Abstract
Geobacillus stearothermophilus T-6 is a thermophilic soil bacterium that has a 38-kb gene cluster for the utilization of arabinan, a branched polysaccharide that is part of the plant cell wall. The bacterium encodes a unique three-component regulatory system (araPST) that includes a sugar-binding lipoprotein (AraP), a histidine sensor kinase (AraS), and a response regulator (AraT) and lies adjacent to an ATP-binding cassette (ABC) arabinose transport system (araEGH). The lipoprotein (AraP) specifically bound arabinose, and gel mobility shift experiments showed that the response regulator, AraT, binds to a 139-bp fragment corresponding to the araE promoter region. Taken together, the results showed that the araPST system appeared to sense extracellular arabinose and to activate a specific ABC transporter for arabinose (AraEGH). The promoter regions of the arabinan utilization genes contain a 14-bp inverted repeat motif resembling an operator site for the arabinose repressor, AraR. AraR was found to bind specifically to these sequences, and binding was efficiently prevented in the presence of arabinose, suggesting that arabinose is the molecular inducer of the arabinan utilization system. The expression of the arabinan utilization genes was reduced in the presence of glucose, indicating that regulation is also mediated via a catabolic repression mechanism. The cluster also encodes a second putative ABC sugar transporter (AbnEFJ) whose sugar-binding lipoprotein (AbnE) was shown to interact specifically with linear and branched arabino-oligosaccharides. The final degradation of the arabino-oligosaccharides is likely carried out by intracellular enzymes, including two α-l-arabinofuranosidases (AbfA and AbfB), a β-l-arabinopyranosidase (Abp), and an arabinanase (AbnB), all of which are encoded in the 38-kb cluster.
INTRODUCTION
The natural degradation of biomass from plants is a key step in the carbon cycle (53, 69, 79). This process is carried out mainly by microorganisms that can be found either free or as part of the digestive system in higher animals (76). The three main polysaccharides in the plant cell wall are cellulose, hemicellulose, and pectin, which are rigidified by lignin, a heterogeneous aromatic polymer (28, 60). Pectin is a complex polysaccharide and may account for up to 30% of the dry weight of the plant cell wall (46). Arabinan is a pectic polysaccharide consisting of a backbone of α-1,5-linked l-arabinofuranosyl units, which are further decorated mainly with α-1,2- and α-1,3-linked arabinofuranosides (46).
Three general strategies are taken by the microbial world for plant cell wall degradation and can be described as follows. Anaerobic bacteria, such as Clostridium spp., have evolved unique multienzyme complexes, named cellulosomes, that integrate many cellulolytic and hemicellulolytic enzymes and mediate both the attachment of the cell to the crystalline polymer and its controlled hydrolysis (9, 16, 23, 65). Aerobic fungi, such as Trichoderma and Aspergillus, secrete a large variety of free cellulases, hemicellulases, and ligninases that work synergistically to completely degrade the polymers into mono- and disaccharides that can also be utilized by the surrounding microorganisms (17). Lastly, aerobic bacteria, such as Bacillus and Cellvibrio spp., secrete only a limited number of endo-type enzymes that degrade the polysaccharide backbone into relatively large oligosaccharides. The final breakdown of these oligosaccharides is carried out by cell-associated or intracellular enzymes (35, 50, 67). The latter strategy, which is presented elegantly in the Geobacillus spp. (67, 68), has an advantage since the extracellular soluble products are not easily available to nonhemicellulolytic competing microorganisms. Although many microbial hemicellulolytic enzymes have been studied extensively (64), the current knowledge of sensing and transporting cellulose, hemicellulose, and pectin degradation products is limited. In Gram-positive bacteria, relatively few ATP-binding cassette (ABC) transporters that are involved in the uptake of hemicellulolytic products have been characterized (19). The ABC transport systems, XynEF and BxlEFG, from G. stearothermophilus (68) and Streptomyces thermoviolaceus (72), respectively, were demonstrated to bind xylo-oligosaccharides. The ABC transporter, AraNPQ, from Bacillus subtilis is involved in the uptake of arabino-oligosaccharides (22). In the anaerobic bacterium Clostridium thermocellum, five sugar ABC transporters were characterized for cellodextrins (CbpB to -D), cellotriose (CbpA), and laminaribiose (Lbp) (52).
