Abstract
Starvation causes cells in a dense population of Myxococcus xanthus to change their gliding movements and construct mounds. Short-range C-signaling between rod-shaped cells within mounds induces gene expression that promotes differentiation into spherical spores. Several C-signal-dependent genes have been shown to be regulated by cooperative binding of two transcription factors to the promoter region. These FruA- and MrpC2-regulated genes (designated fmg) each exhibit a different arrangement of binding sites. Here, we describe fmgE, which appears to be regulated by three sites for cooperative binding of FruA and MrpC2. Chromatin immunoprecipitation analysis showed that association of MrpC2 and/or its longer form, MrpC with the fmgE promoter region, depends on FruA, consistent with cooperative binding of the two proteins in vivo. Electrophoretic mobility shift assays with purified His10-MrpC2 and FruA-His6 indicated cooperative binding in vitro to three sites in the fmgE promoter region. The effects of mutations on binding in vitro and on expression of fmgE-lacZ fusions correlated site 1 (at about position −100 relative to the transcriptional start site) with negative regulation and site 2 (just upstream of the promoter) and site 3 (at about position +100) with positive regulation. Site 3 was bound by His10-MrpC2 alone, or the combination of His10-MrpC2 and FruA-His6, with the highest affinity, followed by site 1 and then site 2, supporting a model in which site 3 recruits MrpC2 and FruA to the fmgE promoter region, site 1 competes with site 2 for transcription factor binding, and site 2 occupancy is required to activate the promoter but only occurs when C-signaling produces a high concentration of active FruA.
INTRODUCTION
Myxococcus xanthus is a Gram-negative bacterium that provides an attractive model to study signaling and gene regulatory mechanisms (59). Flexible, rod-shaped cells move over solid surfaces, forming swarms that seek prey bacteria on which to feed (3). Upon nutrient limitation, cells alter the frequency with which they reverse their direction of gliding and coordinate their movements to construct mounds containing approximately 105 cells (14). During this process of aggregation, some cells undergo programmed cell death (33), others remain outside the nascent fruiting bodies as peripheral rods (36), and others differentiate into dormant, stress-resistant, spherical spores. The mature fruiting body is a mound of spores capable of producing a swarm of cells when a nutrient source becomes available.
Fruiting body development is coordinated by intracellular and extracellular signals. Nutrient limitation triggers a stringent response that leads to production of intracellular (p)ppGpp and induction of early developmental genes (10, 47), including dnaA (perhaps only in cells destined to become spores), which encodes the initiator protein of DNA replication (41). Cells secrete proteases that produce a mixture of amino acids and peptides known as the A-signal (23, 38), which appears to function as a quorum signal, allowing aggregation to begin and additional genes to be expressed if cells are at a sufficiently high density (15, 22, 24). A second extracellular signal called the C-signal is unusual for bacterial signals in that it functions only at short range (2), perhaps only when cells are in contact. Efficient C-signaling requires that cells become aligned (18), as they do in the outer domain of a nascent fruiting body (43). Many studies support a model in which an increasing level of C-signaling governs mound formation and ensures that spores form within mounds (reviewed in references 12, 14, 45, and 49). The mechanism of C-signaling is only partly understood. It involves CsgA (46), a protein associated with the outer membrane, where it is cleaved by a secreted protease to a 17-kDa form that appears to be the C-signal (17, 26, 40); however, a receptor has not been identified. In any case, a csgA mutant fails to form stable aggregates, exhibits reduced or abolished expression of many developmental genes, and forms very few spores (19, 46). Genes normally expressed shortly after early mound formation exhibit reduced expression in a csgA mutant, and genes normally expressed later fail to be expressed (19).
The C-signal-dependent changes in motility behavior and gene expression that lead to aggregation and sporulation are mediated by FruA, which is similar to the response regulators of two-component signal transduction systems (6, 37). It has been proposed that FruA is phosphorylated in response to C-signal (6), but a cognate histidine protein kinase has not been identified, and some evidence suggests that FruA might function without being phosphorylated (29). The C-terminal DNA-binding domain of FruA has been shown to bind to sites in the promoter regions of genes that fail to be expressed in a fruA mutant, suggesting that FruA is a transcriptional activator (52–53, 58, 62). Recently, FruA was shown to bind cooperatively with MrpC2 to the promoter region of the C-signal-dependent fmgA gene (29).
MrpC2 is a truncated form lacking the 25 N-terminal amino acid residues of MrpC (54). MrpC is similar to the proteins in the cyclic AMP receptor protein (CRP) family (50). Its production is inhibited during growth by phosphorylation (34–35). The phosphorylated form of MrpC binds weakly to DNA compared with the unphosphorylated form. Starvation inhibits phosphorylation of MrpC, and some of the unphosphorylated form may be cleaved to MrpC2 or a slightly shorter form lacking 7 more residues at its N terminus (54). MrpC2 binds better than MrpC to sites upstream of the mrpC and fruA promoters, activating transcription so that the concentrations of MrpC, MrpC2, and FruA increase in starving cells (35, 54). MrpC also binds to the mazF promoter region and activates expression of MazF, a toxin that causes programmed cell death during development (33). Interestingly, MrpC binds to MazF and inhibits its activity, allowing some cells in the population to escape programmed cell death and eventually form spores. MrpC2 has not yet been tested for binding to MazF and to the mazF promoter region.
The finding that MrpC2 and FruA bind cooperatively just upstream of the fmgA promoter to activate transcription suggests that positional information through C-signaling and FruA is integrated with starvation sensing and cell death control through MrpC and MrpC2 (29). Such combinatorial regulation was subsequently demonstrated for other C-signal-dependent genes, although different arrangements of MrpC2 and FruA binding sites were discovered. The two proteins bind cooperatively just upstream of the fmgBC and fmgD promoters, but FruA binds proximal to these promoters (25, 30), whereas MrpC2 binds proximal to the fmgA promoter (29). In the fmgD promoter region, a second MrpC2 binding site partially overlaps the FruA binding site and the promoter (25). It was proposed that binding of MrpC2 to the downstream site represses fmgD transcription until C-signaling causes the concentration of active FruA to increase sufficiently to outcompete the downstream MrpC2 for cooperative binding with the upstream MrpC2. The model would explain why fmgD transcription begins later during development and is more dependent on C-signaling than transcription of fmgA and fmgBC.
