Abstract
Salmonella enterica serovar Typhimurium possesses a stimulon of genes that are differentially regulated in response to conditions of low fluid shear force that increase bacterial virulence and alter other phenotypes. In this study, we show that a previously uncharacterized member of this stimulon, ydcI or STM1625, encodes a highly conserved DNA binding protein with related homologs present in a range of Gram-negative bacterial genera. Gene expression analysis shows that ydcI is expressed in different bacterial genera and is involved in its autoregulation in S. Typhimurium. We demonstrate that purified YdcI protein specifically binds a DNA probe consisting of its own promoter sequence. We constructed an S. Typhimurium ΔydcI mutant strain and show that this strain is more sensitive to both organic and inorganic acid stress than is an isogenic WT strain, and this defect is complemented in trans. Moreover, our data indicate that ydcI is part of the rpoS regulon related to stress resistance. The S. Typhimurium ΔydcI mutant was able to invade cultured cells to the same degree as the WT strain, but a strain in which ydcI expression is induced invaded cells at a level 2.8 times higher than that of the WT. In addition, induction of ydcI expression in S. Typhimurium resulted in the formation of a biofilm in stationary-phase cultures. These data indicate the ydcI gene encodes a conserved DNA binding protein involved with aspects of prokaryotic biology related to stress resistance and possibly virulence.
INTRODUCTION
Bacterial growth environments characterized by low fluid shear force have been shown to induce a multitude of phenotypic responses, including altered acid, oxidative, thermal, and osmotic stress resistance (7, 33, 36, 41, 52–55), increased biofilm formation (6, 33, 52), altered protein secretion (14, 15), altered cell surface lipid and polysaccharide profiles (6, 7, 55), and increased survival in cellular and animal hosts (39, 52–54). Notably, the virulence of Salmonella enterica serovar Typhimurium is increased by low fluid shear growth conditions as measured using murine infection assays and tissue culture models (39, 52–54). Low fluid shear force (defined here as approximately <0.01 to 0.2 dynes/cm2) is characterized by a low-turbulence, low-agitation environment, as opposed to high fluid shear (defined here as approximately from 5 to >50 dynes/cm2) where liquid moves with higher velocity over the cellular surface (3–5, 21, 23, 25, 36). Low fluid shear growth environments include spaceflight, ground-based suspension culture models such as the rotating-wall vessel (RWV) bioreactor, and the spaces between cellular microvilli, the last of which is encountered by numerous pathogens during the natural course of infection (21, 23, 25, 30, 40, 41). Previous work has shown that bacteria grown in low fluid shear environments induce a molecular response which includes genome-wide changes in gene expression (the low fluid shear stimulon) (6, 7, 39, 48, 52, 53, 55). Since growth under low fluid shear conditions is able to induce cellular phenotypes which are difficult or impossible to obtain via conventional culture methods, there is much potential for important previously uncharacterized genes and regulatory schemes to be revealed via the study of the low fluid shear response.
The first studies to identify the bacterial genes of the low fluid shear stimulon were performed with S. Typhimurium cultures grown in an RWV bioreactor (39, 55) and during spaceflight (52, 53). Genes of the S. Typhimurium low fluid shear stimulon were found to be distributed across the genome and belong to a wide variety of functional groups, including various transport systems, lipopolysaccharide synthesis pathways, protein secretion mechanisms, various metabolic pathways, and genes of unknown function (52, 53, 55). In addition, several genes encoding previously uncharacterized transcriptional regulatory proteins in S. Typhimurium were also found to be part of the low fluid shear stimulon (52, 53, 55). These genes are of particular interest not only for their potential role in mediating bacterial responses to low fluid shear but also for their potential to allow us to better understand the global circuits that influence bacterial physiology in general. It is highly likely that previously unexplored transcriptional regulators that are identified via the low fluid shear stimulon will be involved in aspects of prokaryotic biology that are able to be studied under “standard conditions” outside the context of low fluid shear.
The S. Typhimurium gene ydcI (also known as STM1625 in the S. Typhimurium genome) belongs to the aforementioned class of potential transcriptional regulators that are members of the low fluid shear stimulon as determined using microarray analysis with total cell RNA with a 2-fold change cutoff (54, 55). However, the ydcI gene is completely uncharacterized and has not been previously studied beyond being identified via genome sequence analysis. This study aimed to characterize the ydcI gene (and the YdcI protein) to answer basic questions regarding its role in S. Typhimurium biology and the function of its protein product. Our results indicate that the S. Typhimurium ydcI gene (i) is conserved across genera, (ii) autoregulates its expression, (iii) encodes a DNA binding protein that binds with specificity, (iv) is required for full resistance of S. Typhimurium to acid stress, (v) appears to be a member of the rpoS regulon, and (vi) when induced, alters S. Typhimurium interactions with host cells and facilitates biofilm formation. To our knowledge, this is the first report to demonstrate that a previously uncharacterized gene identified via study of the low fluid shear stimulon is involved in responses that can be observed outside the low fluid shear environment. This report provides a foundation for further analysis of the ydcI gene and other previously uncharacterized transcriptional regulators of the low fluid shear stimulon. The study of these genes may allow us to engineer bacteria for beneficial purposes by disrupting or inducing certain signals in a controlled manner resulting in improved vaccines and other applications (10, 11, 32).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Please refer to Table 1 for a list of strains and plasmids used in this study. S. Typhimurium strain χ3339 was used as the wild type (WT), and all mutations were analyzed in this background (22). Construction of the S. Typhimurium ΔydcI mutation was performed using lambda Red recombination as previously described (12, 45) and by using the DNA primers listed in Table S1 in the supplemental material. A Cmr cassette from pKD3 (12) was inserted at nucleotide 166 of the S. Typhimurium ydcI open reading frame (ORF) in the same orientation as the ydcI ORF. To obtain the ΔydcI::lacZ fusion, the Cmr cassette at the site of the ΔydcI mutation was deleted using Flp recombinase expressed from pCP20 (12), and then the lacZ plasmid pCE36 (13) was inserted at the single FRT site at this location as previously described (13). To construct the WT ydcI::lacZ fusion, we performed an identical procedure by inserting the Cmr cassette immediately after the stop codon of ydcI and inserting pCE36 at this location. The strain containing Cm::lacZ was obtained by inserting the Cmr-encoding gene from pKD3 immediately upstream of the lacZ gene in the ΔydcI::lacZ mutant strain using Red recombination such that the Cmr-encoding gene promoter replaces the ydcI promoter and drives lacZ expression. All constructions were verified using PCR analysis. The ΔrpoS mutation (which is marked by Apr) was transferred via P22 transduction from donor strain χ4973 (38). To restore WT rpoS in the WT ydcI::lacZ ΔrpoS strain, a strain containing a Cmr marker at gene STM2901 (located approximately 20 kb from the rpoS gene) was used as a donor for P22 transduction, and Cmr recipients were screened for ampicillin sensitivity (50). A Cmr Apr isolate from this transduction was used as a control. Plasmid pBAD18+ydcI was constructed using a synthesized ydcI gene in which a six-histidine tag was fused to the C terminus and cloned into pUC57 (56) and then subcloned into pBAD18 (24) downstream of the arabinose-inducible promoter (Genscript, Inc., Piscataway, NJ). The entire length of the construct was verified via sequencing. Strains were grown using LB (Lennox) medium supplemented with the following antibiotics when necessary: ampicillin, 200 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 50 μg/ml.
