Abstract
The Spx protein of Bacillus subtilis is a global regulator of the oxidative stress response. Spx concentration is controlled at the level of proteolysis by the ATP-dependent protease ClpXP and a substrate-binding protein, YjbH, which interacts with Spx. A yeast two-hybrid screen was carried out using yjbH as bait to uncover additional substrates or regulators of YjbH activity. Of the several genes identified in the screen, one encoded a small protein, YirB (YuzO), which elevated Spx concentration and activity in vivo when overproduced from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible yirB construct. Pulldown experiments using extracts of B. subtilis cells producing a His-tagged YirB showed that native YjbH interacts with YirB in B. subtilis. Pulldown experiments using affinity-tagged Spx showed that YirB inhibited YjbH interaction with Spx. In vitro, YjbH-mediated proteolysis of Spx by ClpXP was inhibited by YirB. The activity of YirB is similar to that of the antiadaptor proteins that were previously shown to reduce proteolysis of a specific ClpXP substrate by interacting with a substrate-binding protein.
INTRODUCTION
Regulated proteolysis plays two fundamental roles in bacterial physiology: it functions in the removal of denatured and damaged protein resulting from stress and interrupted translation, and it is often involved in controlling the concentration of key regulatory factors (17–19). Proteases that take part in proteome quality control and in proteolytic regulation include the ATP-dependent, multicomponent Clp proteases composed of a substrate-binding 100-kDa heat shock protein (HSP100)/Clp ATPase and a peptide-cleaving catalytic component, ClpP (37). In Escherichia coli, two Clp proteases, ClpAP and ClpXP, function in both removal of damaged protein and proteolytic control of gene expression. The substrate selectivity of both proteases is determined in part by substrate recognition factors, or adaptors, that interact with specific substrates and deliver the substrate to the Clp protease (2, 24). This also involves specific contacts between the adaptor and Clp ATPase subunit. An oft-cited example of proteolytic control is the ClpXP-catalyzed turnover of the general stress sigma subunit of enteric bacteria, σS (3, 5, 34, 42, 50). The adaptor protein that facilitates σS proteolysis is RssB, which is a member of the response regulator class of two-component regulatory proteins. RssB serves three roles in ClpXP-catalyzed σS turnover; it contacts the substrate by targeting a specific amino acid motif near the N terminus of σS. While bound to substrate, it also contacts the ClpX subunit. Finally, it renders accessible an N-terminal peptide of σS that is recognized by ClpX. RssB is controlled by phosphorylation, but it is also the target of small proteins termed “antiadaptors” that bind to RssB and inhibit its substrate-binding activity (5–7). The three RssB-binding peptides identified are produced in response to diverse stress conditions, all of which lead to stabilization of σS through inhibition of RssB activity (5).
In Bacillus subtilis, a single protein that fulfills the role of an antiadaptor within the competence developmental pathway was identified, the ComS protein encoded by the srf operon (10, 21). Through a quorum-sensing pathway involving the ComX ComPA signal transduction system (11, 40), comS is induced, and its product inhibits the adaptor activity of MecA, which mediates the ClpCP-catalyzed degradation of the competence transcription factor, ComK (35, 38, 44, 45).
Spx, a global regulator that functions in the B. subtilis stress response (12, 22, 30, 33, 51), is also under proteolytic control that requires the Clp protease, ClpXP (31, 32), and a substrate recognition factor, YjbH (16, 26), which exhibits characteristics of an adaptor protein. YjbH protein accelerates ClpXP-catalyzed Spx degradation in vitro and is necessary to keep the concentration of Spx low in unperturbed cells. The YjbH protein bears a metal-binding His-Cys cluster at its N terminus that is required for its activity and suggests a sensory role involving Cys oxidation and metal ion release (16). Among the YjbH homologs in Gram-positive bacteria, this N-terminal metal binding motif is not highly conserved, which suggests that there might exist alternative mechanisms, aside from oxidative metal ion release, controlling YjbH activity.
In this report, we present the results of a screen for YjbH-interacting proteins designed to uncover alternative YjbH substrates or regulators of YjbH activity. Of the interacting proteins uncovered, one, the product of the yirB (yuzO) gene, was found to inhibit YjbH-mediated activity in vivo and in vitro and to disrupt interaction between YjbH and Spx. The YirB protein might be one of several inhibitory small proteins that disrupt the substrate recognition activity of YjbH.
MATERIALS AND METHODS
Bacterial strains, chemicals, and materials.
B. subtilis strains are listed in Table 1 and are derivatives of JH642. E. coli TOP10 (Invitrogen) and DH5α were used for general cloning procedures. Medium components were from Difco. TSS medium was described previously (14). All restriction/modification enzymes were from New England BioLabs. Oligonucleotide primers were from Invitrogen; plasmid pET-23a and E. coli BL21(DE3)(pLysS) were from Novagen. The Ni2+-nitrilotriacetic acid (NTA) resin, StrepTactin Superflow-Plus, and PCR/plasmid purification kits were from Qiagen (Germany). High-Q anion-exchange prepack columns were procured from Bio-Rad. All analytical-grade chemicals were from Sigma unless otherwise stated.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype |
|---|---|
| Strains | |
| JH642 | trpc2 pheA1 |
| ORB3834 | trpc2 pheA1 spx::neo |
| ORB6824 | trpc2 pheA1 thrC::trxB-lacZ |
| ORB4541 | trpC2 pheA1 trxB::trxB-lacZ |
| ORB4561 | trpC2 pheA1 trxB::trxB-lacZ spx::tet |
| ORB7399 | trpC2 pheA1 amyE::pSK1 (Physpank-yirB) |
| ORB7402 | trpC2 pheA1 amyE::Physpank |
| ORB7403 | trpC2 pheA1 spx::neo amyE::pSK1 (Physpank-yirB) |
| ORB7406 | trpC2 pheA1 spx::neo amyE::Physpank |
| ORB7407 | trpC2 pheA1 thrC::trxB-lacZ amyE::pSK1 (Physpank-yirB) |
| ORB7408 | trpC2 pheA1 thrC::trxB-lacZ amyE::Physpank |
| ORB7430 | trpC2 pheA1 ΔyirB::ermf |
| ORB7431 | trpC2 pheA1 ΔyirB::ermr |
| ORB7437 | trpC2 pheA1 ΔyirB::ermf trxB::trxB-lacZ |
| ORB7556 | trpC2 pheA1 ΔyirB::erm amyE::Physpank-yirB trxB::trxB-lacZ |
| ORB7557 | trpC2 pheA1 ΔyirB::erm amyE::Physpank-yirB-His6trxB::trxB-lacZ |
| Plasmids | |
| pDR111 | Physpank spc bla |
| pUC19 | A derivative of pUC18 |
| pTKlac | Promoter-probe vector; Ampr Cmr |
| pDG646 | Source of erythromycin resistance cassette |
| pSK1 | Phyperspank fused with yirB gene |
| pSK5.1, 5.2 | pUC18 with yirB::erma |
| pSG-3 | yjbH coding sequence in yeast two-hybrid vector pGBKT7 |
| pSG-4 | yjbH(Δ1–24) coding sequence in yeast two-hybrid vector pGBKT7 |
erm, erythromycin-resistant marker.