Biomass is considered a readily viable option as a renewable energy source for liquid fuel (bioethanol) that does not contribute net CO2 to the atmosphere (4, 21, 55). The two main challenges in converting biomass to liquid fuel are the economical solubilization of cellulose, i.e., converting cellulose to glucose, and the utilization of five-carbon sugars by fermenting microorganisms (21, 30, 40, 56). To engineer fermenting microorganisms capable of utilizing pentoses, it is possible to use pentose-related functions taken from hemicellulolytic microorganisms, such as the various transporters and metabolic enzymes. In this regard, recently the yeast Saccharomyces cerevisiae was engineered to express the cellodextrin transport system from Neurospora crassa. In both saccharification and fermentation experiments, the engineered yeast converted cellulose to ethanol more efficiently than did the yeast lacking this system (25).
Geobacillus stearothermophilus T-6 is a thermophilic aerobic bacterium that possesses an extensive hemicellulolytic system for the utilization of xylan, galactan, and arabinan (27, 67). The bacterium secretes a limited number of extracellular polysaccharide backbone-degrading enzymes (e.g., extracellular xylanase, arabinanase, and galactanase) which partially hydrolyze the main backbone to give relatively large decorated oligosaccharides. The uptake of these oligosaccharides is mediated by specific ABC sugar transporters, and the final breakdown is carried out by intracellular enzymes (3, 12–14, 33, 63, 80).
In this study, we characterized the arabinan and arabinose utilization system in G. stearothermophilus and demonstrated the role of specific sensing, transporting, and regulating elements.
MATERIALS AND METHODS
Bacterial strains and plasmids.
G. stearothermophilus T-6 (NCIMB 40222) was isolated following an enrichment procedure for microbial strains capable of producing alkaline-tolerant, extracellular, thermostable xylanases (39, 66). The Escherichia coli strains used were XL-1 Blue (Stratagene, La Jolla, CA) for general cloning and JM109(DE3) (Promega, Madison, WI) and BL21(DE3) (Novagen, Madison, WI) for expression via the T7 RNA polymerase expression system with either pET11d or pET9d (Novagen). The vector pGEM-T Easy was obtained from Promega.
Growth conditions.
The growth medium for G. stearothermophilus was basic salt medium (BSM) supplemented with 0.5% glucose or arabinose. BSM contained the following per liter: KH2PO4, 0.4 g; MgSO4 · 7H2O, 0.1 g; (NH4)2SO4, 2 g; MOPS (N-morpholinepropanesulfonic acid) buffer (pH 7.0), 10 g; trace element solution, 4 ml. Trace element solution contained the following per liter: CaCl2 · 7H2O, 0.92 g; FeSO4 · 7H2O, 1.51 g; MnSO4 · 4H2O, 0.148 g; ZnSO4 · 7H2O, 0.105 g; CuSO4 · 5H2O, 0.156 g. The pH was adjusted to 2.0 with sulfuric acid.
DNA and RNA isolation and manipulation.
G. stearothermophilus T-6 genomic DNA was isolated by the procedure of Marmur (44) as outlined by Johnson (38). Plasmid DNA was purified using the DNA Clean-Up System (Promega). DNA was manipulated by standard procedures (59). Total RNA was isolated with the RNeasy kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol.
Genome sequencing of G. stearothermophilus.
The de novo whole-genome sequencing of G. stearothermophilus T-6 was carried out using the 454 Life Sciences Corp. technology (43) by DYN LABS Ltd. (Caesarea, Israel). Sequence homologies were searched with the FASTA (54) and BLAST (5) algorithms. Protein sequences were analyzed with the Expert Protein Analysis System (ExPASY) proteomics server (http://www.expasy.org) of the Swiss Institute of Bioinformatics.
Expression and extraction of G. stearothermophilus T-6 AraR.
The araR gene was cloned via PCR (Table 1) into T7 expression vector pET11d to give pET11d-araR. Expression of araR was carried out by growing, in 2-liter shake flasks shaken at 230 rpm at 37°C, 200-ml cultures of E. coli JM109(DE3)(pLysS) carrying pET11d-araR in terrific broth (59) supplemented with chloramphenicol (25 μg/ml) and carbenicillin (50 μg/ml). Induction by 4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was carried out at a cell turbidity at 600 nm of 0.6 unit. After 3 h of incubation, the cells were harvested; resuspended in 20 ml of a solution containing 50 mM Tris-Cl (pH 7.5), 100 mM KCl, 10% glycerol, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol; and disrupted by a single passage through a French press (Spectronic Instruments, Inc., Rochester, NY). Following centrifugation of the cell extract (14,000 × g for 15 min), the soluble fraction was used for gel retardation assays.
