Abstract
Previously, we isolated a selenate- and arsenate-reducing bacterium, designated strain SF-1, from selenium-contaminated sediment and identified it as a novel species, Bacillus selenatarsenatis. B. selenatarsenatis strain SF-1 independently reduces selenate to selenite, arsenate to arsenite, and nitrate to nitrite by anaerobic respiration. To identify the genes involved in selenate reduction, 17 selenate reduction-defective mutant strains were isolated from a mutant library generated by random insertion of transposon Tn916. Tn916 was inserted into the same genome position in eight mutants, and the representative strain SF-1AM4 did not reduce selenate but did reduce nitrate and arsenate to the same extent as the wild-type strain. The disrupted gene was located in an operon composed of three genes designated srdBCA, which were predicted to encode a putative oxidoreductase complex by the BLASTX program. The plasmid vector pGEMsrdBCA, containing the srdBCA operon with its own promoter, conferred the phenotype of selenate reduction in Escherichia coli DH5α, although E. coli strains containing plasmids lacking any one or two of the open reading frames from srdBCA did not exhibit the selenate-reducing phenotype. Domain structure analysis of the deduced amino acid sequence revealed that SrdBCA had typical features of membrane-bound and molybdopterin-containing oxidoreductases. It was therefore proposed that the srdBCA operon encoded a respiratory selenate reductase complex. This is the first report of genes encoding selenate reductase in Gram-positive bacteria.
INTRODUCTION
Selenium is the 34th element on the periodic table and has chemical properties resembling those of sulfur. It is obtained in limited ways, such as a by-product of the electric smelting of copper, and is an important material used in photoelectric devices, photosensitive drums used in dry copying, semiconductors, and the colorization and decolorization of glasses. Biologically, selenium is an essential element used for the synthesis of selenocysteine contained in selenoproteins, such as mammalian glutathione peroxidase and bacterial formate dehydrogenase. However, exposure to higher concentrations of selenium is toxic, and therefore it is important to understand how environmental selenium is controlled. Selenium has several oxidation states in the environment, i.e., selenate (+VI), selenite (+IV), elemental selenium (0), selenide (−II), and organic selenium (−II), and it is known that prokaryotes play a major role in its oxidation and reduction (28).
The molecular mechanisms of selenate reduction have been analyzed in some bacteria. The selenate reductase complex of Thauera selenatis has been intensively studied biochemically and genetically. The selenate reductase complex of T. selenatis is a soluble periplasmic protein (20) that consists of three subunits, i.e., a catalytic subunit containing a molybdenum cofactor [Mo(V)], a subunit containing iron-sulfur clusters (one [3Fe-4S] cluster and three [4Fe-4S] clusters), and a subunit containing heme b (6, 25). The genes encoding them were identified as serA, serB, and serC, respectively. They comprise an operon with serD, which encodes a system-specific chaperone (14). Furthermore, identification of cytochrome c4 as an electron mediator from quinols to SerABC revealed the important link between SerABC and selenate respiration in T. selenatis (15).
Another well-studied selenate reductase belongs to Enterobacter cloacae strain SLD1a-1. The selenate reductase complex of strain SLD1a-1 also comprises three subunits containing molybdenum, heme, and nonheme iron (21). In contrast to the protein in T. selenatis, the selenate reductase complex of E. cloacae strain SLD1a-1 is a membrane-bound insoluble protein. In this organism, genetic studies also revealed that the selenate reduction pathway requires the global transcriptional regulator gene fnr (33), the twin-arginine translocation pathway genes tatABC (16), and the menaquinone biosynthetic pathway genes menFDHBCE (17), although the genes encoding the selenate reductase complex remain unknown. In Escherichia coli, two operons, ygfKMN (1) and ynfEFGH (10), have been proposed to encode the selenate reductase, although selenate reduction by E. coli has not been characterized. Thus, the molecular mechanisms of selenate reduction have been studied in a range of Gram-negative bacteria but to date have never been studied in Gram-positive bacteria. Since Gram-positive bacteria have no outer membrane and periplasmic space, the potential differences between the selenate reductases in Gram-negative and Gram-positive bacteria are interesting, such as whether the proteins are soluble or membrane bound and whether they are cytoplasmic or extracytoplasmic.
We previously isolated a selenate-reducing bacterium, strain SF-1, from the effluent sediment of a glass manufacturing plant (8) and identified it as a new species, Bacillus selenatarsenatis (32). Strain SF-1 is able to reduce selenate to selenite, arsenate to arsenite, and nitrate to nitrite, independently, by anaerobic respiration (13, 30). These reductive activities are inhibited by the addition of 1 mM tungstate to the culture medium, implying that these reduction reactions are catalyzed by molybdoenzymes (31), although the molecular and genetic mechanisms of these reactions have not yet been fully analyzed.
The current study analyzed the respiratory selenate reduction mechanism in strain SF-1 using molecular genetic methods. Selenate reduction-defective mutants were generated by random insertion of the conjugative transposon Tn916 (7) into its genome, and three genes related to selenate reduction were identified in the region flanking the insertion site. From analysis of the mutant phenotypes and the observation of selenate reduction in recombinant E. coli cells, it was proposed that the identified genes were specific for respiratory selenate reduction and independent of arsenate and nitrate reduction. Analyses of the deduced amino acid sequences predicted that the genes encode a selenate reductase complex.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. B. selenatarsenatis SF-1SMR is a spontaneous streptomycin-resistant (Smr) mutant isolated from strain SF-1 by selection on LB agar plates containing streptomycin at 1,000 μg/ml. Enterococcus faecalis strain CG110 was a kind gift from M. Morikawa of Hokkaido University, Japan. E. coli DH5α was used to host recombinant DNA vectors.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference(s) or source |
|---|---|---|
| Bacillus selenatarsenatis strains | ||
| SF-1 | Wild type | 8, 32 |
| SF-1SMR | Spontaneous Smr mutant | This work |
| Enterococcus faecalis CG110 | Tn916 donor; Tcr Sms | 9 |
| Escherichia coli DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 23 |
| Plasmids | ||
| pGEM-T Easy vector | TA cloning vector; Apr | Promega |
| pGEMsrdBCA | srdBCA operon on pGEM-T Easy vector | This work |
| pGEMsrdCA | Deletion of srdB, based on pGEMsrdBCA | This work |
| pGEMsrdBA | Deletion of srdC, based on pGEMsrdBCA | This work |
| pGEMsrdBC | Deletion of srdA, based on pGEMsrdBCA | This work |
| pGEMsrdB | Deletion of srdCA, based on pGEMsrdBCA | This work |
| pGEMsrdC | Deletion of srdBA, based on pGEMsrdBCA | This work |
| pGEMsrdA | Deletion of srdBC, based on pGEMsrdBCA | This work |
| pGEMsrdpt | Deletion of srdBCA, based on pGEMsrdBCA | This work |
Growth media and conditions.