Yeast two-hybrid screen for YjbH-interacting proteins.
The plasmids pYJBHAD and pTYJBHAD (16) are derivatives of the vector pGADT7 (Clontech) and contain, respectively, the coding sequences of yjbH and yjbH(Δ1–24) fused to the part of GAL4 that encodes the activation domain (AD). The oligonucleotides to amplify the yjbH and yjbH(Δ1–24) coding sequences in the construction of pYJBHAD and pTYJBHAD (16) were used again to generate the inserts for construction of the DNA-binding domain (BD) fusion plasmids pSG-3 and pSG-4 (Table 1) derived from vector pGBKT7. Plasmid pSN11 is a pGBKT7 derivative carrying DNA specifying the spx-GAL4 DNA-binding domain fusion (31). The B. subtilis genomic DNA-activation domain fusion library in pGADT7 was described previously (31). The fusion library and pSG-3 were used to transform Saccharomyces cerevisiae PJ69-4A (His− Ade− Trp− Leu−), which carries GAL2-ADE2 and GAL2-HIS3 fusions controlled by GAL4. The yeast two-hybrid reporter strain (PJ69-4A) expressing different BD and AD fusion proteins was assayed for growth on an adenine dropout plate or a histidine dropout plate. Yeast two-hybrid experiments were done according to the standard lab protocol, as previously described (16).
Construction of yirB deletion mutation.
A yirB deletion mutant strain was constructed by first introducing a BamHI site into the yirB coding sequence by using primers sk-08-15 and sk-08-16 (Table 2). The yirB gene was amplified in two fragments (5′ yirB and 3′ yirB), each with a BamHI site at one end. The two fragments were joined by ligation at the BamHI ends and then further amplified with the upstream primer of the 5′ yirB fragment (sk-08-14) and downstream primer (sk-08-17) of the 3′ yirB fragment. The amplified fragments had SphI and XmaI ends, which were used to insert the fragment by ligation into plasmid pUC19 that was cleaved with SphI and XmaI. The ligation mixture was used to transform E. coli TOP10 competent cells. A plasmid bearing the insert, pSK4, was cleaved with BamHI and ligated with an erythromycin resistance cassette obtained from pDG646 (20) by digestion of the plasmid with BamHI and then isolation of the drug resistance fragment from low-melting-point agarose gel. Plasmids with the erythromycin cassette in each of both the orientations (pSK5.1 and pSK5.2) were then transformed into JH642 competent cells to generate the yirB null mutants ORB7430 and ORB7431 (Table 1). The insertion of the erythromycin (erm) resistance cassette in the yirB gene was confirmed by PCR analysis. The yirB mutant was also made in the trxB-lacZ background to create strain ORB7437. Western blot analysis of Spx protein with yirB mutant protein extract was performed by a previously described procedure (32) with samples that were untreated or treated with 1 mM diamide. β-Galactosidase activity was also measured in trxB-lacZ ΔyirB and trxB-lacZ yirB+ strains, as described previously (30).
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| sk-08-11 | GCGCAAGCTTAAAAGGTGGTGAACTACTATGAGAGAATTGGACGAAATGAT |
| sk-08-08 | TATAGCTAGCTTATGTGAAAGAATAGCCGTTAA |
| sk-08-12 | TATAGAATTCTCCTTCAACTGCATGC |
| sk-08-13 | ATATAAGCTTGATTCCTCTGTTTCGC |
| sk-08-14 | TACGGCATGCTGATAAAATCTTTGTG |
| sk-08-15 | TATAGGATCCTCTTTTTCGCAAGCG |
| sk-08-16 | GCATGGATCCGGAATCAAAGTGGAG |
| sk-08-17 | ATATCCCGGGAGAAGAAGTCATCGAA |
Construction of yirB-overexpressing B. subtilis strain.
The Physpank plasmid pDR111 (4) was used to construct the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Physpank-yirB construct. Primers sk-08-11 and sk-08-08 (Table 2) were used to amplify the yirB coding sequence. The PCR fragment was cleaved with HindIII and NheI, followed by insertion into cleaved pDR111 by ligation. The resulting plasmid, pSK1, was subjected to nucleotide sequencing to confirm that the insert was oriented so as to be under the control of the vector's IPTG-inducible Physpank promoter. The Physpank-yirB construct was introduced by transformation into JH642 competent cells to obtain strain ORB7399 (amyE::Physpank-yirB). Chromosomal DNA of ORB7399 was used to transform the trxB-lacZ strain, ORB6824, to obtain strain ORB7407. ORB7402 is a control strain that contains the pDR111 vector only (amyE::Physpank). Chromosomal DNA from ORB7402 was used to transform ORB6824 (thrC::trxB-lacZ) to obtain ORB7408, which was used as a control strain in β-galactosidase activity experiments. Thus, the following strains were made to measure β-galactosidase activity: ORB7407 (amyE::Physpank-yirB thrC::trxB-lacZ) and ORB7408 (amyE::Physpank-thrC::trxB-lacZ).
