Abstract
BacA of Sinorhizobium meliloti plays an essential role in the establishment of nitrogen-fixing symbioses with Medicago plants, where it is involved in peptide import and in the addition of very-long-chain fatty acids (VLCFA) to lipid A of lipopolysaccharide (LPS). We investigated the role of BacA in Rhizobium species strain NGR234 by mutating the bacA gene. In the NGR234 bacA mutant, peptide import was impaired, but no effect on VLCFA addition was observed. More importantly, the symbiotic ability of the mutant was comparable to that of the wild type for a variety of legume species. Concurrently, an acpXL mutant of NGR234 was created and assayed. In rhizobia, AcpXL is a dedicated acyl carrier protein necessary for the addition of VLCFA to lipid A. LPS extracted from the NGR234 mutant lacked VLCFA, and this mutant was severely impaired in the ability to form functional nodules with the majority of legumes tested. Our work demonstrates the importance of VLCFA in the NGR234-legume symbiosis and also shows that the necessity of BacA for bacteroid differentiation is restricted to specific legume-Rhizobium interactions.
INTRODUCTION
Symbiotic interactions between rhizobia, Gram-negative soil bacteria, and leguminous plants result in the formation of new plant organs called nodules. Rhizobia penetrate inside nodules and differentiate into bacteroids, which reduce atmospheric nitrogen to compounds the plant can assimilate. A successful interaction requires the correct exchange of molecular signals, which also determine the number of partners with which a legume host or rhizobial strain can develop functional symbioses (6, 27). Certain rhizobia, such as Sinorhizobium meliloti, can nodulate only a few legumes, whereas others, such as Rhizobium sp. strain NGR234, have a broad host range and can nodulate more than 120 genera of legumes (40).
There are several parallels between the mechanisms used to establish and maintain a functional bacteroid and those used by pathogenic bacteria during infection of eukaryotic cells. In fact, a successful symbiosis requires an “acute infection” (rhizobia have to enter cells of the plant root) and also a “chronic infection” (persistence of nitrogen-fixing bacteroids within nodules)—stages often associated with intracellular bacterial pathogens (13, 22). Several molecular signals, such as modifications of bacterial surface components, have been described to be involved in these two phases (12, 14).
BacA was one of the first rhizobial proteins shown to be required for the establishment of a successful chronic infection (23). Initially, bacA was identified by screening S. meliloti mutants for symbiotic deficiencies (34). bacA mutants invade nodules normally but then lyse and die before differentiating into functional nitrogen-fixing bacteroids. This observation led to the suggestion that BacA is required for nodule maturation and persistence (23). BacA is also important for prolonged intracellular survival during host-bacterium pathogenic interactions, such as chronic infection by Brucella abortus (32). S. meliloti bacA mutants display pleiotropic phenotypes. In addition to impaired nodulation with Medicago sativa (19), they are more resistant to the glycopeptide bleomycin and to aminoglycoside antibiotics (e.g., gentamicin). On the other hand, the absence of BacA results in an increased sensitivity to sodium dodecyl sulfate (SDS), deoxycholate (DOC), and ethanol, which suggests that there could be alterations in membrane integrity. Indeed, the S. meliloti bacA mutant was shown to be affected in its lipopolysaccharide (LPS) composition (19).
Bacterial LPS is typically composed of three parts: a hydrophobic domain known as lipid A, a nonrepeating “core” oligosaccharide, and a distal (predominantly repeating) polysaccharide (or O antigen) which may or may not be present. The correct assembly of rhizobial LPS is important for symbiosis, as mutants impaired in LPS synthesis frequently do not form functional nodules (9–11, 28, 39). Furthermore, changes to both the core and O antigen can occur during the symbiotic interaction. Rhizobium leguminosarum LPS becomes more hydrophobic during bacteroid development (15, 29), while Rhizobium etli LPS is also modified in response to plant exudates (38). In the case of NGR234, a new LPS species, characterized by the presence of a rhamnose-rich “rhamnan” O antigen, is synthesized upon induction by flavonoids, the initiating molecular signals of host plants (5, 21, 44).
Rhizobial lipid A contains a very-long-chain fatty acid (VLCFA; 27-OH-28:0), attached as the secondary fatty acid of an acyloxyacyl substituent on the distal glucosamine residue (24, 42, 43), which is important for symbiosis. For S. meliloti, R. etli, and R. leguminosarum, two genes involved in the addition of VLCFA to lipid A have been identified: lpxXL, which encodes a specific long-chain acyltransferase, and acpXL, which encodes a dedicated acyl carrier protein. VLCFA is synthesized attached to AcpXL in the cytoplasm and transferred onto the lipid A precursor by the inner membrane-associated LpxXL (2, 8, 17, 48). The VLCFA moiety is also found in a number of facultative intracellular pathogens that form chronic infections (e.g., Bartonella henselae, Brucella abortus, and Legionella pneumophila) (4, 54).
C28 fatty acids are not present in the lipid A fractions of S. meliloti lpxXL and acpXL mutants. As a consequence, the outer membrane is altered, and the cells are more sensitive to detergents (e.g., SDS and DOC). When these mutants are used to inoculate Medicago plants, they have reduced competitive abilities and take longer to induce nodules. Nevertheless, the nodules elicited on M. sativa and Medicago truncatula roots are normal and fix nitrogen, suggesting that VLCFA is important but not essential for chronic infection by S. meliloti (17, 48). LPS alterations in S. meliloti bacA mutants manifest as a significant decrease (about 50%) in the VLCFA content of lipid A (16, 19). This reduction in VLCFA on its own, however, cannot explain the symbiotic deficiency of this mutant, as acpXL, lpxXL, and acpXL lpxXL mutants are able to establish successful symbioses with alfalfa. Moreover, S. meliloti acpXL bacA and lpxXL bacA double mutants lack VLCFA but are as symbiotically defective as the bacA mutant, suggesting that BacA must have additional functions required for chronic infection (17).
BacA is a homologue of SbmA of Escherichia coli, a transporter of peptide antibiotics, and thus could be involved in peptide uptake in rhizobia. Like SbmA, BacA is an integral inner membrane protein with seven transmembrane domains. Involvement of S. meliloti BacA in peptide uptake was recently confirmed using a truncated form of the eukaryotic antimicrobial peptide Bac7 (35). BacA is essential for Bac7(1-16)-mediated cell death and for intracellular accumulation of fluorescently labeled Bac7(1-16). This phenotype is independent of the effect of BacA on lipid A. It is not known whether BacA is directly or indirectly involved in peptide uptake (35). Peptides are produced in developing nodules of legumes, and it has been suggested that these peptides have a key function in S. meliloti bacteroid development (51). It has been proposed that BacA-dependent peptide uptake could play a critical role in the chronic infection process of symbiotic and pathogenic bacteria (36).
BacA is essential for development of indeterminate pea nodules formed by R. leguminosarum bv. viciae but not of determinate bean nodules formed by R. leguminosarum bv. phaseoli and R. etli, although the bacA mutants of these strains show similar phenotypes under free-living conditions (30). Since the broad host range of NGR234 includes plants that form both determinate and indeterminate nodules, we tested the symbiotic phenotype of an NGR234 bacA mutant, as well as the role of BacA in LPS synthesis and peptide transport in NGR234.
MATERIALS AND METHODS
General microbiological and molecular techniques.
