Abstract
Spores of Bacillus subtilis require the GerAA, GerAB, and GerAC receptor proteins for l-alanine-induced germination. Mutations in gerAA, both random and site directed, result in phenotypes that identify amino acid residues important for receptor function in broad terms. They highlight the functional importance of two regions in the central, integral membrane domain of GerAA. A P324S substitution in the first residue of a conserved PFPP motif results in a 10-fold increase in a spore's sensitivity to alanine; a P326S change results in the release of phase-dark spores, in which the receptor may be in an “activated” or “quasigerminated” state. Substitutions in residues 398 to 400, in a short loop between the last two likely membrane-spanning helices of this central domain, all affect the germination response, with the G398S substitution causing a temperature-sensitive defect. In others, there are wider effects on the receptor: if alanine is substituted for conserved residue N146, H304, or E330, a severe defect in l-alanine germination results. This correlates with the absence of GerAC, suggesting that the assembly or stability of the entire receptor complex has been compromised by the defect in GerAA. In contrast, severely germination-defective mutants such as E129K, L373F, S400F, and M409N mutants retain GerAC at normal levels, suggesting more local and specific effects on the function of GerAA itself. Further interpretation will depend on progress in structural analysis of the receptor proteins.
INTRODUCTION
In endospore-forming bacteria, the gerA family of operons generally encode three proteins that are likely to interact to form specific receptors for individual chemical germinants (15, 23). Most species encode a number of such operons, which have evolved to provide responses to different germinants by a process of gene duplication and divergence. The first example of this type of operon described was the gerA operon of Bacillus subtilis; the GerA proteins are required for germination in l-alanine as the sole germinant (18, 21, 28), whereas the GerB and GerK receptor proteins are required for an alternative germination in a mixture of l-asparagine, d-glucose, d-fructose, and K+ (AGFK). The gerA operon is expressed at a low level during sporulation, in the developing forespore, and is under the control of the forespore-specific sigma factor sigma G; the levels of expression are downregulated by AbrB-like protein SpoVT (3) and dependent on activity of the PrpE phosphatase (10).
The proteins encoded (GerAA, GerAB, and GerAC) are all predicted to be targeted to the spore inner membrane, and direct evidence for this has been obtained for GerAA and GerAC (11). The GerAA protein is predicted to contain an integral membrane domain of five predicted membrane spans, following a large hydrophilic N-terminal domain, and followed in turn by a relatively short hydrophilic C-terminal domain. As gerA is expressed in the forespore, it is likely that the large N-terminal domain of GerAA would be located on the inside of the inner spore membrane. Germination-defective point mutants were isolated, following enrichment for spores that failed to lose heat or chemical resistance following exposure to alanine as the germinant, and were mapped to the gerA locus (18, 21, 25). Cloning of the adjacent citG gene (16) allowed classification of a selection of point mutations into three complementation groups (27), corresponding to the three operons gerAA, gerAB, and gerAC (28). In general, most of the point mutants examined had an extreme defect in germination in l-alanine without seriously interfering with germination in the AGFK combination of germinants. With the exception of the two gerAB mutations that reduced the sensitivity of germination to l-alanine (17, 21), the amino acid changes in the point mutants had not been examined.
In an attempt to explore functionally important residues, albeit in the absence of any direct structural information, point mutations in several gerA random mutants have been identified and additional site-directed mutants constructed. These studies have defined several regions of the GerAA protein important in receptor function and distinguish mutations that disrupt receptor organization from those likely to have a more specific defect.
MATERIALS AND METHODS
Strains and culture conditions.
B. subtilis mutant strains used in this study are listed in Table 1. The routine culture media were L broth for Escherichia coli and Oxoid nutrient broth for B. subtilis. Antibiotic resistance was selected by adding the following antibiotics to the nutrient agar: ampicillin (50 μg ml−1) for E. coli and kanamycin (5 μg ml−1) and chloramphenicol (3 μg ml−1) for B. subtilis. Spores of B. subtilis were generally prepared in CCY medium (24) containing 20 μg ml−1 tryptophan and washed as previously described (18). For two strains, those containing gerAA56 and gerAA107 alleles, sporulation in CCY medium was asynchronous, with some lysis; therefore, SG liquid medium (13) containing 20 μg ml−1 tryptophan, in which they sporulated to completion, was used instead. Spores of the parental strain 1604 prepared in both media showed similar germination behavior, although the rate of germination in AGFK was a little slower for spores prepared in SG liquid medium.
Table 1.
Mutant strains used in this studya
| Strain | gerAA alleleb | AA changeb | Mutation type(s)c | Phenotypic groupd | GerAC statuse |
|---|---|---|---|---|---|
| BH4951 | 80 | E129K | R | III | Present |
| AM1671 | 108 | N146A | D | III | None |
| AM1666 | 103 | V252A | D | I | |
| AM1667 | 104 | S294P | D | II | |
| AM1672 | 109 | H304A | D | III | None |
| AM1673 | 110 | P324S | D | I* | |
| AM1674 | 111 | F325A | D | I | |
| AM1681 | 113 | P326S | D | ||
| AM1683 | 114 | E330A | D | III | None |
| AM1684 | 115 | E335A | D | III | 5% |
| BH4819 | 7 | P349S | R | II | |
| AM1878 | 84 | V369 | R, FS | III | None |
| AM1668 | 105 | L373F | D | III | Present |
| BH4818 | 16 | G398S | R | III | Present |
| AM1675 | 112 | L399N | D | II | |
| BH4810 | 56 | S400F | R | III | Present |
| AM1669 | 106 | M409N | D | III | Present |
| AM1670 | 107 | H430A | D | II |
Strains from the mutant collection of D. A. Smith contained random mutations. Strain AM1681 had phase-dark spores, and strain BH4818 was temperature sensitive.