Western blot analysis.
JH642, ORB7402, and ORB7399 were grown in culture to mid-log phase in Difco sporulation medium (DSM). The culture was then split into two flasks, one untreated and other treated with 1 mM IPTG. Cultures were incubated further at 37°C, and cells were harvested by centrifugation after 30 and 60 min of induction. The cells were lysed by French press, and the lysates were cleared by centrifugation at 17,000 × g for 30 min at 4°C (16). The cleared extracts were applied to a 15% SDS-PAGE gel. The gel-resolved proteins were electrotransferred to a nitrocellulose membrane for Western blot analysis. For Western blotting, polyclonal anti-Spx was used as a primary antibody (32).
Spx-Strep-II tag construction and purification.
To generate the Strep-II-tagged Spx, the spx coding sequence was amplified from genomic DNA of B. subtilis JH642 with primers 5′ TTA TAT CAT ATG GTT ACA CTA TAC ACA TCA C 3′ (forward) and 5′ATA TGG ATC CGT TTG CCA AAC GCT GTG 3′ (reverse). (The restriction sites are underlined.) The PCR product was inserted into vector pMMN771 (recombinant vector constructed in the lab of M. M. Nakano for C-terminal Strep-II tag fusion construction) at the NdeI and BamHI sites to produce plasmid pSG25, which was introduced by transformation into E. coli BL21(DE3)(pLysS). The transformed cells were grown in a 500-ml culture at 37°C in LB broth containing 50 μg ml−1 ampicillin and 5 μg ml−1 chloramphenicol to an A600 of 0.5 to 0.6, and expression was induced with 0.3 mM IPTG for 3 h at 30°C. Cells were harvested by centrifugation in a Super ST21 centrifuge with an ST-H750 rotor at 4,000 rpm for 30 min at 4°C. The cells were suspended in 2 ml of buffer A (20 mM HEPES [pH 7.6], 100 mM KCl, 10% glycerol, 5 mM dithiothreitol [DTT]). One tablet of EDTA-free protease inhibitor cocktail (Roche Applied Science, Germany) was added per 10 ml of resuspended cells. The cells were subjected to freeze-thaw cycles and disrupted by a French press. The lysates were cleared by centrifugation at 17,000 × g for 30 min at 4°C. The clear lysate was applied to a StrepTactin column equilibrated with buffer A and washed with W buffer (100 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM DTT) and eluted with 2.5 mM d-desthiobiotin. The fractions were dialyzed against buffer B (25 mM sodium phosphate [pH 6.8], 100 mM KCl, 5% glycerol, 5 mM DTT), applied to a High-S column (Bio-Rad), and eluted with a gradient of buffer B from 100 to 300 mM KCl. Spx-containing fractions were pooled and dialyzed against a mixture of 25 mM sodium phosphate (pH 8.0), 100 mM KCl, and 5% glycerol, and aliquots were stored at −80°C.
Construction and purification of His6-tagged YirB.
To obtain YirB-expressing E. coli lysate, the yirB coding sequence was PCR amplified from genomic DNA of B. subtilis JH642 with primers 5′ TTA TAT CAT ATG GTT ACA CTA TAC ACA TCA C 3′ (forward) and 5′ ATATCT CGAGTG TGA AAG AAT AGC CG 3′ (reverse). (The restriction sites are underlined.) The PCR product was inserted into vector pET-23a at the NdeI and XhoI sites to produce plasmid pSG23. The pSG23-transformed E. coli BL21(DE3)(pLysS) cells were grown at 37°C in LB broth containing 50 μg ml−1 ampicillin and 5 μg ml−1 chloramphenicol to an A600 of 0.5 to 0.6, and expression was induced with 0.3 mM IPTG for 3 h at 30°C. Cells from a 1-liter culture were harvested by centrifugation, as described above, and suspended in 10 ml of buffer E (50 mM Tris-Cl, 300 mM NaCl [pH 8.0]). One tablet of EDTA-free protease inhibitor cocktail (Roche Applied Science, Germany) was added per 10 ml of resuspended cells. The cells were subjected to freeze-thaw cycles and disrupted by French press. The lysate was cleared by centrifugation as described above. The soluble fraction from the whole-cell lysate was applied to a preequilibrated 2-ml Ni2+-nitrilotriacetic acid (NTA) affinity matrix (Qiagen, Germany). The column was washed with 25 column volumes (CV) of buffer E at a flow rate of 1.0 ml/min and then with 10 column volumes of buffer F (50 mM Tris-Cl, 100 mM KCl, 30 mM imidazole [pH 8.0]). The bound protein was eluted in buffer G (50 mM Tris-Cl, 100 mM KCl, 200 mM imidazole [pH 8.0]). The fractions containing YirB were identified by 10 to 20% gradient SDS-PAGE gel and were concentrated by an Amicon filtration device. The concentrate was dialyzed against a mixture containing 50 mM HEPES-KOH (pH 7.6), 100 mM KCl, 5% glycerol, and 5 mM DTT. The protein concentration was measured using protein concentration dye reagent (Bio-Rad), and bovine serum albumin (BSA) was used to generate the standard curve.
Complementation of yirB with a yirB allele encoding a His6-tagged YirB protein.
For construction of the yirB-His-expressing strain, DNA was PCR amplified with the forward primer sk-08-13, with the reverse primer 5′ ATA TGC TAG CTC AGT GGT GGT GGT GGT G 3′ and pSG23 DNA as a template. PCR products were digested with HindIII and NheI and inserted into plasmid pDR111, which was cleaved with the same enzymes. Ligation generated plasmid pSG24, which was used to transform strain ORB7557 (JH642 ΔyirB::erm) with selection on DSM agar containing spectinomycin and erythromycin. To check the integration of yirB-His at the amyE locus, clones were plated on an LB-starch plate and grown for 16 h at 37°C. The resulting amyE mutant strain, ORB7557, was examined to determine if yirB-His complements the yirB null mutation. To determine the relative enhancement of Spx function, a reporter assay with trxB-lacZ fusion was conducted as described above.
Expression and purification of B. subtilis His6-tagged YjbH.