Standard molecular cloning techniques were used throughout this study (1, 45). All strains and plasmids used are listed in Table 1, and all oligonucleotide primers are listed in Table 2. E. coli strains were grown at 37°C with Luria-Bertani medium (45). NGR234 and its derivatives were grown at 27°C with TY medium (3) or rhizobial minimal medium supplemented with succinate (RMS) (7). Ampicillin (Ap), gentamicin (Gm), kanamycin (Km), rifampin (Rif), and spectinomycin (Sp) were added at concentrations of 100, 20 (10 for E. coli), 50, 100, and 50 μg/ml, respectively. Where appropriate, the flavonoid apigenin was added at 10−6 M to induce NGR234 strains.
Table 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | NBRC no. | Reference or source |
|---|---|---|---|
| Strains | |||
| Escherichia coli | |||
| DH5α | supE44 ΔlacU169(φ80dlacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | BRL, Bethesda, MD | |
| Rhizobium strainsa | |||
| NGR234 | Broad-host-range bacterium isolated from nodule of Lablab purpureus; Rifr | 101917 | 49 |
| NGRΔacpXL | NGR234 derivative with acpXL deleted; Rifr | 106209 | This work |
| NGRΩbacA | NGR234 derivative containing an Ω cassette inserted into the SmaI site of bacA; Rifr Spr | 106210 | This work |
| Plasmids | |||
| pBluescript II KS(+) | High-copy-number ColEI-based phagemid; Apr | Stratagene, La Jolla, CA | |
| pKS-acpXL | pBluescript KS(+) derivative carrying a 1.4-kb PCR fragment containing the acpXL gene; Apr | This work | |
| pKS-ΔacpXL | pKS-acpXL derivative in which the acpXL gene has been deleted; Apr | This work | |
| pKS-acpXLpro | pBluescript KS(+) derivative carrying a 0.5-kb PCR fragment containing the acpXL promoter; Apr | This work | |
| pKS-bacApro | pBluescript KS(+) derivative carrying a 0.5-kb PCR fragment containing the bacA promoter; Apr | This work | |
| pK18mobsacB | Suicide vector used to generate directed mutagenesis; Kmr | 46 | |
| pK18-ΔacpXL | pK18mobsacB derivative containing the acpXL deletion; Kmr | This work | |
| pJQ200SK+ | Suicide vector used to generate directed mutagenesis; Gmr | 41 | |
| pJQ-bacA | pJQ200SK+ derivative carrying bacA of NGR234; Gmr | This work | |
| pJQ-bacAΩSp | pJQ200SK derivative carrying the mutated bacAΩSp fragment; Gmr Spr | This work | |
| pRK2013 | Tra+ helper plasmid | 20 | |
| pPROBE-GT and pPROBE-GT′ | Broad-host-range promoter probe vectors containing the GFP open reading frame; Gmr | 37 | |
| pGT′-acpXL | pPROBE-GT′ derivative containing the acpXL promoter region; Gmr | This work | |
| pGT′-bacA | pPROBE-GT′ derivative containing the bacA promoter region; Gmr | This work | |
| pGT-fixF | pPROBE-GT derivative containing the fixF promoter region; Gmr | Antoine le Quéré, unpublished data |
NGR234 strains have been deposited at the NITE Database of Biological Resources (NBRC; http://www.nite.go.jp/) for long-term storage and distribution.
Table 2.
Oligonucleotide sequences used for this study
| Name | Sequence (5′–3′)a |
|---|---|
| acpXLfor | AAAGGATCCCACGCCGCGAATGATGTTTGC |
| acpXLrev | AAAAGCTTGGGGCCATCAATCCCACTTC |
| acpXLdel UP | AAAGAATTCATGCACCGCAAAAGCCAATGC |
| acpXLdel DOWN | AAAGAATTCAAGGCCTGATCGCCTCTGC |
| acpXLprom rev | AAAGGTACCCGCAAAAGCCAATGCCTCCC |
| bacAfor | CCAATCCTTCTTCCCGCAACC |
| bacArev | GGCCTTGATGAACTGCTGGTC |
| bacAprom for | AAAGGATCCGCTGCTGGTCGAGGCTGTC |
| bacAprom rev | AAAGGTACCGGGACGGCACTCGCTTTTCT |
Restriction sites are underlined.
Mutation of acpXL and bacA.
To delete acpXL, a 1.4-kb fragment containing acpXL and its flanking regions was amplified by PCR from NGR234 genomic DNA, using primers acpXLfor and acpXLrev (Table 2), and cloned into pBluescript II KS(+) to create pKS-acpXL. A PCR using primers acpXLdelUP and acpXLdelDOWN, with pKS-acpXL as a template, resulted in a PCR product deleted of acpXL. This product was digested with EcoRI and then circularized to create pKS-ΔacpXL. The accuracy of the PCR and the acpXL deletion were verified by sequencing of this plasmid. The 1.1-kb fragment containing the acpXL deletion was subcloned from pKS-ΔacpXL into the suicide vector pK18mobsacB (46) by use of BamHI and HindIII, generating pK18-ΔacpXL.
To mutate bacA, a 1.4-kb fragment containing bacA was amplified by PCR from genomic DNA, using primers bacAfor and bacArev (Table 2), digested with BamHI and XhoI, and cloned into the corresponding sites of pJQ200SK+ to create pJQ-bacA. An Ω cassette conferring resistance to spectinomycin was inserted into the unique SmaI site located within the cloned bacA open reading frame (ORF), generating pJQ-bacAΩSp.
Triparental matings using the helper plasmid pRK2013 (20) were used to transfer the resulting suicide plasmids, pK18-ΔacpXL and pJQ-bacAΩSp, into NGR234. Mutants with double recombination were selected by plating bacteria onto TY plates containing 5% sucrose and appropriate antibiotics. Putative mutants were confirmed by PCR and Southern blots of digested DNA, using standard procedures.
Transcriptional analyses with promoter-green fluorescent protein (GFP) fusions.
The promoter regions of bacA and acpXL were amplified by PCR from genomic DNA of NGR234, using the primers bacApromfor and bacApromrev for bacA and acpXLfor and acpXLpromrev for acpXL (Table 2). The fragments obtained were cloned into pBluescript II KS(+) and sequenced to verify PCR fidelity. The promoter regions were then subcloned into the broad-host-range promoter probe vector pPROBE-GT′ (37). Plasmids were mobilized from E. coli DH5α into NGR234 and mutant derivatives by triparental matings, using the helper plasmid pRK2013 (20).
Bacterial strains carrying pPROBE constructs were grown in RMS supplemented with the appropriate antibiotics for 48 h. These precultures were used to inoculate 10 ml of RMS to an optical density at 600 nm (OD600) of 0.1; to test gene induction by flavonoids or stress conditions, apigenin (10−6 M), polymyxin B (0.1 or 0.5 μg/ml), or NaCl (1% [wt/vol]) was added when appropriate. The optical density (600 nm) and fluorescence (excitation filter at 485 nm and emission filter at 528 nm) from 100-μl aliquots were recorded at 0, 6, 24, 48, and 72 h postinoculation, using a Synergy 2 multimode microplate reader (BioTek Instruments, Winooski, VT). At least three transcriptional assays were performed for each strain, and the fluorescence values obtained were normalized to the average optical density at each time point.
Nodulation tests.
