Abstract
The heparin-binding hemagglutinin adhesin (HBHA) is a surface adhesin on the human pathogen Mycobacterium tuberculosis. Previously, it has been shown that HBHA exists as a dimer in solution. We investigated the detailed nature of this dimer using circular dichroism spectroscopy and analytical ultracentrifugation techniques. We demonstrate that the heparan sulfate (HS) binding region does not play a role in dimerization in solution, while the linker region between the predicted N-terminal coiled-coil and the C-terminal HS binding region does affect dimer stability. The majority of contacts responsible for dimerization, folding, and stability lie within the predicted coiled-coil region of HBHA, while the N-terminal helix preceding the coiled-coil appears to trigger the folding and dimerization of HBHA. Constructs lacking this initial helix or containing site-specific mutations produce nonhelical monomers in solution. Thus, we show that HBHA dimerization and folding are linked and that the N-terminal region of this cell surface adhesin triggers the formation of an HBHA coiled-coil dimer.
INTRODUCTION
Roughly one-third of the world's population has been infected by the bacterial pathogen Mycobacterium tuberculosis, the causative agent of tuberculosis (TB) (6). There has been a resurgence in both the developed and developing worlds of TB cases in AIDS patients, whose immune-compromised systems are good hosts for M. tuberculosis. Due to its significant impact on public health, the pathogenesis of Mycobacterium tuberculosis has been studied for many years. It appears that both human epithelial cells and the extracellular matrix around these cells serve as scaffolds upon which M. tuberculosis binds during infection and dissemination (12). One protein responsible for adhesion of the bacterium to lung epithelial cells is heparin-binding hemagglutinin adhesin (HBHA) (11).
HBHA plays an important role in infection, especially during extrapulmonary dissemination to other systems in the human host (15). HBHA has also been investigated as a potential diagnostic tool for latent tuberculosis (7), as a possible vaccine (9), and as a booster of BCG immunity, probably by inducing cytokines that may help with the production of Th1 effector-memory lymphocytes (5). HBHA is known to bind heparin (16), and recently, Esposito et al. have analyzed HBHA dimers in solution (2, 3).
We focused on the regions of HBHA expected by sequence analysis to form coiled-coil interactions. Coiled-coils are a common protein motif and are established protein-protein interfaces involving helices and, thus, relatively short regions of primary structure (10). Coiled-coil alpha-helices wrapped around each other to form left- or right-handed super coils (10). The primary sequence for most coiled-coils consists of heptad repeats (residues are denoted a to g), where a and d are hydrophobic residues and e and g are often charged residues (10). Coiled-coil regions pack against each other in a knobs-in-hole fashion, generating a hydrophobic core while aligning the charge residues so that they face the solvent. Here, we describe the importance of the coiled-coil and other regions of HBHA during protein dimerization.
MATERIALS AND METHODS
Cloning, expression, and purification.
Genes encoding HBHA 1_199, 6_199, 6_156, 6_138, 6_111, 6_99, 6_88, and 20_199 (where the beginning and ending residues are indicated by the first and second numbers, respectively) were amplified from an M. tuberculosis (strain H37Rv) genome via PCR and incorporated into the LIC (ligation-independent cloning) vector pMCSG7 (20). This vector incorporates a hexa-His tag at the N terminus of the HBHA construct, linked via a tobacco etch virus (TEV) protease cleavage site. L14A and L15A mutants of HBHA 6_199 and 6_111 were introduced using site-directed mutagenesis. Sequence-confirmed constructs were inserted into pMCSG7 and then transformed into chemically competent E. coli BL21-AI cells (Invitrogen). Transformed cells grew to an optical density of 0.8 in terrific broth (TB) at 37°C with shaking. An l-arabinose solution was added to the growth at a final concentration of 0.2% vol/vol, and the temperature was reduced to 18°C for 30 min. Protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and cells were allowed to grow for 16 h. Cells were then centrifuged at 4,500 × g for 30 min at 4°C. Pellets were suspended in 50 ml lysis buffer (50 mM potassium phosphate, 500 mM NaCl, 5% glycerol, and 20 mM imidazole, pH 7.4) per 4 liters of culture. The suspended pellet was frozen dropwise in a liquid nitrogen bath. Pellets were resuspended in the lysis buffer plus protease inhibitor tablets (Roche) and DNase. Lysis was achieved using a Branson sonic dismembrator at 30% power for 3 cycles of 1 min at 4°C. Constructs were purified over a HisTrap column (GE Healthcare). The hexa-His affinity tag was removed using TEV protease with further purification over a Superdex 200 column (GE Healthcare) preequilibrated in buffer (10 mM potassium phosphate, pH 7.5, 200 mM potassium fluoride). Fractions containing HBHA were combined and used in the following experiments.