Here, we report that a second gene in the late class that depends completely on C-signaling is under combinatorial control of MrpC2 and FruA, which bind cooperatively to three sites in the promoter region. The gene, herein designated fmgE, was first identified as a developmentally regulated gene by an insertion of the transposon Tn5 lac at the Ω4406 locus (21). Developmental lacZ expression was abolished in a csgA mutant (19). The gene at the Ω4406 locus (MXAN3464) (8) is predicted to code for a protein annotated as hypothetical (27), which a BLAST search (1) shows is similar to hypothetical proteins in diverse bacteria and a few eukaryotes, and all of which share a conserved domain found in zinc-dependent metalloproteases. fmgE is likely monocistronic (27). The Tn5 lac insertion in fmgE delays and reduces aggregation and reduces sporulation by about 8-fold (S. Mittal and L. Kroos, unpublished data). Deletion analyses revealed unusual regulation of fmgE. A negative regulatory element was localized between positions −533 and −100, relative to the transcriptional start site, and a positive regulatory element was localized between +50 and +140 (55). On the other hand, mutational analysis highlighted the importance of sequences just upstream of the promoter, as in other C-signal-dependent promoter regions (56, 57, 60, 61), including two 5-bp elements (consensus sequence GAACA) at positions −69 to −65 and positions −64 to −60 and a C-box-like sequence, CATCGTG, at positions −55 to −49 (the C-box consensus sequence is CAYYCCY, in which Y means C or T) (55). Using a combination of in vitro and in vivo approaches, we found that MrpC2 and FruA bind cooperatively not only immediately upstream of the fmgE promoter but also slightly farther upstream at about position −100 and downstream at about +100, accounting for the negative and positive regulatory elements, respectively. The different affinities of the three sites for the combination of MrpC2 and FruA support a model in which a rising concentration of active FruA produced in response to C-signaling during development results in occupancy of the downstream site first, followed by occupancy of the upstream site, and finally, occupancy of the site just upstream of the promoter, which activates transcription.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are listed in Table 1. To construct plasmids, pKV45 was used as the template for PCR amplification of fmgE DNA segments, using upstream primers each containing an XhoI site and downstream primers each containing a BamHI site (Table 2 lists primers). Each PCR product was cloned using pCR2.1-TOPO and Escherichia coli strain TOP10, as described by the manufacturer. Each DNA insert was sequenced at the Michigan State University Genomics Technology Support Facility to ensure that the correct sequence was obtained. Each pCR2.1-TOPO derivative was digested with XhoI and BamHI, the DNA insert was gel purified, and it was cloned into XhoI-BamHI-digested pREG1727 using E. coli strain DH5α and standard methods (44).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| TOP10 | λ− F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 9 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm with DE3, a λ prophage carrying the T7 RNA polymerase gene | Novagen |
| SMhisMrpC2 | BL21(DE3) containing pET16b/His10-MrpC2 | 29 |
| SMFruAhis | BL21(DE3) containing pET11km/FruA-His6 | 29 |
| M. xanthus strains | ||
| DK1622 | Wild type | 13 |
| DK5285 | fruA::Tn5lac Ω4491 | 20 |
| MKV6 | attB::pKV6 (pREG1727 with 150-bp XhoI-BamHI fragment from pKV3)a | 55 |
| MKV24 | attB::pKV24 (pREG1727 with 351-bp XhoI-BamHI fragment from pPV251) | This study |
| MKV54 | attB::pKV54 (pREG1727 with 398-bp XhoI-BamHI fragment from pKV45) | This study |
| MBS03 | attB::pBS08 (pREG1727 with 197-bp XhoI-BamHI fragment from pBS02) | This study |
| MBS04 | attB::pBS07 (pREG1727 with 216-bp XhoI-BamHI fragment from pBS01) | This study |
| MBS06 | attB::pBS21 (pREG1727 with 193-bp XhoI-BamHI fragment from pBS15) | This study |
| MBS07 | attB::pBS18 (pREG1727 with 170-bp XhoI-BamHI fragment from pBS13) | This study |
| MBS08 | attB::pBS19 (pREG1727 with 160-bp XhoI-BamHI fragment from pBS14) | This study |
| Plasmids | ||
| pET16b/His10-MrpC2 | pET16b carrying a gene encoding His10-MrpC2 under the control of a T7 RNA polymerase promoter | 35 |
| pET11km/FruA-His6 | pET11km carrying a gene encoding FruA-His6 under the control of a T7 RNA polymerase promoter | S. Inouye |
| pREG1727 | Apr Kmr; P1-incattP ′lacZ | 7 |
| pCR2.1-TOPO | Apr Kmr; lacZα | Invitrogen |
| pKV3 | pCR2.1-TOPO with fmgE DNA from positions −100 to +50 | 55 |
| pKV4 | pREG1727 with 1.0-kb XhoI-BamHI fragment from pPV4406-1.0 | 55 |
| pKV12 | pCR2.1-TOPO with fmgE DNA from positions −100 to + 320 | 55 |
| pKV28 | pKV12 with CATCGTG-to-ACGATGT mutation from positions −55 to −49 | 55 |
| pKV29 | pKV12 with GGACA-to-TTCAC mutation from positions −69 to −65 | 55 |
| pKV30 | pKV12 with GAACC-to-TCCAA mutation from positions −64 to −60 | 55 |
| pKV45 | pCR2.1-TOPO with fmgE DNA from positions −150 to +251, generated by PCR using LK0926 and LK1245 as primers and pKV4 as template | This study |
| pKV47 | pKV12 with A-to-C mutation at position −54 | 55 |
| pPV251 | pCR2.1-TOPO with fmgE DNA from positions −100 to +251 | 55 |
| pBS01 | pCR2.1-TOPO with fmgE DNA from positions −100 to +116, generated by PCR using LK1328 and LK2527 as primers and pKV45 as template | This study |
| pBS02 | pCR2.1-TOPO with fmgE DNA from positions −150 to +50, generated by PCR using LK0926 and LK1034 as primers and pKV45 as template | This study |
| pBS13 | pCR2.1-TOPO with fmgE DNA from positions −120 to +50, generated by PCR using LK2577 and LK1034 as primers and pKV45 as template | This study |
| pBS14 | pCR2.1-TOPO with fmgE DNA from positions −110 to +50, generated by PCR using LK2578 and LK1034 as primers and pKV45 as template | This study |
| pBS15 | pCR2.1-TOPO with fmgE DNA from positions −100 to +93, generated by PCR using LK1328 and LK2606 as primers and pKV45 as template | This study |
Where possible, the plasmid description is given in parentheses after the strain description.
Table 2.
Primers used in this study
| Primer | Sequencea | Description or referenceb |
|---|---|---|
| LK0926 | GGCTCGAGGTTTTTCTCTGTGAAAGAGGAC | Position −150 forward with XhoI site |
| LK1034 | CCGGATCCGTTGTTACCGGCATTGGTGC | Position +50, reverse with BamHI site |
| LK1245 | CCGGATCCACTGGCGCCGGCTGTAGGC | Position +251, reverse with BamHI site |
| LK1328 | GGCTCGAGCATTCTGTGCGGCGTTTCAG | Position −100, forward with XhoI site |
| LK1861 | CCTTGAGCGCGATGGAGATA | 60 |
| LK1862 | CTCGGCGGCCTCATCGAC | 60 |
| LK2442 | GAGGGCGCACGAATCGTCC | Position −25, reverse |
| LK2464 | CAATCCCATGTCCTCATCTG | Position −25, forward |
| LK2487 | TTTCTCTGTGAAAGAGGACCC | Position −147, forward |
| LK2512 | CCTGAAACGCCGCACAGAATG | Position −80, reverse |
| LK2527 | GGATCCCGATTGCGTTCCTGCTCCATG | Position +116, reverse with BamHI site |
| LK2577 | GGCTCGAGCATTAACGGGCGATCTTCCTC | Position −120, forward with XhoI site |
| LK2578 | GGCTCGAGCGATCTTCCTCATTCTGTGCG | Position −110, forward with XhoI site |
| LK2606 | GGATCCGGTGAGGACAGCGGTCAG | Position +93, reverse with BamHI site |
Restriction sites are underlined.