B. selenatarsenatis SF-1 and its mutants were cultivated in Bacto Trypticase soy broth (TSB) (Becton-Dickinson, Franklin Lakes, NJ) supplemented with 24 g/liter NaCl, 7 g/liter MgSO4 · 7H2O, 5.3 g/liter MgCl2 · 6H2O, 0.7 g/liter KCl, and 0.1 g/liter CaCl2 (pH 7.5) (34) or on Difco LB broth (Lennox) (Becton-Dickinson) containing 1.5% (wt/vol) agar, at 37°C, unless otherwise stated. Basal salt medium (BSM), used for phenotypic analysis of the mutant strain SF-1AM4, contained 0.1 g/liter NaCl, 0.1 g/liter KH2PO4, 0.2 g/liter CaCl2 · 2H2O, 0.24 g/liter NH4Cl, 0.12 g/liter MgCl2 · 6H2O, 0.6 mg/liter H3BO3, 0.17 mg/liter CoCl2 · 6H2O, 0.07 mg/liter CuCl2, 0.22 mg/liter ZnCl2, and 1.0 g/liter yeast extract in 50 mM Tris-HCl buffer (pH 8.0). For aerobic cultivation, 10 g/liter of glucose was added to BSM as a carbon source. For anaerobic cultivation, 20 mM sodium lactate and 1 mM sodium selenate, sodium selenite, sodium arsenate, or sodium nitrate were used as the electron donor and the electron acceptor, respectively, to supplement BSM. E. faecalis CG110 and E. coli strains were cultivated on LB broth containing 1.5% (wt/vol) agar at 37°C. When necessary, streptomycin (Sm) (500 μg/ml), tetracycline (Tc) (10 μg/ml), and ampicillin (Ap) (30 μg/ml) were supplemented into the media.
DNA manipulation.
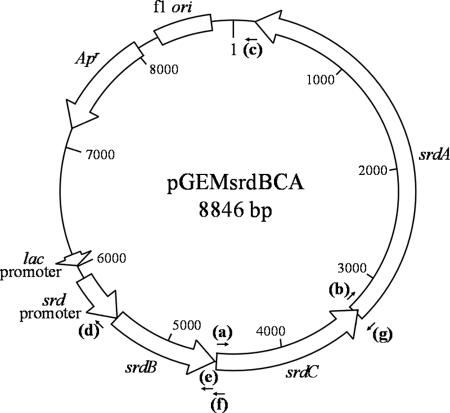
Restriction enzymes (TaKaRa Bio, Shiga, Japan, or Toyobo, Osaka, Japan) and T4 DNA ligase (TaKaRa Bio) were used according to the manufacturer's instructions. PCR amplification was performed as previously described (23) using a GeneAmp PCR system 9700 (Life Technologies Japan, Tokyo, Japan), with the primers listed in Table 2. KOD-plus DNA polymerase (Toyobo) was used for inverse PCR and the construction of plasmid vectors, and Ex Taq DNA polymerase (TaKaRa Bio) was used for all other PCR applications and for the A-tailing of DNA fragments. An AquaPure genomic DNA kit (Bio-Rad Laboratories, Tokyo, Japan) was used for the preparation of genomic DNA and the Quantum Prep Plasmid MiniPrep kit (Bio-Rad) was used for the preparation of plasmid DNA; both were used according to the manufacturer's instructions. DNA fragments were extracted from agarose gels using the Illustra GFX PCR DNA and gel band purification kit (GE Healthcare, Buckinghamshire, United Kingdom). Nucleotide sequences were determined using the BigDye Terminator kit and an ABI3100 system (Life Technologies Japan). Agarose gel electrophoresis and the transformation of E. coli were performed as previously described (23). To construct pGEMsrdBCA plasmid DNA (Fig. 1), a DNA fragment including srdBCA with its original promoter region was amplified by PCR using the primers SRDBCAF and SRDBCAR and inserted into the pGEM-T Easy vector (Promega, Tokyo, Japan). Plasmids lacking one or two of the open reading frames (ORFs) from srdBCA were constructed by self-ligation of DNA amplified using the following primer sets (Table 2): SRDCF and SRDPR for pGEMsrdCA, SRDAF and SRDBR2 for pGEMsrdBA, SRDTF and SRDCR for pGEMsrdBC, SRDTF and SRDBR1 for pGEMsrdB, SRDTF and SRDCR for pGEMsrdC, SRDAF and SRDPR for pGEMsrdA, and SRDTF and SRDPR for pGEMsrdpt. The annealing sites for primers on pGEMsrdBCA are shown in Fig. 1. Plasmid pGEMsrdBCA was used as a template for the construction of vectors lacking in each ORF, except for pGEMsrdC, where pGEMsrdCA was used as the template.
Table 2.