Circular dichroism (CD) experiments.
Wavelength spectra were recorded in CD buffer (10 mM potassium phosphate, pH 7.5, and 200 mM potassium fluoride) from 260 to 185 nm using an Aviv 62 DS CD spectrophotometer. HBHA constructs were analyzed at a concentration of 10 μM in quartz cuvettes (Hellma) with a path length of 0.1 cm. Each sample was scanned three times at 20°C with a 10-s averaging time. Spectra were averaged, and the buffer signal subtracted. Values were converted from ellipticity to mean residue ellipticity (MRE) (degree × cm2 × dmol−1) by using the following equation:
| (1) |
where θ is ellipticity in millidegrees, Mr is the molecular weight divided by the number of peptide bonds, C is the concentration in grams per liter, and l is the path length in cm.
Thermal denaturation.
HBHA constructs were heated from 4°C to 85°C using 1-degree steps with an equilibration time of 2 min for each temperature step. Measurements were taken at 222 nm with a 10-s averaging time for each degree step within a 0.3-degree dead band. Transition curves were normalized to the fraction folded (F) using the following equation:
| (2) |
where [θ]obs is the observed signal at a given temperature, [θ]u is the value when the peptide is unfolded, and [θ]n is the value when the peptide is in its native state. Reversibility was checked by rapid cooling to 25°C, followed by a wavelength scan collected from 260 to 185 nm.
Analytical ultracentrifugation sedimentation equilibrium (AUC-SE).
Sedimentation equilibrium experiments were performed on HBHA constructs at 20°C in a Beckman XL-I analytical ultracentrifuge using 6-sector cells and an An-50 Ti rotor. Samples at three different concentrations (20, 10, and 5 μM) were spun at three velocities (3.22 × 104, 7.24 × 104, and 1.30 × 105 × g) until equilibrium was reached. Absorbance was monitored every 2 h at 230 nm and 280 nm. Absorbance offset values were determined using the meniscus depletion method by overspeeding the samples at 1.63 × 105 × g for 6 h. The program SEDNTERP (John Philo, Thousand Oaks, CA, and RASMB) was used to calculate solvent density and viscosity along with molecular mass and partial specific volume for each HBHA construct. Data sets were initially analyzed using SEDFIT (19) in order to generate the proper file type for analysis with SEDPHAT (18). Using SEDPHAT, a species analysis model was applied to each construct data set to determine the solution makeup for each construct. Standard curves for each HBHA construct were generated at 230 nm in order to derive the extinction coefficients using the Beer-Lambert law. Data for each construct were also plotted using the following equation:
| (3) |
where Cr is the concentration at radius r, Cr0 is the concentration at the reference point, M is the molecular weight, v̄ is the partial specific volume of the molecule, ρ is the density of the solution, ω is the angular velocity of the experiment, R is the gas constant, T is the temperature (K) of the experiment, r is radius, and r0 is the reference radius. The resulting plot is linear if the molecule population contains a single species that is monodisperse, while a curved lined indicates the polydispersity of the sample. The slope of the line is proportional to the mass of a single species.
RESULTS
Design of HBHA constructs for analysis.
