Abstract
In Selenomonas ruminantium, a strictly anaerobic and Gram-negative bacterium, cadaverine covalently linked to the peptidoglycan is required for the interaction between the peptidoglycan and the S-layer homologous (SLH) domain of the major outer membrane protein Mep45. Here, using a series of diamines with a general structure of NH3+(CH2)nNH3+ (n = 3 to 6), we found that cadaverine (n = 5) specifically serves as the most efficient constituent of the peptidoglycan in acquiring the high resistance of the cell to external damage agents and is required for effective interaction between the SLH domain of Mep45 and the peptidoglycan, facilitating the correct anchoring of the outer membrane to the peptidoglycan.
TEXT
The cell surface structure of Selenomonas ruminantium has a typical Gram-negative three-layer organization comprised by the cytoplasmic membrane, the peptidoglycan layer, and the outer membrane (7) but is characterized by the exceptional feature of the absence of murein lipoprotein, which is widely distributed among Gram-negative bacteria and serves as a structural protein that anchors the outer membrane to the peptidoglycan (2, 11). Instead, the peptidoglycan of this bacterium contains cadaverine covalently linked to the α-carboxy group of the d-glutamic acid residue (5, 6). In our previous study, it was revealed that peptidoglycan-linked cadaverine is required for the interaction between the peptidoglycan and the periplasm-exposed S-layer homologous (SLH) domain of Mep45, a major outer membrane protein, and it was shown that the structural connection between the outer membrane and the peptidoglycan depends on the presence of peptidoglycan-linked cadaverine (9). This cadaverine-mediated interaction system was considered a functional counterpart of murein lipoprotein. The anchoring mechanism between the outer membrane and the peptidoglycan in Gram-negative bacteria is implicated in maintaining cell surface integrity and thus is responsible for cell resistance to external damage agents, such as antibiotics, detergents, and dyes (1, 2, 3, 11). Here, we focused on the biological significance of the cadaverine-mediated anchoring mechanism in S. ruminantium and the structural specificity of the diamine requirement for this mechanism, since cadaverine is not the conventional polyamine found in most eubacteria, as their major polyamines are usually putrescine and spermidine (12). In this bacterium, cadaverine is synthesized by the decarboxylation of l-lysine catalyzed by lysine/ornithine decarboxylase (LDC/ODC) (13). However, this enzyme shows significant homology to eukaryotic ornithine decarboxylase (ODC), an enzyme responsible for putrescine synthesis through the decarboxylation of l-ornithine, rather than to LDC (14, 15). In our previous study, it was found that five amino acid residues located in the catalytic domain of LDC/ODC are responsible for this notable substrate preference for lysine, and substituting these residues for the corresponding residues of ODC resulted in the conversion of substrate specificity toward l-ornithine (15). These facts lead us to assume that cadaverine may possess some inherent biological significance compared to putrescine or other polyamines, as one of the possible explanations for this characteristic molecular conversion of ODC to LDC. Furthermore, our previous study clarified that the cellular level and activity of LDC/ODC are tightly regulated in an antizyme-mediated manner, in which ribosomal protein L10 binds to LDC/ODC and triggers ATP-dependent proteolysis of LDC/ODC (16, 17). This antizyme-mediated proteolysis essentially occurs at the early stationary phase but is also shown to be inducible by the presence of putrescine, suggesting that the activity of LDC/ODC is highly regulated to focus on the exclusive synthesis of cadaverine. These findings tempt us to investigate the specific biological significance of cadaverine covalently linked to the peptidoglycan. To this end, we have examined the structural specificity of the diamine requirement for cell growth, maintenance of cell surface integrity, and resistance of the cell to external damage agents such as antibiotics, detergents, or dyes, using a series of diamines with the structure NH3+(CH2)nNH3+ (n = 3 to 6). For convenience, we designate each diamine by its carbon chain length, such as C5 and C4 for cadaverine [NH3+(CH2)5NH3+] and putrescine [NH3+(CH2)4NH3+], respectively.
C3, C4, and C6 all incorporate into the peptidoglycan in vivo at the same level as C5 but are insufficient for maintaining cell resistance to external damage agents.