Primers used in this study
| Primer | Nucleotide sequence (5′ to 3′) |
|---|---|
| TN916F | ATACCATTCACATCGAAGTGCCGCCA |
| TN916R | TGGCAAACAGGTTCACCGGTACTAACA |
| SRDBCAF | CCAGAAACAGCAAAGTCCTTGTCG |
| SRDBCAR | GCAGCTTCCCTTTCGCACAAAGTT |
| SRDAF | ATGGAAAACCAACACCAGAAATTC |
| SRDBF | CTTATGGAGGTGAAATAAATGG |
| SRDCF | ATGTTAAAAAAATTATATTTTACAGTG |
| SRDTF | TCTTTAAAAGATCTATTTAACAGCAAC |
| SRDPR | TTATTTCACCTCCATAAGAATTAAAC |
| SRDBR1 | TATTCAGCACCTCCTCTTTATG |
| SRDBR2 | TTATGTTAAGTAATATACATTTGGTTCAG |
| SRDCR | TTACGCCTTGATATGAATTTCTG |
Fig. 1.
Genetic map of pGEMsrdBCA. The srdBCA operon with its own promoter region was inserted into the pGEM-T Easy vector under the direction of the lac promoter. Annealing sites of primers used for the construction of derivative vectors are indicated: a, SRDCF; b, SRDAF; c, SRDTF; d, SRDPR; e, SRDBR1; f, SRDBR2; g, SRDCR.
Transposon mutagenesis and screening of mutants defective for selenate reduction.
E. faecalis CG110 was used as the donor strain of transposon Tn916, and strain SF-1SMR was used as the recipient. Strain SF-1SMR was cultivated in 3 ml of TSB supplemented with Sm at 37°C for 20 h, and E. faecalis CG110 was streaked and cultivated on an LB agar plate at 37°C for 20 h. The culture of strain SF-1SMR was centrifuged and washed twice with 3 ml of LB medium and then resuspended in 3 ml of LB medium. A 0.2-ml suspension of strain SF-1SMR was spread on the LB agar plate on which strain CG110 had already grown, as described above, and the plate containing both strains was incubated at 37°C overnight. The cells on the agar plate were recovered as a suspension in 10 ml of TSB, diluted 100-fold, and then spread onto an LB agar plate containing Sm, Tc, and 0.5 mM sodium selenate. The plates were incubated at 37°C overnight and then incubated at 30°C for several additional hours to allow the colonies to reduce selenate and develop the red color indicative of elemental selenium. White colonies were isolated and incubated on LB agar plates containing Sm, Tc, and 1 mM sodium selenate at 37°C overnight and then further incubated at 30°C for more than 2 days under anaerobic conditions using an AnaeroPouch-Anaero (Mitsubishi Gas Chemical Company, Japan) to confirm selenate reduction. Clones that grew stably and formed white colonies were identified as potential selenate reduction-defective mutants.
Southern blotting to detect integrated transposons.
The transfer of HindIII-digested genomic DNAs of strain SF-1SMR and selenate reduction-defective mutant strains to Hybond-N+ (GE Healthcare) was performed as previously described (23). The DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Diagnostics, Japan) was used for probe labeling, hybridization, and detection. A partial DNA fragment of the tet(M) gene of Tn916 was PCR amplified using primers TN916F and TN916R and used as a probe.
Cloning of the flanking region of the transposon insertion site.
The flanking region of the Tn916-insertion site was amplified by inverse PCR (23) using HindIII-digested genomic DNA. Amplified DNA fragments were separated by agarose gel electrophoresis and purified. The DNA fragments were subcloned into the pGEM-T Easy vector in E. coli DH5α, and the inserted DNA sequence was determined. An LA PCR in vitro cloning kit (TaKaRa Bio) was used for cloning entire genes.
Phenotypic analysis of mutant strain SF-1AM4.
To analyze the phenotype of strain SF-1AM4, cells of strain SF-1SMR or SF-1AM4 were grown in 20 ml of BSM containing glucose in a 50-ml bottle at 30°C for 12 h. The cells were harvested by centrifugation (6,000 × g, 10 min, 4°C) and resuspended in a small volume of 50 mM Tris-HCl buffer (pH 8.0). The cell suspensions were inoculated into 20 ml of BSM containing 20 mM lactate and an appropriate electron acceptor (1 mM selenate, selenite, arsenate, or nitrate) in a 50-ml bottle. The bottles were sealed with butyl rubber septa and aluminum crimp seals. The headspace was replaced with N2 gas, and the bottles of broth were incubated on a rotary shaker at 30°C for 12 h (selenate, arsenate, and nitrate) or 48 h (selenite). The concentrations of selenate, selenite, arsenate, and nitrate were determined by ion-exchange chromatography (HIC-SP system; Shimadzu, Japan) with an IonPac AS4A-SL column (Dionex) for selenate, selenite, and nitrate or with a HIC-SA3 column (Shimadzu, Japan) for arsenate. The mobile phase was a 3 mM Na2CO3 solution prepared with ultrapure water from a DIRECT-Q system (Nihon Millipore, Japan).
Phenotypic analysis of recombinant E. coli.
Selenate reduction in the recombinant E. coli strains was analyzed on agar plates. E. coli DH5α strains carrying pGEMsrdBCA and its derivative plasmids were cultivated aerobically in 3 ml of LB medium containing Ap at 37°C for 6 h. Each broth culture was spotted onto LB agar plates containing Ap and 0.5 mM selenate or selenite. The plates were incubated at 37°C for 60 h, and the color of the colonies was observed.
Computational analysis.
All sequences were analyzed with the GENETYX-WIN Program (version 3.2; Genetyx, Japan). The similarity of the sequences was analyzed using BLASTN searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) based on the nucleotide collection database. The deduced amino acid sequences were annotated using BLASTX searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) based on the Swiss-Prot database, and their domain architectures were predicted by SMART (http://smart.embl-heidelberg.de/) (26). All of the analyses based on databases were performed on 27 October 2009. Multiple-alignment and phylogenetic analyses of the catalytic subunits, including SrdA, were performed using the ClustalW software (version 1.83), as previously described (22). All amino acid sequences except for that of SrdA were obtained from the Swiss-Prot database (http://au.expasy.org/sprot/) on 27 October 2009. The tree was generated using the NJplot program (19).
Nucleotide sequence accession number.
The nucleotide sequence of the srdBCA operon is available in the DNA Data Bank of Japan (DDBJ) under accession number AB534554.