Legume seeds were obtained from the suppliers listed by Pueppke and Broughton (40). Nodulation tests were performed in Magenta jars as described by Skorpil et al. (48a). Plants were grown at a day temperature of 28°C and a night temperature of 18°C, with a photoperiod of 16 h. Two plants were grown per Magenta jar, and each was inoculated with 107 bacteria. At harvest (6 weeks after inoculation for all plants, with the exception of Leucaena leucocephala, which was harvested at 7 weeks postinoculation), the aerial portion of the plant was removed and its dry weight recorded. Functional nodules were identified visually by their pink coloration due to the presence of leghemoglobin, an indicator of nitrogenase activity. The total number of active (pink) nodules and their fresh weight were determined for each replicate.
Polysaccharide analysis by SDS-PAGE and silver staining.
Strains were grown in RMS for 40 h and supplemented with 10−6 M apigenin where required. Polysaccharides were obtained from cells collected by centrifuging 4 ml of culture, as described previously (25). Briefly, the cell pellets were resuspended in 30 μl lysis buffer (1 M Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 4% [vol/vol] β-mercaptoethanol, 10% [vol/vol] glycerol, 0.03% [wt/vol] bromophenol blue) and boiled for 10 min. Lysed cells were treated with 10 μl protease K (2.5 mg/ml) at 60°C for 1 h, and then the samples were diluted by adding 80 μl of sample buffer (120 mM Tris-HCl [pH 6.8], 3% [wt/vol] SDS, 9% [vol/vol] β-mercaptoethanol, 30% [vol/vol] glycerol, 0.03% [wt/vol] bromophenol blue). Polysaccharides were separated by SDS-PAGE (18% acrylamide), and the gels were stained specifically for LPS or capsular polysaccharides (KPS) as described previously (31).
LPS extraction and analysis of hydroxy fatty acids.
Strains were grown in 1 liter of RMS for 40 h. Cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C and then washed twice with 1% (wt/vol) NaCl and twice with distilled H2O. Pellets were resuspended in 20 ml H2O, heated at 65°C, and then mixed with the same volume of hot phenol (at 65°C). Samples were incubated at 65°C for 15 min, and then the water and phenol phases were separated by centrifugation at 13,000 rpm for 15 min at 10°C. A second extraction with the same volume (20 ml) of H2O was performed, and the two water phases were combined. The combined water phases were then dialyzed against H2O and lyophilized. To determine the content of lipid A hydroxy fatty acids, this material was incubated at 80°C for 16 h in methanolic 1 M HCl, followed by trimethyl-silylization of the hydroxyl groups (53). The resulting derivatives were analyzed by gas chromatography with a 30-m SPB-1 column (Supelco; Sigma-Aldrich, St. Louis, MO). After injection, the oven temperature was raised 2°C min−1 from 150 to 180°C, 1°C min−1 from 180 to 200°C, and 5°C min−1 from 200 to 305°C and then maintained at 305°C for 25 min. The identity and relative molar response of each fatty acid were determined by comparison to parallel analyses of purified LPS of Salmonella enterica and R. etli strains whose lipid A structures and average compositions are known.
Sensitivity assays.
To test membrane integrity, bacteria were grown in liquid RMS for 48 h. The cells were then centrifuged, washed, and resuspended in fresh RMS to an OD600 of 0.2. For the filter disk assay, 500 μl of the resuspended culture was spread onto RMS plates. After 30 min, a paper disk (6-mm diameter; bioMérieux, Nürtingen, Germany) was applied to the center of the plate, and 5 μl of 10% (wt/vol) SDS was applied. At least three plates were prepared for each test. The plates were incubated for 72 to 96 h at 27°C, and then the diameters of the growth inhibition zones were recorded. For the DOC assay, RMS plates containing 0 mM, 2 mM, or 10 mM DOC were prepared, and 20 μl of culture was streaked for each strain to be tested. The plates were incubated at 27°C for 72 h. To test the sensitivity of strains to polymyxin B, the strains were precultured for 48 h in RMS and then washed and resuspended to an OD600 of 0.05 in RMS containing polymyxin B at different final concentrations. Growth was monitored by measuring the OD600 for the next 72 to 96 h.
To test the sensitivity of NGR234 to Bac7, strains were grown in liquid TY to early stationary phase and then diluted to an OD600 of 0.05 in fresh TY medium containing Bac7 at different final concentrations. Growth was monitored by measuring the OD600 for the next 72 to 96 h. The tests were repeated at least three times for each strain and stress condition.
RESULTS
Identification and mutation of acpXL and bacA in NGR234.
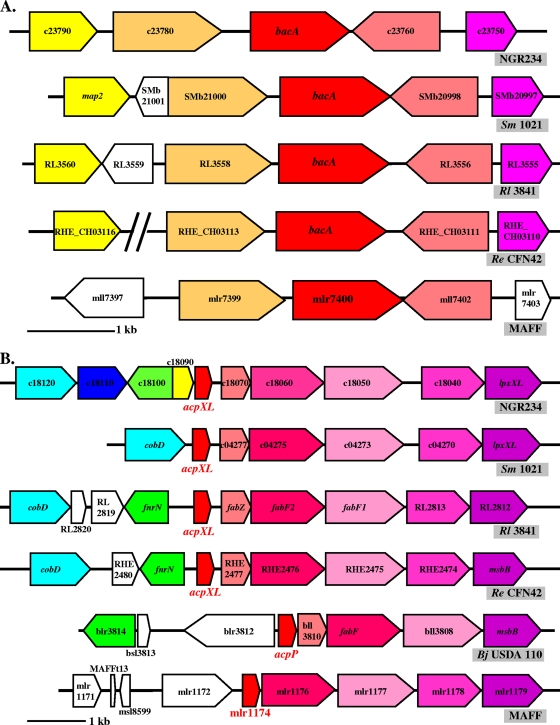
Homology searches using bacA of S. meliloti 1021 located a single bacA locus on the chromosome of NGR234 (Fig. 1A). bacA of NGR234 encodes a predicted protein of 378 amino acids belonging to a subfamily of the ABC transporter family (SbmA-BacA). The bacA loci from S. meliloti 1021, R. leguminosarum bv. viciae 3841, R. etli CFN42, Bradyrhizobium japonicum USDA 110, and Mesorhizobium loti MAFF303099 and the homology (to NGR234) of proteins encoded by the ORFs are shown in Fig. 1A and Table 3, respectively. The bacA region is well conserved in the rhizobia we examined, with the exception of B. japonicum USDA110 (Fig. 1A and Table 3). Despite this conservation, predicted gene functions give little insight into any possible symbiotic roles of the proteins encoded by this locus (Table 3). To mutate bacA, an ΩSpec cassette was inserted into the unique SmaI site of bacA (see Materials and Methods) to create NGRΩbacA.
Fig. 1.
bacA and acpXL loci of various rhizobia. Conservation of the bacA (A) and acpXL (B) loci is shown for NGR234, S. meliloti (Sm) 1021, R. leguminosarum bv. viciae (Rl) 3841, R. etli (Re) CFN42, and M. loti MAFF303099. In B. japonicum (Bj) USDA110, only a fragment of bacA is present and is surrounded by ORFs encoding putative transposases. Homologous genes are indicated by the same color. The genes are named according to their annotation in the published genomes (S. meliloti pSymB, GenBank accession no. NC_003078; R. leguminosarum bv. viciae 3841, NC_008380; R. etli CFN42, NC_007761; B. japonicum USDA110, NC_004463; and M. loti MAFF303099, NC_002678).
Table 3.