Table 1.
Strains and plasmids used in this study
| Species and strain or plasmid | Comment(s) |
|---|---|
| Salmonella Typhimurium | |
| χ3339 | 22 |
| χ3339 ΔydcI | This study |
| χ3339 WT ydcI::lacZ | This study |
| χ3339 ΔydcI::lacZ | This study |
| χ3339 WT ydcI::lacZ ΔrpoS | This study |
| χ3339 Cm::lacZ | This study |
| χ3339 Cm::lacZ ΔrpoS | This study |
| χ3339 WT ydcI::lacZ ΔrpoS | |
| STM2901::Cm | This study |
| χ3339 WT ydcI::lacZ WT rpoS | |
| STM2901::Cm | This study |
| χ3339 ΔSPI-1 | 50 |
| Escherichia coli TOP10 | Invitrogen |
| Salmonella Typhi Ty2 | 17 |
| Proteus mirabilis ATCC 7002 | American Type Culture Collection |
| Serratia marcescens ATCC 14041 | American Type Culture Collection |
| Klebsiella pneumoniae ATCC 13883 | American Type Culture Collection |
| Citrobacter koseri ATCC 27156 | American Type Culture Collection |
| Plasmids | |
| pKD3 | 12 |
| pCE36 | 13 |
| pCP20 | 12 |
| pBAD18 | 24 |
| pBAD18+ydcI | This study |
| pBAD18+lacZ | 42, this study |
Sequence analysis.
The YdcI homologs from the indicated Gram-negative species were identified using BLAST search analysis with S. Typhimurium YdcI (GenBank accession no. AAL20543) as the query (1). Homologs were aligned using CLUSTAL W analysis as previously described (46, 51).
Gene expression analysis.
Reverse transcription (RT)-PCR was performed as described previously (51, 55), using approximately 4 to 6 μg RNA from late-log-phase cultures (optical density at 600 nm [OD600] = 0.9 to 1.4). RNA was isolated using RNA Protect reagent and RNeasy spin prep kits (Qiagen, Inc., Valencia, CA). LacZ assays were performed with late-log-phase cultures using standard protocols as described previously (35), and the data presented represent the mean and standard deviation of three or four independent experiments each assayed in duplicate or triplicate reactions.
Expression and purification of YdcI protein.
Escherichia coli strain TOP10 containing pBAD18+ydcI was grown to an OD600 of approximately 0.5 to 0.8, induced with 0.1% arabinose, and then grown for an additional 3 h. Cells were harvested, lysed using B-Per reagent (Pierce/Thermo-Fisher, Rockford, IL), and processed for isolation of YdcI protein (with a six-histidine tag) using nickel-agarose spin columns in accordance with the manufacturer's instructions (Zymo Research, Orange, CA). Coomassie staining and Western blot analysis with anti-six-histidine antibody (Immunology Consultants Lab, Inc., Newberg, OR) were performed using standard protocols as described previously (2, 49, 50).
Gel shift assays.
Gel shift reactions were performed in the presence of 10 mM Tris-HCl [pH 7.5], 50 mM KCl, 10% glycerol, 1 mM dithiothreitol, 0.5 mM EDTA, 1 μg salmon sperm DNA, and 0.1 μg/ml bovine serum albumin with approximately 1 ng of probe. The reaction mixtures were incubated for 20 min at room temperature before being loaded onto a prerun 8% acrylamide gel in 0.5× Tris-borate-EDTA (TBE) buffer at 150 V. Probe labeling and detection were performed using a nonradioactive kit in accordance with the manufacturer's instructions (Pierce/Thermo-Fisher, Rockford, IL) with positively charged nylon membrane (GE Healthcare, Piscataway, NJ). The 180-bp probe H3 consisted of DNA extending upstream from 38 bp downstream of the ydcI ATG codon (S. Typhimurium coordinates 1714755 to 1714935) that was PCR amplified using the primers indicated in Table S1 in the supplemental material and purified using a spin column in accordance with the manufacturer's instructions (Zymo Research, Orange, CA). The 182-bp probe H5 consisted of DNA upstream of the H3 probe from coordinates 1714573 to 1714754, and the lacZ probe consisted of 152 bp amplified from the lacZ gene using the primers indicated in Table S1 in the supplemental material. Competition assays were performed under the binding conditions described above with increasing amounts of a molar excess of unlabeled DNA that was either specific (the 180-bp ydcI promoter fragment) or nonspecific (lacZ fragment), as indicated.
Thermal, osmotic, and acid stress assays.
Stress assays were performed as described previously, using shaking broth cultures grown to stationary phase (39, 52–54). The data presented represent the mean and standard deviation of four independent experiments each plated in triplicate.
Tissue culture invasion and intracellular survival assays.
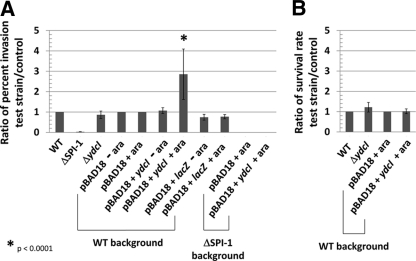
Invasion of Int407 intestinal epithelial cells and survival in J774 macrophages were assayed as described previously with a multiplicity of infection of approximately 30 using late-log-phase broth shaking cultures (49–52, 54). The data (see Fig. 7) are from four to six independent experiments each performed in triplicate tissue culture wells, except for the pBAD18+lacZ strains, which are from three independent experiments performed in triplicate.
Fig. 7.