RESULTS
Generation of selenate reduction-defective mutants.
Selenate reduction-defective mutants of strain SF-1 were generated by insertion of Tn916, which was transferred by conjugation with E. faecalis CG110. To facilitate the screening of mutant strains from donor strains, the spontaneous Smr strain SF-1SMR was used as the recipient. By random insertion into the genome of strain SF-1SMR, a mutant library consisting of 400,000 colonies which exhibited both Sm- and Tc-resistant phenotypes was obtained on selective agar plates containing selenate, Sm, and Tc. From this library, 110 white colonies were screened. Screened colonies were further confirmed on the selective plates, and 17 clones that were able to grow stably and form white colonies were defined as selenate reduction-defective mutants and were designated strains SF-1AM1 to SF-1AM17.
The copy numbers of Tn916 integrated into each clone were identified by Southern hybridization analysis using the partial tetM gene contained in Tn916 as a probe. SF-1AM2, SF-1AM8, SF-1AM9, and SF-1AM12 were found to contain several copies of the transposon (five, four, two, and four copies, respectively), and the others contained only one copy. The parental strain SF-1SMR did not contain any Tn916 sequences.
The nucleotide sequences of the flanking regions of the Tn916 insertion site in the 13 mutants containing a single Tn916 insertion were isolated by inverse PCR, and their determined sequences were analyzed with the BLASTX program (see Table S1 in the supplemental material). Tn916 was inserted into the same genomic position in eight mutants (SF-1AM4, -7, -11, -13, -14, -15, -16, and -17), indicating that they are the same clone, and the disrupted gene was believed to encode a putative thiosulfate reductase. The remaining mutants possessed Tn916 in the genome but at different positions. In mutant strains SF-1AM1, -3, -5, -6, and -10, Tn916 likely affected the sequences encoding diguanylate cyclase, a molybdate metabolism regulator, 2-keto-4-pentenoate hydratase, a membrane protein of unknown function, and an uncharacterized ATP binding cassette (ABC) transporter permease, respectively.
Identification of the srdBCA genes, encoding a putative selenate reductase.
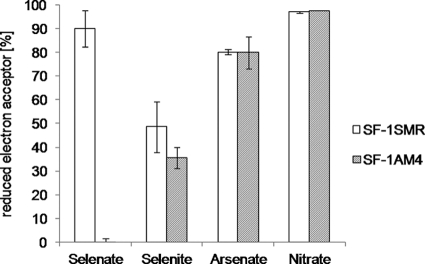
The gene disrupted by Tn916 in mutant strain SF-1AM4 and the other seven mutants may encode a reductase related to selenate reduction. To confirm the relationship between the disrupted gene and selenate reduction, the reduction of selenate, selenite, arsenate, and nitrate by strain SF-1AM4 in liquid medium was analyzed using ion-exchange chromatography (Fig. 2). Selenate reduction was abolished in strain SF-1AM4, whereas arsenate and nitrate were reduced to the same extent as in strain SF-1SMR. Selenite reduction was decreased in strain SF-1AM4 but not significantly (the error bars, indicating standard deviations, overlapped). Strain SF-1AM4 did not grow with selenate as a sole electron acceptor (data not shown). These results suggested that the gene disrupted by Tn916 in the mutant strains is involved mainly in respiratory selenate reduction and has little effect on selenite reduction.
Fig. 2.
Selenate, selenite, arsenate, and nitrate reduction by strains SF-1SMR and SF-1AM4. The ratios of reduced electron acceptors against their initial concentrations (1 mM) are indicated as percentages. Values represent the mean electron acceptor reduction in three independent cultures, and error bars represent the standard deviation (SD).
To obtain the entire gene sequence, approximately 6 kb of sequence flanking the Tn916 insertion site in strain SF-1AM4 was cloned and its sequence was analyzed (Fig. 3; Table 3). Three ORFs were located in this region and designated srdB, srdC, and srdA. Tn916 was inserted into srdA. The sizes of srdB, srdC, and srdA were 876 bp, 1,278 bp, and 3,144 bp, respectively. Therefore, it was predicted that the proteins encoded by srdBCA comprise 292, 426, and 1,048 amino acids, respectively. The start codon for srdC was located 18 bp downstream of the stop codon for srdB, and the start codon for srdA was located 35 bp upstream of the stop codon for srdC. The BLASTN program revealed that the nucleotide sequences of srdBCA shared no significant homology with sequences in any other bacteria, suggesting that the srdBCA genes are phylogenetically novel genes. Through BLASTX searches, these ORFs were predicted to encode subunits of a membrane-anchored oxidoreductase containing an iron-sulfur cluster and a molybdenum cofactor. This composition of subunits is generally known as a respiratory oxidoreductase (22). The adjacent locations of the ORFs implies that they comprise an operon that may be related to anaerobic respiration.
Fig. 3.
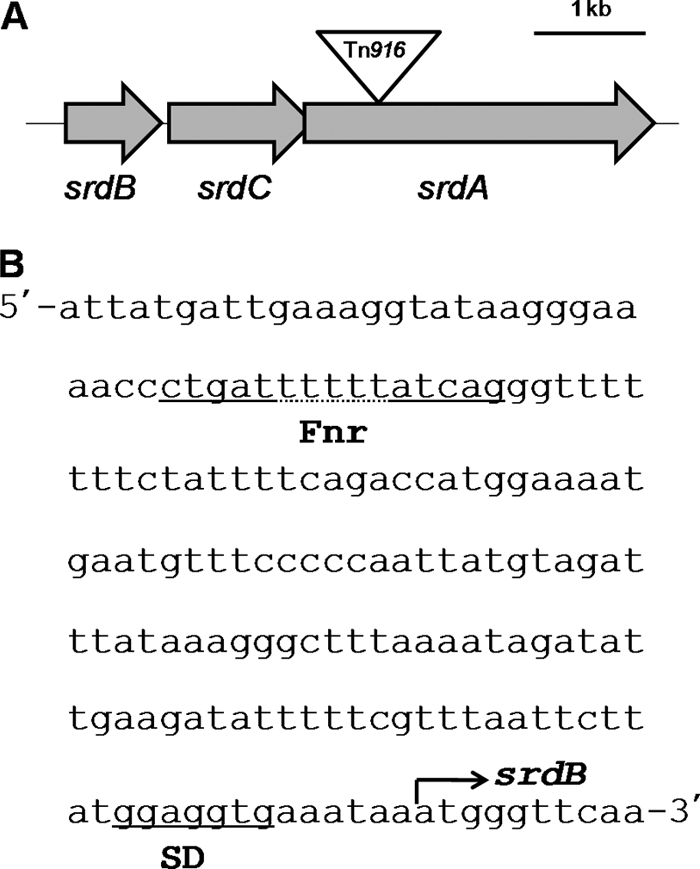
(A) Schematic representation of the srdBCA operon. The inverted triangle indicates the Tn916 insertion site. (B) Nucleotide sequence of the promoter region in the srdBCA operon. A putative Shine-Dalgarno (SD) sequence and Fnr binding motif are located upstream of srdB.