Homology of translated ORFs in the bacA loci of various rhizobia
| Rhizobium NGR234 ORF (no. of aa encoded) | Putative function of encoded protein |
S. meliloti 1021 |
R. leguminosarum 3841 |
R. etli CFN42 |
M. loti MAFF303099 |
||||
|---|---|---|---|---|---|---|---|---|---|
| ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ||
| c23790 (257) | Methionine aminopeptidase (Map2) | SMb21002 (249) | 92 (95) | RL3560 (252) | 75 (88) | RHE_CH03116 (252) | 74 (87) | ||
| c23780 (414) | Major facilitator superfamily (MSF) protein | SMb21000 (377) | 84 (88) | RL3558 (404) | 59 (73) | RHE_CH03113 (372) | 58 (71) | mlr7400 (401) | 49 (65) |
| bacA (378) | Cell envelope modification; peptide uptake | bacA (420) | 60 (75) | bacA (421) | 65 (79) | bacA (421) | 64 (77) | mlr7399 (419) | 53 (69) |
| c23760 (334) | Hypothetical protein | SMb20998 (334) | 83 (93) | RL3556 (328) | 62 (75) | RHE_CH03111 (329) | 60 (74) | mlr7402 (335) | 53 (67) |
| c23750 (192) | Putative transmembrane protein | SMb20997 (194) | 85 (93) | RL3555 (194) | 60 (77) | RHE_CH03110 (194) | 62 (78) | ||
We also mutated acpXL to create a VLCFA-negative strain to use as a comparison with NGRΩbacA. The acpXL-lpxXL locus of NGR234 was identified by homology searches using acpXL and lpxXL of S. meliloti 1021 (Fig. 1B). The region between acpXL and lpxXL includes four other ORFs that are also (based upon their homologies) probably involved in lipid A synthesis (Table 4). acpXL-lpxXL loci show preserved synteny, and the ORFs therein are highly conserved among different rhizobia, whereas the region upstream of acpXL is variable (Fig. 1B and Table 4). An in-frame deletion of acpXL was created, generating NGRΔacpXL (see Materials and Methods).
Table 4.
Homologies of translated ORFs in the acpXL loci of various rhizobia
| Rhizobium NGR234 ORF (no. of aa encoded) | Putative function of encoded protein |
S. meliloti 1021 |
R. leguminosarum 3841 |
R. etli CFN42 |
B. japonicum USDA110 |
M. loti MAFF303099 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ORF (no. of aa encoded) | % Identity (% similarity) | ||
| c18120 (326) | Cobalamin biosynthesis protein | cobD (327) | 78 (86) | cobD (326) | 65 (73) | cobD (326) | 63 (73) | ||||
| c18110 (264) | Hypothetical protein (linked to cobalamin synthesis?) | ||||||||||
| c18100 (246) | Transcriptional regulator (Fnr/CRP family) | fnrN (240) | 62 (80) | fnrN (239) | 64 (81) | blr3814 (282) | 24 (44) | ||||
| c18090 (113) | Hypothetical protein | ||||||||||
| acpXL (92) | Acyl carrier protein | acpXL (95) | 98 (100) | acpXL (92) | 95 (97) | acpXL (92) | 95 (97) | acpP (96) | 76 (90) | mlr1174 (93) | 82 (92) |
| c18070 (158) | Hydroxy-myristoyl acyl carrier protein dehydratase | SMc04277 (158) | 90 (95) | fabZ (160) | 80 (87) | RHE_CH2477 (160) | 81 (87) | bll3810 (156) | 48 (63) | ||
| c18060 (399) | Beta-ketoacyl-ACP synthase II | SMc04275 (400) | 90 (95) | fabF2 (401) | 73 (84) | RHE_CH2476 (401) | 73 (84) | fabF (401) | 44 (62) | mlr1176 (396) | 55 (67) |
| c18050 (427) | Beta-ketoacyl-ACP synthase II | SMc04273 (427) | 96 (98) | fabF1 (428) | 85 (91) | RHE_CH2475 (428) | 86 (92) | bll3808 (425) | 58 (72) | mlr1177 (416) | 67 (79) |
| c18040 (342) | Alcohol dehydrogenase protein | SMc04273 (342) | 95 (98) | RL2813 (342) | 93 (97) | RHE_CH2474 (342) | 94 (97) | mlr1178 (342) | 84 (92) | ||
| lpxXL (c18030) (309) | Acyltransferase | lpxXL (312) | 91 (94) | RL2812 (311) | 74 (86) | msbB (288) | 77 (86) | msbB (310) | 38 (56) | mlr1179 (328) | 59 (76) |
Measurement of acpXL and bacA expression in response to flavonoids.
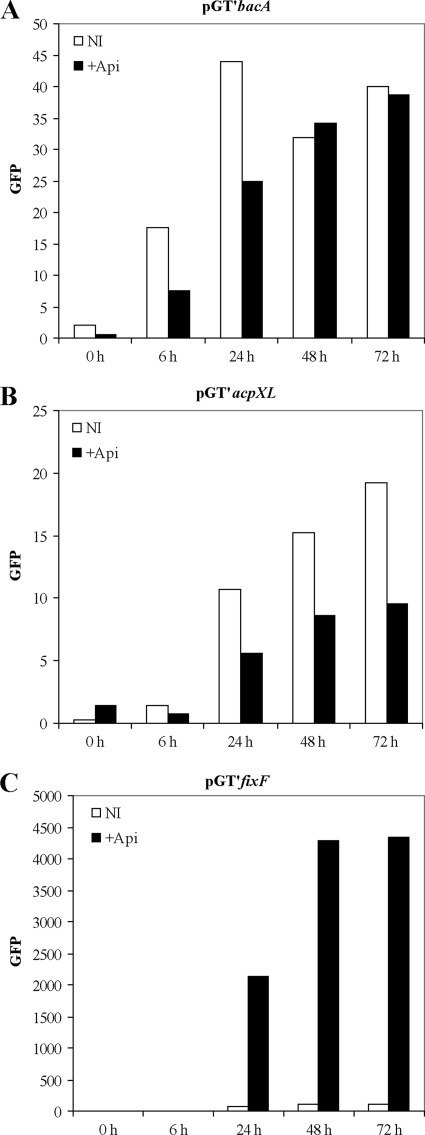
To determine whether the expression of acpXL and bacA is regulated by flavonoids, the promoter regions of the two genes were cloned upstream of GFP in the broad-host-range vector pPROBE-GT′ (see Materials and Methods). Fluorescence of NGR234 strains carrying the promoter-GFP fusions was measured in the presence or absence of flavonoids. The promoter region of a gene (fixF) similarly cloned upstream of GFP in pPROBE-GT′ (and known to be induced by flavonoids) was used as a positive control. Expression of both acpXL and bacA was low and constitutive (Fig. 2). Fluorescence of each mutant was also measured under stressful conditions (osmotic stress or polymyxin B), but no changes in expression were observed for either gene (data not shown).
Fig. 2.
Measurement of bacA and acpXL expression. Expression analyses were performed with the bacA (A), acpXL (B), and fixF (C) promoter regions. The promoter regions were fused to a promoterless gfp gene and assayed at different time points in RMS and RMS supplemented with 10−6 M apigenin. The values for the empty vector control were subtracted from the actual readings, and data were normalized according to the OD600. Numbers shown are the averages for three replicate experiments.
Nodulation tests.