Invasion of Int407 cells and survival in J774 macrophages. (A) The indicated S. Typhimurium strains were tested for invasion of Int407 cells via gentamicin protection assay. The ratio of percent invasion of each strain to that of the control was calculated and graphed. The ΔSPI-1 strain contains a deletion of the SPI-1 genes required for full invasion of Int407 cells. The ΔSPI-1 and ΔydcI mutant strains were compared to the WT for ratio calculation. The pBAD18, pBAD18+ydcI, and pBAD18+lacZ strains were compared to the WT pBAD18 strain under the corresponding arabinose condition for ratio calculation. (B) The indicated strains were tested for survival in J774 macrophages over a period of 18 to 20 h via gentamicin protection assay. The ratio of the percent survival of each strain to that of the control was calculated and graphed. For ratio calculations, the ΔydcI mutant strain was compared to the WT and the pBAD18+ydcI strain was compared to the pBAD18 strain.
Biofilm formation.
Strain χ3339 containing pBAD18, pBAD18+ydcI, or pBAD18+lacZ was grown overnight in the absence of arabinose, diluted 1:50 in fresh medium, grown to an OD600 of approximately 0.6 to 0.8, supplemented with arabinose to 0.1%, and grown for an additional 12 to 16 h. Crystal violet staining of biofilms and subsequent quantitation were performed as described previously (9, 43).
Low fluid shear studies.
To test S. Typhimurium responses to low fluid shear culture, the RWV bioreactor was used to grow cultures of each strain in the low fluid shear and control orientations as described previously (references 52 and 53 for acid stress and reference 54 for oxidative stress). The cultures were subjected to acid stress (pH 3.5 with citric acid) and oxidative stress (70 mM H2O2) and assayed for survival as described previously (references 52 and 53 for acid stress and 54 for oxidative stress). The results are from three to six independent RWV experiments each plated in triplicate.
RESULTS
The S. Typhimurium ydcI gene encodes a conserved protein.
We used the S. Typhimurium YdcI protein sequence (GenBank accession no. AAL20543) as a query in a BLAST search for homologs in the protein database. The results of this search indicated that the YdcI protein sequence belongs to the LysR family of transcriptional regulators and is highly conserved among Gram-negative genomes, including the genera Escherichia, Shigella, Citrobacter, Enterobacter, Klebsiella, and Serratia (Fig. 1). The e values for the alignments in Fig. 1 are all less than 10−140, except that for the Serratia protein, which is 10−92. We also found that the YdcI protein is encoded by Ralstonia, Pseudomonas, Burkholderia, Rhizobium, Agrobacterium, Brucella, and Ruegeria at a somewhat lower level of homology (e values between 10−71 and 10−49) but still representing a high degree of conservation in these genera (see Fig. S1 in the supplemental material). The genomic organization of the location of the ydcI gene in the genera from Fig. 1 is also conserved (see Fig. S2 in the supplemental material). These results indicate that the ydcI gene is conserved across multiple Gram-negative genera and suggest evolutionary selection for the function of this gene in prokaryotic biology.
Fig. 1.
Alignment of the S. Typhimurium YdcI protein with homologs present in a range of other Gram-negative genera. Yellow highlighting indicates amino acid identity (in at least five of the proteins). The e values for the aligned proteins are less than 10−140, except for the Serratia protein, whose e value is 10−92.
Analysis of ydcI gene expression.
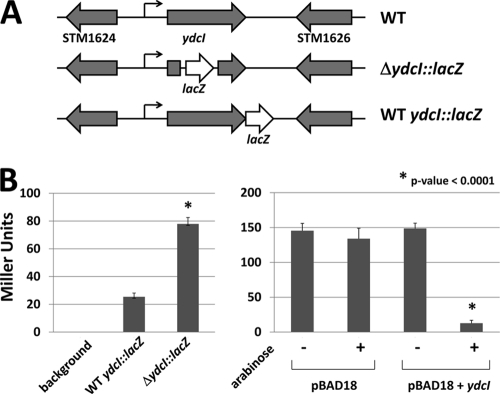
To test if the ydcI gene is expressed in different bacterial genera, we isolated total RNA from cultures of S. Typhimurium, Salmonella Typhi, Proteus mirabilis, Serratia marcescens, Klebsiella pneumoniae, and Citrobacter koseri and performed an RT-PCR analysis using primers against the ydcI homolog in each strain. We detected ydcI homolog expression via this assay indicating that this gene is able to be transcribed across genera (Fig. 2). To further analyze ydcI gene expression, we constructed lacZ transcriptional fusions in the chromosomal copy of the S. Typhimurium ydcI gene. Two such fusions were constructed; the WT ydcI::lacZ and ΔydcI::lacZ alleles contain the promoterless lacZ gene fused immediately after the ydcI stop codon and at nucleotide 166 of the 912-bp ydcI gene, respectively (Fig. 3A). The latter construct is predicted to abolish normal YdcI protein function, thereby allowing comparison of lacZ reporter expression in the presence and absence of WT YdcI protein. We found that the WT ydcI::lacZ fusion was expressed 3-fold less than the ΔydcI::lacZ construct (Fig. 3B). We introduced a plasmid expressing the S. Typhimurium ydcI gene from an arabinose-inducible promoter (pBAD18+ydcI) into the ΔydcI::lacZ mutant strain. In the presence of arabinose, this strain expressed significantly less lacZ than the control strain containing the vector only (Fig. 3B). Collectively, these results indicate that the YdcI protein negatively autoregulates its expression under the conditions tested here and is expressed at relatively low levels when this autoregulation is intact.
Fig. 2.
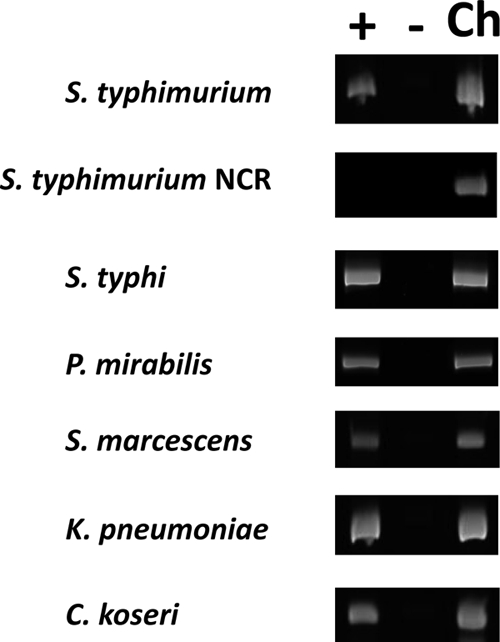
RT-PCR of ydcI gene homologs in a range of Gram-negative genera. The plus and minus lanes contain samples with and without reverse transcriptase, respectively. The Ch lane contains samples where chromosomal DNA isolated from the indicated strain was used in the PCR. NCR refers to a noncoding region in the S. Typhimurium genome between two divergent promoters that is not predicted to be transcribed.