Table 3.
Summary of genes in the srdBCA operon
| ORF | Start (base) | Stop (base) | Length (bp) | Accession no. | Organism | Putative function | Score | E value | % Identity |
|---|---|---|---|---|---|---|---|---|---|
| srdB | 496 | 1,371 | 876 | P31076 | Wolinella succinogenes | Polysulfide reductase chain B | 135 | 4.00E−31 | 36 |
| srdC | 1,392 | 2,669 | 1,278 | O29750 | Archaeoglobus fulgidus | Heterodisulfide reductase-like menaquinol oxidoreductase integral membrane subunit | 126 | 3.00E−28 | 27 |
| srdA | 2,635 | 5,778 | 3,144 | P46448 | Haemophilus influenzae | Formate dehydrogenase subunit alpha | 62.4 | 2.00E−08 | 25 |
Putative Shine-Dalgarno sequences were found upstream of the srdB, srdC, and srdA genes (5′-GGAGGTG-3′, 5′-GGAGGTG-3′, and 5′-GGAGGTA-3′, respectively). Upstream of the srdB gene, a conserved sequence (5′-CTGATNNNNNATCAG-3′) was identified, which was similar to the consensus sequence (5′-TTGATNNNNATCAA-3′) for the Fnr protein binding site that positively regulates genes under anaerobic conditions (Fig. 3B) (29).
Domain structure analysis of the deduced amino acid sequences of the srdBCA operon.
The deduced amino acid sequences of the srdBCA operon were analyzed to predict the domain structures and the functions of the encoding proteins. The SMART program was employed for the prediction of conserved domains. It was predicted that SrdA has four conserved domains: a twin-arginine translocation (TAT) signal on the N terminus (amino acids 8 to 33) (2), a [4Fe-4S] cluster domain of molybdopterin oxidoreductase (amino acids 56 to 127), and two domains for molybdopterin (amino acids 130 to 717 and 892 to 1041). In the SrdB sequence, three domains were predicted: a TAT signal on the N terminus (amino acids 1 to 43) and two [4Fe-4S] cluster binding domains (amino acids 86 to 109 and 163 to 186). According to the consensus sequence for the [4Fe-4S] cluster binding domain, CAX2CBX2-11CCX3CDP (22), the sequence of SrdB apparently has two more [4Fe-4S] iron-sulfur cluster binding motifs located at amino acids 138 to 151 and 230 to 250, in addition to two predicted [4Fe-4S] domains. The SMART program predicted that SrdC has a signal peptide (amino acids 1 to 28) and nine transmembrane regions (amino acids 39 to 61, 74 to 96, 116 to 138, 187 to 209, 224 to 246, 259 to 281, 301 to 323, 328 to 350, and 383 to 405).
Selenate reduction by recombinant E. coli strains containing the srdBCA operon.
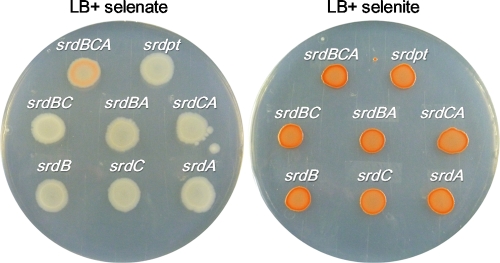
To confirm the necessity of the ORFs in the srdBCA operon for selenate reduction, E. coli DH5α was transformed with pGEMsrdBCA and its derivative plasmids containing the ORFs of the srdBCA operon in several combinations (Fig. 1; Table 1). Since a transformation method for B. selenatarsenatis has not been established, E. coli was used as the host, with expression of srdBCA being expecting to occur via its own promoter because the promoter region has a sequence similar to the −35 and −10 sequences for σ70 of E. coli. Selenate and selenite reduction was examined on agar plates containing selenate and selenite, respectively (Fig. 4). In a preliminary experiment, agar plates were incubated anaerobically; however, this resulted in selenate reduction in all strains despite the presence or absence of the srdBCA genes. This may be due to the expression of E. coli genes, such as ygfKMN (1) and ynfEFGH (10), whose functions are related to anaerobic selenate reduction. Therefore, we selected aerobic conditions in subsequent experiments. Under aerobic conditions, DH5α/pGEMsrdBCA formed red spots of cells in the presence of selenate, whereas other recombinant strains formed white spots of cells. Observing the spots more precisely, the generation of elemental selenium in DH5α/pGEMsrdBCA occurred only in the subsurface of the thick rim of the spot of cells. This indicated that the expression of srdBCA occurred microaerobically in recombinant E. coli. All strains formed red spots of cells on agar plates containing selenite. These results suggested that all ORFs of srdBCA are essential for selenate reduction.
Fig. 4.
Plate assay for selenate reduction. E. coli DH5α strains harboring pGEMsrdBCA and its derivatives were spotted onto LB agar plates containing 0.5 mM selenate (left) or 0.5 mM selenite (right). All plates were incubated at 37°C for 60 h. The red color indicates the presence of elemental selenium.