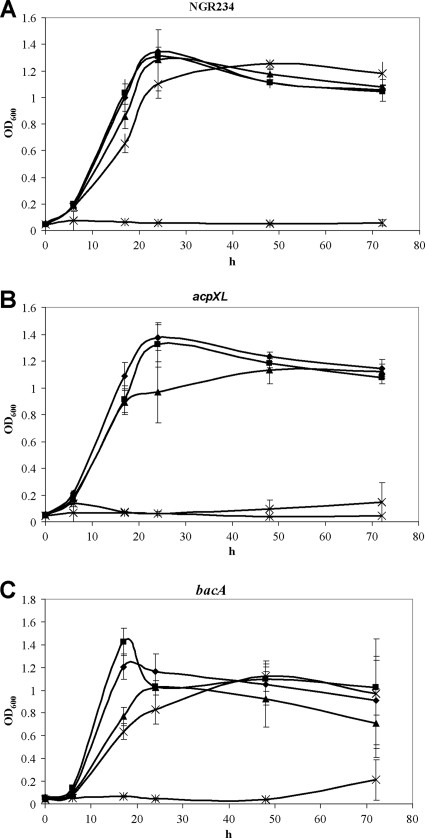
To investigate their symbiotic proficiency, the acpXL and bacA mutants were inoculated onto various plants belonging to different tribes and forming determinate or indeterminate nodules. No symbiotic differences were observed between the bacA mutant and NGR234 after inoculation onto Lablab purpureus (Phaseoleae), Leucaena leucocephala (Mimoseae), Tephrosia vogelii (Millettieae), or Vigna unguiculata (Phaseoleae) (Table 5). We tested more NGR234 host plants—Cajanus cajan, Macroptilium atropurpureum, Pachyrhizus tuberosus (all Phaseoleae), Lotus japonicus (Loteae), and Crotalaria juncea (Crotalarieae)—but again found that the bacA mutant caused no impairment in symbiosis with these legumes (relative to the wild type) (data not shown). In contrast, the acpXL mutant did not form functional (pink) nodules on L. purpureus, T. vogelii, or V. unguiculata, although numerous white (pseudo-)nodules were induced. L. leucocephala was efficiently nodulated by the acpXL mutant (Table 5), however.
Table 5.
Nodulation of Lablab purpureus, Leucaena leucocephala, Tephrosia vogelii, and Vigna unguiculata by NGR234 and derivative mutants
| Plant (nodule type)a and strain | Mean ± SDb |
||
|---|---|---|---|
| No. of nodules | Nodule wt (mg) | Dry wt (mg) | |
| Lablab purpureus (D) | |||
| Control | 0 | 0 | 700 ± 190 |
| NGR234 | 10 ± 4 | 520 ± 160 | 1,020 ± 280 |
| NGRΔacpXL | 0* ± 0 | 0* ± 0 | 650* ± 140 |
| NGRΩbacA | 10 ± 5 | 450 ± 220 | 1,020 ± 350 |
| Leucaena leucocephala (I) | |||
| Control | 0 | 0 | 90 ± 15 |
| NGR234 | 15 ± 3 | 140 ± 30 | 240 ± 40 |
| NGRΔacpXL | 11 ± 3 | 130 ± 20 | 230 ± 50 |
| NGRΩbacA | 13 ± 2 | 140 ± 60 | 240 ± 90 |
| Tephrosia vogelii (I) | |||
| Control | 0 | 0 | 150 ± 30 |
| NGR234 | 5 ± 4 | 200 ± 110 | 260 ± 100 |
| NGRΔacpXL | 0* ± 0 | 0* ± 0 | 150* ± 30 |
| NGRΩbacA | 4 ± 2 | 120 ± 80 | 200 ± 70 |
| Vigna unguiculata (D) | |||
| Control | 0 | 0 | 90 ± 20 |
| NGR234 | 80 ± 18 | 740 ± 150 | 1,320 ± 270 |
| NGRΔacpXL | 0* ± 0 | 0* ± 0 | 60* ± 10 |
| NGRΩbacA | 85 ± 28 | 820 ± 130 | 1,240 ± 350 |
D, determinate nodules; I, indeterminate nodules.
The figures listed are per plant. Only pink (i.e., functional) nodules were scored. Nodulation tests were repeated at least twice, with 6 to 8 plants being tested each time. Statistical analyses (Student's t test) were performed to compare each mutant to the wild-type strain. *, significant difference (P ≤ 0.01).
LPS and KPS profiles.
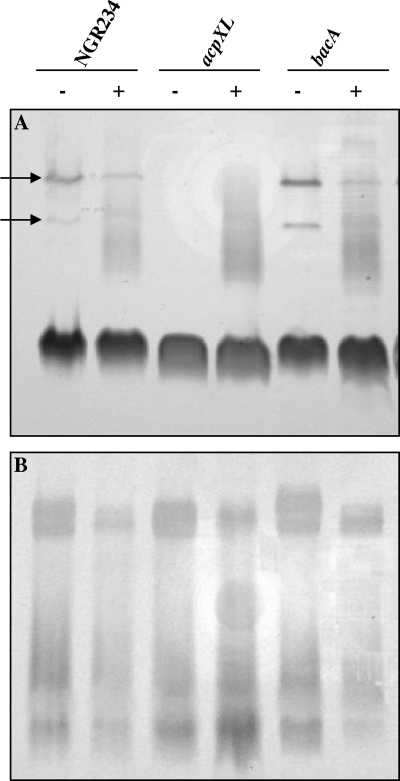
To check whether the production of rhamnan, an O-antigen species produced upon flavonoid induction of NGR234, was affected in the mutant strains, all three (NGR234 as well as the acpXL and bacA mutants) strains were grown in minimal (RMS) medium (with or without flavonoids), and their polysaccharide (LPS and KPS) profiles were analyzed by SDS-PAGE (see Materials and Methods). LPS and KPS profiles of the bacA mutant were indistinguishable from those of NGR234 (Fig. 3). Similarly, the acpXL mutant showed the same KPS profile as NGR234 and made rhamnan in the presence of flavonoids, but the low-mobility bands usually seen in the absence of apigenin (indicated by arrows in Fig. 3A) were absent.
Fig. 3.
LPS and KPS profiles of NGR234 and its mutants. SDS-PAGE analysis was performed to detect LPS (A) and KPS (B) synthesized by NGR234 and the acpXL and bacA mutants grown in the absence (−) or presence (+) of apigenin. The arrows indicate the low-mobility bands present in noninduced samples from NGR234 and NGRΩbacA that are absent in NGRΔacpXL.
Fatty acid composition of LPS.
Crude LPS extracts of NGR234, NGRΔacpXL, and NGRΩbacA were analyzed to determine the contents of hydroxy fatty acids characteristic of lipid A. Only traces of VLCFA (27-OH-28:0) were present in the acpXL mutant, but its content of 3-hydroxy fatty acids (Fig. 4 and Table 6) was similar to that of NGR234. No differences in the VLCFAs as well as the major 3-hydroxy fatty acids of NGRΩbacA and NGR234 were found (Fig. 4 and Table 6).
Fig. 4.
