Abstract
Staphylococcus aureus RN4220, a cloning intermediate, is sometimes used in virulence, resistance, and metabolic studies. Using whole-genome sequencing, we showed that RN4220 differs from NCTC8325 and contains a number of genetic polymorphisms that affect both virulence and general fitness, implying a need for caution in using this strain for such studies.
TEXT
Staphylococcus aureus is a versatile pathogen that is responsible for the majority of nosocomial and community-acquired infections. One of the biggest challenges in treating staphylococcal infections is that many S. aureus strains have developed resistance against various antibiotics. In contrast to clinical isolates, RN4220 is a commonly used laboratory strain that is characterized by a mutation in the sau1 hsdR gene, making it restriction deficient and hence an ideal intermediate cloning host. RN4220 was originally derived from NCTC8325-4 using UV and chemical mutagenesis (10). NCTC8325-4, in turn, was derived from an early clinical isolate, NCTC8325 (also known as PS47 or RN1), cured of three prophages (16). Indeed, RN4220 is not a suitable candidate for the study of antibiotic resistance, because newer methicillin-resistant S. aureus (MRSA) lineages have evolved [e.g., ST239 (hospital), ST80, and ST59 (community)], presumably due to recombination events between lineages. RN4220 is also known to harbor a small deletion in rsbU, a gene within the stress-induced sigB operon, which renders it deficient in σB expression. Additionally, RN4220 shows a Δagr mutant phenotype and does not produce α-hemolysin despite producing small amounts of RNAIII in late log phase (24). Recent studies revealed that a deletion of cvfB (encoding conserved virulence factor B) in RN4220 resulted in diminished agr expression, with a reduction in hla expression, protease production, and virulence, in a silkworm model of systemic infection (14), but the loss of hemolytic activity in the cvfB mutant of RN4220 was found to be due to a defective agr locus and not attributable to the cvfB mutation (14). In another study, SrrAB, a two-component regulatory system, was found to repress the transcription of RNAIII of the agr locus in RN4220 (20, 27). However, the interpretation of virulence and of the associated regulatory data in these studies with RN4220 is suspect due to an inherent agr defect in this strain (24). Finally, O'Neill showed by comparative sequencing that NCTC8325-4, which was thought to be identical to its parent NCTC8325 except for the deletion of three prophages, possesses previously undescribed polymorphisms that may influence the virulence and pathogenicity of NCTC8325-4 (18). As a result of these issues, it is extremely important to delineate an accurate picture of the mutations in RN4220, given the polymorphisms in this strain which can have an impact upon virulence and resistance phenotype.
Using Illumina Solexa-based whole-genome sequencing (paired end) (P. Mayer, L. Farinelli, and E. Kawashima, U.S. patent application WO98/44151), we obtained the whole-genome sequence of strain RN4220 and subsequently identified the polymorphisms in RN4220 compared with the released NCTC8325 genome. Briefly, RN4220 was grown with aeration at 37°C in tryptic soy broth to log phase (optical density at 620 nm [OD620], 0.7). Genomic DNA, isolated with a phenol-chloroform extraction method (6), was sent to Ambry Genetics (California) for library preparation and sequencing. The library preparation was carried out by shearing genomic DNA and blunting it, followed by the addition of adenine at the 3′ end. A specific adapter with bar coding was then ligated to these DNA fragments, followed by PCR amplification (Illumina). Fragments of ∼76 bases were generated and assembled.