YjbH protein is insoluble when produced in E. coli (16), but it can be produced in B. subtilis and purified as a soluble protein. A B. subtilis strain ORB7364 (JH642 yjbH::tet amyE::Physpank-yjbH-His6) was grown at 37°C in 4 liters (1 liter in each flask) of 2XYT medium to an A600 of 0.5. YjbH-His expression was induced with 0.5 mM IPTG for 5 h at 30°C. Cells were harvested by centrifugation in a Sorvall Super T21 with a ST-H750 rotor at 4,000 rpm for 30 min. Purification with an Ni column was conducted as follows. The cell pellet was suspended in 50 ml buffer A (20 mM Tris-HCl [pH 7.6], 200 mM NaCl) with one tablet of EDTA-free protease inhibitor cocktail (Roche Applied Science, Germany) and lysed by 3× passage through a French press. The cell lysate was centrifuged at 17,000 × g for 30 min at 4°C, and the supernatant was applied to a 5-ml Ni2+ affinity column (Bio-Rad) equilibrated with Ni2+ buffer A. The protein was first washed with 5 column volumes of 10% of buffer B (Ni buffer A containing 500 mM imidazole) and then was eluted with 13 column volumes of a gradient formed with 10 to 100% buffer B. Eluted fractions that contain YjbH-His were pooled and diluted with High Q buffer A (20 mM Tris-HCl [pH 8.5], 2% glycerol, 1 mM DTT) to lower the salt concentration to 40 mM. The diluted sample was applied to a High Q column equilibrated with High Q buffer A. The bound proteins were eluted with High Q buffer B in a gradient (High Q buffer A containing 1 M NaCl). Fractions containing YjbH-His were concentrated and stored in buffer (20 mM Tris-HCl [pH 8.5], 50 mM NaCl, 5% glycerol, 1 mM DTT) at −80°C.
YjbH interaction with Spx using C-terminal Strep-II-tagged Spx.
Purified Spx-Strep and YjbH-His were mixed together and incubated for 30 min at room temperature in buffer W (100 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM DTT), with final concentrations of 4 and 1.5 μM, respectively. The mixed sample then was loaded to StrepTactin column that was blocked with 0.1 mg/ml BSA. The column was washed with buffer W three times, each time with 3 CV. The bound proteins were eluted with three times with 1 column volume of buffer W containing 2.5 mM d-desthiobiotin.
Purified samples of Spx-Strep, YjbH-His, and YirB-His were mixed together and incubated for 30 min at room temperature in buffer W at final concentrations of 4, 1.5, and 2.5 μM, respectively. The StrepTactin column chromatography with bound Spx-Strep was performed as described above. YjbH protein was detected with polyclonal anti-YjbH antibody.
In vitro proteolysis reactions.
In vitro proteolysis reaction mixtures were assembled under conditions described previously (16, 49), with some modifications. The reactions were carried out in 50 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM Mg acetate, 5 mM DTT, 5 mM ATP, 5 mM creatine phosphate, and 0.05 U/ml creatine kinase (Sigma). Reaction mixtures (100 μl) were incubated at 30°C in the presence of ClpP, ClpX, and Spx proteins with or without YjbH and YirB (at concentrations indicated in the figure legend). At indicated time intervals, a 12-μl sample from each reaction mixture was collected and treated with 3 μl of stop buffer (SDS loading dye in 0.1 M DTT). The proteins were then resolved on a 12% SDS-PAGE gel, followed by staining with Coomassie brilliant blue. Levels of Spx were defined as ratios of Spx band intensity to ClpP band intensity since the ClpP concentrations in each set of reactions were equal. The Spx/ClpP ratio in a reaction mixture containing no ClpX was given the value 100%.
RESULTS
YjbH-interacting proteins identified by yeast two-hybrid screen.
Previously, evidence was presented that YjbH interacted with Spx and enhanced its proteolytic degradation by ClpXP in vitro, but not the degradation of another ClpXP substrate (16). It was still possible that YjbH could serve as a recognition factor for other Clp substrates, just as MecA targets more than one substrate for ClpCP-catalyzed degradation (32, 35). In addition to other ClpXP substrates, YjbH could also contact factors that regulate its activity, such as redox sensors that affect its Zn-binding center, or proteins that inhibit its activity as a substrate recognition factor for ClpXP. BLAST alignments of YjbH orthologs indicated that the N-terminal metal binding region was not conserved and is absent altogether in the YjbH ortholog of Staphylococcus aureus, suggesting that there may be additional modes of YjbH regulation besides redox control involving metal release. Hence, we undertook a yeast two-hybrid screen for YjbH-interacting proteins.
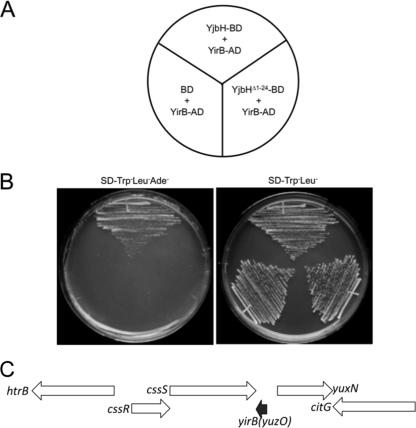
The yjbH coding region was fused to the DNA-binding domain of Gal4 by using the yeast two-hybrid bait vector pGBKT7. The yeast strain bearing the bait plasmid was transformed with the B. subtilis genomic-activation domain fusion library we had previously used (31). Screening for His+ and Ade+ transformants uncovered several clones encoding potential YjbH-binding proteins. Clones showing no activity in the presence of the bait vector pGBKT7 were sequenced, and the coding regions were identified with the BLAST function in Subtilist (http://genolist.pasteur.fr/SubtiList/) and NCBI. The genes are listed in Table S1 in the supplemental material. Two of the proteins, YosL and YqaH, were phage related: the former was encoded by the prophage SPβ (27), and the latter was encoded by the skin element (41, 43). Two proteins identified among the YjbH-interacting clones are transcriptional regulators CtsR and GabR, which could be potential substrates of YjbH-mediated proteolysis. Another protein identified as interacting with YjbH was the product of the yirB (yuzO) gene, which encodes a 54-amino-acid basic polypeptide that is conserved in B. subtilis, Bacillus pumilus, Bacillus licheniformis, and Bacillus amyloliquefaciens. The two activation domain fusion clones that were isolated contained the entire yirB coding sequence, including the starting Met codon, fused to the C-terminal coding end of the activation domain open reading frame. The product of the yirB-AD fusion clone showed no interaction with the Gal4 DNA-binding domain of the pGBKT7 vector or its fusion derivative bearing the product of the mutant yjbH(Δ1–24) allele coding region that lacks the Zn-binding domain (16) (Fig. 1A and B).