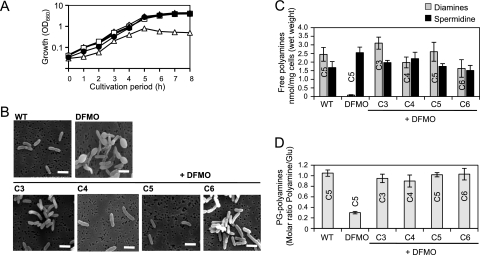
In S. ruminantium, the incorporation of cadaverine into the peptidoglycan is catalyzed by the particulate enzyme, which was designated lipid intermediate:diamine transferase (8). The substrate specificity of this enzyme was examined in vitro, and it shows broad substrate specificity toward a series of diamines, including C3, C4, C5, and C6 (4). We started to examine the effect of C3, C4, and C6 on cell growth and compare the levels of incorporation into the peptidoglycan in vivo to that of C5. Selenomonas ruminantium subsp. lactilytica TAM6421 was cultured in Trypticase peptone-yeast extract-glucose (TYG) medium (9) supplemented with both 1 mM respective diamines and 10 mM dl-α-difluoromethylornithine (DFMO), an irreversible inhibitor for LDC/ODC (13). In the presence of exogenous C3, C4, C5, or C6 and DFMO, the cells completely restored growth rates to rates similar to those of the wild-type cells (Fig. 1A). Their morphologies were also similar (Fig. 1B), indicating that the exogenous C3, C4, and C6 all are sufficient for normal growth similar to that of the C5-grown cells and the wild-type cells. To examine the levels of free and peptidoglycan-linked polyamines in the C3-, C4-, C5-, and C6-grown cells, the cells were harvested from 4-h-cultivated culture and the amounts of polyamines were quantified. The cellular free polyamines were extracted directly by suspension in 5% trichloroacetic acid and subjected to high-pressure liquid chromatography (HPLC) analysis as described previously (16). For quantification of peptidoglycan-linked polyamines, the peptidoglycans from each cell were prepared, hydrolyzed, and analyzed as described previously (9). The cellular free polyamines in the C3-, C4-, C5-, and C6-grown cells were comprised of respective diamines and spermidine, which is similar to that in the wild-type cells except for the structural variations of diamines (Fig. 1C). The concentration of these free polyamines was around 2.0 nmol/mg (wet weight) of cells. The amount of spermidine was not affected by the presence of DFMO, suggesting that spermidine biosynthesis occurs independently of the LDC/ODC activity. It was previously shown that putrescine could be synthesized through the agmatine pathway (10), which probably serves as the precursor-supplying pathway for spermidine synthesis. However, since there was no detectable amount of putrescine in wild-type cells and no complementation was observed by endogenous putrescine for cadaverine deficiency in the presence of DFMO, this pathway is assumed to be exclusively involved in spermidine synthesis, possibly due to the rapid conversion of putrescine into spermidine. The peptidoglycan-linked polyamines in these cells were completely replaced by the respective diamines, which were incorporated into the peptidoglycan almost to the same extent as was cadaverine in the wild-type cells (Fig. 1D). These results indicate that S. ruminantium equally utilizes C3 to C6 as peptidoglycan-linked forms. The incorporation of diamines into the peptidoglycan is expected to take place at the internal side of the cells since it occurs at the intermediate stage of the peptidoglycan biosynthesis and this reaction requires ATP (8), suggesting that the availability of cellular free diamine contents is the primary determinant of the structural diversity of the peptidoglycan-linked polyamine.
Fig. 1.
Effects of exogenous diamines on cell growth (A), cell morphology (B), cellular free polyamine contents (C), and peptidoglycan (PG)-linked polyamine contents (D) in S. ruminantium. To examine the growth and morphology of the cells, S. ruminantium was cultured at 37°C in TYG medium (wild type [WT], □), TYG medium containing 10 mM DFMO (▵), or TYG medium containing 10 mM DFMO and 1 mM C3 (○), C4 (●), C5 (■), or C6 (▴). The cell growth was measured by monitoring the optical density at 660 nm (OD660). For microscopic observation of the cells, cells from 4-h-cultivated culture were adsorbed onto an Isopore membrane filter (Millipore) and then submerged in 2% glutaraldehyde for 1 h at room temperature to fix the cells. After that, the filter was dehydrated by being submerged in ethanol at increasing concentrations (50, 60, 70, 80, 90, and 100%, 30 min for each) and finally submerged in ter-butanol and freeze-dried. Observation was done by scanning electron microscopy (S-4200; Hitachi, Japan) at an accelerating voltage of 15 kV. Bars, 2.5 μm.