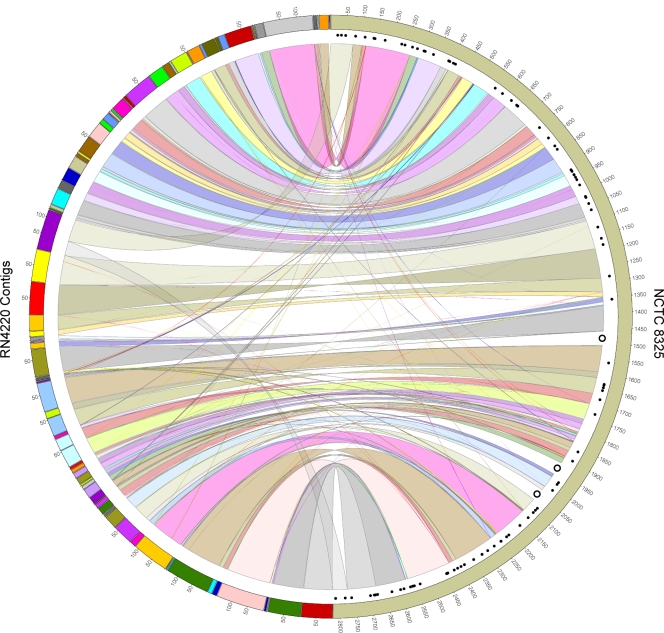
Genome assembly, single-nucleotide polymorphism (SNP) calling, and annotation were done as follows. The obtained 3.5 million paired reads, each of which is 68 nt, were de novo assembled using an Edena assembler (9), development version 3.0. The assembly has been slightly refined using the Minimus assembler (23). Final assembly resulted in 179 contigs (sum = 2.67 Mb, N50 = 80.5 kb [N50 is the contig size such that 50% of the entire assembly is contained in contigs equal to or larger than that size], max = 148 kb). Contigs were annotated using the RAST server (2). Comparison, SNP calling, SNP annotation, and graphical mapping were performed using the MUMmer software package (12), the CIRCOS visualization engine (11), and applications developed in-house. A map of the genome showing the polymorphisms in RN4220 is shown in Fig. 1. The entire genomic sequence of RN4220 can be found in the supplemental material.
Fig. 1.
Graphical mapping of the strain RN4220 contigs onto the strain NCTC8325. The RN4220 contigs are shown on the left-hand side of the circle, while the complete genome sequence of the strain NCTC8325 is shown on the right-hand side. Homologous sequences between the two strains are linked by colored ribbons. The black dots along the NCTC8325 genome sequence indicate the SNP positions, as described in Table S2 in the supplemental material. The three open circles indicate the large regions in NCTC8325 that are deleted in RN4220.
Compared with the published NCTC8325 genome, we identified 121 SNPs and 4 large-scale deletions (see Tables S1 and S2 in the supplemental material). Among the SNPs, 14 were synonymous. The remaining SNPs involve 80 nonhomologous substitutions in coding regions and 27 substitutions in the intergenic regions. As anticipated, three of the four large deletions were associated with the absence of Φ11, Φ12, and Φ13 (Table S1). We also confirmed a subset of 29 nonsynonymous mutations (those that might affect virulence or metabolism) by PCR amplification followed by DNA sequencing (Table 1). This subset included nine nonsynonymous mutations that were also identified by O'Neill (18).
Table 1.
Nonsynonymous, PCR-verified SNPs identified in RN4220 relative to NCTC8325
| Genome position | Putative gene product and function | Nucleotide change |
Amino acid changea | Locus tag | |
|---|---|---|---|---|---|
| NCTC8325 | RN4220 | ||||
| 174867 | HsdR family type I site-specific DNase | G | A | W197* | SAOUHSC_00162 |
| 281533 | EssC (DNA segregation FtsK/SpoIIIE, S-DNA-T family) | G | A | W52* | SAOUHSC_00262 |
| 292106 | Hypothetical protein | G | K | SAOUHSC_00274 | |
| 292107 | C | Y | |||
| 292179 | C | Y | |||
| 292199 | A | R | |||
| 292328 | G | R | |||
| 388693 | Conserved hypothetical protein | C | T | P134S | SAOUHSC_00383 |
| 590402 | Conserved hypothetical protein | G | Frameshift | SAOUHSC_00591 | |
| 751285 | SecA preprotein translocase subunitb | A | T | E449V | SAOUHSC_00769 |
| 795429 | Clumping factor ClfA | C | T | S815L | SAOUHSC_00812 |
| 827849 | Hypothetical proteinb | A | T | T164S | SAOUHSC_00859 |
| 939304 | Competence transcription factor, putativeb | C | T | E53K | SAOUHSC_00961 |
| 1016979 | Spermidine/putrescine ABC transporter, putativeb | G | A | E220K | SAOUHSC_01048 |
| 1020577 | Manganese transport protein MntHb | G | T | S286* | SAOUHSC_01053 |
| 1063555 | UvrC, excinuclease ABC subunit C | C | T | P331S | SAOUHSC_01102 |
| 1123048 | Orotate phosphoribosyltransferaseb | G | A | G42S | SAOUHSC_01 |
| 1160531 | RimM, 16S rRNA-processing protein | G | A | A106T | SAOUHSC_01209 |
| A | G | ||||
| 1358230 | Kgd, alpha-ketoglutarate decarboxylaseb | C | T | D590N | SAOUHSC_01418 |
| 1632629 | (5-Methylaminomethyl-2-thiouridylate)methyltransferase | A | Frameshift | SAOUHSC_01726 | |
| 1733572 | Septation ring formation regulator EzrA | G | T | T73N | SAOUHSC_01827 |
| A | G | F54S | |||
| 2087725 | GroEL chaperone | A | T | F218I | SAOUHSC_02254 |
| 2096628 | AgrA | A | Frameshift | SAOUHSC_02265 | |
| 2106539 | ABC transporter, ATP-binding protein, putative | A | T | L602F | SAOUHSC_02274 |
| 2446162 | Phosphotransferase system sucrose-specific IIBC component, putative | C | T | A211T | SAOUHSC_02661 |
| C | G | ||||
| C | A | ||||
*, truncation.