Fig. 3.
Analysis of ydcI::lacZ fusions in S. Typhimurium. (A) A promoterless lacZ gene was transcriptionally fused to the WT and ΔydcI mutant alleles of the S. Typhimurium ydcI gene in separate strains. The maps of the WT, ΔydcI::lacZ, and WT ydcI::lacZ gene loci in the S. Typhimurium strain genomes are shown. (B) The LacZ activity of the strains in panel A is shown. In addition, the ΔydcI::lacZ strain was transformed with either pBAD18 or pBAD18+ydcI and assayed for LacZ activity in the presence or absence of arabinose.
Purification and DNA binding activity of YdcI protein.
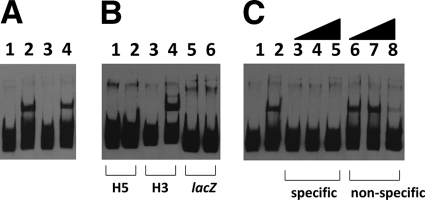
To test the YdcI protein for DNA binding activity, we cloned the S. Typhimurium ydcI gene downstream of an arabinose-inducible promoter on the plasmid vector pBAD18 (24). In this construct, we also fused the coding sequence for a six-histidine epitope immediately after the last codon such that the YdcI protein would be expressed with a C-terminal tag. Arabinose-dependent expression of the YdcI protein was demonstrated via Western blot analysis, and we then purified the fusion protein using nickel agarose columns (see Fig. S3 in the supplemental material). To test the DNA binding activity of YdcI, we incubated YdcI protein from either cell extracts or purified preparations with a 180-bp DNA probe (named H3) consisting of the sequence immediately upstream of the S. Typhimurium ydcI ORF as indicated in Materials and Methods. Both sources of YdcI protein shifted the labeled H3 probe in this assay (Fig. 4A). To test the specificity of the YdcI DNA binding activity, we used a 152-bp probe consisting of lacZ gene DNA, and YdcI did not shift this probe (Fig. 4B). In addition, we also synthesized a 182-bp probe (named H5) consisting of DNA from the ydcI promoter region that extends immediately upstream of the H3 probe and show that YdcI does not shift this probe (Fig. 4B). We also incubated gel shift reaction mixtures with an excess of unlabeled ydcI promoter fragment H3 (specific competitor) or unlabeled lacZ fragment (nonspecific competitor). The results are consistent with a specific YdcI DNA binding activity since the specific competitor abolished binding to the probe and the nonspecific competitor did not (Fig. 4C). Since YdcI is a DNA binding protein that has not been previously characterized, we used the gel shift assay to determine an apparent dissociation constant (KD) for the YdcI binding activity. The concentration of YdcI protein at which 50% binding occurred (and thus indicated the apparent KD under these conditions) was approximately 2.3 to 4.6 nM (see Fig. S4 in the supplemental material). This magnitude of apparent KD is consistent with documented specific DNA binding activity of members of this protein family (34, 44, 47) and is consistent with the conclusion that the ydcI gene encodes a specific DNA binding protein.
Fig. 4.
Gel shift assays with YdcI protein. (A) The labeled H3 DNA probe from the S. Typhimurium ydcI promoter region was incubated with YdcI cell extracts or purified YdcI protein and run in an 8% acrylamide–TBE gel. Lanes: 1, probe alone; 2, TOP10(pBAD18+ydcI) extract, 300 ng; 3, TOP10(pBAD18) extract, 300 ng; 4, purified YdcI protein, approximately 10 nM. (B) Gel shift reaction mixtures with different probes incubated with purified YdcI protein. Lanes: 1, H5 probe alone; 2, H5 probe plus YdcI; 3, H3 probe alone; 4, H3 probe plus YdcI; 5, lacZ probe alone; 6, lacZ probe plus YdcI. (C) YdcI gel shift reaction mixtures with labeled H3 probe were incubated with unlabeled specific (H3 DNA fragment) or nonspecific (lacZ DNA fragment) competitor DNA and then run in an 8% acrylamide gel. Lanes: 1, probe alone; 2, no competitor; 3 to 5, specific competitor at 135×, 270×, and 540× molar excesses, respectively; 6 to 8, nonspecific competitor at 135×, 270×, and 540× molar excesses, respectively.
Since our results indicated that a YdcI DNA binding site is located within the H3 probe, we used the Microfootprinter program (http://bio.cs.washington.edu/MicroFootPrinter.html) to identify possible YdcI binding sites within the H3 fragment (37). This analysis yielded two candidate sites in the H3 fragment, but two separate 40-bp probes containing each site did not display binding to YdcI protein in the gel shift assay (data not shown).
Stress assays reveal an acid resistance defect in an S. Typhimurium ydcI mutant.
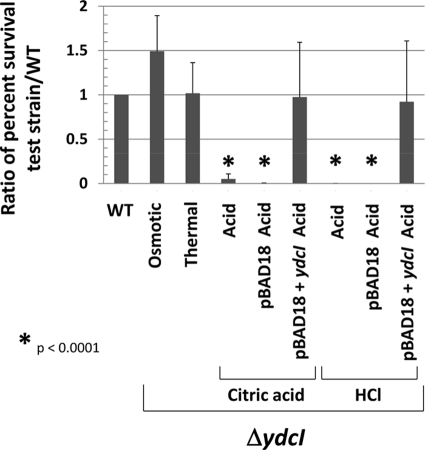
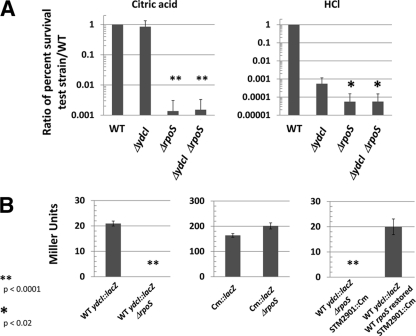
In order to study the role of the ydcI gene in Salmonella biology, we constructed a knockout mutation of the ydcI gene in the chromosome of S. Typhimurium using the lambda Red recombination system (see Materials and Methods for details). We observed no significant growth differences between the WT and ΔydcI mutant strains using OD600 and CFU measurements in a growth curve (see Fig. S5 in the supplemental material). Since the ydcI gene was initially brought to our attention as a member of a stimulon of genes that change expression under conditions that alter bacterial stress resistance, we tested the ydcI mutant for osmotic, thermal, oxidative, and acid stress resistance compared to that of the isogenic WT strain. Under conditions of osmotic and thermal stress, we did not observe a significant difference in survival between the WT and ΔydcI mutant strains (Fig. 5). Under conditions of oxidative stress, we observed inconsistent results that may indicate a survival defect under these conditions of the ΔydcI mutant compared to the WT, but this experiment requires further study to determine a conclusive effect (data not shown). However, under both organic (citric acid) and inorganic (HCl) acid stress (pH 3.5), we observed a significant difference between the survival of the ΔydcI mutant and that of the WT strain (Fig. 5). Both the organic and inorganic acid resistance defects can be complemented by a functional copy of ydcI provided on a plasmid (Fig. 5). These data indicate that the ydcI gene is required for full acid stress resistance of S. Typhimurium under the conditions studied here and likely expresses a protein that serves to regulate genes involved in acid stress resistance.