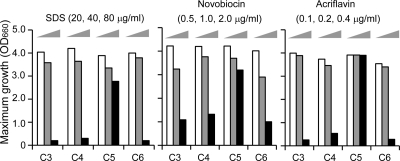
To obtain further insight into the biological functions of C3, C4, and C6 in comparison to that of C5, we focused on the susceptibilities of the cells to external damage agents, such as SDS, novobiocin, and acriflavine. We compared maximum growth levels of C3-, C4-, C5-, and C6-grown cells in the presence of these damage agents (Fig. 2). It was shown that the growth of C3-, C4-, and C6-grown cells was highly inhibited by all of these agents in comparison with that of the C5-grown cells, indicating that susceptibility to these agents was increased in the C3-, C4-, or C6-grown cells. The MICs of these agents for C3-, C4-, C5-, and C6-grown cells were determined, respectively, as follows: SDS (μg/ml), 100, 100, 180, and 100; novobiocin (μg/ml), 8, 10, 18, and 8; acriflavine (μg/ml), 1.0, 1.0, 2.0, and 1.0, respectively. These values indicate that the increased susceptibility is also reflected in the differences of MICs of these agents for each cell. This finding clearly distinguishes the biological function of C5 from those of other diamines of C3, C4, or C6 and suggests that the biological significance of C5 is related to the maintenance of cell resistance to external damage agents.
Fig. 2.
Susceptibilities of S. ruminantium cells cultured in the presence of exogenous diamines and DFMO to external damage agents. S. ruminantium was cultured at 37°C in TYG medium containing 10 mM DFMO and 1 mM C3, C4, C5, or C6 in the presence of increasing concentrations of SDS (20, 40, or 80 μg/ml), novobiocin (0.5, 1.0, or 2.0 μg/ml), or acriflavine (0.1, 0.2, or 0.4 μg/ml). Growth of the cells in each culture was monitored by optical density at 660 nm (OD660), and maximum growth levels were compared. Values are the averages of duplicate experiments.
C3-, C4-, and C6-grown cells display detachment of the outer membrane from the peptidoglycan layer.
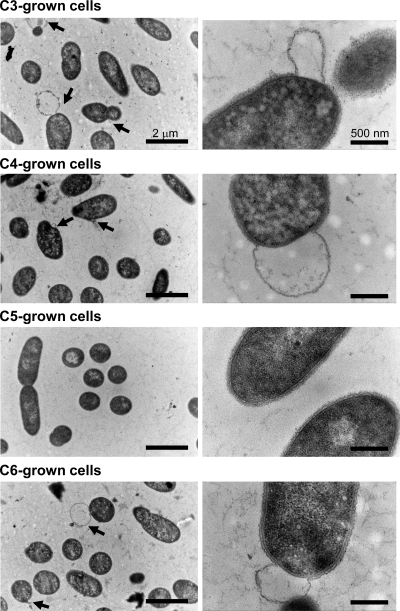
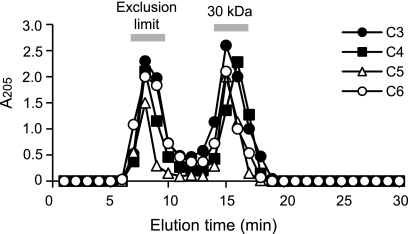
To clarify whether the difference in susceptibilities is linked to a defect in cell surface integrity, we examined the cell surface structure of the C3-, C4-, C5-, and C6-grown cells (Fig. 3). Procedures for ultrathin sectioning and transmission electron microscopy were described previously (9). It was shown that all of the C3-, C4-, and C6-grown cells had an obvious morphological defect in their cell surface structures, displaying detachment of the outer membrane from the peptidoglycan, in contrast to the C5-grown cells, which displayed complete integrity of their cell surface structure. In appearance, the peptidoglycan layer itself displays no defect in C3-, C4-, and C6-grown cells. To further confirm that the basic property of the peptidoglycan is not affected by the structural variation of covalently linked diamines, we examined the sensitivity of each peptidoglycan to lysozyme treatment (Fig. 4). Twenty-five milligrams of the purified peptidoglycan preparation from each cell was suspended in 5 ml of 10 mM sodium phosphate buffer (pH 8.0) containing 20 μg/ml lysozyme (Wako Pure Chemicals, Japan) and digested for 10 h at 37°C. After that, the sample was boiled for 15 min, the supernatant collected by centrifugation (20,000 × g, 15 min) was subjected to gel filtration chromatography using a G3000SW column (Tosoh, Japan), and the digestion patterns were compared. The result showed that all of the peptidoglycans were digested in similar patterns, in which the approximately 30-kDa fragment was predominantly produced to almost the same extent, indicating that the sensitivity to lysozyme was not varied despite the structural variation of diamines. Taken together, these results indicate a direct correlation between the correct maintenance of cell surface integrity and the susceptibility of the cells to external damage agents and suggest that the functional difference between C5 and other diamines is derived from the ability to anchor the outer membrane to the peptidoglycan but not from alteration of the properties of the peptidoglycan itself.