Fig. 1.
Yeast two-hybrid results showing interaction between YirB and YjbH proteins. (A) Schematic of plates shown in panel B. AD denotes activation domain fusion, and BD is the DNA-binding domain fusion. (B) Yeast strains bearing two-hybrid constructs. The plate on the left contains Trp− Leu− Ade− dropout medium; growth without Ade is indicative of YirB-YjbH fusion interaction. On the right is a control plate that selects for two hybrid plasmid-bearing strains. No evidence of interaction is observed when a control BD plasmid or a BD fusion construct made with the N-terminal deletion derivative of YjbH, YjbH(Δ1–24), is propagated in the yeast strain carrying the YirB-AD fusion plasmid. (C) Organization of the chromosomal region of B. subtilis where the yirB (yuzO) gene resides. The black arrow indicates the location and transcriptional orientation of yirB. The yirB gene resides downstream of the cssRS operon (white arrows) encoding the two-component regulatory system controlling the cell's response to secretion stress. Also shown are the location and transcriptional orientation of the htrB gene that is divergently transcribed from cssRS.
The yirB gene is not listed among the genes of B. subtilis strain 168 found in Subtilist but is located in the region between the cssS and yuxN genes (Fig. 1C) of B. subtilis 168 and other strains. The yirB gene is located downstream from the cssRS operon, but convergently oriented, suggesting that cssRS transcription would negatively impact yirB expression. The cssRS operon is induced by secretion stress and by heat shock and is positively autoregulated (9, 46).
YirB expression results in elevated Spx protein concentration and Spx-activated gene expression.
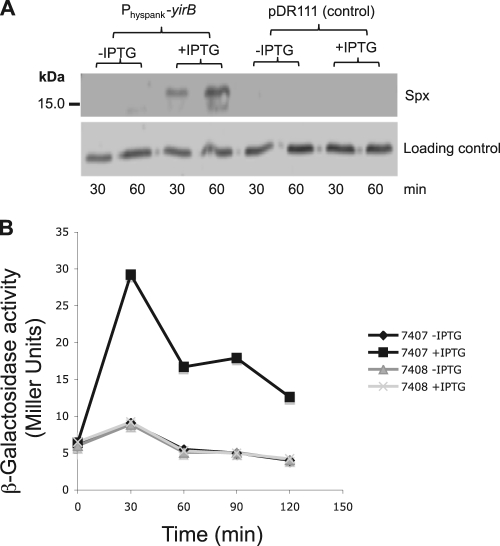
The yqaH, yosL, and yirB coding regions were placed under the control of the IPTG-inducible Physpank promoter of the expression vector pDR111 (4) to determine if induction of each of the genes and the resultant production of YjbH-binding products would lead to changes in Spx concentration and/or activity. Induction of yqaH and yosL expression from the Physpank constructs had no effect on Spx concentration (data not shown). IPTG induction of yirB from the pDR111 construct resulted in an elevated concentration of Spx compared to that of the uninduced control, as determined by Western analysis (Fig. 2A). The control strain bearing only the pDR111 vector showed no elevation of Spx concentration.
Fig. 2.
Expression of IPTG-inducible YirB results in an increase in Spx protein concentration. (A) Western blot of IPTG-induced Physpank-yirB cell (ORB7399) extracts, and those from IPTG-treated cells bearing the vector pDR111 (ORB7402). Cultures were untreated (−) or treated (+) with IPTG for 30 and 60 min before cells were harvested for Western blot analysis. (B) Induction of YirB production increases Spx-dependent expression of trxB-lacZ. A graph shows β-galactosidase activity encoded by the trxB-lacZ fusion in cells bearing the Physpank-yirB construct (ORB7407) or the pDR111 vector (ORB7408). Activity is plotted versus time of culture incubation. β-Galactosidase activity is expressed as Miller units (see Materials and Methods).
To determine if the induction of yirB and resulting increase in Spx concentration elevated Spx-dependent gene expression, the activity of a lacZ fusion with the promoter region of trxB (thioredoxin reductase gene), which requires Spx for optimal expression, was examined in induced and uninduced Physpank-yirB cells. Expression of trxB-lacZ was indeed elevated in the IPTG-treated Physpank-yirB fusion-bearing cells (Fig. 2B).
YirB interacts with YjbH in B. subtilis.
The results above suggested that YirB production increased Spx concentration by inhibiting Spx proteolysis through direct interaction with YjbH. Evidence that YirB and YjbH interact in B. subtilis was obtained by performing pulldown experiments using extracts from cells producing the His6-tagged YirB protein from an IPTG-inducible yirB construct. Cells of log-phase cultures grown in minimal TSS medium were harvested and lysed with a French press. Aliquots of lysates were mixed with Ni2+-NTA beads, and bound protein was collected by centrifugation. Eluted protein was subjected to SDS-PAGE and immunoblot analysis using anti-YjbH and anti-His6 antibody. The YjbH protein produced in wild-type cells was not observed to bind to the Ni2+-NTA beads (Fig. 3A), whereas YirB-His6 protein produced in B. subtilis adsorbed to the beads (Fig. 3B). When the fractions containing YirB-His were analyzed by Western blotting with anti-YjbH, the YjbH protein was detected (Fig. 3C). Binding of YjbH to the affinity matrix required the presence of the YirB-His6 protein, indicative of interaction (direct or indirect) between YirB and YjbH in the B. subtilis extract.
Fig. 3.