Fig. 5.
Acid resistance defect in S. Typhimurium ΔydcI mutant. Isogenic WT and ΔydcI S. Typhimurium strains were tested for resistance to osmotic (2.5 M NaCl), thermal (50°C), and acid (pH 3.5 using citric acid or HCl) stress over a time period of 120 min. The percent survival of the WT and ΔydcI mutant strains under the corresponding stress (compared to before addition of stress) was calculated, and a ratio of the percent survival of the indicated ΔydcI mutant test strain to that of the WT strain is shown. The ΔydcI mutant strain containing pBAD18 or pBAD18+ydcI was grown in the presence of arabinose.
Relationship between the ydcI and rpoS genes.
The S. Typhimurium rpoS gene encodes a highly conserved sigma factor that is required for the expression of a wide array of genes involved in resistance to a range of environmental stresses and has been termed the “master regulator” of bacterial stress resistance (16, 26, 29). Since the ydcI gene is required for full acid stress resistance of S. Typhimurium, we compared the phenotype of an S. Typhimurium ΔydcI ΔrpoS double mutant to that of the ΔydcI and ΔrpoS single mutants to distinguish between the possibilities that ydcI and rpoS are part of the same pathway or part of separate, additive pathways. In the citric acid resistance assay, we observed that the ΔrpoS mutant displayed high sensitivity within 30 min, but the ΔydcI mutant did not display its sensitivity until 120 min after the addition of stress (Fig. 5 and Fig. 6A). Thus, within 30 min of citric acid stress exposure, the ΔydcI and WT strains display similar phenotypes (Fig. 6A). However, the ΔydcI ΔrpoS double mutant displayed a citric acid resistance defect that was identical to that of the ΔrpoS single mutant (Fig. 6A). In the HCl acid resistance assay, both the ΔydcI and ΔrpoS single mutants display high sensitivity within 30 min of acid exposure, but the ΔrpoS mutant is significantly more sensitive than the ΔydcI mutant during this time (Fig. 6A). The ΔydcI ΔrpoS double mutant displayed an HCl acid resistance defect identical to that of the ΔrpoS single mutant. These results suggest that the ydcI and rpoS genes act via the same pathway and do not act in an additive manner in separate pathways to provide WT levels of acid resistance.
Fig. 6.
Relationship between ydcI and rpoS genes. (A) The indicated strains were subjected to acid stress (pH 3.5) with citric acid or HCl as indicated over a 30-min time period. The percent survival of the strains (compared to before the addition of stress) was calculated, and a ratio of the percent survival of the test strain to the WT strain is shown. Note that the phenotype of the ΔydcI mutant is similar to the WT for citric acid stress within 30 min (the data in Fig. 5 are from 120 min of acid stress). The P values were calculated as comparisons of the indicated strains and the ΔydcI single mutant strain. (B) Cultures of the indicated strains were assayed for LacZ activity and plotted for Miller units. All strains are in the χ3339 background, as indicated in Table 1. Cm::lacZ indicates that the lacZ gene in this strain is transcribed via the Cmr gene promoter. STM2901::Cm indicates the gene location of a Cmr marker linked to the WT rpoS gene within approximately 20 kb of DNA in the S. Typhimurium genome.
To test if rpoS plays a role in the expression of the ydcI gene, we constructed a WT ydcI::lacZ ΔrpoS strain and compared the LacZ activity of this strain to that of the WT ydcI::lacZ strain. Remarkably, the WT ydcI::lacZ ΔrpoS strain completely lost LacZ activity, in contrast to the control (Fig. 6B). A control strain in which the same lacZ reporter gene was driven by the Cmr-encoding gene promoter was not affected by the presence of the ΔrpoS mutation (Fig. 6B). To demonstrate that the loss of WT ydcI::lacZ activity was not due to a random artifact of strain construction, we restored the WT rpoS allele to the WT ydcI::lacZ ΔrpoS strain using P22 transduction of a Cmr marker linked to the WT rpoS allele as described in Materials and Methods. This strain displayed WT levels of WT ydcI::lacZ activity, in contrast to the control, confirming that the ΔrpoS mutation was responsible for the expression defect (Fig. 6B). Taken together, the double mutant studies and lacZ reporter analysis provide significant evidence that the ydcI gene is part of the rpoS regulon in S. Typhimurium.
Tissue culture invasion assays.
To test for a role for ydcI in S. Typhimurium invasion of cultured intestinal epithelial cells, we compared the ΔydcI mutant strain to the WT for entry into Int407 cells. The results indicated that the ΔydcI mutant did not differ significantly from the WT in Int407 invasion (Fig. 7A). However, we also tested the invasion phenotype of strains in which ydcI expression had been induced or “turned on” as an alternative to analysis of the deletion mutant (in which the gene is removed). Since expression of the ydcI gene from the pBAD18+ydcI construct restored the repression of a chromosomal ΔydcI::lacZ fusion to WT levels, expressed a protein displaying specific DNA binding activity as predicted, and complemented the ΔydcI acid resistance defect back to WT levels, we reasoned that this construct would be an appropriate reagent for this purpose. WT strain χ3339 containing pBAD18+ydcI invaded Int407 cells to a level 2.8 times higher than that of controls in the presence of arabinose (Fig. 7A). Control strains containing the pBAD18 vector alone or pBAD18+lacZ did not display increased cell entry in either the presence or the absence of arabinose (Fig. 7A). The latter strain expresses copious amounts of LacZ protein (several thousand Miller units) in the presence of arabinose (reference 42 and data not shown). This indicates that this phenotype is specific to ydcI induction and not merely due to general overexpression of protein in this strain. The pBAD18 and pBAD18+ydcI strains displayed virtually identical levels of adherence to Int407 cells under these conditions (see Fig. S6 in the supplemental material). The SPI-1 genes encode a type III secretion system used to facilitate invasion of nonphagocytic cells like those of the Int407 line (18–20). To test if induction of ydcI could serve to reverse the invasion defect of the ΔSPI-1 strain, we introduced the pBAD18+ydcI plasmid into this strain, induced ydcI expression via arabinose, and tested invasion of the culture. We did not observe any reversal of the ΔSPI-1 invasion defect under these conditions compared to the control strain containing the vector alone (Fig. 7A). This may suggest that the increased invasion upon ydcI induction in the WT strain background occurs via the SPI-1 pathway.