The position number is relative to the start site of fmgE transcription, and the orientation (forward or reverse) is relative to the direction of fmgE transcription.
Growth and development.
M. xanthus strains were grown at 32°C in CTT (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH, 7.6]) medium (11) or on CTT agar (1.5%) plates. When required, 40 μg of kanamycin sulfate per ml was added. Fruiting body development was performed on TPM (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH, 7.6]) agar (1.5%) plates, as described previously (21).
ChIP.
M. xanthus strains DK1622 and DK5285 were used for chromatin immunoprecipitation (ChIP) as described previously (29, 62), except Dynabeads protein G (100 μl/ml of cell extract; Invitrogen) were used instead of protein A-Sepharose beads for preclearing and immunoprecipitation. The primers used for PCR of the fmgE promoter region were LK1328 and LK1034, and those used for PCR of the rpoC coding region were LK1861 and LK1862 (Table 2).
Preparation of His10-MrpC2 and FruA-His6.
Recombinant proteins were expressed in E. coli and purified as described previously (29, 35).
Preparation of 32P-labeled DNA fragments.
DNA fragments from the fmgE promoter region were generated by PCR using a wild-type or mutant plasmid (Table 1) as the template and the primers listed in Table 2. For electrophoretic mobility shift assays (EMSAs), 32P-labeled DNA was synthesized by PCR after labeling the primers with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs), and the DNA fragment was purified after 15% PAGE (44).
EMSAs.
EMSAs were performed as described previously (62), except that binding reaction mixtures were incubated at 25°C for 15 min.
Construction of M. xanthus strains and determination of lacZ expression during development.
Strains containing a plasmid integrated at the Mx8 phage attachment site attB were constructed by electroporation (16). Transformants were selected on CTT agar plates containing kanamycin sulfate and screened on TPM agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml in order to avoid rare transformants with unusual developmental lacZ expression (56). Three transformants were typically chosen for further analysis, and β-galactosidase activity was measured as described previously (21).
RESULTS
MrpC and FruA associate with the fmgE promoter region in vivo.
ChIP assays using polyclonal antibodies to MrpC, which also recognize MrpC2 (35), and polyclonal antibodies to FruA were performed to determine whether these transcription factors associate with the fmgE promoter region during M. xanthus development. Cells were collected after 18 h and subjected to ChIP with affinity-purified anti-MrpC immunoglobulin G (IgG) (or IgG from a nonimmunized rabbit as a control) or anti-FruA serum (or preimmune serum from the same rabbit as a control). The ChIP DNA was analyzed by PCR with primers designed to amplify the fmgE promoter region (positions −100 to +50) or, as a control, the rpoC coding region (+1780 to +1905) (62). The PCR analysis showed enrichment of the fmgE promoter region by ChIP with anti-MrpC rather than control IgG (Fig. 1A, lanes 5 and 6) and with anti-FruA compared to control preimmune serum (Fig. 1B, lanes 5 and 6). Similar results were observed in two additional experiments. As expected, the PCR analysis with primers designed to amplify the rpoC coding region showed no enrichment with anti-MrpC or anti-FruA compared to with controls. We conclude that MrpC and/or MrpC2 and FruA are present in the vicinity of the fmgE promoter at 18 h into development.
Fig. 1.
Association of MrpC and/or MrpC2 and FruA with the fmgE promoter region during development. ChIP analysis of M. xanthus at 18 h into development. Cells were treated with formaldehyde and lysed. Cross-linked chromatin was immunoprecipitated with antibodies. DNA was amplified with primers for the fmgE promoter region (positions −100 to +50 relative to the start site of transcription) or for the rpoC coding region (positions +1780 to +1905 relative to the predicted translation start) as a control. A 2-fold dilution series of input DNA purified from 0.025, 0.0125, 0.00625, or 0.003125% of the total cellular extract prior to immunoprecipitation was used as a template in parallel PCRs to show that the PCR conditions allow detection of differences in DNA concentration for each primer set. (A) Wild-type strain DK1622 with affinity-purified IgG antibodies against MrpC (α-MrpC) or, as a control, with total IgG (IgG) from nonimmunized rabbits. (B) Wild-type strain DK1622 with antiserum against FruA (α-FruA) or, as a control, preimmune antiserum (Pre). (C) fruA mutant strain DK5285 with antibodies as shown in panel A.
Association of MrpC and/or MrpC2 with the fmgE promoter region depends on FruA.
It was shown previously that association of MrpC and/or MrpC2 with the fmgA and fmgBC promoter regions depends on FruA, presumably due to cooperative binding of the two proteins just upstream of the promoters (29–30). In contrast, association of MrpC and/or MrpC2 with the fmgD promoter region did not depend on FruA, as determined by ChIP-PCR analysis of a fruA mutant at 18 h into development (25). Although MrpC2 and FruA bind cooperatively just upstream of the fmgD promoter in vitro, a second MrpC2 binding site overlaps the FruA binding site, and two molecules of MrpC2 can bind cooperatively to the fmgD promoter region in the absence of FruA in vitro. Presumably either because of this cooperative binding or simply due to the presence of a higher-affinity binding site than those present in the fmgA and fmgBC promoter regions, MrpC and/or MrpC2 was found to be associated with the fmgD promoter region of a fruA mutant. To test whether association of MrpC and/or MrpC2 with the fmgE promoter region depends on FruA, we performed ChIP-PCR analysis of a fruA mutant at 18 h into development. Reproducibly, the PCR analysis showed no enrichment of the fmgE promoter region by ChIP with anti-MrpC compared to control IgG (Fig. 1C, lanes 5 and 6). Likewise, there was no enrichment of the rpoC coding region compared to the control, as expected. Therefore, association of MrpC and/or MrpC2 with the fmgE promoter region at 18 h into development depends on FruA, as seen previously for the fmgA and fmgBC promoter regions (29–30), suggesting that the two proteins bind cooperatively to the fmgE promoter region.
MrpC2 and FruA bind near the fmgE promoter to sequences important for promoter activity.
Previous studies showed that MrpC2 and FruA bind cooperatively just upstream of the fmgA promoter, with MrpC2 proximal to the promoter (29), and the two proteins bind cooperatively just upstream of the fmgBC and fmgD promoters, with FruA proximal to the promoter (25, 30). Since multiple-base-pair changes just upstream of the fmgE promoter were shown to increase or decrease promoter activity (55) (Fig. 2A, top), we tested whether MrpC2 and/or FruA bind near the fmgE promoter. EMSAs were performed with fmgE promoter region DNA from positions −100 to −25 and purified His10-MrpC2 and FruA-His6. Each protein produced a single shifted complex, although the complex produced by His10-MrpC2 was barely detectable (Fig. 2A, lanes 1 and 2). Together, the two proteins produced two shifted complexes, and the total amount of shifted complexes was greater than that expected for additive binding (Fig. 2A, lane 3). This pattern was shown previously to be indicative of cooperative binding (29).