Mutation identified by O'Neill (18).
Besides the deletions of the three phages, there were also deletions of two hypothetical proteins and an 1,195-bp region that codes for the B subunit of excision endonuclease (also called excinuclease) ABC, which catalyzes the processing of DNA lesions by the UvrABC excinuclease complex for DNA repair (25). Mutations in hsdR (SAOUHSC_00162), essC (SAOUHSC_00262), mntH (SAOUHSC_01053), and a hypothetical gene (SAOUHSC_02790) result in a premature stop codon (Table 1). More specifically, the mutation in hsdR, resulting in a lack of restriction, confirmed previous data of Waldron and Lindsay (26). A rhomboid family protein (SAOUHSC_01649) shows an N-terminal deletion. There are 20 indels (insertion/deletions) in coding regions, causing frame-shift mutations in SAOUHSC_00269, SAOUHSC_00270, SAOUHSC_00274, SAOUHSC_00275, and SAOUHSC_00276, all hypothetical proteins. Among the indels, one of the insertions is in agrA and another is in the gene encoding (5-methylaminomethyl-2-thiouridylate)methyltransferase, while the remaining two are in noncoding regions. Of the 27 mutations in noncoding regions, 5 are within 100 bp of the coding regions of putative regulatory loci. Most of these intergenic SNPs were located in untranslated regions or within putative operons, possibly impacting multiple factors. We found some intergenic SNPs in predicted regulatory noncoding RNA such as Sau-25 (1) and within regions potentially affecting the structure of the highly stable small RNA Teg27/RsaX18 (4, 21).
Eleven of the 121 mutations were also found in NCTC8325-4 (18) (Table S2 in the supplemental material). The remaining mutations, unique to RN4220, can be grouped into two categories: (i) those affecting the survival and fitness of the bacteria, which include uvrC (part of the uvrABC system) (25), ezrA, rimM, and genes encoding ribosomal protein S2 and GroEL, and (ii) those affecting virulence factors, namely, agrA, essC, clfA, and a gene encoding superantigen-like protein (Table 1).
UvrC, part of the system involved in DNA repair, is specifically involved in the incision of the 5′ and 3′ sides of the lesion. The P331S mutation in UvrC in RN4220 may result in disruption of the secondary structure of UvrC, thus making RN4220 more prone to spontaneous mutations. The ezrA gene, likely essential to S. aureus, is thought to regulate septum ring formation (7). RimM (ribosome maturation factor), ribosomal protein S2, and GroEL are important proteins collectively involved in protein synthesis and protein folding. More specifically, RimM is an accessory factor required for 30S maturation and assembly in Escherichia coli (13), and deletion of rimM has been known to decrease the growth rate and reduce translational efficiency at 37°C. In S. aureus, treatment with heat or a cell wall-active antibiotic results in increased transcription of groEL, indicating a role for GroEL in protein folding under heat and antibiotic stress (17, 22). Mutations in these three proteins presumably would thus have an effect on the general fitness (slower growth and translational deficiency) of the strain, especially under stress. The mutation of these genes might result in an inadequate response to antibiotics, which would actually be a false-positive effect of the antibiotic in question. With regard to the virulence factors, the agrA mutation results in the insertion of an extra adenine residue at the 3′ end of agrA, leading to a run of eight adenines and a frameshift that adds three amino acids to the C terminus of AgrA. This finding with agrA in RN4220, resulting in delayed activation of agr and a failure to synthesize delta and alpha hemolysins, has been described by Traber and Novick (24). The membrane protein EssC is one product of the eight-gene cluster of the ESAT-6-like secretion system (Ess), which is essential for the secretion of EsxA-EsxB (5). The essC mutation in RN4220 results in a truncated EssC. S. aureus mutants that fail to secrete EsxA and EsxB display significantly reduced virulence, dissemination, and colonization in mice (5). The S. aureus clumping factor ClfA, a surface protein belonging to the MSCRAMM family, binds the γ-chain of fibrinogen (15) and induces platelet aggregation (3); it also mediates adherence of S. aureus to fibrinogen-coated surfaces and contributes to protection against phagocytosis by neutrophils (19). The S815L mutation in ClfA in RN4220 results in the substitution of a polar hydrophilic amino acid with a nonpolar hydrophobic residue. This may conceivably disrupt the protein fold and reduce its affinity for fibrinogen. The superantigen-like protein is similar to exotoxin Set6, which is a virulence-associated protein (8).