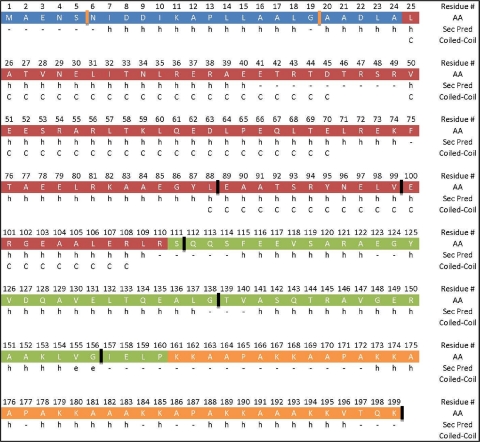
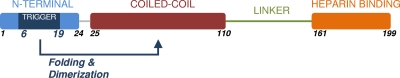
The full-length (HBHA 1_199) and initially designed HBHA construct lacking the first five amino acids (including residues 6 to 199; HBHA 6_199) behaved similarly in our assays (see data below), and thus, both are considered representative of the full-length protein in vitro. We preferred using HBHA 6_199 because the construct expressed well and exhibited an improved half-life in solution, which was important for subsequent analysis. Based upon previously published coiled-coil predictions (3), we anticipated that the coiled-coil would start near Leu-24 and terminate near Arg-107. Thus, construct HBHA 20_199 was generated to examine the putative start of the predicted coiled-coil, while constructs HBHA 6_156 and HBHA 6_111 were created to determine the extent of the coiled-coil into the C-terminal regions of HBHA. Additional constructs (6_88, 6_99, 6_138, 6_111 L14A L15A, and 6_199 L14A L15A) were employed to further clarify the limits of the coiled-coil, heparin binding, and linker regions, as well as to determine the importance of specific residues. As a working model, we classify, using secondary structure (14) and coiled-coil predictions (4), regions of HBHA into N-terminal, predicted coiled-coil, linker, and heparan sulfate (HS) binding regions (Fig. 1).
Fig. 1.
Diagram of the primary sequence for HBHA aligned with a secondary structure prediction and the coiled-coil prediction. The sequence is separated into 4 regions: blue, N terminus; red, predicted coiled-coil; green, linker; and orange, heparin binding region. Starts and ends to constructs are shown by orange and black hashes, respectively. AA represents the amino acid at the residue number above, while Sec Pred is the secondary structure predicted for that amino acid. Finally, if the residue is part of the predicted coiled-coil, a “C” is listed in the Coiled-Coil row.
Circular dichroism spectroscopy.
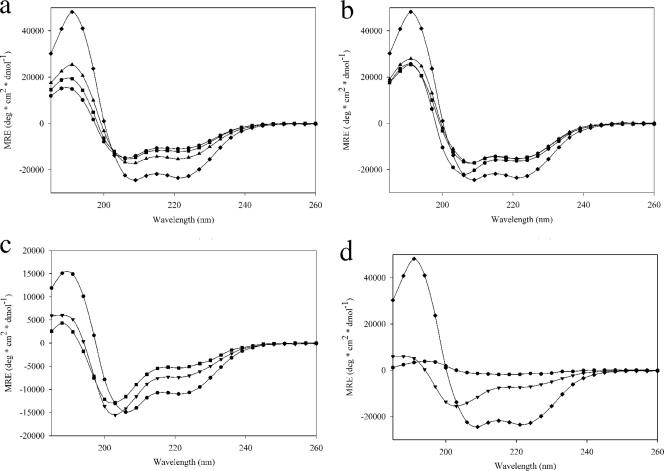
CD spectra from 260 nm to 185 nm for each construct at 10 μM are shown in Fig. 2a to d. The spectra of HBHA 1_199 and 6_199 were identical, indicating that the truncation of the first five residues did not affect the overall structure of the protein (Fig. 2a). The CD spectra of constructs 6_156 and 6_111 were then compared to that of 6_199, which showed that the secondary structure of HBHA became dominated by alpha-helices as the predicted coiled-coil region was isolated. The spectrum from HBHA 6_138 was similar to that of HBHA 6_156, demonstrating that cutting into the linker region had little effect on the overall secondary structure composition (Fig. 2b). The CD spectra from HBHA 6_88 and HBHA 6_99 were similar to one another, and both showed loss of alpha-helical secondary structure due to truncation from the C terminus. Compared to the results for HBHA 6_111 (Fig. 2b), the minima at 208 nm and 222 nm are more positive and the maximum at 190 nm is more negative. These transitions were accompanied by a blue shift of the minimum observed near 208 nm; together, these data support the conclusion that alpha-helical secondary structure is lost in these constructs relative to the structure of the longer version of HBHA examined.
Fig. 2.
Far UV-CD spectra of regional HBHA constructs at 10 μM. (a) ●, 6_199; ▴, 6_156; ♦, 6_111; and ■, 1_199. (b) ■, 6_156; ▴, 6_138; ♦, 6_111; and ●, 6_88. (c) ●, 6_199; ▾, 20_199; and ■, 6_199 L14A L15A. (d) ♦, 6_111; ▾, 20_199; and ●, 6_111 L14A L15A.