Fig. 2.
Binding of MrpC2 and FruA to the fmgE promoter region. (A) Effects of mutations in the fmgE promoter region on fmgE-lacZ expression in vivo and on MrpC2 and FruA binding in vitro. (Top) Summary of the effects of four mutations on developmental fmgE-lacZ expression (55). The mutations are shown, and the numbers indicate the maximum β-galactosidase activity during development, expressed as a percentage of the maximum activity observed for the wild-type promoter. (Bottom) EMSAs with 32P-labeled fmgE DNA (2 nM) spanning from positions −100 to −25 and His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM), as indicated. The probe DNA had the wild-type (WT) sequence or the indicated mutation. The filled arrowhead points to the shifted complex produced by His10-MrpC2 alone, and the open arrowhead points to the complex produced by FruA-His6 alone. (B) Summary of binding sites for MrpC2 and FruA in the fmgE promoter region. The approximate location and relative positions of MrpC2 and FruA at sites 1, 2, and 3 are deduced from the effects of 5′-end deletions, the mutations shown in panel A, and 3′-end deletions, respectively, on binding in vitro, as explained in the text. The position of MrpC2 binding alone, depicted upstream of position +50, is less precisely known but lies between positions −25 and +50.
To test whether mutations that affect fmgE promoter activity also affect the binding of MrpC2 and FruA, EMSAs were performed with fmgE promoter region DNA from positions −100 to −25 containing multiple-base-pair changes. The GGACA-to-TTCAC change, which increased promoter activity by about 3-fold in vivo (55), increased in vitro binding of His10-MrpC2 alone and greatly increased the apparent cooperative binding of the two proteins (Fig. 2A, lanes 4 to 6), suggesting that the increased promoter activity is due to increased binding of MrpC2 and FruA. In contrast, the GAACC-to-TCCAA change, which decreased promoter activity by about 4-fold (55), decreased binding of FruA-His6 alone, and cooperative binding was no longer apparent (Fig. 2A, lanes 8 and 9). Likewise, the CATCGTG-to-ACGATGT change, which decreased promoter activity by about 9-fold (55), decreased binding of FruA-His6 alone, and cooperative binding was no longer apparent (Fig. 2A, lanes 11 and 12). Therefore, for all three mutations, promoter activity in vivo was correlated with cooperative binding of the two transcription factors in vitro. We conclude that cooperative binding of MrpC2 and FruA just upstream of the fmgE promoter is important for promoter activity. Interestingly, both mutations that decreased promoter activity not only decreased the binding of FruA-His6 alone but also increased the binding of His10-MrpC2 alone (Fig. 2A, compare lanes 1, 7, and 10). In the case of the CATCGTG-to-ACGATGT change, not only did binding of His10-MrpC2 alone increase markedly but also a second shifted complex was observed (Fig. 2A, lane 10). We propose that this mutation creates a second MrpC2 binding site and that the other two mutations alter the normal MrpC2 binding site (see Discussion). Since the most upstream mutation tested (GGACA to TTCAC) affected MrpC2 binding but not FruA binding, we infer that MrpC2 binds upstream of FruA (Fig. 2B, site 2), an arrangement similar to that just upstream of the fmgBC and fmgD promoters (25, 30). The binding sites for the two proteins might partly overlap, given that both downstream mutations (GAACC to TCCAA and CATCGTG to ACGATGT) affected the binding of both of the proteins. In previous work, it appeared that MrpC2 and FruA bind to partially overlapping sites just upstream of the fmgA promoter, and it was suggested that the two proteins interact with opposite faces of the DNA in a region of overlap (29).
One mutation that affected fmgE promoter activity did not detectably alter the binding of MrpC2 or FruA, as determined by EMSAs. Changing A to C at position −54 increased promoter activity by about 3-fold (55) but had no effect on shifted complex formation (Fig. 2A, lanes 13 to 15). There are several possible explanations for this (see Discussion).
MrpC2 and FruA bind to a downstream positive regulatory element.
In previous work, deletion of downstream DNA between +50 and +140 reduced fmgE promoter activity by 3-fold, indicating the presence of a positive regulatory element (55). To test whether MrpC2 and/or FruA bind to the downstream region, EMSAs were performed with several DNA fragments. His10-MrpC2 produced a single shifted complex with DNA from positions −25 to +50, but FruA-His6 did not bind detectably (Fig. 3A, lanes 1 to 3). Likewise, FruA-His6 did not bind detectably to DNA from positions −25 to +93; however, His10-MrpC2 produced two shifted complexes, suggesting the presence of two MrpC2 binding sites on this fragment (Fig. 3A, lanes 4 to 6). In contrast, DNA from positions −25 to +116 produced a single shifted complex with FruA-His6, and two shifted complexes with His10-MrpC2, and when both proteins were added, the majority of DNA was shifted to an upper complex, suggestive of cooperative binding (Fig. 3A, lanes 7 to 9). Taken together, the EMSA results suggest that MrpC2 and FruA bind cooperatively between +50 and +116, with MrpC2 proximal to the fmgE promoter (Fig. 2B, site 3), and MrpC2 binds weakly between positions −25 and +50 (Fig. 2B).
Fig. 3.
MrpC2 and FruA bind to a downstream positive regulatory element. (A) Binding of MrpC2 and FruA to site 3. EMSAs with 32P-labeled fmgE DNA (2 nM) spanning from position −25 to the indicated 3′ ends and His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM), as indicated. The filled arrowhead points to the shifted complex produced by His10-MrpC2 alone, and the open arrowhead points to the complex produced by FruA-His6 alone. (B) Effects of 3′-end deletions on developmental fmgE-lacZ expression. β-Galactosidase specific activity during development was measured for lacZ fused to fmgE spanning from positions −100 to +50 (▴), −100 to +93 (○), or −100 to +116 (■). The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Points show the average values of three transformants, and each error bar depicts 1 standard deviation of the data.
Since deletion of DNA between +50 and +140 previously reduced fmgE promoter activity by 3-fold (55), we hypothesized that the positive regulatory element implicated in this region involves cooperative binding of MrpC2 and FruA to site 3, as depicted in Fig. 2B. To test this hypothesis, we compared the fmgE promoter activities of DNA fragments with different 3′ ends. Each fragment was cloned into the transcriptional fusion vector pREG1727, which has the E. coli lacZ gene as a reporter and integrates in the M. xanthus chromosome via site-specific recombination at the Mx8 phage attachment site (7). Consistent with the hypothesis that site 3 is a positive regulatory element, DNA from positions −100 to +116 exhibited higher developmental lacZ expression than DNA from positions −100 to +93 or from −100 to +50 (Fig. 3B). We conclude that MrpC2 and FruA bind cooperatively to a positive regulatory element located about 100 bp downstream of the fmgE promoter.
MrpC2 and FruA bind to an upstream negative regulatory element.