Fig. 3.
Cell surface structure of S. ruminantium cultured in the presence of exogenous diamines and DFMO. S. ruminantium was cultured at 37°C in TYG medium containing 10 mM DFMO and 1 mM C3, C4, C5, or C6. Cells were harvested after 4 h of cultivation and ultrathin sectioned. Sections were stained with 4% uranyl acetate and 0.4% lead citrate. Arrows indicate the sites where the outer membrane was detached from the peptidoglycan. Bars, 2 μm (left) and 500 nm (right).
Fig. 4.
Sensitivity of peptidoglycan to lysozyme treatment. The purified peptidoglycan preparations from C3-, C4-, C5-, or C6-grown cells were digested with lysozyme, and the digestion pattern was analyzed by gel filtration chromatography using a G3000SW column (Tosoh, Japan), monitored by measuring A205.
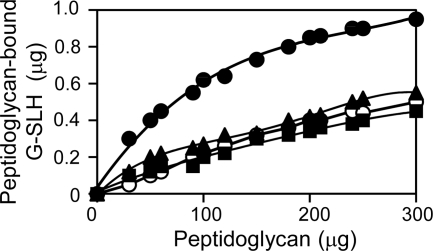
The C5-linked peptidoglycan specifically shows a strong binding affinity to the SLH domain of Mep45.
To examine whether the binding ability of the SLH domain of Mep45 varies in dependence on the structural diversity of the peptidoglycan-linked diamines, we examined the binding affinity of the SLH domain to each peptidoglycan with covalently linked C3, C4, C5, or C6 (Fig. 5). We used the glutathione S-transferase–SLH domain fusion protein (G-SLH) (9) and examined the amount of G-SLH bound to each peptidoglycan (30 to 300 μg). The purified peptidoglycan was suspended with 1 μg of G-SLH in 30 μl of 20 mM sodium phosphate buffer (pH 7.4) containing 150 mM NaCl and was incubated at room temperature for 90 min. The suspension was centrifuged at 20,000 × g for 20 min to separate the peptidoglycan-bound G-SLH (precipitate) from non-peptidoglycan-bound G-SLH (supernatant). The precipitate was subjected to SDS-PAGE and stained with Coomassie brilliant blue, and G-SLH was quantified by measuring band intensity using ImageJ software. To compare the binding affinities of G-SLH to each peptidoglycan, we estimated the amount of peptidoglycan required for the 50% binding of 1 μg of G-SLH. The values for C3-, C4-, C5-, and C6-linked peptidoglycans were approximately 300 μg, 300 μg, 70 μg, and 280 μg, respectively, indicating that the binding ability of the SLH domain depends on the structural diversity of covalently linked diamines, and C5-linked peptidoglycan is the most preferential binding partner. This was considered the direct explanation for the difference in biological functions among C5 and other diamines, representing a basis for the mechanism for anchoring the outer membrane to the peptidoglycan in S. ruminantium, which occurs through efficient binding between the SLH domain of Mep45 and the peptidoglycan in a manner dependent on specific mediation by the peptidoglycan-linked cadaverine.
Fig. 5.
Comparison of the binding affinities of G-SLH to the purified peptidoglycans with covalently linked C3, C4, C5, or C6. Thirty to 300 μg of purified C3 (■)-, C4 (○)-, C5 (●)-, or C6 (▴)-linked peptidoglycan was applied to the binding assay mixture. The amount of G-SLH bound to the peptidoglycan was examined. Values are the averages of triplicate experiments.
Conclusion and discussion.