The gerAA allele and the consequent amino acid (AA) change are shown.
D, site-directed mutation; R, random mutation; FS, frameshift mutagenesis.
The phenotypic groups are based on l-alanine-dependent germination. Group I strains showed normal or near-normal germination. The group I* strain was more sensitive to germinant. Group II strains showed a slower decrease in the optical density, while group III strains showed little or no response to germinant.
The GerAC status is shown only for group III mutants and reports the presence or absence and the level of GerAC protein detected in Western blots of spore extracts.
Transformation of E. coli was accomplished by standard methods, and transformation of B. subtilis was brought about by the method of Kunst and Rapoport (12). Minimal medium (1) contained tryptophan (40 μg ml−1) with sodium d,l-lactate (0.5% [wt/vol]; Fisher Scientific, United Kingdom) as the carbon source for selection of Cit+ transformants. Cells from the transformation mix were washed three times in Spizizen minimal salts before plating on minimal medium containing lactate.
Spore germination.
Spores were heat activated at 70°C for 30 min and cooled in ice before the addition of germinants. Spores were diluted to an optical density at 600 nm (OD600) of 0.6 in 1 mM l-alanine, 20 mM KCl, and 10 mM Tris-HCl (pH 7.6) or in 30 mM l-asparagine, 5.6 mM d-glucose, 5.6 mM d-fructose, 20 mM KCl, and 50 mM Tris-HCl (pH 8.4) for AGFK (l-asparagine, d-glucose, d-fructose, and K+) germination. Germination was carried out at 37°C, and the change in OD490 of the suspension was measured every 2 min for 120 min using a VICTOR2 1420 multilabel counter (Wallac). Spores were germinated in 20-ml volumes, and aliquots were removed for measurement of dipicolinic acid (DPA) release (22). The proportion of heat-resistant organisms during germination was measured after 10−4 dilution in H2O by heating the organisms at 70°C for 60 min. l-Alanine, d-alanine, l-valine, cycloleucine, and γ-aminobutyric acid were from Sigma (>99% purity), and dl-β-aminobutyric acid and l-α-aminobutyric acid (>98%) were from MP Biomedicals Inc. The pH values of the germinants were adjusted before use.
Plasmid templates for mutagenesis.
The 11-kb pAAM53 plasmid (27) contains most of citG, as well as gerAA and gerAB, in the shuttle vector pHV33, a hybrid of pBR322 and pC194. This plasmid was digested with HindIII in order to eliminate the pC194 vector. Digested DNA was diluted, religated, and used to transform E. coli DH5α (dam+) to ampicillin resistance, yielding the 8-kb pAAM201 plasmid. Plasmid pAAM210 was constructed by cloning the 5-kb EcoRI fragment from pAAM3 (27), which contains most of citG, all of gerAA and gerAB, as well as approximately 900 bp of gerAC, into pBR322.
Recipient B. subtilis strains for introduction of site-directed mutations.
The closely linked citG gene provided a selection for introduction of linked gerA alleles from mutagenized plasmids. Three different citG::kan mutants were made at different stages of the work; strain AM1651 carried an insertion in citG, requiring screening of Cit+ transformants for the linked gerA mutation, whereas in strains AM1679 and AM1720, sections of the citG-gerA region were replaced with a kan cassette, requiring simultaneous introduction of citG and the gerA allele to reconstruct the region.
Strain AM1651 (citG::kan).
A kanamycin resistance cassette was amplified from pDG792 (9) by PCR with NcoI sites at both ends and cloned into the unique NcoI site within citG in pAAM201 to generate pAAM202. The pAAM202 (citG::kan) plasmid was linearized with EcoRI and introduced into competent cells of B. subtilis 1604, selecting kanamycin-resistant transformants. A typical transformant, AM1651, failed to grow on minimal medium with lactate as the sole carbon source and, as expected for citG mutants, was asporogenous.
Strain AM1697 (citG-gerAB)::kan.
The NcoI-StuI region of citG-gerA, including gerAA and half of gerAB, in pAAM201 was replaced by the kan cassette from pDG792, as a NcoI-StuI fragment to generate pAAM220. DNA was linearized at the ScaI site in the bla gene and used to transform strain 1604 to kanamycin resistance; transformants were checked for failure to grow on lactate.
Construction of site-directed mutations in the gerAA gene of B. subtilis.
Thirteen gerAA mutations generated by site-directed mutagenesis of pAAM201 were introduced individually into the chromosome of B. subtilis (Table 1). Some changed conserved amino acid residues, often alanine substitutions, while others were changes that introduced an amino acid found at that position in a homolog. For example, the changes in gerAA103(V252A) and gerAA104(S294P) were made to introduce residues present at that position in GerBA, the changes in gerAA110(P324S) and gerAA113(P326S) were in conserved residues, and the changes in gerAA105(L373F) and gerAA112(L399N) introduced the residues at that position in GerXA of Bacillus anthracis and GerIA of Bacillus cereus ATCC 10876, respectively.