At the N terminus of HBHA, there is a marked stretch of hydrophobic residues (Fig. 1). Two leucines within this region, leucines 14 and 15, were mutated to alanines by using site-directed mutagenesis, with the goal of disrupting the coiled-coil. Previously published work had shown that truncation of the first 24 residues of HBHA results in a loss of secondary structure (2). Therefore, a construct starting at residue 20 (alanine) was designed, since a truncation at residue 25 is within a predicted helix and close to the predicted coiled-coil start. The introduction of the N-terminal truncation caused a loss of secondary structure compared to the structure of 6_199, as indicated by the observed shift in the minimum at 208 nm to 200 nm and a loss of CD signal intensity at 222 nm and 190 nm (Fig. 2c). The mutation of leucines 14 and 15 to alanines had a similar disruptive effect on the secondary structure (Fig. 2c). Thus, altering the N-terminal region lying outside the predicted coiled-coil has adverse effects on the alpha-helical secondary structure of HBHA.
The leucine mutations were then incorporated into the most alpha-helical construct, HBHA 6_111 (Fig. 1). However, following TEV protease cleavage during purification of the construct, the protein formed a soluble aggregate as determined by size exclusion chromatography and dynamic light scattering (data not shown). This aggregate was examined on the CD spectrophotometer along with HBHA 6_111 and HBHA 20_199 (Fig. 2d). We observed a total loss of secondary structure signal relative to that in 6_199 when the leucine mutations were introduced into the 6_111 construct, due to the aggregate state of the construct. Taken together, these data demonstrate that the primary structural component for HBHA is the alpha-helix and that the maintenance of this structure is insensitive to C-terminal truncations up to the predicted end of the coiled-coil at residue 111 but highly sensitive to mutation or truncations within the N terminus leading up to residue 20.
AUC-SE.
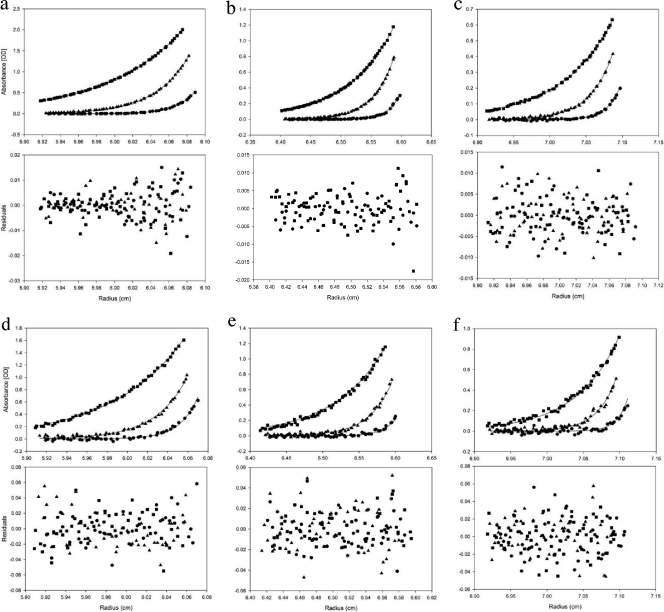
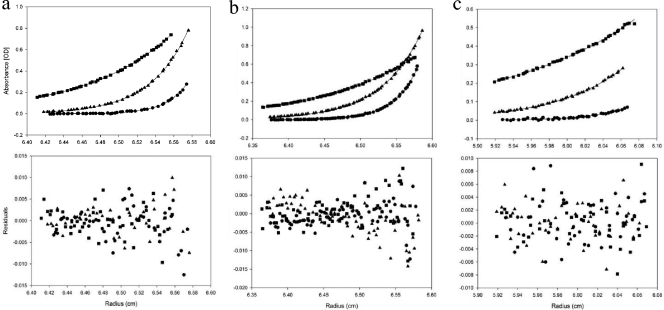
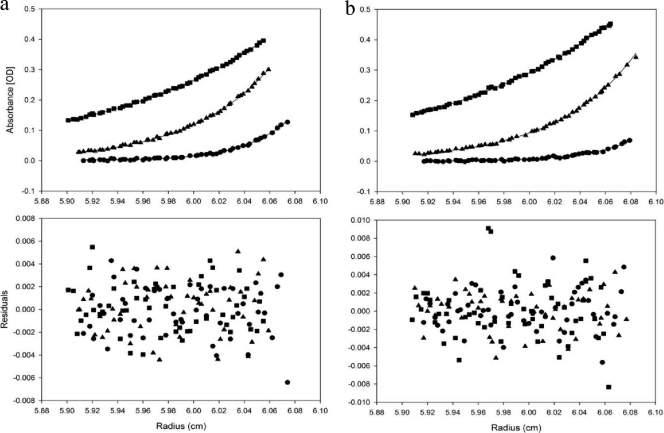
Because HBHA may form an elongated dimer (3), we employed analytical ultracentrifugation sedimentation equilibrium (AUC-SE) to obtain accurate masses of HBHA constructs due to the shape independence of this technique. Comparison of the full-length construct of HBHA 1_199 to HBHA 6_199 confirmed that the deletion of the first 5 residues had no effect on oligomerization (Fig. 3a to f). Regional constructs HBHA 6_156 and HBHA 6_111 were examined at 3 different concentrations, with the 