Based on the above analysis, investigators using RN4220 in virulence studies should proceed with caution, since mutations identified in this paper show that the virulence genes as well as those involved in fitness and numerous stress-associated putative regulators are altered in RN4220.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant AI37142 to A.L.C.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Abu-Qatouseh L. F., et al. 2010. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 88:565–575 [DOI] [PubMed] [Google Scholar]
- 2. Aziz R. K., et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bayer A. S., et al. 1995. Staphylococcus aureus induces platelet aggregation via a fibrinogen-dependent mechanism which is independent of principal platelet GP IIb/IIIa fibrinogen-binding domains. Infect. Immun. 63:3634–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beaume M., et al. 2011. Orientation and expression of methicillin-resistant Staphylococcus aureus small RNAs by direct multiplexed measurements using the nCounter of NanoString technology. J. Microbiol. Methods 84:327–334 [DOI] [PubMed] [Google Scholar]
- 5. Burts M. L., Williams W. A., DeBord K., Missiakas D. M. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U. S. A. 102:1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheung A. L., Bayer M. G., Heinrichs J. H. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Errington J., Daniel R. A., Scheffers D. J. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gill S. R., et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hernandez D., Francois P., Farinelli L., Osteras M., Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 18:802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kreiswirth B. N., et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 11. Krzywinski M., et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurtz S., et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lovgren J. M., et al. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 10:1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsumoto Y., Kaito C., Morishita D., Kurokawa K., Sekimizu K. 2007. Regulation of exoprotein gene expression by the Staphylococcus aureus cvfB gene. Infect. Immun. 75:1964–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McDevitt D., Francois P., Vaudaux P., Foster T. J. 1994. Cloning and sequencing of the clumping factor of Staphylococcus aureus. Mol. Microbiol. 11:237–248 [DOI] [PubMed] [Google Scholar]
- 16. Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166 [DOI] [PubMed] [Google Scholar]
- 17. Ohta T., Honda K., Kuroda M., Saito K., Hayashi H. 1993. Molecular characterization of the gene operon of heat shock proteins HSP60 and HSP10 in methicillin-resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 193:730–737 [DOI] [PubMed] [Google Scholar]
- 18. O'Neill A. J. 2010. Staphylococcus aureus SH1000 and 8325-4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett. Appl. Microbiol. 51:358–361 [DOI] [PubMed] [Google Scholar]
- 19. Palmqvist N., Patti J. M., Tarkowski A., Josefsson E. 2004. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 6:188–195 [DOI] [PubMed] [Google Scholar]
- 20. Pragman A. A., Yarwood J. M., Tripp T. J., Schlievert P. M. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberts C., et al. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188:2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh V. K., Jayaswal R. K., Wilkinson B. J. 2001. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiol. Lett. 199:79–84 [DOI] [PubMed] [Google Scholar]
- 23. Sommer D. D., Delcher A. L., Salzberg S. L., Pop M. 2007. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Traber K., Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol. Microbiol. 59:1519–1530 [DOI] [PubMed] [Google Scholar]
- 25. Truglio J. J., Croteau D. L., Van H. B., Kisker C. 2006. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106:233–252 [DOI] [PubMed] [Google Scholar]
- 26. Waldron D. E., Lindsay J. A. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yarwood J. M., McCormick J. K., Schlievert P. M. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.