Mutations in gerAA were introduced into pAAM201 using a QuikChange II XL site-directed mutagenesis kit (Stratagene) and appropriate complementary oligonucleotide primers (primer details available on request). Mutagenized plasmids were isolated from E. coli transformants, the mutation was checked by sequencing, and the plasmid DNA, linearized at the unique ScaI site in the bla gene, was introduced into B. subtilis AM1651 (citG::kan) competent cells by transformation (12). CitG+ transformants were selected on minimal medium with 0.5% (wt/vol) d,l-lactate (Fisher) and then tested for kanamycin resistance. The CitG+ Kans transformants were then examined for their spore germination phenotype on rich medium by the tetrazolium (Tzm) overlay test (18), and PCR-amplified DNA was sequenced to check for the presence of the mutation; cotransformation of the gerAA mutation in most cases with citG+ was 20 to 40%, depending on the position of the mutation in gerAA. In the case of group I mutants, displaying near-wild-type germination and red in the tetrazolium test, direct sequencing of PCR products from random transformants was the only screening approach possible.
The revised more efficient strategy used pAAM210 as the template for mutagenesis and used linearized DNA from sequenced, mutagenized plasmids to transform citG mutant strains AM1679 and AM1720, selecting for growth on lactate. All Cit+ transformants in this case carried the ger mutation, which was also checked by sequencing. One checked colony of each mutant was retained, and spores were prepared and stored at 4°C.
Western blotting of proteins from crude spore extracts.
Spores (10 mg [dry weight]) were suspended in 0.5 ml prechilled breakage buffer (50 mM Tris-HCl [pH 7.5] containing 0.5 mM EDTA and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Spore breakage, preparation of extracts, SDS-PAGE, and Western blotting were performed by the method of Cooper and Moir (6a).
RESULTS
Identification of random point gerA mutants.
Eleven gerA mutant strains of B. subtilis from the laboratory collection of D. A. Smith, Birmingham University, Birmingham, United Kingdom, that were isolated by enrichment techniques after in vivo mutagenesis with UV or N-methyl-N′-nitro-N-nitrosoguanidine (NTG) (18, 25), and one (gerAA84) mutant strain from a frameshift mutagenesis (S. Taylor and D. A. Smith, unpublished data) were available for sequencing of PCR-amplified chromosomal DNA. The mutations (gerA2, gerA7, gerA12, gerA16, and gerA61) in several mutant strains were already classified by complementation analysis as mutations in the gerAA gene (27). The mutations all proved to be G-to-A transitions, except for the gerAA84 frameshift mutation. Most of these mutations were in the gerAA gene, but the gerA87 mutation (18, 25) results in a G87D substitution in likely membrane-spanning helix 3 in GerAB, and the G17E substitution in gerAC1 (18) would affect the prelipoprotein LSGC consensus cleavage site for the prelipoprotein-specific signal peptidase.
Five gerA mutant alleles (gerA2, gerA12, gerA16, gerA61, and gerA86) giving a germination phenotype that is at least partially temperature sensitive (18) were found to be identical; at least three of the five mutants had been isolated from entirely independent mutagenesis experiments, followed by specific enrichments for temperature-sensitive defects, so they could not be siblings. These data suggest that the temperature-sensitive phenotype results from a very specific amino acid change, in a likely loop between two membrane-spanning helices; the gerAA16 allele was used as a single representative of the group for further study. The germination behavior of the gerAA16 mutant was identical to that of the gerAA12 mutant, presented in reference 18. The other random mutant alleles gerAA7, gerAA56, gerAA80, and gerAA84 were all distinct. All mutants had a single missense mutation in gerAA (Fig. 1), except for the gerAA84 mutant, which had a one-base deletion, causing a frameshift in gerAA and introducing a stop 46 codons further downstream.
Fig. 1.
Amino acid sequence of B. subtilis GerAA protein. An alignment was generated using 10 proteins: B. subtilis GerBA (NP_391461), GerKA (NP_388252), YndD (NP_389658), YfkQ (NP_388660), B. cereus GerLA (NP_977103), GerIA (AAD03541), B. anthracis GerAA (NP_845469), B. thuringiensis GerAA (YP_037228), and Clostridium tetani GerAA (NP_780991) from the NCBI Protein Database. An asterisk above the position indicates that the residue was identical in all. Predicted transmembrane regions (by TOPCONS [4], including the sequences of confirmed functional GerAA homologs GerAA, GerBA, and GerKA from B. subtilis and GerLA, GerXA, GerHA, and GerSA from B. anthracis) are shown as black lines above the sequence. Amino acid substitutions in gerAA mutants are indicated by single underline (random mutations) and double underlines (site-directed mutations). A summary of the effects of the changes is shown in Table 1.
Consequences of individual gerAA point mutations on spore germination.