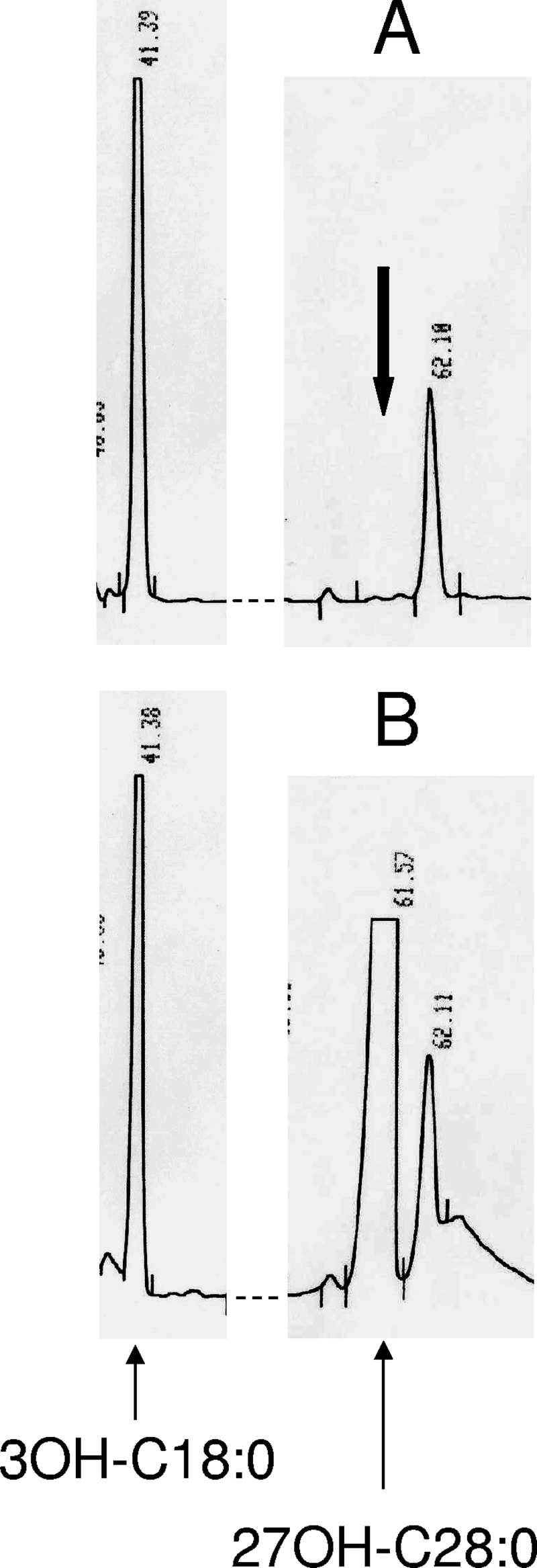
Detection of VLCFA in NGRΔacpXL and NGRΩbacA. Gas chromatograms for NGRΔacpXL (A) and NGRΩbacA (B) LPS extracts show where 27OH-C28:0 emerges and, as a loading control, where 3OH-C18:0 peaks. The relative absence of 27OH-C28:0 in NGRΔacpXL is indicated by the arrow in panel A.
Table 6.
Hydroxy fatty acid compositions of NGR234 and derivatives normalized to glucosamine
| Strain | Relative amt of hydroxy fatty acida |
|||
|---|---|---|---|---|
| 14:0 | 16:0 | 18:0 | 28:0 | |
| NGR234 | 2.09 | 0.35 | 0.63 | 0.81 |
| NGRΔacpXL | 2.95 | 0.20 | 0.55 | 0.01 |
| NGRΩbacA | 2.82 | 0.30 | 0.50 | 0.83 |
The relative molar contents of 3-hydroxymyristic acid (14:0), 3-hydroxypalmitic acid (16:0), 3-hydroxystearic acid (18:0), and 27-hydroxyoctacosanoic acid (28:0) per 2.0 mol of glucosamine are shown.
Sensitivity assays.
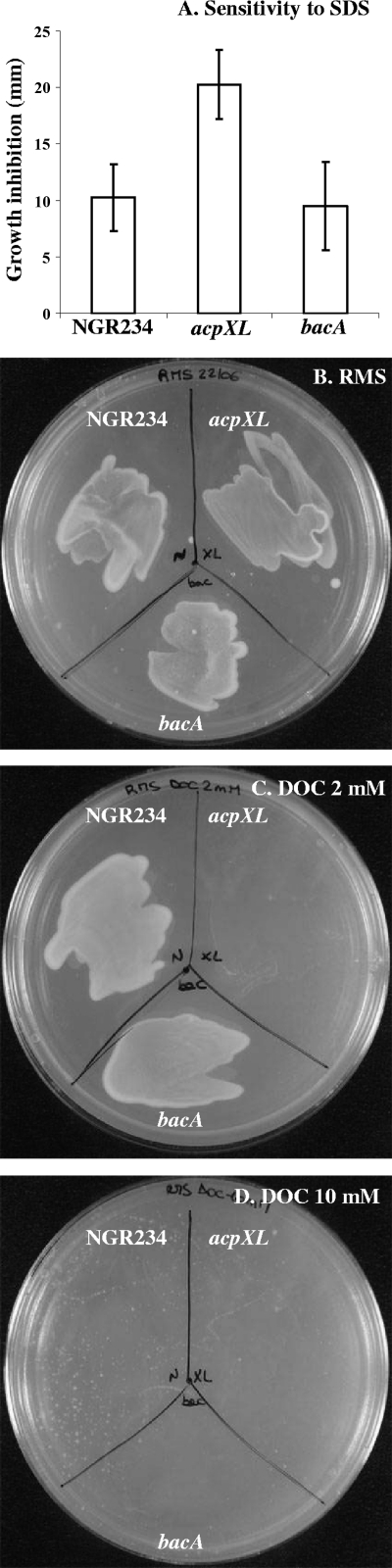
Bacterial mutants affected in LPS in general, and lipid A in particular, are generally sensitive to various stresses. As an example, agents that affect membranes, such as detergents or polymyxin B (an antimicrobial peptide which binds to LPS and permeabilizes the cell membrane), can be used to assay membrane integrity. The sensitivity of NGR234, NGRΔacpXL, and NGRΩbacA to SDS and DOC was tested on RMS plates. In the presence of detergents, the growth of the acpXL mutant was significantly inhibited compared to that of NGR234, whereas the bacA mutant was not affected (Fig. 5). Sensitivity to polymyxin B was tested in liquid cultures. In the presence of 0.25 μg/ml polymyxin B, the growth of the acpXL mutant was significantly slower than the growth of NGR234, and it stopped completely at concentrations of ≥0.5 μg/ml, while the growth of NGR234 was only slightly reduced at these concentrations. As in the case of detergents, the bacA mutant displayed the same phenotype as NGR234 (Fig. 6).
Fig. 5.
Sensitivity to detergents. (A) NGR234, NGRΔacpXL, and NGRΩbacA were plated on RMS agar, and 5 μl of 10% (wt/vol) SDS was added on a paper disk. The diameters of growth inhibition were recorded after 72 to 96 h at 27°C. (B to D) Sensitivities of NGR234, NGRΔacpXL, and NGRΩbacA to RMS (B), RMS + 2 mM DOC (C), and RMS + 10 mM DOC (D).
Fig. 6.
Sensitivity to polymyxin B. Growth of NGR234 (A), NGRΔacpXL (B), and NGRΩbacA (C) was measured in liquid RMS (♦) and RMS supplemented with polymyxin B at different final concentrations (■, 0.1 μg/ml; ▴, 0.25 μg/ml; ×, 0.5 μg/ml; *, 1 μg/ml). The values shown are the averages for three replicate experiments.
BacA as a peptide transporter.