We also tested if the ΔydcI mutation or induction of ydcI expression had any effect on S. Typhimurium survival in J774 macrophages. Neither the deletion nor the induction of the ydcI gene had any effect on S. Typhimurium macrophage survival (Fig. 7B).
Biofilm formation.
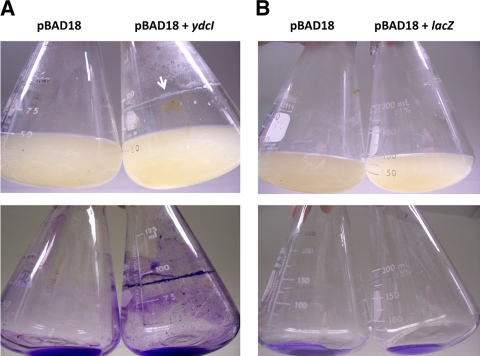
When cultures of strain χ3339 containing pBAD18 or pBAD18+ydcI were supplemented with arabinose and allowed to grow for approximately 12 to 16 h postsupplementation, we observed that the strain containing pBAD18+ydcI formed a biofilm which was especially prominent at the medium-air interface (Fig. 8A). This biofilm was readily stained with crystal violet, indicating significant cell adhesion (Fig. 8A), and this observation was confirmed via microscopy (data not shown). Biofilm staining was readily observed in areas below the medium-air interface, suggesting that biofilm-mediated adhesion was not dependent on the interface (Fig. 8A). The control strain containing pBAD18 did not form a biofilm after arabinose supplementation (Fig. 8A). In addition, both strains did not form a biofilm under the same growth conditions in the absence of arabinose (data not shown). To confirm that biofilm formation was not the result of general protein expression from the pBAD18 arabinose-inducible promoter, we performed the same assay with strain χ3339 containing pBAD18+lacZ. This strain had a phenotype identical to that of the pBAD18 control and did not form a biofilm upon arabinose supplementation (Fig. 8B). Quantitation of crystal violet staining of the biofilm confirmed the qualitative differences observed between the strains in Fig. 8 (see Fig. S7 in the supplemental material).
Fig. 8.
Induction of ydcI expression results in S. Typhimurium biofilm formation. (A) S. Typhimurium strain χ3339 containing either pBAD18 or pBAD18+ydcI was grown in the presence of arabinose for approximately 16 h in LB medium. The white arrow indicates significant biofilm accumulation at the medium-air interface. The upper and lower panels show culture appearance and crystal violet-stained flasks (with cultures removed), respectively. (B) The same as panel A, except that strain χ3339 containing pBAD18+lacZ was tested.
Low fluid shear studies.
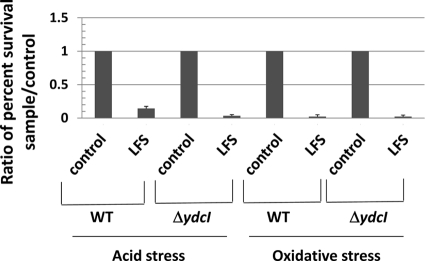
To test if the ydcI gene plays a role in transmission of the low fluid shear environmental signal in S. Typhimurium, we tested both the WT and ΔydcI mutant strains for two separate phenotypes induced by low fluid shear growth in the RWV apparatus. Previous studies have established that low fluid shear conditions alter the acid and oxidative stress resistance profiles of WT S. Typhimurium (39, 41, 52–55). We grew cultures of the WT and ΔydcI mutant strains under low fluid shear and control conditions in the RWV and subjected these cultures to separate acid (pH 3.5 using citric acid) and oxidative (70 mM H2O2) stresses. In both tests, the ΔydcI mutant strain displayed the same low fluid shear-induced phenotype as the WT strain (Fig. 9). This indicates that the ydcI gene is not required for these low fluid shear responses in S. Typhimurium under these conditions.
Fig. 9.
Low fluid shear phenotypes of WT and ΔydcI mutant strains via RWV culture. The S. Typhimurium WT and ΔydcI mutant strains were grown in the RWV apparatus in low fluid shear (LFS) or control conditions, and the cultures were tested for acid stress resistance (pH 3.5 with citric acid) or oxidative stress resistance (70 mM H2O2) in separate assays. The percent survival of the WT and ΔydcI mutant strains for each culture condition in each stress was calculated, and a ratio of the percent survival of each sample to the control condition for each stress is shown. Low fluid shear culture alters stress resistance compared to the control for both the WT and ΔydcI mutant strains.
DISCUSSION
Growth conditions such as spaceflight and RWV culture that are characterized by low fluid shear force have been shown to increase bacterial virulence (39, 52, 53). In addition, an array of other phenotypes is induced or altered via this growth environment (6, 7, 40, 41). Interestingly, these fluid shear levels are relevant to those encountered by certain pathogens in the human host during the course of infection (23, 36, 40, 41). However, this environmental cue is not well characterized, and a number of uncharacterized genes with unknown function are part of a stimulon associated with low fluid shear force (7, 52, 53, 55). Unexplored genes with potentially important, observable roles in prokaryotic biology could be part of this stimulon, but they may not have been previously analyzed because they have not been identified as part of any established regulatory networks or phenotypes. One such gene, S. Typhimurium ydcI, was discovered to be a member of the S. Typhimurium low fluid shear stimulon and has been predicted be homologous to the LysR family of transcriptional regulators (54, 55). However, the ydcI gene was previously uncharacterized before this study.