Figure 1 shows the protein sequence of GerAA and the residues altered by random and site-directed mutagenesis. The amino acid substitutions involved and their consequences for germination are summarized in Table 1. The analysis of a variety of mutations provides the opportunity for preliminary analysis of structure-function relationships in GerAA, though interpretation is limited by the absence of structural information on this protein and the absence of significant homology to GerAA proteins outside the endospore formers.
Topology predictions for GerAA.
GerAA and its homologs all contain a central, largely hydrophobic, integral membrane domain that includes a number of potential membrane-spanning helices, although their organization and precise number are not clear; predictions vary from three to six, depending on the program applied and the homolog tested, but the most probable number is five. The TOPCONS suite (4) at topcons.net, comparing a set of GerAA homologs from B. subtilis and B. anthracis, and memsat-svm (19), which incorporates a PSI-BLAST stage in the analysis, both predicted the general arrangement indicated in Fig. 1, with some small differences in the end positions of the helices. For the purpose of describing amino acid changes in this article and without implying that this topology is any more likely than other predictions, a hypothetical transmembrane organization with 5 transmembrane spans has been indicated.
Characterization of spore germination phenotype.
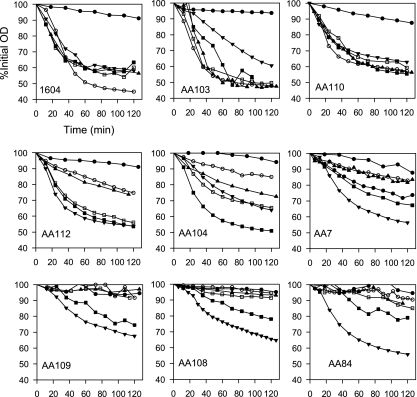
Spore germination in l-alanine and the combination of asparagine, glucose, fructose, and KCl (AGFK) were tested for random and site-directed mutations, and the resulting mutants could be classified into three groups based on germination in l-alanine. Examples are shown in Fig. 2. Group I includes mutants having normal or near-normal phenotype (gerAA103, gerAA110, and gerAA111 mutants), group II comprises mutants with some residual germination function in l-alanine (gerAA7, gerAA104, gerAA107, and gerAA112 mutants), and group III contains mutants showing an extreme germination defect (gerAA56, gerAA80, gerAA84, gerAA105, gerAA106, gerAA108, and gerAA109 mutants and the special case of the temperature-sensitive gerAA16 mutant). Spores were also examined for germination with sugars (5.6 mM d-glucose and/or d-fructose) as cogerminants (Fig. 2). The alanine-GFK response is mediated by both GerA/K and GerB/K receptor combinations (2, 14). Group I mutants still have GerA receptor function (although modification to that function is described below for gerAA110 and gerAA111 mutants), and the addition of either glucose or fructose does not significantly stimulate germination rates in alanine under these conditions, where germination is already efficient. Glucose significantly increased spore germination in l-alanine in most of the group II mutants, in some cases (AA107 and AA112 mutants) giving maximum rates of germination without fructose addition, whereas in others, e.g., the AA104 mutant, the addition of both glucose and fructose was required for maximal germination rates.
Fig. 2.
Effects of mutations on spore germination. Group I mutants, represented by gerAA103 (AA103) and AA110 mutants, showed an approximately wild-type phenotype. Group II mutants, represented by gerAA112 (AA112), AA104, and AA7 mutants, had a partial defect in l-Ala germination. Group III mutants, represented by gerAA109 (AA109), AA108, and AA84 mutants, had extreme defects in l-Ala germination. For each graph, time (in minutes) is shown on the x axis, and the percentage of the initial optical density is shown on the y axis. Symbols: ●, buffer; ○, l-alanine; □, l-alanine plus Glc; ▴, l-alanine plus Fru; ■, l-alanine plus Glc plus Fru; ▾, AGFK.
Group III mutants have minimal GerA receptor function. In this group, spores germinate only slowly and fractionally in l-alanine with added glucose and fructose, presumably via the GerB/K receptor combination only; they do not attain the wild-type rates or extent seen for some of the point mutants in group II. This suggests that the response to l-alanine of the Ala-GFK receptor is not maximal, in the absence of some contribution from the GerA receptor. The absence of functional GerA receptor may slow asparagine-GFK germination rates a little, but as there was more variability between germination rates in wild-type spore preparations in this combination, this was difficult to define.
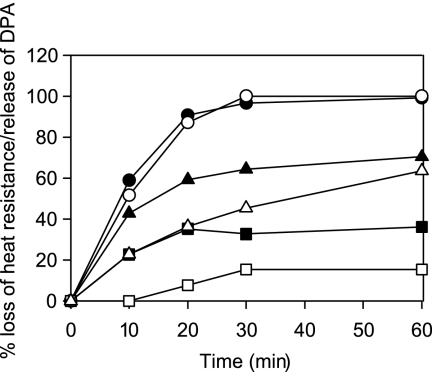
In two mutants with intermediate levels of germination of spore suspensions, measurements were made of other germination parameters—loss of heat resistance and release of dipicolinic acid (DPA) (Fig. 3). For both the gerAA111(F325A) mutant (group I but with a lower maximal rate; Fig. 4) and the gerAA107(H430A) mutant (group II), heat resistance loss was incomplete and significantly preceded release of DPA, within the time scale of the experiment. The data suggest that those spores that did respond by losing heat resistance took longer to release their DPA than was the case for wild-type spores. The significance of this is not clear and would require analysis of DPA release from single spores.