DISCUSSION
In this study, we isolated the srdBCA operon, which is essential for selenate reduction in strain SF-1. Each gene of this operon encoded a subunit of oxidoreductase, and the operon conferred the phenotype of selenate reduction on E. coli DH5α. Therefore, our results strongly suggest that the srdBCA operon encodes a selenate reductase complex. Through domain structure analyses, the features of the molybdopterin-containing oxidoreductase were predicted from the SrdBCA amino acid sequences.
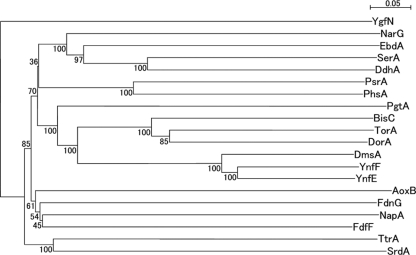
SrdA exhibits the typical features of molybdenum-containing catalytic subunits of bacterial respiratory oxidoreductases. This was consistent with other findings, such as the facts that selenate reduction in the mutant strain SF-1AM3 was affected by insertion of Tn916 into the locus encoding the molybdate metabolism regulator (see Table S1 in the supplemental material) and selenate reduction was inhibited by tungstate, as shown in our previous study (31). To further characterize SrdA, phylogenetic analysis was performed using representative catalytic subunits (Fig. 5). SrdA exhibited the highest similarity to TtrA, which is subunit A of the tetrathionate reductase complex of Salmonella enterica serovar Typhimurium (11), although it was only 22% (see Fig. S1 in the supplemental material). Based on an amino acid sequence alignment between SrdA and TtrA, five cysteine residues were found to be conserved. Four of them were assumed to consist of a [4Fe-4S] cluster binding motif of molybdopterin oxidoreductase (Fig. 6A) (17). The remaining cysteine residue was assumed to be the ligand for molybdenum in TtrA, according to sequence alignments with PhsA of Salmonella enterica serovar Typhimurium and PsrA of Wolinella succinogenes (11) (Fig. 6B). These findings imply that SrdA is a type I molybdoenzyme and therefore differs from the selenate reductase SerA of T. selenatis, which belongs to the type II molybdoenzymes characterized by the aspartate residue which is a ligand for molybdenum (12).
Fig. 5.
Neighbor-joining phylogenetic tree of representative catalytic subunits, including SrdA. Multiple-alignment and phylogenetic analyses were performed using the ClustalW program. The tree was generated using NJplot. Five percent substitution of the sequence is indicated by a bar. Bootstrap values were calculated from 100 replicates. All protein sequences were obtained from the Swiss-Prot database: YgfN, putative hypoxanthine oxidase in Escherichia coli K-12; FdnG, formate dehydrogenase in E. coli K-12; AoxB, arsenate oxidase in Alcaligenes faecalis; NapA, periplasmic nitrate reductase in E. coli K-12; FdhF, formate dehydrogenase in E. coli K-12; PsrA, polysulfide reductase in Wolinella succinogenes; PhsA, thiosulfate reductase in Salmonella enterica serovar Typhimurium; NarG, nitrate reductase in E. coli K-12; EbdA, ethylbenzene dehydrogenase in Azoarcus sp. strain EB1; SerA, selenate reductase in Thauera selenatis; DdhA, dimethylsulfide dehydrogenase in Rhodovulum sulfidophilum; PgtL, pyrogallol hydroxytransferase in Pelobacter acidigallici; BisC, biotin sulfoxide reductase in E. coli K-12; TorA, trimethylamine-N-oxide reductase in E. coli K-12; DorA, DMSO/trimethylamine N-oxide reductase in Rhodobacter capsulatus; DmsA, DMSO reductase in E. coli K-12; YnfF, probable DMSO reductase in E. coli K-12; YnfE, putative DMSO reductase in E. coli K-12; TtrA, tetrathionate reductase in S. enterica serovar Typhimurium; SrdA, selenate reductase in Bacillus selenatarsenatis SF-1.
Fig. 6.
Amino acid sequence alignment of SrdB or SrdA with related molybdoenzymes. (A) Alignment of the Fe-S cluster binding sites of SrdA and TtrA from S. enterica serovar Typhimurium. (B) Multiple alignments of the molybdenum cofactor binding regions of SrdA, TtrA, and PhsA from S. enterica serovar Typhimurium and PsrA from Wolinella succinogenes. (C) Multiple alignments of DmsB from E. coli, SrdB from Bacillus selenatarsenatis SF-1, SerB from Thauera selenatis, and NarH from E. coli. All amino acid sequences were obtained from the Swiss-Prot database.
SrdB is proposed to be a four-cluster protein of molybdopterin oxidoreductase, which participates in electron transfer between quinones and the catalytic subunit (22). To date, two types of four-cluster proteins are known: one contains four [4Fe-4S] clusters, similar to DmsB of the dimethyl sulfoxide (DMSO) reductase complex DmsABC from E. coli (3), and the other contains three [4Fe-4S] clusters and one [3Fe-4S] cluster, similar to NarH of the nitrate reductase complex NarGHI from E. coli (4). SrdB apparently belongs to the former type of four-cluster protein, as all of its motifs for Fe-S cluster binding contain four cysteine residues. In contrast, SerB, a subunit of the selenate reductase complex from T. selenatis, belongs to the latter type of four-cluster proteins (Fig. 6C) (14).
The hydrophobic structure of SrdC suggests that it may be a membrane anchor protein subunit of molybdoenzyme (22). Membrane anchor protein subunits of molybdoenzymes are known to have diverse numbers of transmembrane regions (22). Given that SrdC has nine transmembrane regions, its structure may most resemble that of TtrC, which is subunit C of the tetrathionate reductase complex in S. enterica serovar Typhimurium (11). TtrC likely belongs to the DmsC/NrfD/PsrC family (22). Enzymes belonging to the DmsC/NrfD/PsrC family have a menaquinol binding site localized toward the periplasmic side, which plays an important role in menaquinol binding and oxidation, allowing electrons to pass to an adjacent subunit of the respiratory reductase complex (27, 35). SrdC is likely to have a similar function.