10 μM traces being displayed in Fig. 4a and b. These constructs demonstrated no loss of dimerization with truncations from the C terminus. It further appears that the gain of secondary structure in comparison to the structure of HBHA 6_199 was not due to a change in oligomeric state (Table 1). With construct 6_88 (Fig. 4c), the HBHA molecule was truncated into the predicted coiled-coil region. This construct was treated the same as previous constructs and was also found to be a dimer in solution (Table 1). N-terminal truncation and leucine mutant constructs were treated like the above-described constructs, with the 10 μM traces shown in Fig. 5a (HBHA 20_199) and Fig. 5b (HBHA 6_199 L14A L15A), along with the corresponding residuals. These two constructs were calculated to have the mass of a monomer (Table 1). Indeed, at all three concentrations, HBHA 20_199 and HBHA 6_199 L14A L15A exist as monomers in solution. Together with the CD spectra, these data indicate a link between a major loss in secondary structure signal and the conversion to monomer in solution. In contrast, constructs with alpha-helical characteristics existed as dimers in solution.
Fig. 3.
Absorbance-detected analytical ultracentrifugation sedimentation equilibrium traces with residual plots. (a to c) Plot of AUC-SE for HBHA 1_199 at 3 concentrations, 5 μM (a), 10 μM (b), and 20 μM (c), at three velocities, 3.22 × 104 × g (■), 7.24 × 104 × g (▴), and 1.30 × 105 × g (●). (d to f) Plot of AUC-SE for HBHA 6_199 at 3 concentrations, 5 μM (d), 10 μM (e), and 20 μM (f), at three velocities, 3.22 × 104 × g (■), 7.24 × 104 × g (▴), and 1.30 × 105 × g (●). OD, optical density.
Fig. 4.
Absorbance-detected analytical ultracentrifugation sedimentation equilibrium traces with residual plots. (a to c) HBHA constructs HBHA 6_156 (a), HBHA 6_111 (b), and HBHA 6_88 (c) at 10 μM at three velocities, 3.22 × 104 × g (■), 7.24 × 104 × g (▴), and 1.30 × 105 × g (●). OD, optical density.
Table 1.
Calculated and buoyant masses of HBHA constructs
| Construct | Calculated mass (Da) | Buoyant mass (Da) | Oligomer |
|---|---|---|---|
| 1_199 | 21,675 | 43,406 | 2.0 |
| 6_199 | 21,274 | 49,117 | 2.3 |
| 6_156 | 16,973 | 34,537 | 2.0 |
| 6_111 | 12,240 | 26,377 | 2.2 |
| 6_88 | 9,867 | 19,795 | 2.0 |
| 6_199 L14A L15A | 21,190 | 25,350 | 1.2 |
| 20_199 | 19,926 | 23,323 | 1.2 |
Fig. 5.
Absorbance-detected analytical ultracentrifugation sedimentation equilibrium traces with residual plots. (a, b) HBHA constructs HBHA 20_199 (a) and HBHA 6_199 L14A L15A (b) at 10 μM at three velocities, 3.22 × 104 × g (■), 7.24 × 104 × g (▴), and 1.30 × 105 × g (●). OD, optical density.