Fig. 3.
Effects of the gerAA107 and gerAA111 mutant alleles on loss of heat resistance and DPA release during germination. Spores germinating at 37°C in 10 mM Tris-HCl (pH 7.5) containing 1 mM l-alanine and 20 mM KCl were sampled at intervals, and loss of heat resistance and release of DPA were measured. Values for the loss of heat resistance (black symbols) and release of DPA (white symbols) in strain 1604 (ger+) (circles), gerA111 mutant (triangles), and gerAA107 mutant (squares) are shown. The percentages of OD loss at 60 min were 27 for the gerAA111 mutant and 20 for the gerAA107 mutant.
Fig. 4.
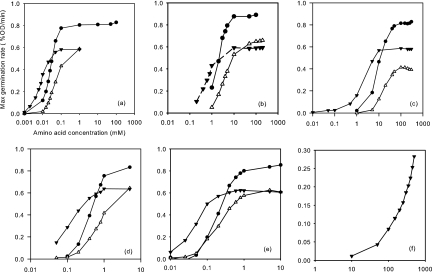
Dependence of maximum germination rate on germinant concentration for the wild type and two mutants. l-alanine (a), l-valine (b), cycloleucine (c), β-aminobutyrate (d), α-aminobutyrate (e), and γ-aminobutyrate (f) were used as germinants for the wild type (●), gerAA110 mutant (▾), and gerAA111 mutant (▵). In each graph, the amino acid concentration (in millimolar) is shown on the x axis, and the maximum germination rate (percent optical density per minute) is shown on the y axis. Only the gerAA110 mutant gave a significant rate in panel f.
An amino acid change in a proline-rich loop (PFPP) of GerAA can increase the responsiveness to germinants.
The gerAA110 and AA111 mutants (group I; P324S and F325A, respectively), germinate well in l-alanine, but they do show some important differences from wild-type behavior. Spore germination of these gerAA mutants was tested in various concentrations of l-alanine and analogues, including l-valine, cycloleucine, β-aminobutyrate, α-aminobutyrate, and γ-aminobutyrate. As described previously (21), the maximum slope of each curve (germination rate) was determined. The germination rate was then plotted against the concentration of germinant on a logarithmic scale (Fig. 4), and C50, the concentration for half-maximal germination rate, was calculated.
Spores of the gerAA110 mutant were more responsive than the wild type to low concentrations of alanine and its analogues. However, the maximum germination rate of the gerAA110 mutant was lower than the wild type, reflecting a less synchronous response. This suggests that the amino acid substitution in this mutant alters the receptor protein so that it will respond to a smaller germination stimulus, but paradoxically, the average spore takes longer to germinate, even in excess germinant (see Fig. 2 for an alanine germination curve). Spores of the gerAA110 mutant also germinated in very high concentrations of γ-aminobutyrate as the sole germinant, whereas the wild type will only do so in combination with sugars (26). In contrast, spores of the gerAA111 mutant germinated slightly slower than the wild type in l-alanine and its analogues; even at high concentrations of germinants, the maximum rate of germination was at least 25% lower than the wild type, and the concentration of germinant required for a half-maximal response was increased (Fig. 4). Like the wild type, gerAA111 spores did not germinate in γ-aminobutyrate up to 0.5 M. This amino acid substitution has only a minor effect on spore germination, but the spores are consistently less responsive to l-alanine or its analogues than those of the parent.
d-Alanine inhibition.
d-Alanine is a competitive inhibitor of germination initiated by l-alanine and its analogues (26), though it does not inhibit germination in l-alanine plus sugar adjuncts. Spore germination of the wild type and gerAA110 and gerAA111 mutants was determined in the presence of l- and d-alanine. l-Alanine was used at 1 mM, as this would give the maximum germination rate for the wild type and mutants (Fig. 4a), and the ratio of l- to d-alanine was varied by reducing the d-alanine concentration (Table 2). At a 1:1 ratio, germination of wild-type and gerAA111 spores was inhibited by d-alanine by approximately 90%. The gerAA111 mutant is more sensitive to d-alanine inhibition, still showing some inhibition at an l-alanine/d-alanine ratio of 20:1. The affinity of this mutant for binding to d-alanine was probably not reduced, unlike the situation for l-alanine. In contrast, spores of the gerAA110 mutant were more resistant to d-alanine inhibition; a 1:1 ratio of l- to d-alanine gave only 40% inhibition. This is consistent with the observation that these spores are more responsive than the wild type to l-alanine. Overall, the data for the gerAA110 and gerAA111 mutants suggest that although the responsiveness to l-alanine is altered, the sensitivity to the competitive inhibitor d-alanine remains unaffected.
Table 2.
