broeck W., Cox E., Oudega B., Goddeeris B. M. 2000. The F4 fimbrial antigen of Escherichia coli and its receptors. Vet. Microbiol. 71:223–244 [DOI] [PubMed] [Google Scholar]
- 88. van der Woude M. W., Low D. A. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol. Microbiol. 11:605–618 [DOI] [PubMed] [Google Scholar]
- 89. Wang Y., et al. 2010. GGDEF and EAL proteins play different roles in the control of Sinorhizobium meliloti growth, motility, exopolysaccharide production, and competitive nodulation on host alfalfa. Acta Biochim. Biophys. Sin. (Shanghai) 42:410–417 [DOI] [PubMed] [Google Scholar]
- 90. Weening E. H., et al. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wu C. C., Huang Y. J., Fung C. P., Peng H. L. 2010. Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156:1983–1992 [DOI] [PubMed] [Google Scholar]
- 92. Wu H., Fives-Taylor P. M. 2001. Molecular strategies for fimbrial expression and assembly. Crit. Rev. Oral Biol. Med. 12:101–115 [DOI] [PubMed] [Google Scholar]
- 93. Wu Y., Outten F. W. 2009. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J. Bacteriol. 191:1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yeh K. S., Hancox L. S., Clegg S. 1995. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J. Bacteriol. 177:6861–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yeh K. S., Tinker J. K., Clegg S. 2002. FimZ binds the Salmonella typhimurium fimA promoter region and may regulate its own expression with FimY. Microbiol. Immunol. 46:1–10 [DOI] [PubMed] [Google Scholar]
- 96. Zhao H., Li X., Johnson D. E., Blomfield I., Mobley H. L. 1997. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol. Microbiol. 23:1009–1019 [DOI] [PubMed] [Google Scholar]