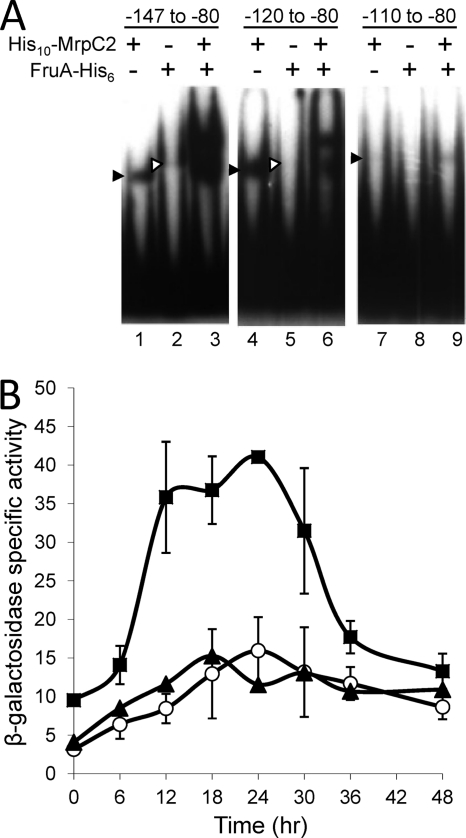
In previous work, deletion of upstream DNA between positions −533 and −100 increased fmgE promoter activity by 4-fold, indicating the presence of a negative regulatory element (55). Comparison of fmgE promoter activity of DNA fragments spanning from positions −150 to +251 and from positions −100 to +251, using the lacZ transcriptional fusion vector described above, localized the negative regulatory element to the region between positions −150 and −100 (data not shown). To test whether MrpC2 and/or FruA binds to this upstream region, EMSAs with several DNA fragments were performed. Although a 5′-end deletion to position −100 eliminated negative regulation, we reasoned that a binding site might include sequences slightly farther downstream, so we tested fragments with different 5′ ends and the same 3′ end at position −80 (to avoid the MrpC2 and FruA binding sites located just upstream of the fmgE promoter) (Fig. 2B, site 2). His10-MrpC2 and FruA-His6 each produced a single shifted complex with DNA from positions −147 to −80, and in combination, the two proteins produced at least two shifted complexes in greater abundance, suggestive of cooperative binding (Fig. 4A, lanes 1 to 3). With DNA from positions −120 to −80, His10-MrpC2 produced a single shifted complex, but binding of FruA-His6 was barely detectable; however, in combination, the two proteins produced an upper complex, suggestive of cooperative binding (Fig. 4A, lanes 4 to 6). In contrast, DNA from positions −110 to −80 appeared to bind His10-MrpC2 weakly, binding of FruA-His6 was not detected, and there was no evidence of cooperative binding (Fig. 4A, lanes 7 to 9). These results suggest that MrpC2 and FruA bind cooperatively between positions −120 and −80, with MrpC2 proximal to the fmgE promoter (Fig. 2B, site 1).
Fig. 4.
MrpC2 and FruA bind to an upstream negative regulatory element. (A) Binding of MrpC2 and FruA to site 1. EMSAs with 32P-labeled fmgE DNA (2 nM) spanning from the indicated 5′ ends to position −80 and His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM), as indicated. The filled arrowhead points to the shifted complex produced by His10-MrpC2 alone, and the open arrowhead points to the complex produced by FruA-His6 alone. (B) Effects of 5′-end deletions on developmental fmgE-lacZ expression. β-Galactosidase specific activity during development was measured for lacZ fused to fmgE spanning from positions −150 to +50 (▴), −120 to +50 (○), or −110 to +50 (■). The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Points show the average values of two or three transformants, and each error bars depicts 1 standard deviation of the data.
To see if negative regulation correlated with MrpC2 and FruA binding to site 1, as depicted in Fig. 2B, DNA fragments with 5′ ends at the −150, −120, or −110 position, and the same 3′ end at +50, were tested for fmgE promoter activity using the lacZ transcriptional fusion vector described above. The two fragments with 5′ ends at −150 or −120 exhibited much lower developmental lacZ expression than the fragment with its 5′ end at position −110 (Fig. 4B). Therefore, the same 5′-end deletion to position −110 that eliminated cooperative binding of His10-MrpC2 and FruA-His6 to site 1 (Fig. 4A) also eliminated or greatly reduced negative regulation (Fig. 4B), correlating the upstream negative regulatory element with site 1.
Sites 1, 2, and 3 exhibit different affinities for MrpC2 and for the combination of MrpC2 and FruA in vitro.
Since the 5′-end deletions tested for fmgE-lacZ expression as shown in Fig. 4B had the same 3′ end at +50 and therefore lacked site 3, we conclude that site 1 functions negatively in the absence of site 3 in vivo. Also, site 3 functions positively in the absence of site 1, since the 3′-end deletions tested for fmgE-lacZ expression, as shown in Fig. 3B, as well as others tested previously (55), had the same 5′ end at position −100 and therefore lacked site 1. To explain how sites 1 and 3 function negatively and positively, respectively, we considered the possibility that site 2 must be occupied by MrpC2 and FruA to activate the fmgE promoter and that sites 1 and 3 independently influence the occupancy of site 2 in different ways. Specifically, because our comparison of His10-MrpC2 and FruA-His6 binding to site 1 (Fig. 4A, lane 3), site 2 (Fig. 2A, lane 3), and site 3 (Fig. 3, lane 9) suggested that site 3 exhibits the highest affinity, followed by site 1 and then site 2, we hypothesized that site 3 acts positively by recruiting MrpC2 and FruA to the fmgE promoter region and that site 1 acts negatively by competing with site 2 for binding of the transcription factors. According to this model, the three sites would be occupied in the order of their affinities for the transcription factors, as depicted in Fig. 5A. We further noted that His10-MrpC2 alone appeared to bind site 3 with the highest affinity (Fig. 3A, lane 7), followed by site 1 (Fig. 4A, lane 1) and then site 2 (Fig. 2A, lane 1), whereas FruA-His6 alone appeared to bind each site with similarly low affinity (compare Fig. 2A, lane 2, Fig. 3A, lane 8, and Fig. 4A, lane 2). To confirm these observations, we compared the binding of His10-MrpC2 alone at different concentrations to sites 1, 2, and 3. As expected, His10-MrpC2 bound to site 3 with the highest affinity, followed by site 1 and then site 2 (Fig. 6A). Likewise, we compared the binding of His10-MrpC2 at different concentrations in combination with FruA-His6 and found the same order of binding affinities for sites 1 to 3 (Fig. 6B). We conclude that the affinities of sites 1, 2, and 3 for the combination of MrpC2 and FruA in vitro are correlated with the affinities of the three sites for MrpC2 alone in vitro and that the affinities are consistent with a model in which site 3 would be occupied first in vivo and act positively by recruiting MrpC2 and FruA to the fmgE promoter region, site 1 would be occupied second and act negatively by competing with site 2 for binding of the transcription factors, and finally, site 2 would be occupied when the transcription factors reach a sufficient concentration, activating transcription of fmgE (Fig. 5A).
Fig. 5.