Inhibition of germination of wild-type, gerAA110, and gerAA111 spores by d-alanine a
| Ratio of alanine isomers (l-alanine/d-alanine) | Alanine isomer concns (mM) (l-alanine/d-alanine) | Inhibition (%) |
||
|---|---|---|---|---|
| Wild type | AA110 mutant | AA111 mutant | ||
| 1:1 | 1:1 | >95 | 37 | 88 |
| 2:1 | 1:0.5 | 62 | 12 | 78 |
| 4:1 | 1:0.25 | 24 | <5 | 53 |
| 8:1 | 1:0.125 | 8 | <5 | 31 |
| 10:1 | 1:0.1 | <5 | <5 | 28 |
| 20:1 | 1:0.05 | <5 | <5 | 16 |
All conditions contained 20 mM KCl in 10 mM Tris-HCl (pH 7.5). Germination was measured by the loss of optical density (OD), and the percentage of inhibition was calculated as 100 × [(maximum rate of the decrease in the OD in the presence of d-alanine)/(maximum rate of the decrease in OD in 1 mM l-alanine only)]. Germination was not seen in d-alanine in the absence of l-alanine (OD decrease of ≤5% in 120 min).
Altering the conserved PFPP motif can affect spore maturation.
A P326S change (mutant allele gerAA113) was introduced at the next residue in the PFPP motif. This mutant grows normally, but during sporulation, it lyses prematurely, so that cultures at the end of sporulation contain much cell debris and some free phase-dark spores. No free phase-bright spores are obtained. Only early, partially phase-gray forespores can be seen in mother cells, and these darken rather than becoming more phase bright before release. The forespore fails to mature properly in the mother cell, possibly as a consequence of spontaneous germination via a defective germinant receptor. This phenotype is entirely distinct from that caused by the absence of the receptor, which would not interfere with spore maturation.
Charged residues in a potential amphipathic region.
Immediately after this likely PFPP hinge region is a sequence that could be within the membrane, as suggested in some topology predictions, and has some elements of an amphipathic helix. Two conserved glutamate residues in this region were altered to alanine—E330A and E335A in gerAA114 and gerAA115 alleles, respectively. Both of these changes resulted in loss of alanine germination. As discussed below, this was matched by the complete loss, or 95% reduction, of GerAC protein. This suggests that the region including these charged residues is likely to be of (at least) structural importance to the receptor complex.
Mutations define an important loop between membrane spans 4 and 5.
Multiple isolates of mutants with a temperature-sensitive defect carry the G398S substitution (gerA16 and other mutants [see above]) in a loop between potential membrane spans 4 and 5. The local sequence is not strongly conserved across GerAA homologs in sporeformers, but one of three adjacent residues in the loop is G, A, or P in a variety of homologs. This loop is also the site of a second point mutation, gerA56(S400F), that results in a severe loss of function of GerAA (group III). A site-directed mutation (L399N; gerAA112) introduced the amino acid present at the equivalent position in the loop in GerIA/GerHA, the inosine receptor of B. cereus/B. anthracis. This change resulted in an intermediate-level germination defect (group II); germination in l-alanine was reduced in both rate and extent, but germination was as fast as the wild type when glucose and fructose were also included (Fig. 2).
Conserved residues in the N-terminal domain of GerAA are important for germination.
Altering either of two highly conserved amino acids at the extreme ends of a relatively conserved section within the N-terminal hydrophilic domain of GerAA (E129K in gerAA80; N146A in gerAA108) prevented germination in l-alanine (a group III phenotype).
Some changes to GerAA prevent assembly of GerAC in the spore, providing strong evidence for a germination receptor complex.
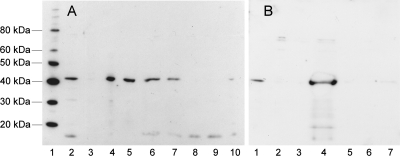
If a gerAA mutant fails to germinate in l-alanine (group III), that phenotype could result from the loss of function of a crucial amino acid involved in the germination process, or in a more general sense a loss of receptor function because of a loss of stability of the GerAA protein, either individually or in a receptor complex. A GerAA antipeptide antibody (11) was no longer active enough to detect GerAA protein in spores, so no direct test of levels of GerAA in spores was possible. Instead, the more effective anti-GerAC polyclonal antibody raised against an overexpressed GerAC protein (11) was used to detect the GerAC protein in wild-type and gerAA mutant spores (Fig. 5). Some of the group III gerAA point mutants that show an extreme defect in alanine germination lack GerAC entirely, at least at this limit of detection.
Fig. 5.
Levels of GerAC protein in gerAA mutant spores. Western blot of gerAA mutant broken spore extracts were separated on a Nu-PAGE gel (Invitrogen), probed with anti-GerAC antibody, and detected using ECL Plus (GE Healthcare). The GerAC band runs at 42 kDa under the conditions used. (A) Lanes: 1, MagicMark protein markers (Invitrogen); 2, strain 1604; 3, gerA84 mutant; 4, gerA16 mutant; 5, gerA80 mutant; 6, gerAA105 mutant; 7, gerAA106 mutant; 8, gerAA109 mutant; 9, gerAA114 mutant; 10, gerAA115 mutant. (B) Lanes: 1, strain 160; 2, gerAA108 mutant; 3, gerAA109 mutant; 4, gerA56 mutant; 5, gerAA84 mutant; 6, gerAA114 mutant; 7, gerAA115 mutant.