The goals of this study were to characterize the S. Typhimurium ydcI gene for a role in bacterial biology and to analyze the DNA binding activity of the YdcI protein. In this report, we demonstrate that the ydcI gene (i) is conserved in a range of Gram-negative bacteria, (ii) is expressed in S. Typhimurium and other Gram-negative species, (iii) is autorepressed in S. Typhimurium, (iv) encodes a DNA binding protein that binds specifically to a probe from the ydcI promoter, (v) is required for full resistance of S. Typhimurium to acid stress, (vi) is part of the rpoS regulon, (vii) can increase S. Typhimurium invasion of intestinal epithelial cells when induced, and (viii) can cause biofilm formation in S. Typhimurium when induced. These results indicate that the ydcI gene likely serves as a DNA binding transcriptional regulator that regulates genes involved in aspects of bacterial biology, including stress resistance and possibly virulence. A major goal in the future study of ydcI will be to identify the genes that are members of a potential “YdcI regulon” to understand how genes regulated by YdcI are linked to stress resistance, host cell interactions, biofilm formation, and other prokaryotic functions. In addition, future studies will focus on determining if ydcI has a role in the transmission of the low fluid shear environmental cue to bacterial cells beyond the experiments performed here. Though the results of Fig. 9 show that the ydcI gene is not required for the low fluid shear phenotypes tested as part of this study, this gene may be involved in other low fluid shear responses not yet tested (including in other species beyond S. Typhimurium).
Expression of the ydcI gene is upregulated by low fluid shear growth conditions (54, 55). Interestingly, though the ydcI gene is clearly expressed in bacteria under “standard” culture conditions, the S. Typhimurium ydcI gene appears to be repressed and expressed at relatively low levels under these conditions. One intriguing possibility is that ydcI is expressed at a certain level under one set of conditions and is then upregulated by an environmental signal(s) to induce adaptive cellular changes for another set of conditions. As indicated by our data, these changes could include altering resistance to stresses, interactions with host cells, or physiology related to biofilm formation. Whether these changes function to adapt S. Typhimurium or other bacteria for their roles as pathogens remains to be determined with future studies. However, our data suggest that, at a minimum, the ydcI gene likely regulates genes with the potential for involvement in functions related to stress resistance, growth/survival, and possibly virulence. The observation that ydcI expression involves the RpoS sigma factor provides support to this hypothesis since RpoS controls the expression of a regulon of genes involved in stress resistance and virulence (16, 26, 29, 38). However, we have previously reported that RpoS is not required for a range of low fluid shear responses in S. Typhimurium (54). Therefore, a possible scheme for the regulation of ydcI expression is that RpoS is used for expression under certain conditions and another (yet to be identified) pathway is used for expression under other conditions (such as low fluid shear). Future studies will be aimed at determining how ydcI gene regulation fits into regulatory schemes in a range of bacteria.
This is the first report to demonstrate DNA binding activity of the YdcI protein. We show that both (i) extracts containing YdcI protein and (ii) purified YdcI protein are able to bind a DNA probe (from the S. Typhimurium ydcI promoter region) in a specific manner. The YdcI DNA binding activity displays an apparent dissociation constant with a value consistent with specific DNA binding proteins with biologic functions and consistent with other members of LysR family (to which YdcI belongs) (34, 44, 47). Induced expression of the YdcI protein repressed an S. Typhimurium ydcI::lacZ fusion and complemented the acid resistance defect of an S. Typhimurium ΔydcI mutant. These observations suggest that the YdcI DNA binding activity functions in bacterial cells to affect physiological changes. Future studies will be aimed at determining (i) the specific DNA site that is recognized by YdcI, (ii) the genes associated with YdcI binding sites, and (iii) whether these sites are used by YdcI in other bacterial species. We identified potential binding sites within the H3 probe fragment, but smaller DNA fragments containing these individual sites did not display YdcI binding activity. Identification of the actual YdcI binding site will require additional, systematic study that is beyond the scope of this report. However, this report provides a foundation for analyzing the previously uncharacterized YdcI DNA binding activity via a number of approaches.
Though induced ydcI expression affects S. Typhimurium intestinal epithelial cell invasion and biofilm formation, we do not know how the YdcI protein functions in these phenotypes. We demonstrate that the host cell interaction phenotype only affects the invasion of intestinal epithelial cells and not survival in macrophages. One possibility for this may be that ydcI can alter the expression of genes involved in SPI-1 type III secretion-mediated invasion of nonphagocytic cells or other genes involved in specific interactions with epithelial cells. Indeed, the induction of ydcI did not increase invasion by a ΔSPI-1 strain, suggesting that ydcI possibly acts through the SPI-1 pathway for increased invasion. We tested a SPI-1 invI::lacZ fusion for altered expression in the presence of pBAD18+ydcI and found that ydcI induction produced a 7.1-fold decrease in reporter expression (data not shown). This indicates that YdcI may potentially alter SPI-1 gene expression as part of its function, but how YdcI would do this in the context of the many different SPI-1 genes and the other regulators of SPI-1 gene expression remains to be determined. Systematic identification of the genes regulated by YdcI and elucidation of how YdcI controls these genes as part of an overall regulatory scheme will require further controlled study. Biofilm formation in Salmonella spp. appears to be a complex process and has been shown to involve a number of different genes involved in flagella and curli fiber formation, cellulose production, and O-antigen synthesis (8, 9, 27, 28, 31). In addition, there are likely at least two different types of biofilms formed by Salmonella spp. that are utilized on either cholesterol-rich surfaces (like gallstones) or on material surfaces like glass or plastic (8, 9). We do not know how ydcI affects Salmonella genes involved in biofilm formation, but studies aimed at determining the nature of the ydcI-induced biofilm and the genes involved in this process are currently being pursued.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the support of NASA grant NCC2-1362, NASA grant NNX09AH40G, and the Department of Biology and ORSP, Villanova University.