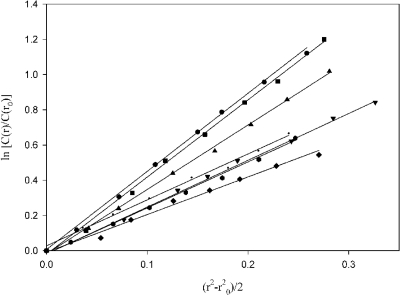
Constructs were also plotted using equation 3, described in Materials and Methods. The resulting plots for each construct were linear, indicating a single species in solution (Fig. 6). The molecular weights of each macromolecule influence the slope of the line, resulting in the construct with the largest molecular weight, HBHA 1_199, having the greatest slope and the construct with the smallest molecular weight, HBHA 6_88, having the smallest slope. The resultant molecular weights from these plots corroborated the molecular weights achieved using the fitting software.
Fig. 6.
AUC data at 7.24 × 104 × g of HBHA constructs plotted in relation to molecular weight. HBHA 1_199 (●); HBHA 6_199 (■); HBHA 6_156 (▴); HBHA 6_111 (▾); HBHA 6_88 (♦); HBHA 6_199 L14A L15A ( ); and HBHA 20_199 (·).
); and HBHA 20_199 (·).
Thermal denaturation.
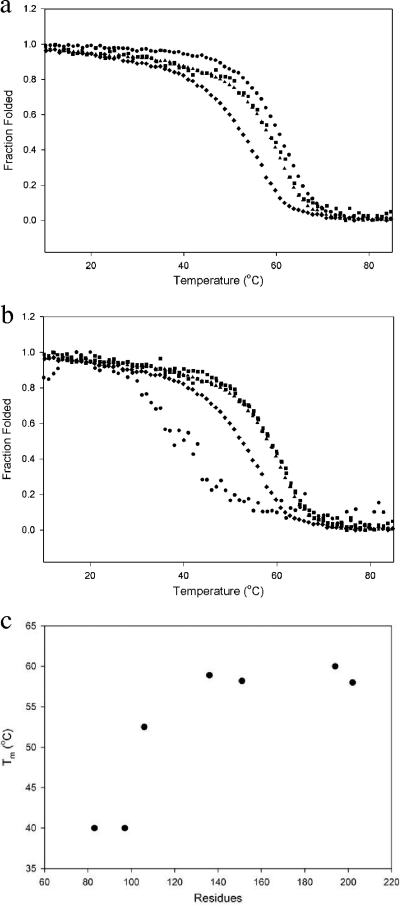
The CD signal at 222 nm is generally accepted as an accurate measure of the alpha-helical content of a protein (17), and because HBHA is composed of primarily alpha-helices, 222 nm was considered an appropriate measure of HBHA folding and stability. The constructs 6_199, 6_156, and 6_111 were analyzed along with the full-length construct, HBHA 1_199 (Fig. 7a). Melting temperatures (Tm) were determined and are listed in Table 2. As reported above, the signal generated by HBHA 1_199 was similar to that of 6_199 (Fig. 7a). No difference in protein stability was detected upon the removal of the HS binding domain (e.g., see the data for HBHA 6_156 in Fig. 7a). However, the Tm began to decrease once the full linker region was removed from the HBHA construct (e.g., HBHA 6_111 in Fig. 7a). Truncating the linker region increased the alpha-helical secondary structure and had an effect on stability, as seen by a decrease in the Tm (Fig. 7b). Once the predicted coiled-coil region was disrupted, significant reductions in Tm were observed (Table 2). It was noted that when transitioning from 60°C to 40°C, the data for HBHA 6_88 were noisier than the data for the other constructs (Fig. 7b), probably due to the disordered nature of the construct.
Fig. 7.
Thermal denaturation of HBHA constructs monitored at 222 nm. (a) Constructs 6_199 (●), 1_199 (■), 6_156(▴), and 6_111 (♦) were all at 10 μM. (b) Constructs 6_156 (▴), 6_138 (■), 6_111 (♦), and 6_88 (●) were all at 10 μM. (c) Plot of construct size (number of amino acids) versus calculated melting temperature.
Table 2.