Table 1.
Oligonucleotides used in this study
| Application and primer | Sequence (5′–3′)a |
|---|---|
| Cloning into T7 expression vectors | |
| araP N-ter | CAGAATCCATGGCGCATCACCATCACCATCACTATGCCTACTATTATCAACAA |
| araP C-ter | TGATCAGGATCCTCATTTGGCATAAAAATGGTCGAC |
| araT N-ter | GTTCTCAAATCCATGGATCATCATCATCATCATCATTGGAAGGTGCTGATTGC |
| araT C-ter | GGTGCTTGGATCCTCAATCTGTATATTCACTGATC |
| araR N-ter | GGAATTCAAGCTTCATGAAAGAGAAAACGCTG |
| araR C-ter | CGTCTAGACTGCAGGGATCCTATTAAATTTCCTTGTGTGA |
| Primer extension analysis | |
| araD | GCAAGTTGGCCTCTAATACG |
| araR | CATCCGGCTTCATTTGTC |
| 5′ RACE analysis | |
| abnE_GSP1 | GGAATTCACATAAGCGCGAAGTTCTCCG |
| abnE_GSP2 | CGCATCTGTCGGCAATGGCAATG |
| araE_GSP2 | GGGACTTGGTGCAAACGGCGGAAAAG |
| araE_GSP1 | CGACTACGTCCTCAGCATATTGC |
| Real-time RT-PCR analysis | |
| Fwr abfA | ATTGCCCACGGCTACAAAGA |
| Rev abfA | CACGGACCGTCCATCTCATT |
| Fwr araP | ATTATTGGAAACGGTGCCTCAA |
| Rev araP | TTAATGATTCCGCCGCATCT |
| Fwr araJ | ATCCGCATGAACCCTAAGCTT |
| Rev araJ | CTGGTCGAATGGACGCAATA |
| Fwr araN | GCTGTCCGCCGAACTTAGC |
| Rev araN | TGCGTCCGATGTTTGCAA |
| Fwr 16sRNA | AAGTGCGCGTCATTTCG |
| Rev 16sRNA | CACCGTGACTTCAATCG |
| DNA-binding assays | |
| 1IR_araD | TTATAGAAAAATTGTACGTACAATAGTATAAT |
| 2IR_araD | TTATTATACTATTGTACGTACAATTTTTCTAT |
| 1IR_abnE | TTTTGACATGTACGAACAATTAGATTAATATG |
| 2IR_abnE | TTCATATTAATCTAATTGTTCGTACATGTCAA |
| ProaraE2 | GAATCAATATCGAAGAATTCGCATGATCAGTGAATATAC |
| ProaraE3 | CTATATCATCGAAGGATCCGTTCGTCTTTTCAA |
Boldface bases indicate engineered restriction sites.
Production and purification of His6-tagged AraP, AbnE, and AraT.
The AraP, AbnE, and AraT open reading frames were all cloned into the pET9d vector (Novagen), yielding plasmids pET9d-araP, -abnE, and -araT, respectively. The lipoproteins AraP and AbnE were cloned without their 21 and 16 N-terminal lipoprotein coding sequences (CDSs), respectively. All the primers were designed to allow in-frame cloning of the genes into the T7 polymerase expression vector using an NcoI restriction site at the 5′ terminus and a BamHI restriction site at the 3′ terminus (Table 1). The N-terminal primers contained six histidine codons to provide His6-fused products. For protein production, E. coli BL21(DE3) cultures containing the appropriate vector (pET9d-araP, -abnE, or -araT) were grown overnight in terrific broth (59) with kanamycin (25 μg/ml) (0.5 liter in 2-liter baffled shake flasks shaken at 230 rpm at 37°C) to a final turbidity at 600 nm of 15 to 20 units. The cultures were harvested, resuspended in 30 ml of buffer (20 mM imidazole, 20 mM phosphate buffer, 500 mM NaCl, pH 7.0), disrupted by two passages through a French press, and centrifuged (14,000 × g for 15 min) to obtain soluble extracts. The His-tagged, fused proteins were isolated using a 5-ml Histrap column (GE Health Care) and mounted on an AKTAexplorer fast protein liquid chromatography system (GE Health Care) according to the manufacturer's instructions. Proteins were stored at −80°C before use. In the case of the purified response regulator AraT, the protein was dialyzed overnight against 2 liters of buffer containing 50 mM Tris-HCl (pH 7.0) and 100 mM KCl, followed by the addition of EDTA and glycerol to final concentrations of 1 mM and 10%, respectively. The protein was stored at −80°C in 100-μl aliquots.