We thank John Friede for providing S. marcescens, Dennis Wykoff for providing reagents and advice, and Jason Scheidel for technical support.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Ausubel F., et al. 1996. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 3. Bannister S. R., et al. 2002. Shear force modulates osteoblast response to surface roughness. J. Biomed. Mater. Res. 60:167–174 [DOI] [PubMed] [Google Scholar]
- 4. Beeson J. G., et al. 2000. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat. Med. 6:86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai Z., Xin J., Pollock D. M., Pollock J. S. 2000. Shear stress-mediated NO production in inner medullary collecting duct cells. Am. J. Physiol. Renal Physiol. 279:F270–F274 [DOI] [PubMed] [Google Scholar]
- 6. Crabbé A., et al. 2008. Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ. Microbiol. 10:2098–2110 [DOI] [PubMed] [Google Scholar]
- 7. Crabbé A., et al. 2010. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 12:1545–1564 [DOI] [PubMed] [Google Scholar]
- 8. Crawford R. W., Gibson D. L., Kay W. W., Gunn J. S. 2008. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 76:5341–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crawford R. W., Reeve K. E., Gunn J. S. 2010. Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 192:2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Curtiss R., III, et al. 2009. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 77:1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curtiss R., III, et al. 2010. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit. Rev. Immunol. 30:255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ellermeier C. D., Janakiraman A., Slauch J. M. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161 [DOI] [PubMed] [Google Scholar]
- 14. Fang A., Pierson D. L., Koenig D. W., Mishra S. K., Demain A. L. 1997. Effect of simulated microgravity and shear stress on microcin B17 production by Escherichia coli and on its excretion into the medium. Appl. Environ. Microbiol. 63:4090–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fang A., Pierson D. L., Mishra S. K., Demain A. L. 2000. Growth of Streptomyces hygroscopicus in rotating-wall bioreactor under simulated microgravity inhibits rapamycin production. Appl. Microbiol. Biotechnol. 54:33–36 [DOI] [PubMed] [Google Scholar]
- 16. Fang F. C., et al. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89:11978–11982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Felix A., Pitt R. M. 1951. The pathogenic and immunogenic activities of Salmonella typhi in relation to its antigenic constituents. J. Hyg. (Lond.) 49:92–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galán J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53–86 [DOI] [PubMed] [Google Scholar]
- 19. Galán J. E., Collmer A. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322–1328 [DOI] [PubMed] [Google Scholar]
- 20. Galán J. E., Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573 [DOI] [PubMed] [Google Scholar]
- 21. Gao H., Ayyaswamy P. S., Ducheyne P. 1997. Dynamics of a microcarrier particle in the simulated microgravity environment of a rotating-wall vessel. Microgravity Sci. Technol. 10:154–165 [PubMed] [Google Scholar]
- 22. Gulig P. A., Curtiss R., III 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55:2891–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo P., Weinstein A. M., Weinbaum S. 2000. A hydrodynamic mechanosensory hypothesis for brush border microvilli. Am. J. Physiol.-Renal Physiol. 279:F698–F712 [DOI] [PubMed] [Google Scholar]
- 24. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammond T. G., Hammond J. M. 2001. Optimized suspension culture: the rotating-wall vessel. Am. J. Physiol. Renal Physiol. 281:F12–25 [DOI] [PubMed] [Google Scholar]
- 26. Hengge-Aronis R. 2000. The general stress response in Escherichia coli, p. 161–168 In Storz G., Hengge-Aronis R. (ed.), Bacterial stress responses. ASM Press, Washington, D.C [Google Scholar]
- 27. Jonas K., et al. 2007. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim S. H., Wei C. I. 2009. Molecular characterization of biofilm formation and attachment of Salmonella enterica serovar typhimurium DT104 on food contact surfaces. J. Food Prot. 72:1841–1847 [DOI] [PubMed] [Google Scholar]
- 29. Klauck E., Typas A., Hengge R. 2007. The sigmaS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci. Prog. 90:103–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klaus D. M., Todd P., Schatz A. 1998. Functional weightlessness during clinorotation of cell suspensions. Adv. Space Res. 21:1315–1318 [DOI] [PubMed] [Google Scholar]
- 31. Latasa C., et al. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322–1339 [DOI] [PubMed] [Google Scholar]
- 32. Li Y., et al. 2009. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc. Natl. Acad. Sci. U. S. A. 106:593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lynch S. V., Mukundakrishnan K., Benoit M. R., Ayyaswamy P. S., Matin A. 2006. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl. Environ. Microbiol. 72:7701–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. MacLean A. M., Anstey M. I., Finan T. M. 2008. Binding site determinants for the LysR-type transcriptional regulator PcaQ in the legume endosymbiont Sinorhizobium meliloti. J. Bacteriol. 190:1237–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Nauman E. A., et al. 2007. Novel quantitative biosystem for modeling physiological fluid shear stress on cells. Appl. Environ. Microbiol. 73:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neph S., Tompa M. 2006. MicroFootPrinter: a tool for phylogenetic footprinting in prokaryotic genomes. Nucleic Acids Res. 34:W366–W368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nickerson C. A., Curtiss R., III 1997. Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect. Immun. 65:1814–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nickerson C. A., et al. 2000. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 68:3147–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nickerson C. A., et al. 2003. Low-shear modeled microgravity: a global environmental regulatory signal affecting bacterial gene expression, physiology, and pathogenesis. J. Microbiol. Methods 54:1–11 [DOI] [PubMed] [Google Scholar]
- 41. Nickerson C. A., Ott C. M., Wilson J. W., Ramamurthy R., Pierson D. L. 2004. Microbial responses to microgravity and other low-shear environments. Microbiol. Mol. Biol. Rev. 68:345–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Sullivan L. E., Nickerson C. A., Wilson J. W. 2010. A series of IncQ-based reporter plasmids for use in a range of Gram negative genera. J. Microbiol. Biotechnol. 20:871–874 [PubMed] [Google Scholar]
- 43. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 44. Parsek M. R., Ye R. W., Pun P., Chakrabarty A. M. 1994. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region catBC promoter activation by CatR. J. Biol. Chem. 269:11279–11284 [PubMed] [Google Scholar]
- 45. Quick L. N., Shah A., Wilson J. W. 2010. A series of vectors with alternative antibiotic resistance markers for use in lambda Red recombination. J. Microbiol. Biotechnol. 20:666–669 [DOI] [PubMed] [Google Scholar]
- 46. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tralau T., Mampel J., Cook A. M., Ruff J. 2003. Characterization of TsaR, an oxygen-sensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 69:2298–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tucker D. L., et al. 2007. Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol. 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilson J. W., Coleman C., Nickerson C. A. 2007. Cloning and transfer of the Salmonella pathogenicity island 2 type III secretion system for studies of a range of gram-negative genera. Appl. Environ. Microbiol. 73:5911–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wilson J. W., Nickerson C. A. 2006. Cloning of a functional Salmonella SPI-1 type III secretion system and development of a method to create mutations and epitope fusions in the cloned genes. J. Biotechnol. 122:147–160 [DOI] [PubMed] [Google Scholar]
- 51. Wilson J. W., Nickerson C. A. 2006. A new experimental approach for studying bacterial genomic island evolution identifies island genes with bacterial host-specific expression patterns. BMC Evol. Biol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson J. W., Ott C. M., Honer zu Bentrup K., et al. 2007. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl. Acad. Sci. U. S. A. 104:16299–16304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilson J. W., Ott C. M., Quick L., Davis R., Honer zu Bentrup K., et al. 2008. Media ion composition controls regulatory and virulence response of Salmonella in spaceflight. PLoS One 3:e3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilson J. W., et al. 2002. Low-shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS-independent manner. Appl. Environ. Microbiol. 68:5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson J. W., et al. 2002. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc. Natl. Acad. Sci. U. S. A. 99:13807–13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.