Cadaverine specifically serves as the most efficient constituent of the peptidoglycan in maintaining cell resistance to external damage agents and is required for effective interaction between the SLH domain of Mep45 and the peptidoglycan, facilitating the correct anchoring of the outer membrane to the peptidoglycan. According to these findings, we describe a correlation between structural anchoring of the outer membrane to the peptidoglycan in vivo and the binding strength of the SLH domain of Mep45 to the peptidoglycan in vitro, and we propose that part of the inherent biological significance of cadaverine is its efficiency in correct maintenance of cell surface integrity in S. ruminantium. Since this bacterium lacks the murein lipoprotein which is responsible for anchoring the outer membrane to the peptidoglycan in other Gram-negative bacteria such as Escherichia coli, the cadaverine-mediated anchoring mechanism may represent a subclass of anchoring systems of Gram-negative bacteria. However, the potential difference in fundamental roles between the two anchoring systems remains to be investigated, since the inhibitory effect of DFMO directly influenced the viability of S. ruminantium cells, in contrast to the fact that the defect in the structural connection between the outer membrane and the peptidoglycan, which was found in a murein lipoprotein-lacking mutant of E. coli, does not immediately affect cell growth (2). It is yet unknown whether the binding strength of the SLH domain of Mep45 to the peptidoglycan directly affects cell viability or not, and this needs to be clarified in further research.
Footnotes
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Cascales E., Bernadac A., Gavioli M., Lazzaroni J. C., Lloubes R. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fung J., MacAlister T. J., Rothfield L. I. 1978. Role of murein lipoprotein in morphogenesis of the bacterial division septum: phenotypic similarity of lkyD and lpo. J. Bacteriol. 133:1467–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Godlewska R., Wiśniewska K., Pietras Z., Jagusztyn-Krynicka E. K. 2009. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298:1–11 [DOI] [PubMed] [Google Scholar]
- 4. Kamio Y. 1987. Structural specificity of diamines covalently linked to peptidoglycan for cell growth of Veillonella alcalescens and Selenomonas ruminantium. J. Bacteriol. 169:4837–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamio Y., Itoh Y., Terawaki Y., Kusano T. 1981. Cadaverine is covalently linked to peptidoglycan in Selenomonas ruminantium. J. Bacteriol. 145:122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamio Y., Pösö H., Terawaki Y., Paulin L. 1986. Cadaverine covalently linked to a peptidoglycan is an essential constituent of the peptidoglycan necessary for the normal growth in Selenomonas ruminantium. J. Biol. Chem. 261:6585–6589 [PubMed] [Google Scholar]
- 7. Kamio Y., Takahashi H. 1980. Outer membrane proteins and cell surface structure of Selenomonas ruminantium. J. Bacteriol. 141:899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamio Y., Terawaki Y., Izaki K. 1982. Biosynthesis of cadaverine-containing peptidoglycan in Selenomonas ruminantium. J. Biol. Chem. 257:3326–3333 [PubMed] [Google Scholar]
- 9. Kojima S., et al. 2010. Cadaverine covalently linked to the peptidoglycan mediates the interaction between the peptidoglycan and periplasm-exposed SLH domain of major outer membrane protein Mep45 in Selenomonas ruminantium. J. Bacteriol. 192:5953–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao S., et al. 2008. Occurrence of agmatine pathway for putrescine synthesis in Selenomonas ruminantium. Biosci. Biotechnol. Biochem. 72:445–455 [DOI] [PubMed] [Google Scholar]
- 11. Suzuki H., et al. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167:1–9 [DOI] [PubMed] [Google Scholar]
- 12. Tabor C. W., Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takatsuka Y., et al. 1999. Novel characteristics of Selenomonas ruminantium lysine decarboxylase capable of decarboxylating both L-lysine and L-ornithine. Biosci. Biotechnol. Biochem. 63:1063–1069 [DOI] [PubMed] [Google Scholar]
- 14. Takatsuka Y., Tomita T., Kamio Y. 1999. Identification of the amino acid residues conferring substrate specificity upon Selenomonas ruminantium lysine decarboxylase. Biosci. Biotechnol. Biochem. 63:1843–1846 [DOI] [PubMed] [Google Scholar]
- 15. Takatsuka Y., Yamaguchi Y., Ono M., Kamio Y. 2000. Gene cloning and molecular characterization of lysine decarboxylase from Selenomonas ruminantium delineate its evolutionary relationship to ornithine decarboxylase from eukaryotes. J. Bacteriol. 182:6732–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamaguchi Y., Takatsuka Y., Kamio Y. 2002. Identification of a 22-kDa protein required for the degradation of Selenomonas ruminantium lysine decarboxylase by ATP-dependent protease. Biosci. Biotechnol. Biochem. 66:1431–1434 [DOI] [PubMed] [Google Scholar]
- 17. Yamaguchi Y., Takatsuka Y., Matsufuji S., Murakami Y., Kamio Y. 2006. Characterization of a counterpart to mammalian ornithine decarboxylase antizyme in prokaryotes. J. Biol. Chem. 281:3995–4001 [DOI] [PubMed] [Google Scholar]