The gerA84 mutant, in which a frameshift mutation truncates the GerAA protein at residue 369, part way through the membrane-embedded domain, contains no GerAC protein. This could result from a failure of assembly of the entire complex as a result of a truncated GerAA protein or from some polar effect of the frameshift on downstream gene expression in the operon. However, a number of group III mutations that result in an amino acid substitution also lack GerAC protein; no polarity would be invoked in such a situation. The significance of this observation is considered in more detail and also in the context of GerAB mutant proteins in Cooper and Moir (6a), but briefly, these data demonstrate that some changes in the GerAA protein can result in a failure to assemble a receptor complex that includes GerAC; this provides strong evidence for protein interaction to form a receptor complex. Such changes in GerAA that result in the absence of GerAC from the spore are likely to have destabilized the overall structure of the GerAA protein, affecting receptor complex assembly, but changes in GerAA that allow normal assembly of GerAC into the complex are likely to have more specific effects. For example, consider the two amino acid substitutions in the N-terminal hydrophilic domain that give group III type (severe) defects in alanine germination. The gerAA108 mutant spores are likely to lack the GerA receptor on the basis of the absence of GerAC protein in Western blot analysis (Fig. 5); any failure to assemble the receptor would give a severe phenotype. In contrast, the spores with the E129K change in the N-terminal domain still contained approximately wild-type levels of GerAC, suggesting that the structure of the receptor may be intact, despite loss of its function. Substituting alanine for the conserved H304, E330, and E335 residues results in a severe depletion of GerAC, suggesting that they affect structural stability. In contrast, residue N146 discussed above and residues L373, G398, S400, and M409 do not destabilize complex assembly and are therefore likely to have a more specific, but unknown, functional role in the germination process.
DISCUSSION
It has so far proved impossible to overexpress the integral membrane proteins that form major components of the family of spore germinant receptors, of which the GerA receptor is the paradigm, and as a result, there is no structural information to guide functional analysis. One potential source of information relevant to receptor function, however, is an analysis of strains bearing point mutations. In the mutagenesis described in this work, the receptor operon was retained at its normal location, at single copy, but this meant that the time-consuming substitution approach adopted limited the number of mutations that could be made.
The data presented above are somewhat diverse but establish the usefulness of defining the consequences of individual amino acid substitutions on germinant receptor function. The major conclusions follow. (i) A region including the PFPP conserved motif beginning at residue 324, and including the following conserved E330 and E335 residues, is particularly important; changes in this region result in a variety of phenotypes, including increasing receptor sensitivity, a spore maturation failure, or a loss of receptor. This region might represent either an atypical membrane helix or an exposed loop between membrane helices, as there is currently no structural information on this protein. (ii) A loop between likely transmembrane spans 4 and 5 is also important for function; substitutions in this region can give a loss of function of the receptor or a reversible temperature-sensitive germination phenotype, suggesting the importance of conformation in this region (S400F or G398S, respectively). (iii) The loss of function in some gerAA mutants, including not just null but also some missense mutations, is accompanied by loss of the GerAC protein from the spore. This demonstrates that the incorporation of the GerAC protein is dependent on the GerAA protein (as well as on the GerAB protein, as described in the accompanying paper [6a]) and distinguishes GerAA amino acid substitutions resulting in a total functional defect, such as E129K and M409N, from those in which the overall receptor complex is destabilized, such as N146A or E330A.
The GerA proteins form a specialized receptor for l-alanine and its analogues, which bind to initiate germination, but the molecular details of the signal transduction process mediated by the membrane-associated receptor complex that results in germination-associated changes, including release of ions, small molecules, and DPA, and activation of lytic events in cortex and coats (15, 23), remain largely unclear. Receptors may function as a single type or in concert with others of different specificity (2). It is not clear whether a single subunit of the receptor binds germinants or whether multiple subunits might. The GerAB component, a homolog of the amino acid-polyamine-organocation (APC) family of single-component membrane transporters, is most likely to bind germinant. Two mutant alleles (gerAB38 and gerAB44) with amino acid changes in predicted membrane-spanning helices of GerAB alter dramatically the responsiveness of spores to l-alanine (17, 21), and tellingly, the alternate GerAB-related proteins GerVB and GerUB of Bacillus megaterium QM B1551 confer somewhat different germinant specificities on a single receptor (5, 6).
The range of phenotypes seen on changing amino acids in the local proline-rich hinge-like region emphasizes its importance to the function of the GerAA protein. Previously, the equivalent amino acid substitution to gerAA110(P324S) in the homologous GerBA protein of B. subtilis (2, 20) allowed a slow germination response to l-alanine as the sole germinant in the GerB receptor, which normally requires additional GerK receptor-mediated stimulation. This may also be the case in gerAA110 for the GerA receptor and γ-aminobutyrate, although it is also possible that trace contamination with l-alanine, at <0.001%, could give this result, given the increased sensitivity of the mutant to l-alanine and its analogues. Overall, the increased sensitivity suggests that the conformation of this proline-rich bend region may influence the responsiveness of the overall receptor complex to germinant, perhaps by affecting germinant binding or by altering the likelihood of a conformational change following germinant binding. The absence of structural information makes this hard to interpret further, although one might speculate that changing this first proline in the PFPP motif renders the receptor more easily converted to a germinant-bound “activated” conformation, and that changing the next proline residue, as in the gerAA113 mutant, results in a GerA receptor that is permanently activated, at least in the developing sporangium, leading to autogermination before spore maturation (7).