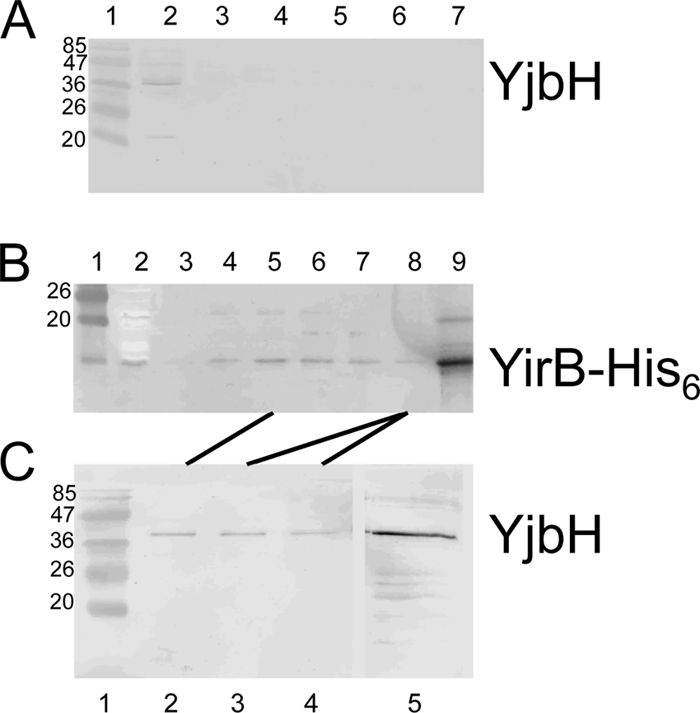
YirB interacts with YjbH in vivo. (A) Western blot of fractions eluted from a Ni2+-NTA column to which was applied extracts of B. subtilis strain ORB7437 (ΔyirB trxB-lacZ). Western blotting was performed with anti-YjbH antibody. Lane 1, marker with molecular mass (kDa) of proteins indicated; lane 2, extract prior to nickel affinity chromatography; lanes 3 to 7, eluted fractions from affinity column. (B) Western blot of fractions eluted from an Ni2+-NTA column to which was applied extracts of strain ORB7557 (ΔyirB Physpank-yirB-His6). Western blotting was performed with anti-His tag antiserum. Lane 1, molecular mass marker; lane 2, extract prior to affinity chromatography; lanes 3 to 8, eluted fractions from nickel affinity column; lane 9, purified YirB-His protein. (C) Western blot of the eluted fractions of panel B performed with anti-YjbH antiserum. Lane 1, molecular mass marker; lanes 2 to 4, fractions of lanes 5 and 8 in panel B; lane 5, purified YjbH. See Materials and Methods for details.
YirB prevents Spx interaction with YjbH.
The yeast two-hybrid and YirB-His6 pulldown results provided evidence that YirB interacts with YjbH and that the putative metal-binding domain of YjbH plays an essential role in this interaction. Furthermore, our previous studies (16) showed that the putative metal-binding domain of YjbH is also important for association with Spx, suggesting that Spx and YirB may be sharing the same interacting surfaces of YjbH. Moreover, we showed above that overexpression of YirB in B. subtilis resulted in the accumulation of Spx in vivo, confirming that YirB enhances the stability of Spx, likely through inhibition of YjbH-mediated proteolytic degradation by ClpXP. Therefore, we sought to determine whether YirB can prevent the interaction between Spx and YjbH.
To test this hypothesis, we first produced and purified YirB (see Fig. S1 in the supplemental material). The yirB gene was inserted into a pET-23a vector with the aim of overexpressing His6-tagged YirB protein in E. coli. YirB was purified by metal affinity chromatography. On SDS-PAGE, recombinant YirB corresponds to a molecular mass of ∼6 kDa, which is in agreement with predicted theoretical molecular mass of YirB with a 6×His tag (see Fig. S1A in the supplemental material). The His6-tagged version of YirB was produced in a B. subtilis strain bearing a yirB::erm null mutation (ORB7557). The production of active YirB-His with the Physpank-yirB-His construct in the ΔyirB background was evident by the enhancement of Spx function (trxB-lacZ expression) (see Fig. S1B in the supplemental material). Thus, it was shown that the YirB-His protein is active in vivo.
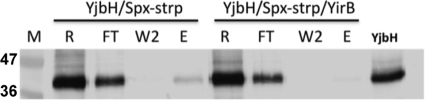
To test whether YirB can prevent the interaction of Spx and YjbH, a pulldown assay was carried out as described in Materials and Methods. Strep-II-tagged Spx was mixed with YjbH-His purified from B. subtilis. YjbH-His was found in the wash fractions, but a significant amount coeluted with Spx-Strep (Fig. 4). YjbH did not coelute with Spx-Strep when YirB-His was included in the reaction mixture (Fig. 4). The results obtained provide evidence that YirB, perhaps through a direct interaction with YjbH, prevented or disrupted YjbH binding to Spx-Strep.
Fig. 4.
YirB blocks interaction between Spx and YjbH in vitro. Data from an in vitro pulldown assay of YjbH-Spx-Strep (strp) protein interaction in the absence and presence of purified YirB-His. R, reaction; FT, column flowthrough fraction; W2, second wash fraction from column; E, elution. YjbH protein is used as marker in the far right lane. M, prestained molecular mass (kDa) marker.
YirB inhibits YjbH-mediated proteolysis of Spx by ClpXP.