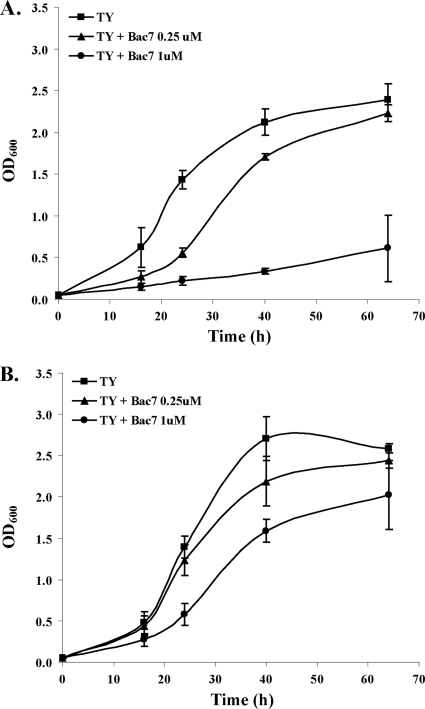
BacA of S. meliloti is involved in peptide transport, and a bacA mutant is more resistant than the parent strain to the antimicrobial peptide Bac7 (which has been shown to inhibit DnaK as well as other unknown targets [47]) and to the glycopeptide antibiotic bleomycin (18, 26, 32). Since NGR234 is naturally resistant to bleomycin, this assay could not be used, but the bacA mutant was similarly resistant, implying that bleomycin resistance is expressed through a mechanism that does not involve BacA (data not shown). On the other hand, Bac7 inhibited the growth of NGR234, but NGRΩbacA was significantly less susceptible. The sensitivity of NGR234 and NGRΩbacA to Bac7 was tested by measuring the growth inhibition in liquid TY medium in the presence of different concentrations of Bac7 (Fig. 7). Furthermore, the sensitivity of the acpXL mutant to Bac7 was also tested, but this mutant showed the same phenotype as NGR234 (data not shown).
Fig. 7.
NGRΩbacA has increased resistance to Bac7. Growth was measured for NGR234 (A) and the bacA mutant NGRΩbacA (B) in liquid TY containing different concentrations of Bac7. The graphs represent the means (± SD) for three different experiments.
DISCUSSION
Our data show that bacA influences neither the VLCFA content nor the LPS of NGR234. This is in stark contrast to the S. meliloti bacA mutant, which induces only nonfunctional nodules and produces lipid A with reduced quantities of VLCFA. Since S. meliloti acpXL and lpxXL mutants lack VLCFA but remain symbiotically proficient (16, 17, 33), BacA must have other functions critical for chronic infection in nodules. BacA of S. meliloti is involved in the uptake of synthetic peptides (35). Thus, BacA may have a role in the recognition/transport of plant peptides required for bacteroid differentiation within the indeterminate nodules formed specifically by galegoid legumes (e.g., Medicago and Vicia species), but not with phaseoloid legumes forming determinate nodules (30, 51). BacA is important in the symbiosis between Mesorhizobium huakuii 7653R and Astragalus sinicus, which also belongs to the Galegeae. An M. huakuii 7653R bacA mutant is defective in nitrogen fixation during symbiosis. The same mutant under free-living conditions is also sensitive to cell envelope-disrupting agents, is resistant to bleomycin and has reduced amounts of VLCFA in its lipid A (50). R. leguminosarum bv. phaseoli and R. etli bacA mutants display similar phenotypes in their free-living states, but they are able to establish functional symbioses (30). Our data show that BacA of NGR234 might be involved uniquely in peptide uptake (but not to permit bacteroid differentiation) and has no role in VLCFA synthesis or transport.
The NGR234 acpXL mutant did not possess VLCFA in its lipid A. Although we cannot rule out the possibility that the nature of the mutation affected downstream genes, this absence of VLCFA had detrimental effects on the ability of this strain to nodulate several legumes. The fact that the NGR234 acpXL mutant was not able to nodulate most of the plants tested was also unexpected, since acpXL mutants of S. meliloti and R. leguminosarum, although less efficient, still induce functional nodules (17, 48, 53). In the case of R. leguminosarum, the acpXL mutant lacks VLCFA in vitro, but lipid A extracted from acpXL mutant-containing bacteroids of R. leguminosarum possessed VLCFA. This suggests that in this strain, there is another acp gene that is activated during symbiosis which is able to replace AcpXL function and add VLCFA to lipid A (52). One possible reason for the disparity in phenotypes between the NGR234 and R. leguminosarum acpXL mutants is that NGR234 does not possess functional acp homologues.
Why is VLCFA important for symbiosis, and particularly so for NGR234, even though expression of acpXL in NGR234 is independent of flavonoids? Possible explanations were revealed by studying the acpXL mutant under free-living conditions. The absence of VLCFA compromises membrane integrity, as shown by the mutant's heightened sensitivities to membrane-disrupting agents (detergents and polymyxin B). The fact that VLCFA is needed for maximal outer membrane stability is likely to be particularly acute during rhizobial uptake into plant cells as well as in rhizobial persistence within the cortical cells as bacteroids. At these stages of the symbiosis, rhizobia, especially at their outer surfaces, are subjected to osmotic and possibly oxidative stresses as well as plant defense reactions. In a similar vein, it has also been suggested that VLCFA can span the whole outer bacterial membrane, significantly contributing to its stability (53).
We have shown that BacA is not necessary for bacteroid development by NGR234 in nongalegoid legumes that form either determinate or indeterminate nodules. BacA is responsible for resistance to Bac7 but does not affect the VLCFA content of LPS or membrane stability. On the other hand, the presence of VLCFA in the lipid A of NGR234 is essential for successful symbiotic interactions with most plants tested.
ACKNOWLEDGMENTS
We are very grateful to Yin-Yin Aung for nodulation tests and plant handling. We thank Dora Gerber for her unstinting help. We also acknowledge the comments of the three anonymous reviewers of this work.
This work was supported by the Fonds National Suisse de la Recherche Scientifique (projects 3100AO-104097 and 3100A0-116858 to W.J.B. and W.J.D.), the Département de l'Instruction Publique du Canton de Genève (W.J.B. and W.J.D.), the Université de Genève (to W.J.B.), grant DE-FG02-98ER-20307 from the U.S. Department of Energy (to K.D.N.), National Institutes of Health grant GM31010 (to G.C.W.), and the MIT Center for Environmental Health Sciences NIEHS (grant P30 ES002109). G.C.W. is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 25 February 2011.