Receptors may function as a single type or in concert with others of different specificity (2). Mutants in group II have a significant defect in l-alanine germination; spores are much delayed in their response to l-alanine, and only one-third to one-half of the population germinate in l-alanine within 2 h. Nevertheless, a proportion of the spores germinate with a microlag well within the normal range. This fractional germination reflects some heterogeneity in the properties of individual spores (8). However, all the spores will respond at near-normal rates if both glucose and fructose are added, invoking GerK and GerB receptors (Fig. 2). The levels of GerAC protein have not been screened in all these mutants, so it is not known whether this slow and limited individual response reflects lower overall levels of GerA receptor or whether it is the direct consequence of a change in side chain interactions within the GerAA protein. Those group III mutations that cause a major germination defect but still contain GerAC protein in the spore at near-normal levels are easier to interpret; presumably, they identify changes that affect GerAA function rather than gross receptor structure. The next challenge is to define what such changes mean for the signal transduction process and the role of the GerAA domains in the germination process. The current work highlights the need for topology analysis, and an appreciation of the requirements for formation of a stable germinant receptor complex in the spore, as well as for structural information on the receptor complex itself.
ACKNOWLEDGMENTS
This work was funded by a scholarship from the Royal Thai Government (W.M.), a University of Sheffield Krebs studentship (G.R.C.) and a U.S. Air Force European Office of Aerospace Research & Development (EOARD) grant FA8655-07-1-3057 (A.M. and R.N.A.).
Footnotes
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Anagnostopoulos C., Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atluri S., Ragkousi K., Cortezzo D. E., Setlow P. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagyan I., Hobot J., Cutting S. 1996. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 178:4500–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernsel A., Viklund H., Hennerdal A., Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christie G., Lazarevska M., Lowe C. R. 2008. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J. Bacteriol. 190:2014–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christie G., Lowe C. R. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 189:4375–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a. Cooper G. R., Moir A. 2011. Amino acid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis 168. J. Bacteriol. 193:2261–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dion P., Mandelstam J. 1980. Germination properties as marker events characterizing later stages of Bacillus subtilis spore formation. J. Bacteriol. 141:786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh S., Setlow P. 2009. Isolation and characterization of superdormant spores of Bacillus species. J. Bacteriol. 191:1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guérout-Fleury A. M., Shazand K., Frandsen N., Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 10. Hinc K., et al. 2006. Expression of genes coding for GerA and GerK spore germination receptors is dependent on the protein phosphatase PrpE. J. Bacteriol. 188:4373–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hudson K. D., et al. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 183:4317–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunst F., Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leighton T. J., Doi R. H. 1971. Stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 246:3189–3195 [PubMed] [Google Scholar]
- 14. McCann K. P., Robinson C., Sammons R. L., Smith D. A., Corfe B. M. 1996. Alanine germination receptors of Bacillus subtilis. Lett. Appl. Microbiol. 23:290–294 [DOI] [PubMed] [Google Scholar]
- 15. Moir A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526–530 [DOI] [PubMed] [Google Scholar]
- 16. Moir A. 1983. The isolation of lambda transducing phages carrying the citG gene and gerA gene of Bacillus subtilis. J. Gen. Microbiol. 129:303–310 [DOI] [PubMed] [Google Scholar]
- 17. Moir A., Corfe B. M., Behravan J. 2002. Spore germination. Cell. Mol. Life Sci. 59:403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moir A., Lafferty E., Smith D. A. 1979. Genetic analysis of spore germination mutants of Bacillus subtilis 168 - correlation of phenotype with map location. J. Gen. Microbiol. 111:165–180 [DOI] [PubMed] [Google Scholar]
- 19. Nugent T., Jones D. T. 2010. Predicting transmembrane helix packing arrangements using residue contacts and a force-directed algorithm. PLoS Comput. Biol. 6:e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paidhungat M., Setlow P. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J. Bacteriol. 181:3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sammons R. L., Moir A., Smith D. A. 1981. Isolation and properties of spore germination mutants of Bacillus subtilis 168 deficient in the initiation of germination. J. Gen. Microbiol. 124:229–241 [Google Scholar]
- 22. Scott I. R., Ellar D. J. 1978. Study of calcium dipicolinate release during bacterial spore germination by using a new, sensitive assay for dipicolinate. J. Bacteriol. 135:133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556 [DOI] [PubMed] [Google Scholar]
- 24. Stewart G. S. A. B., Johnstone K., Hagelberg E., Ellar D. J. 1981. Commitment of bacterial spores to germinate - a measure of the trigger reaction. Biochem. J. 198:101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trowsdale J., Smith D. A. 1975. Isolation, characterization, and mapping of Bacillus subtilis 168 germination mutants. J. Bacteriol. 123:83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woese C. R., Morowitz H. J., Hutchinson C. A. 1958. Analysis of action of L-alanine analogues in spore germination. J. Bacteriol. 76:578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zuberi A. R., Feavers I. M., Moir A. 1985. Identification of three complementation units in the gerA spore germination locus of Bacillus subtilis. J. Bacteriol. 162:756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuberi A. R., Moir A., Feavers I. M. 1987. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene 51:1–11 [DOI] [PubMed] [Google Scholar]