Models for C-signal-dependent regulation of fmgE and fmgD. As development proceeds, C-signaling causes the concentration of active FruA to rise (triangles). (A) In the fmgE promoter region, three sites for cooperative binding of MrpC2 (light gray ovals) and FruA (dark gray ovals) are occupied in the order of their affinities for the two transcription factors, first site 3, then site 1, and finally site 2, resulting in activation of transcription. (B) In the fmgD promoter region, cooperative binding of two MrpC2 initially represses transcription, but eventually FruA outcompetes the downstream MrpC2 for binding to the upstream MrpC2, activating transcription. Activation of fmgD might occur at a slightly lower concentration of active FruA than activation of fmgE (see Discussion).
Fig. 6.
Relative affinities of MrpC2, and the combination of MrpC2 and FruA, for sites 1, 2, and 3 in the fmgE promoter region. (A) Binding of MrpC2 to sites 1, 2, and 3. EMSAs with 32P-labeled fmgE DNA (2 nM) spanning from positions −147 to −80 (site 1), from positions −100 to −25 (site 2), or from positions −25 to +116 (site 3) and His10-MrpC2 at decreasing concentrations (1, 0.5, 0.25, and 0.125 μM). The asterisk denotes a band observed in lanes 9 to 12, which was also observed in the absence of His10-MrpC2 (data not shown), and therefore appears to be a minor contaminant of this probe preparation rather than a shifted complex. (B) Binding of MrpC2 and FruA to sites 1, 2, and 3. EMSAs with 32P-labeled fmgE DNA and His10-MrpC2 at decreasing concentrations, as shown in panel A, and with FruA-His6 (3 μM).
DISCUSSION
The novel contribution of this study is that multiple cooperative binding sites for MrpC2 and FruA can be used to regulate a gene during M. xanthus development. Not only do the two transcription factors bind immediately upstream of the fmgE promoter, as observed for other fmg genes (25, 29–30), they bind slightly farther upstream to regulate negatively and downstream at about position +100 to regulate positively. It is interesting that fmgE exhibits complex combinatorial control by MrpC2 and FruA. This could be a general characteristic of genes that are induced late during M. xanthus development and depend strongly on C-signaling for expression, since fmgD falls into this category, and its regulation was more complex than those of fmgA and fmgBC, which are expressed earlier and depend only in part on C-signaling. Whereas MrpC2 and FruA bind cooperatively just upstream of the fmgA, fmgBC, and fmgD promoters to activate transcription (25, 29–30), a second MrpC2 binding site partially overlaps both the FruA binding site and the fmgD promoter −35 region (25) (Fig. 5B). The position of the downstream MrpC2 binding site and the effects of mutations on binding in vitro and on promoter activity in vivo suggest that binding of MrpC2 to the downstream site represses fmgD transcription. Derepression presumably depends on C-signal-dependent activation of FruA, which outcompetes the downstream MrpC2 for cooperative binding with the upstream MrpC2. Likewise, we propose that cooperative binding of MrpC2 and FruA to site 2 (Fig. 2B) just upstream of the promoter activates fmgE transcription but that occupancy of site 2 occurs only when C-signaling produces a high enough concentration of active FruA (Fig. 5A). According to our model, site 1 acts negatively by competing with site 2 for cooperative binding of MrpC2 and FruA. Consistent with the model, not only did loss of site 1 (due to a 5′-end deletion to position −100) increase promoter activity, but also activity no longer depended on C-signaling (55). Further, we propose that the high-affinity site 3 acts positively by recruiting MrpC2 and FruA to the vicinity of sites 1 and 2, effectively increasing the local concentration of the two transcription factors as they dynamically bind and are released from many sites in the chromosome.
Our ChIP analysis provides evidence that association of MrpC and/or MrpC2 with the fmgE promoter region depends on FruA (Fig. 1), consistent with the cooperative binding of the two proteins in vivo. Sites 1, 2, and 3 exhibited enhanced binding by the combination of His10-MrpC2 and FruA-His6, relative to the binding of either protein alone in EMSAs (Fig. 2A, 3A, and 4A), a pattern shown previously to be indicative of cooperative binding (29). EMSAs also provided evidence for binding of His10-MrpC2 alone between positions −25 and +50 (Fig. 3A); however, the binding was weak in vitro, and ChIP analysis of a fruA mutant suggested that none of the MrpC2 sites in the fmgE promoter region are occupied appreciably in the absence of FruA, at least at 18 h into development. In contrast, MrpC and/or MrpC2 are associated with the fmgD promoter region in a fruA mutant at 18 h into development, based on a comparable ChIP experiment (25). This difference between fmgD and fmgE supports the notion that the two genes are regulated differently (Fig. 5), despite their similar late induction during development and strong dependence on C-signaling.
The effects of mutations in site 2, just upstream of the fmgE promoter, showed a correlation between the cooperative binding of His10-MrpC2 and FruA-His6 in vitro and fmgE-lacZ expression during development (Fig. 2A). The GGACA-to-TTCAC mutation that increased cooperative binding also increased fmgE-lacZ expression, and the GAACC-to-TCCAA mutation that decreased cooperative binding also decreased expression. These results indicate that cooperative binding of MrpC2 and FruA to site 2 is important for fmgE promoter activity. Interestingly, both mutations increased the binding of His10-MrpC2 alone. A possible explanation is that both mutations create a sequence that matches the (A/G)TTT(C/G)A(A/G) consensus sequence for MrpC2 binding (35) at 5 out of 7 positions (i.e., the GGACA-to-TTCAC mutation creates ACTTCAC from positions −71 to −65, and the GAACC-to-TCCAA mutation creates ACTTGGA at positions −58 to −64 on the opposite strand), but the GAACC-to-TCCAA mutation positions MrpC2 inappropriately for cooperative binding with FruA. The wild-type sequence does not match the consensus noted above, but it does match a second consensus for MrpC2 binding, GTGTC(N8)GACAC (35), at 6 out of 10 positions (i.e., GGGAA(N8)GACAG at positions −81 to −64). Like the GAACC-to-TCCAA mutation, the CATCGTG-to-ACGATGT mutation reduced cooperative binding and promoter activity, and it increased binding of His10-MrpC2 alone, but in this case, two shifted complexes were observed, as if a second MrpC2 binding site was created by the mutation (Fig. 2A). Indeed, the mutation creates part of the sequence ATGTC(N8)GATTC from positions −52 to −35, which matches the second MrpC2 binding consensus sequence noted above at 7 out of 10 positions. If MrpC2 binds to this sequence, it might repress fmgE transcription by blocking the binding of FruA and/or the RNA polymerase, analogous to the proposed role of the downstream MrpC2 binding site in the fmgD promoter region (25) (Fig. 5B). The A-to-C mutation at position −54 did not affect binding of His10-MrpC2 and/or FruA-His6 to site 2 in the fmgE promoter region (Fig. 2A, lanes 13 to 15), yet the mutation increased fmgE-lacZ expression by nearly 3-fold (55). A simple explanation for the discrepancy is that the binding reaction conditions present in vitro do not reflect the conditions present in vivo. The mutation occurs within the sequence CATCG(N5)CGGG from positions −55 to −42, which matches the consensus sequence for binding of the FruA DNA-binding domain, GGG(C/T)(A/G)(N4–6)(C/T)GGG (58), at 6 out of 9 positions, but the mutation does not improve the match to the consensus. Perhaps the mutation alters the interaction of FruA with its site in a subtle way that does not increase affinity but, nevertheless, permits greater activation of transcription. We cannot rule out the possibility that the mutation affects the binding of a protein other than FruA and/or MrpC2; however, there is no evidence for such a protein.