REFERENCES
- 1. Ausubel F. M., et al. (ed.). 1991. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 2. Basu S. S., Karbarz M. J., Raetz C. R. 2002. Expression cloning and characterization of the C28 acyltransferase of lipid A biosynthesis in Rhizobium leguminosarum. J. Biol. Chem. 277:28959–28971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beringer J. E. 1974. R-factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188–198 [DOI] [PubMed] [Google Scholar]
- 4. Bhat U. R., Carlson R. W., Busch M., Mayer H. 1991. Distribution and phylogenetic significance of 27-hydroxy-octacosanoic acid in lipopolysaccharides from bacteria belonging to the alpha-2 subgroup of Proteobacteria. Int. J. Syst. Bacteriol. 41:213–217 [DOI] [PubMed] [Google Scholar]
- 5. Broughton W. J., et al. 2006. Flavonoid-inducible modifications to rhamnan O antigens are necessary for Rhizobium sp. strain NGR234-legume symbioses. J. Bacteriol. 188:3654–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Broughton W. J., Jabbouri S., Perret X. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Broughton W. J., et al. 1986. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J. Cell Biol. 102:1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brozek K. A., Carlson R. W., Raetz C. R. 1996. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J. Biol. Chem. 271:32126–32136 [DOI] [PubMed] [Google Scholar]
- 9. Campbell G. R., et al. 2003. Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J. Bacteriol. 185:3853–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell G. R. O., Reuhs B. L., Walker G. C. 2002. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. U. S. A. 99:3938–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cava J. R., Elias P. M., Turowski D. A., Noel K. D. 1989. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J. Bacteriol. 171:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deakin W. J., Broughton W. J. 2009. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7:312–320 [DOI] [PubMed] [Google Scholar]
- 13. Den Herder G., Parniske M. 2009. The unbearable naivety of legumes in symbiosis. Curr. Opin. Plant Biol. 12:491–499 [DOI] [PubMed] [Google Scholar]
- 14. D'Haeze W., Holsters M. 2004. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12:555–561 [DOI] [PubMed] [Google Scholar]
- 15. D'Haeze W., Leoff C., Freshour G., Noel K. D., Carlson R. W. 2007. Rhizobium etli CE3 bacteroid lipopolysaccharides are structurally similar but not identical to those produced by cultured CE3 bacteria. J. Biol. Chem. 282:17101–17113 [DOI] [PubMed] [Google Scholar]
- 16. Ferguson G. P., et al. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. U. S. A. 101:5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferguson G. P., Datta A., Carlson R. W., Walker G. C. 2005. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol. 56:68–80 [DOI] [PubMed] [Google Scholar]
- 18. Ferguson G. P., Jansen A., Marlow V. L., Walker G. C. 2006. BacA-mediated bleomycin sensitivity in Sinorhizobium meliloti is independent of the unusual lipid A modification. J. Bacteriol. 188:3143–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferguson G. P., Roop R. M., Walker G. C. 2002. Deficiency of a Sinorhizobium meliloti bacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184:5625–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Figurski D. H., Helinski D. R. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fraysse N., Jabbouri S., Treilhou M., Couderc F., Poinsot V. 2002. Symbiotic conditions induce structural modifications of Sinorhizobium sp. NGR234 surface polysaccharides. Glycobiology 12:741–748 [DOI] [PubMed] [Google Scholar]
- 22. Gibson K. E., Kobayashi H., Walker G. C. 2008. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42:413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Glazebrook J., Ichige A., Walker G. C. 1993. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7:1485–1497 [DOI] [PubMed] [Google Scholar]
- 24. Gudlavalleti S. K., Forsberg L. S. 2003. Structural characterization of the lipid A component of Sinorhizobium sp. NGR234 rough and smooth form lipopolysaccharide. J. Biol. Chem. 278:3957–3968 [DOI] [PubMed] [Google Scholar]
- 25. Hitchcock P. J., Brown T. M. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ichige A., Walker G. C. 1997. Genetic analysis of the Rhizobium meliloti bacA gene: functional interchangeability with the Escherichia coli sbmA gene and phenotypes of mutants. J. Bacteriol. 179:209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones K. M., Kobayashi H., Davies B. W., Walker G. C. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanipes M. I., Ribeiro A. A., Lin S., Cotter R. J., Raetz C. R. 2003. A mannosyl transferase required for lipopolysaccharide inner core assembly in Rhizobium leguminosarum. Purification, substrate specificity, and expression in Salmonella waaC mutants. J. Biol. Chem. 278:16356–16364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kannenberg E. L., Carlson R. W. 2001. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39:379–391 [DOI] [PubMed] [Google Scholar]
- 30. Karunakaran R., et al. 2010. BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 192:2920–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le Quéré A. J.-L., et al. 2006. Structural characterization of a K-antigen capsular polysaccharide essential for normal symbiotic infection in Rhizobium sp. NGR234. J. Biol. Chem. 281:28981–28992 [DOI] [PubMed] [Google Scholar]
- 32. Le Vier K., Phillips R. W., Grippe V. K., Roop R. M., Walker G. C. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492–2493 [DOI] [PubMed] [Google Scholar]
- 33. Le Vier K., Walker G. C. 2001. Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J. Bacteriol. 183:6444–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Long S., McCune S., Walker G. C. 1988. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J. Bacteriol. 170:4257–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marlow V. L., et al. 2009. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol. 191:1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mergaert P., et al. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U. S. A. 103:5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller W. G., Leveau J. H., Lindow S. E. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 38. Noel K. D., Box J. M., Bonne V. J. 2004. 2-O-methylation of fucosyl residues of a rhizobial lipopolysaccharide is increased in response to host exudate and is eliminated in a symbiotically defective mutant. Appl. Environ. Microbiol. 70:1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noel K. D., Vandenbosch K. A., Kulpaca B. 1986. Mutations in Rhizobium phaseoli that lead to arrested development of infection threads. J. Bacteriol. 168:1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pueppke S. G., Broughton W. J. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host-ranges. Mol. Plant-Microbe Interact. 12:293–318 [DOI] [PubMed] [Google Scholar]
- 41. Quandt J., Hynes M. F. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 42. Que N. L., Lin S., Cotter R. J., Raetz C. R. 2000. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. Demonstration of a conserved distal unit and a variable proximal portion. J. Biol. Chem. 275:28006–28016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Que N. L., Ribeiro A. A., Raetz C. R. 2000. Two-dimensional NMR spectroscopy and structures of six lipid A species from Rhizobium etli CE3. Detection of an acyloxyacyl residue in each component and origin of the aminogluconate moiety. J. Biol. Chem. 275:28017–28027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reuhs B. L., et al. 2005. Structural characterization of a flavonoid-inducible Pseudomonas aeruginosa A-band-like O antigen of Rhizobium sp. strain NGR234, required for the formation of nitrogen-fixing nodules. J. Bacteriol. 187:6479–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sambrook J., Fritsch E. F., Maniatis T. (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Schäfer A., et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 47. Scocchi M., et al. 2009. The proline-rich antibacterial peptide Bac7 binds to and inhibits in vitro the molecular chaperone DnaK. Int. J. Pept. Res. Ther. 15:147–155 [Google Scholar]
- 48. Sharypova L. A., Niehaus K., Scheidle H., Holst O., Becker A. 2003. Sinorhizobium meliloti acpXL mutant lacks the C28 hydroxylated fatty acid moiety of lipid A and does not express a slow migrating form of lipopolysaccharide. J. Biol. Chem. 278:12946–12954 [DOI] [PubMed] [Google Scholar]
- 48a. Skorpil, et al. 2005. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of modulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57:1304–1317 [DOI] [PubMed] [Google Scholar]
- 49. Stanley J., Dowling D. N., Broughton W. J. 1988. Cloning of hemA from Rhizobium sp. NGR234 and symbiotic phenotype of a gene-directed mutant in diverse legume genera. Mol. Gen. Genet. 215:32–37 [Google Scholar]
- 50. Tan X. J., Cheng Y., Li Y. X., Li Y. G., Zhou J. C. 2009. BacA is indispensable for successful Mesorhizobium-Astragalus symbiosis. Appl. Microbiol. Biotechnol. 84:519–526 [DOI] [PubMed] [Google Scholar]
- 51. Van de Velde W., et al. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327:1122–1126 [DOI] [PubMed] [Google Scholar]
- 52. Vedam V., et al. 2006. The pea nodule environment restores the ability of a Rhizobium leguminosarum lipopolysaccharide acpXL mutant to add 27-hydroxyoctacosanoic acid to its lipid A. J. Bacteriol. 188:2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vedam V., et al. 2003. A Rhizobium leguminosarum acpXL mutant produces lipopolysaccharide lacking 27-hydroxyoctacosanoic acid. J. Bacteriol. 185:1841–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zahringer U., et al. 1995. The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog. Clin. Biol. Res. 392:113–139 [PubMed] [Google Scholar]