Transcriptional analyses.
Transcriptional analyses were performed on total RNA extracted from exponentially growing cells. Northern blot analysis was conducted as described by Moran (47). Primer extension reactions were carried out as described previously (47), with avian myeloblastosis virus reverse transcriptase (Promega), 40 μg of total RNA, and the primers listed in Table 1. The rapid amplification of cDNA ends (RACE) technique was used to amplify portions of the putative transcripts at their 5′ ends (Clontech, Mountain View, CA). Briefly, reverse transcription (RT) was generated using random hexamers with a reverse transcriptase that exhibits terminal transferase activity. The cDNA was amplified with universal primers (UPM), as well as specific primers antiparallel and complementary to a sequence downstream of the araE and abnE genes (Table 1). When required, the PCR products were first cloned into the pGEM vector, electrotransformed into E. coli XL1-Blue, and subsequently sequenced.
Real-time RT-PCR analysis.
RT of RNA was performed with the Verso cDNA kit by following the manufacturer's protocol (Thermo Fisher Scientific), with 1 μg of total RNA and random hexamers as primers. To check for DNA contamination, control reactions were carried out in the absence of reverse transcriptase. Real-time RT-PCR primers for the abfA, araJ, araM, araP, and 16S rRNA genes were designed with the aid of the Primers Express 2.0 software (Applied Biosystems) (Table 1). Gene relative quantification was performed with the Applied Biosystems 7300 real-time PCR system. Each 20-μl reaction mixture included template cDNA, 300 nM each primer, and Power SYBR green PCR Master Mix (Applied Biosystems). cDNA was amplified with two primers for each gene using Power SYBR green PCR master mix (Applied Biosystems) and the Applied Biosystems 7300 real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. The amplification conditions for all reactions were 1 cycle of 95°C for 15 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 15 s. Melt curves were analyzed to ensure specificity of primer annealing and lack of primer secondary structure. Data analysis was carried out with the 7300 system software (Applied Biosystems) using 16S rRNA for normalization.
Mobility shift DNA-binding assays.
Three unique radioactive DNA probes were used for gel retardation assays. For His6-AraT, a 139-bp DNA probe (from position −162 to position −23 relative to the transcriptional start site of the araE gene) was generated via PCR with the primers listed in Table 1. This PCR product was purified with a Wizard SV gel PCR cleanup system (Promega), double digested with EcoRI and BamHI, purified again, and end labeled using [α-32P]dATP with Klenow fragment (Fermentas) as described by the manufacturer. For binding analysis with AraR, two double-stranded probes (corresponding to the araD and abnE promoter regions) were composed of two synthetic complementary oligonucleotides (Table 1). The double-stranded probes were composed of two noncomplementary T nucleotides at the 5′ end for end labeling with Klenow fragment (Fermentas) in the presence of [α-32P]dATP. The binding reaction mixture (30-μl total volume) contained 20 μl of a solution containing 50 mM Tris-Cl (pH 7.5), 100 mM KCl, 10% glycerol, 1 mM EDTA, 2 μg of salmon sperm DNA, 0.66 mM dithiothreitol, 33 μg of bovine serum albumin, and 0.08 ng of labeled probe and the indicated concentration of protein. The binding mixture was incubated for 30 min at 45°C and then separated on a 6.6% nondenaturing polyacrylamide gel prepared in Tris-borate-EDTA buffer (59) and run for 1 h. Gels were dried under vacuum and exposed to a phosphorimager screen before analysis with a Fuji BAS2000 phosphorimager.
Preparation of branched arabino-oligosaccharides.