Cooperative binding of MrpC2 and FruA to site 3, located about 100 bp downstream of the fmgE transcriptional start site, appears to upregulate fmgE transcription (55) (Fig. 3). Positive regulation by transcription factors that bind downstream of promoters is unusual in bacteria. Other instances have been described in M. xanthus, but the mechanisms are not understood (28, 58). Downstream positive regulators in other bacteria have been proposed to function by a variety of mechanisms, such as antagonizing a repressor (4), recruiting an activator (48), or directly facilitating productive interaction of the RNA polymerase with the promoter (32, 39, 51). Our results do not exclude any of these potential mechanisms to explain how cooperative binding of MrpC2 and FruA to site 3 might activate fmgE transcription. However, ChIP followed by sequencing (ChIP-seq) indicates that MrpC and/or MrpC2 binds to a large number (>1,700) of sites in the M. xanthus chromosome (M. Robinson, B. Son, and L. Kroos, unpublished data), and site 3 exhibits high affinity for His10-MrpC2 alone or the combination of His10-MrpC2 and FruA-His6 (Fig. 6), so we favor the simple model that site 3 acts positively by increasing the local concentration of the two transcription factors in the vicinity of the fmgE promoter. We note that the complement of the sequence CAGAACT from positions +71 to +77 (i.e., AGTTCTG) matches the (A/G)TTT(C/G)A(A/G) consensus sequence for MrpC2 binding (35) at 5 out of 7 positions. Also, the complement of the sequence GCTG(N4)CACCC from positions +83 to +95 (i.e., GGGTG(N4)CAGC) matches the GGG(C/T)(A/G)(N4–6)(C/T)GGG consensus sequence for the binding of the FruA DNA-binding domain (58) at 6 out of 9 positions, and the loss of 2 bp from the end of this sequence might explain why FruA-His6 failed to bind to DNA from positions −25 to +93 (Fig. 3A).
Another unusual feature of fmgE regulation is the presence of an upstream cis-regulatory element that acts negatively (55), which we found is located about 100 bp upstream of the transcriptional start site and is correlated with site 1 for cooperative binding of MrpC2 and FruA (Fig. 4). Typically, upstream negative regulation involves cooperative binding of a transcription factor to an upstream site and to a site near or within the promoter, forming a repression loop (5, 31). We considered the possibility that MrpC2 bound to site 1 forms a repression loop with MrpC2 bound somewhere between positions −25 and +50 and that C-signal-dependent activation of FruA leads to cooperative binding with MrpC2 at site 1, breaking the repression loop. However, our ChIP analysis does not support this model since MrpC and/or MrpC2 does not appear to be associated with the fmgE promoter region in the absence of FruA (Fig. 1C). Since site 1 exhibits slightly higher affinity for His10-MrpC2 alone or for the combination of His10-MrpC2 and FruA-His6 than site 2 (Fig. 6), we favor the idea that site 1 acts negatively by competing with site 2 for cooperative binding of the two transcription factors. We note that the sequence GGGCG(N4)TCCT from positions −113 to −101 matches the GGG(C/T)(A/G)(N4–6)(C/T)GGG consensus sequence for binding of the FruA DNA-binding domain (58) at 6 out of 9 positions and could account for the loss of cooperative binding of FruA-His6 and His10-MrpC2 to DNA from positions −110 to −80 (Fig. 4A). Also, the sequence GTTTCAG from positions −87 to −81 matches the (A/G)TTT(C/G)A(A/G) consensus sequence for MrpC2 binding (35) at all 7 positions.
Our results demonstrate that multiple cooperative binding sites for MrpC2 and FruA are used to regulate fmgE during M. xanthus development, raising the question of why such elaborate control is observed. Like fmgD, expression of fmgE depends strongly on C-signaling and occurs late during development. However, regulation of fmgD appears to be simpler, involving cooperative binding of two MrpC2 proteins just upstream of the promoter in the off state and FruA outcompeting the downstream MrpC2 to activate the promoter (25) (Fig. 5B). We note two differences between fmgD and fmgE that might explain the more streamlined regulation of fmgD. First, fmgE appears to be a recent addition to the M. xanthus genome (27). Its coding region does not exhibit the strong bias toward usage of guanine or cytosine at the third codon position typical of M. xanthus genes. Also, the overall G+C content of fmgE and its upstream intergenic region is 59% (27), much lower than that of the whole genome (68.9%) (8). In contrast, fmgD exhibits third-codon-position G+C bias typical for M. xanthus coding regions, so it likely has resided longer in the genome than fmgE, perhaps allowing more streamlined regulation of fmgD to evolve. A second difference between fmgD and fmgE is their spatial regulation. fmgD-lacZ is expressed in the outer domain of nascent fruiting bodies (42), where rod-shaped cells become aligned and presumably engage in efficient C-signaling (43). fmgE-lacZ is expressed primarily in the inner domain of fruiting bodies (42), where spores are observed (43), and β-galactosidase specific activity reaches about 8-fold-higher levels in spores than in rod-shaped cells, whereas it reaches only 2-fold higher levels in spores than in rods for fmgD-lacZ (21). Therefore, the more elaborate control of fmgE might ensure that it is expressed predominantly after cellular differentiation. This raises the intriguing possibility that the concentration of active FruA continues to rise in cells that are forming spores. These cells lose their ability to move independently and presumably lose their ability to engage in C-signaling. It has been proposed that sporulating cells are passively transported to the inner domain of nascent fruiting bodies by the circling movements of undifferentiated rod-shaped cells in the outer domain (42). In any case, our finding that fmgE is under combinatorial control of MrpC2 and FruA, and involves cooperative binding to three sites, underscores the importance and versatility of this regulatory mechanism. As proposed for other fmg genes (25, 29–30), positional information through C-signaling and FruA is presumably integrated with starvation sensing and cell death control through MrpC and MrpC2 to govern fmgE expression, and the particular arrangement and affinity of binding sites for the two transcription factors dictate the spatiotemporal pattern of expression.
ACKNOWLEDGMENTS
We are grateful to Sumiko Inouye for providing plasmids, protocols, and antibodies. We thank Kartik Viswanathan and Poorna Viswanathan for constructing the plasmids used in this study, and we thank Sheenu Mittal for technical advice.
This research was supported by NSF grant MCB-0744343 and by the Michigan Agricultural Experiment Station.