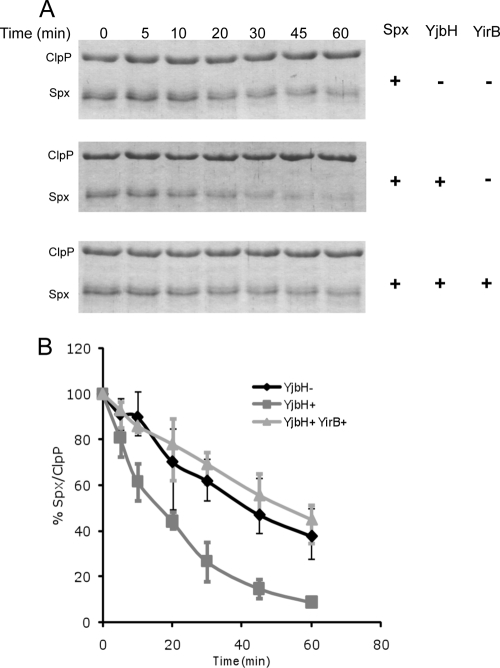
Based on the above results, the inhibition of Spx-YjbH interaction by YirB in the pulldown experiment along with accumulation of Spx and enhancement of its activity upon overexpression of YirB in Bacillus subtilis strongly suggested that YirB serves in a capacity that is reminiscent of antiadaptor proteins (5, 35) in that it prevents substrate interaction with a protein functioning as an adaptor necessary for Clp-catalyzed proteolysis. To test this assumption directly, we assembled an in vitro proteolytic system as described in Materials and Methods. YjbH catalyzed the Spx degradation by ClpXP in a time-dependent manner (Fig. 5), as reported previously (16). However, the rate of Spx degradation was comparable to that in the control reaction lacking YjbH when YirB was present. An ∼10-fold excess of Spx over YirB was added to the YjbH-catalyzed proteolytic reaction, suggesting that the affinity of YirB to YjbH is greater than that of Spx. This supports the model that YirB is an inhibitor of Spx proteolytic control and functions by modulating the YjbH activity by direct interaction.
Fig. 5.
YirB inhibits YjbH-enhanced proteolysis of Spx by ClpXP. (A) Spx (6 μM), ClpX (6 μM), and ClpP (12 μM) were incubated at 30°C in the presence of ATP, an ATP-generating system (creatine kinase), and 10 mM DTT with or without YjbH (6 μM) or YirB (0.5 μM). The top panel shows an SDS-PAGE gel profile of reactions without YjbH. The second panel shows a gel profile of reaction mixtures containing YjbH, and the bottom panel is a gel profile of reaction mixtures containing YirB. (B) A plot of Spx band intensities with the ClpP band as an internal control is shown as the standard error of the mean from three independent preparations.
DISCUSSION
A yeast two-hybrid screen for YjbH-interacting proteins uncovered a small protein encoded by the yirB (yuzO) gene of B. subtilis. YirB is a highly basic 54-amino-acid protein with a pI of 9.92. In addition to the yeast two-hybrid results, pulldown experiments using Ni2+-NTA beads and His6-tagged YirB protein showed that YirB and YjbH form a complex in B. subtilis extracts. In vitro, this interaction results in reduced binding of YjbH with Spx and a loss of YjbH-enhanced proteolysis of Spx by ClpXP protease. The results provide further confirmation of the proposed role of YjbH, as put forth by Larsson et al. (26), that YjbH accelerates Spx proteolysis catalyzed by ClpXP and also provide clues to how YjbH activity might be controlled. Our previous work (16) suggested that YjbH is under redox control that targets its Zn-binding domain, which upon oxidant treatment, releases its Zn ion. However, N termini among YjbH orthologs show striking sequence divergence, suggesting that oxidation of metal-binding domains might not be the only mechanism of YjbH control. The discovery of a small protein, YirB, that negatively affects YjbH activity indicates that YjbH is likely controlled by other cellular factors and, perhaps, under diverse environmental and stress conditions besides oxidative stress.
A deletion of the yirB (yuzO) gene does not show a phenotype with respect to trxB-lacZ fusion expression under unperturbed growth conditions or with respect to Spx protein levels under normal conditions or under disulfide stress. At present, we do not know the conditions under which YirB might be functional. Spx activity and concentration are known to be induced by a variety of harsh treatments and encounters with toxic compounds (1, 12, 30, 33, 36, 47), and a function for YirB within a stress response network has yet to be discovered. A possible explanation for yirB (yuzO) control relates to its chromosomal position, residing immediately downstream from the cssRS operon and convergently oriented with respect to cssRS (Fig. 1C). The CssRS system controls the expression of the stress-induced, protease/chaperone-encoding genes htrA and htrB (9, 23), and in this respect is similar to the CpxAR system of E. coli (8). Secretion stress, a condition that leads to the accumulation of misfolded proteins at the cytoplasmic membrane and cell wall, induces the transcription of cssRS. Transcriptional induction is also observed upon heat stress in an htrA mutant background (9). This transcription requires positive autoregulation by CssR, the response regulator of the CssRS pair. Transcription likely proceeds beyond the end of the cssRS operon and into the convergently oriented yirB (yuzO) coding sequence, which lies just 28 bp downstream from the stop codon of the cssS gene. While a sequence resembling a factor-independent terminator is detected at the end of the cssRS operon, part of its base-paired stem structure overlaps with the C-terminal coding region of cssS, suggesting that translation could prevent efficient terminator secondary structure formation. Thus, a speculative model of yirB control could involve the production of antisense RNA functioning in the downregulation of yirB expression. At present, there is no other supporting evidence that such a mechanism of yirB control operates in B. subtilis.
If YirB were downregulated by such a mechanism, YjbH activity would be upregulated, leading to, presumably, heightened turnover of Spx. Because Spx is known to interfere with activator-stimulated transcription (31, 48), the resulting reduction in Spx concentration would lessen Spx-dependent interference of CssR-stimulated transcription. Interestingly, induction of stress-induced transcription is most often the result of reversal of negative control exerted by repressors, such as HrcA, CtsR, PerR, OhrR, and YodB (15, 25, 28, 29, 39), or activation of transcription by the alternative RNA polymerase form bearing σB. Under these conditions, increases in Spx concentration and activity would not be expected to interfere with transcriptional induction. In contrast, the stress-induced CssRS system might be accompanied by a mechanism to reduce Spx concentration, so as to enhance CssR-stimulated transcription.
Several fusion products were found to test positive for YjbH interaction in the yeast two-hybrid screen. Two of the proteins, YosL and YqaH, are phage related, suggesting that YjbH might play a role in prophage induction control. Notably, one of the potential binding partners of YjbH is the heat shock negative transcriptional regulator CtsR. CtsR serves as a thermal sensor in the control of heat-induced genes (13). One outcome of the aforementioned control of yirB by cssRS operon transcription would be the heightened interaction of YjbH with CtsR, which could elevate the turnover rate of heat-denatured CtsR. Such a scenario would be compatible with physiological goals achieved by induction of the CssRS regulon and repression of yirB expression. The possible involvement of YjbH in the proteolytic control of CtsR and control of phage-related functions are areas of ongoing investigation.