Arabino-oligosaccharides branched with α-l-arabinofuranosyl residues at the C-2 and/or C-3 positions were prepared as follows. Branched sugar beet arabinan (1%, wt/vol, aqueous solution; Megazyme, Wicklow, Ireland) was partially digested with 0.1 mg/ml recombinant extracellular endo-α-1,5-arabinanase (AbnA) from G. stearothermophilus T-6 and incubated for 30 min at 60°C. The enzyme was inactivated by boiling the reaction mixture for 10 min. Following centrifugation (14,000 × g for 10 min) and lyophilization, the resulting soluble products included a wide range of branched oligosaccharides. These sugars were separated using a BioGel P-2 (Bio-Rad, Richmond, CA) gel filtration column (100 by 2 cm) running with water at room temperature. The high-molecular-weight pectic side chain was eluted in the void volume. Fractions (1.5 ml) were collected and detected by thin-layer chromatography analysis using ethyl acetate-acetic acid-water (2:1:1) as the running solvent. To determine the average size of the various saccharides, solutions of 0.25 to 5 mg/ml of selected sugar fractions were treated with 45 and 17.5 μg/ml recombinant arabinofuranosidases AbfA and AbfB, respectively, and 12 μg/ml intracellular α-1,5-arabinanase AbnB, all from G. stearothermophilus T-6, for 48 h at 60°C until they were fully hydrolyzed into arabinose. The increase in reducing power after hydrolysis was measured using the bicinchoninic acid assay for reducing sugars (18), and the values were used to calculate the average size of the arabino-oligosaccharides in each fraction. The size range of the eluted sugars was 2 to 14 arabinose units.
Microcalorimetry titration studies.
Titration calorimetry measurements were performed with a VP-ITC calorimeter (MicroCal, Northampton, MA) as described by Wiseman et al. (78). Protein solutions for isothermal titration calorimetry (ITC) were dialyzed extensively overnight against a buffer containing 50 mM Tris-HCl (pH 7.0), 100 mM NaCl, and 0.02% NaN3. Ligand solutions of arabinobiose (A2), arabinotriose (A3), arabinotetraose (A4), arabinopentaose (A5), arabinohexaose (A6) (Megazyme, Wicklow, Ireland), and branched arabino-oligosaccharides composed of 2 to 14 arabinose units were prepared by dilution with the actual protein dialysis buffer. Aliquots (10 μl) of the ligand solution at 8.5 to 20 times the molar concentration of the binding site were added to the reaction cell containing 1.41 ml of 0.02 to 0.1 mM protein solution by the controlled action of a 250-μl rotating stirrer-syringe. The heat of dilution was determined to be negligible in separate titrations of the ligand into the buffer solution. Calorimetric data analysis was carried out with the ORIGIN 7.0 software (MicroCal). Binding parameters, including the number of binding sites (n), the binding constant (KB, [M−1]), and the binding enthalpy (ΔHB, [kcal/mol]) of bound ligand, were determined by fitting the experimental binding isotherms. KB was determined by the slope of the isotherm at the equivalence point (20).
Nucleotide sequence accession number.
The 77,747-bp sequence containing the xylan, xylose, arabinan, and arabinose utilization region from G. stearothermophilus T-6 has been deposited in the GenBank under accession number DQ868502.
RESULTS
The genome of G. stearothermophilus T-6 has been sequenced using the 454 Life Sciences technology, resulting in a genome coverage of 40-fold. The uncompleted draft genome includes 3,388,716 bases with a coding capacity of 82.6%, providing 3,269 predicted CDSs. The resulting sequence revealed a 38-kb gene cluster that we postulated to encode enzymes for the utilization of l-arabinan and arabinose and that is adjacent to a 40-kb segment for the utilization of xylan (67, 68). The arabinan utilization cluster contains a potential three-component sensing system (araPST), a putative ABC transport system (araEGH), an apparent repressor (araR), putative l-arabinose utilization genes (araDBA-abp-abnB), and the l-arabinan utilization genes (abnEFJ-abnA-abfBA-araJKLMN) (Fig. 1). Based on the locations of potential transcription terminators, the genes appeared to be organized into five transcriptional units. Table 2 lists the genes and their proposed biological functions.
Fig. 1.
Genetic map of the 38-kb segment containing the arabinose and arabinan utilization genes. The letter P indicates the proposed promoter site, and Ω indicates the position of the putative rho-independent transcription termination site. DNA probes for the Northern blot analyses are shown as open boxes. Note that the abp gene is interrupted by an ∼1.4-bp insertion element, IS5377.
Table 2.