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bassler B. L., Losick R. 2006. Bacterially speaking. Cell 125:237–246 [DOI] [PubMed] [Google Scholar]
- 3. Berleman J. E., Kirby J. R. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 33:942–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cowan J. M., Urbanowski M. L., Talmi M., Stauffer G. V. 1993. Regulation of the Salmonella typhimurium metF gene by the MetR protein. J. Bacteriol. 175:5862–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunn T. M., Hahn S., Ogden S., Schleif R. F. 1984. An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. U. S. A. 81:5017–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellehauge E., Norregaard-Madsen M., Sogaard-Andersen L. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807–817 [DOI] [PubMed] [Google Scholar]
- 7. Fisseha M., Gloudemans M., Gill R., Kroos L. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goldman B. S., et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 10. Harris B. Z., Kaiser D., Singer M. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hodgkin J., Kaiser D. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaiser D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45–54 [DOI] [PubMed] [Google Scholar]
- 13. Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaiser D., Robinson M., Kroos L. 2010. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2:a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaplan H. B., Plamann L. 1996. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol. Lett. 139:89–95 [DOI] [PubMed] [Google Scholar]
- 16. Kashefi K., Hartzell P. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthux frzF− defect. Mol. Microbiol. 15:483–494 [DOI] [PubMed] [Google Scholar]
- 17. Kim S. K., Kaiser D. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19–26 [DOI] [PubMed] [Google Scholar]
- 18. Kim S. K., Kaiser D. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926–928 [DOI] [PubMed] [Google Scholar]
- 19. Kroos L., Kaiser D. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840–854 [DOI] [PubMed] [Google Scholar]
- 20. Kroos L., Kuspa A., Kaiser D. 1990. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J. Bacteriol. 172:484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kroos L., Kuspa A., Kaiser D. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252–266 [DOI] [PubMed] [Google Scholar]
- 22. Kuspa A., Kroos L., Kaiser D. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267–276 [DOI] [PubMed] [Google Scholar]
- 23. Kuspa A., Plamann L., Kaiser D. 1992. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 174:3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuspa A., Plamann L., Kaiser D. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360–7369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J.-S., Son B., Viswanathan P., Luethy P. M., Kroos L. 2011. Combinatorial regulation of fmgD by MrpC2 and FruA during Myxococcus xanthus development. J. Bacteriol. 193:1681–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lobedanz S., Sogaard-Andersen L. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loconto J., Viswanathan P., Nowak S. J., Gloudemans M., Kroos L. 2005. Identification of the Ω4406 regulatory region, a developmental promoter of Myxococcus xanthus, and a DNA segment responsible for chromosomal position-dependent inhibition of gene expression. J. Bacteriol. 187:4149–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez-Argudo I., Ruiz-Vazquez R. M., Murillo F. J. 1998. The structure of an ECF-sigma-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol. Microbiol. 30:883–893 [DOI] [PubMed] [Google Scholar]
- 29. Mittal S., Kroos L. 2009. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:1965–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mittal S., Kroos L. 2009. Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development. J. Bacteriol. 191:2753–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mossing M. C., Record M. T., Jr 1986. Upstream operators enhance repression of the lac promoter. Science 233:889–892 [DOI] [PubMed] [Google Scholar]
- 32. Munson G. P., Scott J. R. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391–1402 [DOI] [PubMed] [Google Scholar]
- 33. Nariya H., Inouye M. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55–66 [DOI] [PubMed] [Google Scholar]
- 34. Nariya H., Inouye S. 2005. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol. Microbiol. 58:367–379 [DOI] [PubMed] [Google Scholar]
- 35. Nariya H., Inouye S. 2006. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol. Microbiol. 60:1205–1217 [DOI] [PubMed] [Google Scholar]
- 36. O'Connor K. A., Zusman D. R. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 173:3318–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ogawa M., Fujitani S., Mao X., Inouye S., Komano T. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757–767 [DOI] [PubMed] [Google Scholar]
- 38. Plamann L., Kuspa A., Kaiser D. 1992. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J. Bacteriol. 174:3311–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qi Y., Hulett F. M. 1998. PhoP-P and RNA polymerase σA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187–1197 [DOI] [PubMed] [Google Scholar]
- 40. Rolbetzki A., Ammon M., Jakovljevic V., Konovalova A., Sogaard-Andersen L. 2008. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev. Cell 15:627–634 [DOI] [PubMed] [Google Scholar]
- 41. Rosario C. J., Singer M. 2010. Developmental expression of dnaA is required for sporulation and timing of fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 76:1322–1333 [DOI] [PubMed] [Google Scholar]
- 42. Sager B., Kaiser D. 1993. Spatial restriction of cellular differentiation. Genes Dev. 7:1645–1653 [DOI] [PubMed] [Google Scholar]
- 43. Sager B., Kaiser D. 1993. Two cell-density domains within the Myxococcus xanthus fruiting body. Proc. Natl. Acad. Sci. U. S. A. 90:3690–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sambrook J., Fritsch E., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Shimkets L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525–549 [DOI] [PubMed] [Google Scholar]
- 46. Shimkets L. J., Gill R. E., Kaiser D. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. U. S. A. 80:1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singer M., Kaiser D. 1995. Ectopic production of guanosine penta-and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633–1644 [DOI] [PubMed] [Google Scholar]
- 48. Slominska M., et al. 2003. SeqA-mediated stimulation of a promoter activity by facilitating functions of a transcription activator. Mol. Microbiol. 47:1669–1679 [DOI] [PubMed] [Google Scholar]
- 49. Sogaard-Andersen L., et al. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1–8 [DOI] [PubMed] [Google Scholar]
- 50. Sun H., Shi W. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 183:4786–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szalewska-Palasz A., Wegrzyn A., Blaszczak A., Taylor K., Wegrzyn G. 1998. DnaA-stimulated transcriptional activation of oriλ: Escherichia coli RNA polymerase β subunit as a transcriptional activator contact site. Proc. Natl. Acad. Sci. U. S. A. 95:4241–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ueki T., Inouye S. 2005. Activation of a development-specific gene, dofA, by FruA, an essential transcription factor for development of Myxococcus xanthus. J. Bacteriol. 187:8504–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ueki T., Inouye S. 2005. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J. Biol. Chem. 280:32279–32284 [DOI] [PubMed] [Google Scholar]
- 54. Ueki T., Inouye S. 2003. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:8782–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Viswanathan K., Viswanathan P., Kroos L. 2006. Mutational analysis of the Myxococcus xanthus Ω4406 promoter region reveals an upstream negative regulatory element that mediates C-signal dependence. J. Bacteriol. 188:515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Viswanathan P., Kroos L. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 185:1405–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Viswanathan P., Murphy K., Julien B., Garza A. G., Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 189:3738–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Viswanathan P., Ueki T., Inouye S., Kroos L. 2007. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc. Natl. Acad. Sci. U. S. A. 104:7969–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Whitworth D. E. (ed.). 2008. Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 60. Yoder D., Kroos L. 2004. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J. Bacteriol. 186:661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoder D., Kroos L. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J. Bacteriol. 186:3766–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoder-Himes D., Kroos L. 2006. Regulation of the Myxococcus xanthus C-signal-dependent Ω4400 promoter by the essential developmental protein FruA. J. Bacteriol. 188:5167–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]