The SMART program also predicted TAT signal peptides on the N termini of SrdA and SrdB. The TAT system is a pathway for secreting enzymes containing cofactors (2). Its signal peptides are generally composed of three regions, a polar N-terminal region (n region), a moderately hydrophobic region (h region), and a C-terminal region (c region), and the n region often contains basic residues and a twin-arginine motif, SRRXFLK (24). SrdA and SrdB have similar amino acid sequences at their N terminus. Thus, SrdA and SrdB are extracytoplasmic proteins transported by the TAT system. This also supports the idea that SrdA contains a molybdenum cofactor and a [4Fe-4S] cluster and that SrdB contains four [4Fe-4S] clusters. In contrast, SrdC has a Sec-type signal on its N terminus. The Sec system is a general translocation pathway to the plasma membrane for membrane proteins, and the Sec signal peptides are also composed of positively charged n, h, and c regions, often containing proline and glycine at the cleavage site (18). The amino acid sequence at the N terminus of SrdC fulfills these properties. TAT-targeted SrdAB and membrane-inserted SrdC would therefore form a membrane-bound selenate reductase facing the extracytoplasmic side of the cell. Because Gram-positive bacteria lack an outer membrane and periplasmic space, association with the membrane or cell wall is an issue in these organisms (5). Rothery and coworkers (22) demonstrated that soluble periplasmic molybdoenzymes are rare, and most molybdoenzymes are membrane bound or cytoplasmic in organisms lacking an outer membrane. Therefore, it is reasonable to assume that SrdBCA in the Gram-positive organism B. selenatarsenatis is membrane bound, in contrast with the periplasmic soluble SerABC complex in the Gram-negative organism T. selenatis.
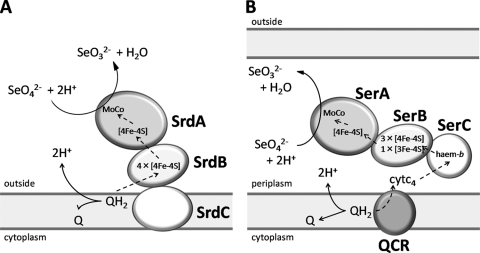
Taken together, the evidence presented here suggests that SrdBCA is a membrane-bound, molybdopterin-containing oxidoreductase belonging to the DMSO reductase family (22). Based on amino acid sequence analyses, we propose the mechanisms of selenate reduction by SrdBCA (Fig. 7A). Selenate is reduced to selenite, releasing oxide, with two electrons provided by the SrdBCA complex, and two protons receive oxide released from selenate and are converted to water. This reductive transformation is coupled to the oxidation of quinols. Quinol (QH2) bound to SrdC is oxidized to quinone (Q), releasing two protons to the outside of the cell membrane and providing two electrons to SrdB. Electrons pass through the [4Fe-4S] clusters of SrdB and transfer to the [4Fe-4S] cluster of SrdA, and then selenate receives electrons via the molybdenum cofactor. These sequential reactions do not directly affect the proton gradient between the inside and outside of the cell membrane but contribute by maintaining the redox loop of Q/QH2.
Fig. 7.
Comparative schematic representations of selenate reduction by SrdBCA from Bacillus selenatarsenatis SF-1 and SerABC from Thauera selenatis. (A) Predicted model of selenate reduction by SrdBCA. Selenate reduction is coupled with quinol oxidation. SrdC mediates quinol oxidation, providing two electrons to SrdB. Electrons pass through the [4Fe-4S] clusters of SrdB and SrdA, and selenate is reduced after receiving electrons via a molybdenum cofactor. (B) Known components in selenate reduction by SerABC (14). In contrast to SrdBCA, SerABC is a periplasmic soluble enzyme. SerABC receives electrons from cytochrome c4, which is reduced by quinol-cytochrome c oxidoreductase coupled with quinol oxidation. The dashed arrows represent electron flow. QCR, quinol-cytochrome c oxidoreductase; Q, quinones; QH2, quinols; cytc4, cytochrome c4; [4Fe-4S], [4Fe-4S] iron-sulfur cluster; [3Fe-4S], [3Fe-4S] iron-sulfur cluster; MoCo, molybdenum cofactor; SeO42−, selenate; SeO32−, selenite.
To date, biochemical analyses have revealed an important part of the selenate reduction mechanism in T. selenatis (Fig. 7B) (15). As described above, SerABC is a soluble, periplasmic, type II molybdoenzyme, containing three [4Fe-4S] clusters and one [3Fe-4S] cluster in SerB and also containing heme b in SerC. SerABC receives electrons from cytochrome c4, which is reduced by quinol-cytochrome c oxidoreductase coupled with quinol oxidation (15). Therefore, SrdBCA is quite different from SerABC.
Selenate reductase has also been studied in E. cloacae SLD1a-1. This selenate reductase exhibited features similar to those of SrdBCA, as it was a membrane-bound protein composed of three subunits containing molybdenum and iron (21), related to the FNR and TAT systems (16, 33). However, the phylogenetic relationships and domain structures of these proteins remain unclear due to the absence of amino acid sequence data. In E. coli, two operons, ygfKMN (1) and ynfEFGH (10), were reported to encode molybdenum-containing oxidoreductases related to selenate reduction. However, selenate reduction by E. coli has not been well characterized. Thus, this study is the first to report the genes encoding a membrane-bound, respiratory selenate reductase in Gram-positive bacteria. Further studies with strain SF-1 are required to elucidate the complete mechanism of selenate reduction, the findings of which will contribute to understanding the physiology of selenate-respiring bacteria.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 25 February 2011.