Supplementary Material
ACKNOWLEDGMENTS
The work reported herein was supported by grant GM045898 from the National Institutes of Health.
We thank Ann A. T. Lin for purification of Spx-Strep.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Antelmann H., Hecker M., Zuber P. 2008. Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev. Proteomics 5:77–90 [DOI] [PubMed] [Google Scholar]
- 2. Baker T. A., Sauer R. T. 2006. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31:647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becker G., Klauck E., Hengge-Aronis R. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. U. S. A. 96:6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ben-Yehuda S., Rudner D. Z., Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 5. Bougdour A., Cunning C., Baptiste P. J., Elliott T., Gottesman S. 2008. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68:298–313 [DOI] [PubMed] [Google Scholar]
- 6. Bougdour A., Gottesman S. 2007. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc. Natl. Acad. Sci. U. S. A. 104:12896–12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bougdour A., Wickner S., Gottesman S. 2006. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev. 20:884–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Danese P. N., Snyder W. B., Cosma C. L., Davis L. J., Silhavy T. J. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387–398 [DOI] [PubMed] [Google Scholar]
- 9. Darmon E., et al. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Souza C., Nakano M. M., Zuber P. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 91:9397–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubnau D., Lovett C. M., Jr 2002. Transformation and recombination, p. 453–471 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 12. Eiamphungporn W., Helmann J. D. 2008. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elsholz A. K., Michalik S., Zuhlke D., Hecker M., Gerth U. 2010. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 29:3621–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fouet A., Jin S. F., Raffel G., Sonenshein A. L. 1990. Multiple regulatory sites in the Bacillus subtilis citB promoter region. J. Bacteriol. 172:5408–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuangthong M., Atichartpongkul S., Mongkolsuk S., Helmann J. D. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garg S. K., Kommineni S., Henslee L., Zhang Y., Zuber P. 2009. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 191:1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gottesman S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565–587 [DOI] [PubMed] [Google Scholar]
- 18. Gottesman S. 1999. Regulation by proteolysis: developmental switches. Curr. Opin. Microbiol. 2:142–147 [DOI] [PubMed] [Google Scholar]
- 19. Gottesman S., Maurizi M. R. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guerout-Fleury A. M., Shazand K., Frandsen N., Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 21. Hamoen L. W., Eshuis H., Jongbloed J., Venema G., van Sinderen D. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55–63 [DOI] [PubMed] [Google Scholar]
- 22. Hochgrafe F., et al. 2008. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 190:4997–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hyyrylainen H. L., et al. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159–1172 [DOI] [PubMed] [Google Scholar]
- 24. Kirstein J., Moliere N., Dougan D. A., Turgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7:589–599 [DOI] [PubMed] [Google Scholar]
- 25. Kruger E., Hecker M. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180:6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larsson J. T., Rogstam A., von Wachenfeldt C. 2007. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol. Microbiol. 66:669–684 [DOI] [PubMed] [Google Scholar]
- 27. Lazarevic V., et al. 1999. Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPβc2. Microbiology 145:1055–1067 [DOI] [PubMed] [Google Scholar]
- 28. Lee J. W., Helmann J. D. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367 [DOI] [PubMed] [Google Scholar]
- 29. Leelakriangsak M., et al. 2008. Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis. Mol. Microbiol. 67:1108–1124 [DOI] [PubMed] [Google Scholar]
- 30. Nakano S., Küster-Schöck E., Grossman A. D., Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakano S., Nakano M. M., Zhang Y., Leelakriangsak M., Zuber P. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakano S., Zheng G., Nakano M. M., Zuber P. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nguyen T. T., et al. 2009. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR). Mol. Microbiol. 71:876–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pratt L. A., Silhavy T. J. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U. S. A. 93:2488–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prepiak P., Dubnau D. 2007. A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP. Mol. Cell 26:639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rukmana A., Morimoto T., Takahashi H., Giyanto, Ogasawara N. 2009. Assessment of transcriptional responses of Bacillus subtilis cells to the antibiotic enduracidin, which interferes with cell wall synthesis, using a high-density tiling chip. Genes Genet. Syst. 84:253–267 [DOI] [PubMed] [Google Scholar]
- 37. Sauer R. T., et al. 2004. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schlothauer T., Mogk A., Dougan D. A., Bukau B., Turgay K. 2003. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. U. S. A. 100:2306–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schulz A., Schumann W. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Solomon J. M., Magnuson R., Srivastava A., Grossman A. D. 1995. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 9:547–558 [DOI] [PubMed] [Google Scholar]
- 41. Stragier P., Kunkel B., Kroos L., Losick R. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507–512 [DOI] [PubMed] [Google Scholar]
- 42. Studemann A., et al. 2003. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 22:4111–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takemaru K., Mizuno M., Sato T., Takeuchi M., Kobayashi Y. 1995. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology 141:323–327 [DOI] [PubMed] [Google Scholar]
- 44. Turgay K., Hahn J., Burghoorn J., Dubnau D. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turgay K., Hamoen L. W., Venema G., Dubnau D. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:119–128 [DOI] [PubMed] [Google Scholar]
- 46. Westers H., et al. 2006. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS Lett. 273:3816–3827 [DOI] [PubMed] [Google Scholar]
- 47. You C., et al. 2008. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiol. 8:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Nakano S., Choi S. Y., Zuber P. 2006. Mutational analysis of the Bacillus subtilis RNA polymerase α C-terminal domain supports the interference model of Spx-dependent repression. J. Bacteriol. 188:4300–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y., Zuber P. 2007. Requirement of the zinc-binding domain of ClpX for Spx proteolysis in Bacillus subtilis and effects of disulfide stress on ClpXP activity. J. Bacteriol. 189:7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou Y., Gottesman S. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.