Calculated melting temperatures for HBHA constructs
| Construct | No. of amino acids | Mean Tm ± SD (°C)a |
|---|---|---|
| 1_199 | 202 | 58.5 ± 0.20 |
| 6_199 | 197 | 60.0 ± 0.07 |
| 6_156 | 154 | 58.2 ± 0.17 |
| 6_138 | 136 | 58.0 ± 0.16 |
| 6_111 | 109 | 52.5 ± 0.16 |
| 6_99 | 97 | 40.0 ± 0.51 |
| 6_88 | 86 | 40.0 ± 1.0 |
| 6_199 L14A L15A | 194 | NA |
| 6_111 L14A L15A | 109 | NA |
| 20_199 | 183 | NA |
NA, not applicable.
The change in Tm as a function of peptide length is plotted in Fig. 7c. As residues are truncated from the C terminus, only a small decrease in the Tm of the HBHA constructs is observed. However, as constructs disrupted the predicted coiled-coil region, a more significant decrease in Tm was observed (Table 2). The limited stability of constructs HBHA 20_199, HBHA 6_199 L14A L15A, and HBHA 6_111 L14A L15A did not allow for successful analysis of these proteins by thermal denaturation. Collectively, though, these data indicate that disrupting the coiled-coil domain from the C terminus weakens the fold of the HBHA dimer.
DISCUSSION
Heparin-binding hemagglutinin adhesin is a surface protein of Mycobacterium tuberculosis employed for adhesion to epithelial cells; in addition, HBHA has been observed to aggregate and agglutinate in solution (13). Recent studies have shown that HBHA exists in a dimeric state in a range of concentrations in solution. This dimer was the focus of our work. With no atomistic model available for HBHA, we employed secondary structure predictions to guide the design of constructs. These data were overlaid with the primary sequence, along with the published coiled-coil prediction (Fig. 1). From the schematic of HBHA, we divided the protein into four regions: the N-terminal region (blue), predicted coiled-coil (red), linker region (green), and heparin sulfate binding region (orange). Constructs were designed to test the role each region may have in dimerization. Most constructs were started at residue 6, asparagine (N), rather than the starting methionine. Two factors led to the decision to start at residue 6: first, the constructs that began with this N expressed well, and second, they exhibited an improved half-life in solution, which was important for subsequent analytical ultracentrifugation studies. However, as shown in our data, constructs that began with either residue 1 or 6 behaved similarly throughout our biophysical characterizations.
Constructs 6_199, 6_156, 6_111, and 20_199 were initially characterized by circular dichroism spectroscopy. The trace for HBHA 6_199 appeared similar to previously published data (3). However, as we started truncating HBHA from the C terminus, an increase in the alpha-helical signal was observed, with HBHA 6_111 being the most alpha-helical (Fig. 2a). These data suggest that, in vitro, a majority of the residues after 111 are either disordered or in a non-alpha-helical conformation and that a vast proportion of residues 1 through 111 are in an alpha-helical state.
HBHA 6_199, HBHA 6_156, and HBHA 6_111 were analyzed by AUC-SE and all were determined to have a buoyant mass corresponding to a dimer (Table 1). Unlike the previous constructs, HBHA 20_199 had a decrease in alpha-helical signal and the protein adopted a more random coil conformation, seen by a shifting of the minimum from 208 to 200 nm. HBHA 20_199 was further characterized by AUC-SE (Fig. 5a and 6), and the resultant buoyant mass was equivalent to a monomer (Table 1). These AUC-SE and CD data indicate a link between protein folding and dimerization, with the N-terminal region being responsible for forming and maintaining the HBHA dimer. HBHA 6_199, HBHA 6_156, and HBHA 6_111 all contain the N-terminal region and have alpha-helical CD spectra. Removal of the N terminus causes the loss of a characteristic alpha-helical signal.
Mutation of Leu-14 and Leu-15 to alanines resulted in a construct that produced a CD spectrum similar to that of HBHA 20_199. And, like HBHA 20_199, this construct was a monomer in solution. These data implicate the N-terminal region, in particular, the predicted helix from residue 7 to 18 (Fig. 1), in dimer formation and folding. However, while the N-terminal region is important for the formation of the dimer and folding of the protein, the coiled-coil is still important for the maintenance of the dimer. When residues are eliminated from the C terminus of the coiled-coil, there is weakening of protein stability. From the thermal denaturation experiments, HBHA 6_88 is not as stable as either HBHA 6_111 or HBHA 6_156 (Fig. 7b). These observations support the conclusion that while contacts present in the coiled-coil are important for stability and preservation of the fold, it is the N-terminal region that triggers folding and dimerization.