REFERENCES
- 1. Bébien M., Kirsch J., Méjean V., Verméglio A. 2002. Involvement of putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148:3865–3872 [DOI] [PubMed] [Google Scholar]
- 2. Berks B. C., Palmer R., Sargent F. 2005. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8:174–181 [DOI] [PubMed] [Google Scholar]
- 3. Bilous P. T., Cole S. T., Anderson W. F., Weiner J. H. 1988. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol. Microbiol. 2:785–795 [DOI] [PubMed] [Google Scholar]
- 4. Blasco F., Iobbi C., Giordano G., Chippaux M., Bonnefoy V. 1989. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol. Gen. Genet. 218:249–256 [DOI] [PubMed] [Google Scholar]
- 5. Desvaux M., Dumas E., Chafsey I., Hébraud M. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256:1–15 [DOI] [PubMed] [Google Scholar]
- 6. Dridge E. J., et al. 2007. Investigation of the redox centres of periplasmic selenate reductase from Thauera selenatis by EPR spectroscopy. Biochem. J. 408:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flannagan S., Zitzow L. A., Su Y. A., Clewell D. B. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350–354 [DOI] [PubMed] [Google Scholar]
- 8. Fujita M., Ike M., Nishimoto S., Takahashi K., Kashiwa M. 1997. Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp. SF-1. J. Ferment. Bioeng. 83:517–522 [Google Scholar]
- 9. Gawron-Burke C., Clewell D. B. 1982. A transposon in Streptococcus faecalis with fertility properties. Nature 300:281–284 [DOI] [PubMed] [Google Scholar]
- 10. Guymer D., Maillard J., Sargent F. 2009. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 191:519–528 [DOI] [PubMed] [Google Scholar]
- 11. Hensel M., Hinsley A. P., Nikolaus T., Sawers G., Berks B. C. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32:275–287 [DOI] [PubMed] [Google Scholar]
- 12. Jormakka M., Richardson D., Byrne B., Iwata S. 2004. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 12:95–104 [DOI] [PubMed] [Google Scholar]
- 13. Kashiwa M., Nishimoto S., Takahashi K., Ike M., Fujita M. 2000. Factors affecting soluble selenium removal by a selenate-reducing bacterium Bacillus sp. SF-1. J. Biosci. Bioeng. 89:528–533 [DOI] [PubMed] [Google Scholar]
- 14. Krafft T., Bowen A., Theis F., Macy J. M. 2000. Cloning and sequencing of the genes encoding the periplasmic-cytochrome B-containing selenate reductase of Thauera selenatis. DNA Seq. 10:365–377 [DOI] [PubMed] [Google Scholar]
- 15. Lowe E. C., et al. 2010. Quinol-cytochrome c oxidoreductase and cytochrome c4 mediate electron transfer during selenate respiration in Thauera selenatis. J. Biol. Chem. 285:18433–18442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma J., Kobayashi D. Y., Yee N. 2007. Chemical kinetic and molecular genetic study of selenium oxyanion reduction by Enterobacter cloacae SLD1a-1. Environ. Sci. Technol. 41:7795–7801 [DOI] [PubMed] [Google Scholar]
- 17. Ma J., Kobayashi D. Y., Yee N. 2009. Role of menaquinone biosynthesis genes in selenate reduction by Enterobacter cloacae SLD1a-1 and Escherichia coli K12. Environ. Microbiol. 11:149–158 [DOI] [PubMed] [Google Scholar]
- 18. Martoglio B., Dobberstein B. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410–415 [DOI] [PubMed] [Google Scholar]
- 19. Perrière G., Gouy M. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369 [DOI] [PubMed] [Google Scholar]
- 20. Rech S. A., Macy J. M. 1992. The terminal reductase for selenate and nitrate respiration in Thauera selenatis are two distinct enzymes. J. Bacteriol. 174:7316–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ridley H., Watts C. A., Richardson D. J., Butler C. S. 2006. Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl. Environ. Microbiol. 72:5173–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rothery R. A., Workun G. J., Weiner J. H. 2008. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 1778:1897–1929 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Sargent F. 2007. The twin-arginine transport system: moving folded proteins across membranes. Biochem. Soc. Trans. 35:835–847 [DOI] [PubMed] [Google Scholar]
- 25. Schröder I., Rech S., Krafft T., Macy J. M. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 272:23765–23768 [DOI] [PubMed] [Google Scholar]
- 26. Schultz J., Milpetz F., Bork P., Ponting C. P. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simon J., Kern M. 2008. Quinone-reactive proteins devoid of haem b form widespread membrane-bound electron transport modules in bacterial respiration. Biochem. Soc. Trans. 36:1011–1016 [DOI] [PubMed] [Google Scholar]
- 28. Stolz J. F., Oremland R. S. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23:615–627 [DOI] [PubMed] [Google Scholar]
- 29. Unden G., et al. 1995. O2-sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch. Microbiol. 164:81–90 [PubMed] [Google Scholar]
- 30. Yamamura S., Ike M., Fujita M. 2003. Dissimilatory arsenate reduction by a facultative anaerobe, Bacillus sp. strain SF-1. J. Biosci. Bioeng. 96:454–460 [DOI] [PubMed] [Google Scholar]
- 31. Yamamura S., Terashi S., Ike M., Yamashita M., Fujita M. 2004. Characterization of arsenate-, selenate-, and nitrate-reducing activities in Bacillus sp. SF-1. Jpn. J. Water Treat. Biol. 40:161–168 [Google Scholar]
- 32. Yamamura S., et al. 2007. Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int. J. Syst. Evol. Microbiol. 57:1060–1064 [DOI] [PubMed] [Google Scholar]
- 33. Yee N., Ma J., Dalia A., Boonfueng T., Kobayashi D. Y. 2007. Se(VI) reduction and the precipitation of Se(0) by the facultative bacterium Enterobacter cloacae SLD1a-1 are regulated by FNR. Appl. Environ. Microbiol. 73:1914–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoon J., et al. 2001. Bacillus jeotgali sp. nov., isolated from jeotgal, Korean traditional fermented seafood. Int. J. Syst. Evol. Microbiol. 51:1087–1092 [DOI] [PubMed] [Google Scholar]
- 35. Zhao Z., Weiner J. H. 1998. Interaction of 2-n-heptyl-4-hydroxyquinoline-N-oxide with dimethyl sulfoxide reductase of Escherichia coli. J. Biol. Chem. 273:20758–20763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.