Thus, we propose that the first helix acts as a trigger sequence (Fig. 8) for HBHA dimerization. Trigger sequences have been identified in other coiled-coils, where they have been shown to control folding and oligomerization (8). HBHA is expressed predominantly on the Mycobacterium tuberculosis surface, facing epithelial cells (1). The heparan sulfate ligand recognized by HBHA is presented on epithelial cells in grouped brush structures (21). Thus, we further propose that the HBHA dimer triggered by the N-terminal residues 6 to 19 of the protein serves to increase the local concentration of this adhesin such that optimal contacts with heparan sulfate brush structure can be formed. Such contacts would be expected to be critical to the initiation of pathogenesis by Mycobacterium tuberculosis.
Fig. 8.
Schematic of HBHA model.
Footnotes
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Dupres V., et al. 2005. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2:515–520 [DOI] [PubMed] [Google Scholar]
- 2. Esposito C., et al. 2010. Dimerisation and structural integrity of heparin binding hemagglutinin A from Mycobacterium tuberculosis: implications for bacterial agglutination. FEBS Lett. 584:1091–1096 [DOI] [PubMed] [Google Scholar]
- 3. Esposito C., et al. 2008. Evidence for an elongated dimeric structure of heparin-binding hemagglutinin from Mycobacterium tuberculosis. J. Bacteriol. 190:4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gruber M., Soding J., Lupas A. N. 2006. Comparative analysis of coiled-coil prediction methods. J. Struct. Biol. 155:140–145 [DOI] [PubMed] [Google Scholar]
- 5. Guerrero G. G., Debrie A. S., Locht C. 2010. Boosting with mycobacterial heparin-binding haemagglutinin enhances protection of Mycobacterium bovis BCG-vaccinated newborn mice against M. tuberculosis. Vaccine 28:4340–4347 [DOI] [PubMed] [Google Scholar]
- 6. Hingley-Wilson S. M., Sambandamurthy V. K., Jacobs W. R., Jr 2003. Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nat. Immunol. 4:949–955 [DOI] [PubMed] [Google Scholar]
- 7. Hougardy J. M., et al. 2007. Heparin-binding-hemagglutinin-induced IFN-gamma release as a diagnostic tool for latent tuberculosis. PLoS One 2:e926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kammerer R. A., et al. 1998. An autonomous folding unit mediates the assembly of two-stranded coiled coils. Proc. Natl. Acad. Sci. U. S. A. 95:13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Locht C., Hougardy J. M., Rouanet C., Place S., Mascart F. 2006. Heparin-binding hemagglutinin, from an extrapulmonary dissemination factor to a powerful diagnostic and protective antigen against tuberculosis. Tuberculosis (Edinb.). 86:303–309 [DOI] [PubMed] [Google Scholar]
- 10. Lupas A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375–382 [PubMed] [Google Scholar]
- 11. Menozzi F. D., Bischoff R., Fort E., Brennan M. J., Locht C. 1998. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc. Natl. Acad. Sci. U. S. A. 95:12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Menozzi F. D., et al. 2006. Mycobacterium tuberculosis heparin-binding haemagglutinin adhesin (HBHA) triggers receptor-mediated transcytosis without altering the integrity of tight junctions. Microbes Infect. 8:1–9 [DOI] [PubMed] [Google Scholar]
- 13. Menozzi F. D., et al. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184:993–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mooij W. T., Mitsiki E., Perrakis A. 2009. ProteinCCD: enabling the design of protein truncation constructs for expression and crystallization experiments. Nucleic Acids Res. 37:W402–W405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pethe K., et al. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190–194 [DOI] [PubMed] [Google Scholar]
- 16. Pethe K., et al. 2000. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. 275:14273–14280 [DOI] [PubMed] [Google Scholar]
- 17. Scholtz J. M., Qian H., York E. J., Stewart J. M., Baldwin R. L. 1991. Parameters of helix-coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers 31:1463–1470 [DOI] [PubMed] [Google Scholar]
- 18. Schuck P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320:104–124 [DOI] [PubMed] [Google Scholar]
- 19. Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stols L., et al. 2002. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25:8–15 [DOI] [PubMed] [Google Scholar]
- 21. Varki A., Esko C. R. J., Hudson F., Hart G., Marth J. 1999. Essentials of glycobiology, 1st ed., vol. 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]