The arabinan utilization genes in G. stearothermophilus T-6
| Category and gene | Proposed biological function |
|---|---|
| Glycoside hydrolases | |
| abp | β-l-Arabinopyranosidase (GH27)a |
| abnA | Extracellular arabinanase (GH43) |
| abnB | Intracellular arabinanase (GH43) |
| abfB | α-l-Arabinofuranosidase (GH51) |
| abfA | α-l-Arabinofuranosidase (GH51) |
| Arabinose-sensing system | |
| araP | Arabinose sensor protein |
| araS | Histidine protein kinase |
| araT | Response regulator |
| Sugar metabolism | |
| araJ | Putative oxidoreductase |
| araK | Aldose 1-epimerase |
| araL | Putative sugar phosphatase |
| araM | Putative glycerol-1-phosphate dehydrogenase |
| araN | Hypothetical protein |
| araD | l-Rribulose-5-phosphate 4-epimerase |
| araB | Ribulose kinase |
| araA | l-Arabinose isomerase |
| Sugar ABC transporters | |
| araE | Arabinose-binding protein |
| araG | Arabinose transport ATP binding |
| araH | Arabinose membrane permease |
| abnE | Arabino-oligosaccharide-binding protein |
| abnF | ABC transport permease |
| abnJ | ABC transport permease |
| Specific regulator araR | Master repressor of the arabinose/arabinan utilization genes |
GH, glycoside hydrolase family.
Sequence analysis of the araPST and araEGH operons.
Based on sequence analysis, the araP gene product shows 34% identity (over a 284-residue span) to the periplasmic glucose-binding protein from Thermotoga maritima (70). The putative araS gene product exhibits features characteristic of bacterial histidine kinase proteins, including two transmembrane helices (TM1 residues 15 to 37 and TM2 residues 219 to 312) flanking an extracellular domain (residues 38 to 289) and a conserved C-terminal cytoplasmic region containing the ATP-binding kinase domain. In addition, AraS contains a motif designated CaChe (Ca transport and chemotaxis, residues 151 to 241) (6). Downstream of araS lies araT, which encodes a protein with strong similarity to response regulators with a predicted two-domain architecture and an N-terminal signal receiver domain linked to a C-terminal effector domain (58). The N-terminal signal receiver domain (residues 7 to 122) shares homology with the CheY superfamily (74), whereas the C-terminal domain, from position 351 to position 395, contains a putative helix-turn-helix motif that resembles the AraC-type DNA-binding domain (26). The intergenic spacer regions between the araPST genes lack any obvious promoter or terminator, suggesting that the araPST cluster constitutes a polycistronic operon. Based on the above analysis, we conclude that araPST constitutes an operon whose products might act together to respond to arabinose availability. The araPST operon is positioned just upstream of a putative ABC transport system (araEGH). A similar arrangement is found in G. stearothermophilus T-6 for the xylotriose ABC transporter, which is adjacent to, and regulated by, a two-component system (68), suggesting that the putative araPST operon regulates the expression of the ABC transport system (araEGH).
AraE appears to correspond to an extracellular sugar-binding protein with features characteristic of signal peptides of bacterial lipoproteins. Its N-terminal region contains a putative cleavage site with an almost perfect match to the consensus cleavage site sequence for the lipoprotein signal peptidase II (Leu-Ala-Gly/Ala↓Cys) (75). araG encodes a putative ATP-binding protein involved in energy coupling of the sugar uptake system. Hydropathy analyses of AraH show patterns of hydrophobic and hydrophilic regions that suggest that this protein is likely to have membrane-spanning regions and 10 transmembrane helices. AraH shows homology to several integral cytoplasmic membrane proteins involved in sugar transport and contains a conserved hydrophilic segment with the consensus sequence EAAX3GX9IXLP; this sequence is typical of integral membrane proteins from binding protein-dependent transport systems (32).
AraP is an arabinose-binding protein.
Based on sequence homology, araP encodes a sugar-binding protein. In Gram-negative bacteria, substrate-binding proteins are located in the periplasmic space, whereas in Gram-positive bacteria, they were found to be linked to the cytoplasmic membrane by a lipid anchor (29, 73). The ability of AraP to bind arabinose or arabino-oligosaccharides was demonstrated using ITC. ITC provides a direct measurement of binding enthalpy, ΔHB, and allows the simultaneous determination of the binding parameters, including the binding constant (KB), entropy (ΔSB), the free energy of binding (ΔGB), and the binding stoichiometry (n). ITC measurements indicated that AraP binds strongly to arabinose (KD = 0.6 μM at 30°C) (Fig. 2), with enthalpy and entropy values of ΔHB = −7.7 ± 0.1 kcal/mol and TΔSB = 0.9 kcal/mol. The titration curve fit very well into a single binding model with a calculated n (stoichiometry) of 1. AraP failed to interact with glucose, galactose, xylose, or arabino-oligosaccharides (data not shown). Thus, the effector of the three-component sensing system, AraPST, appears to be arabinose.
Fig. 2.
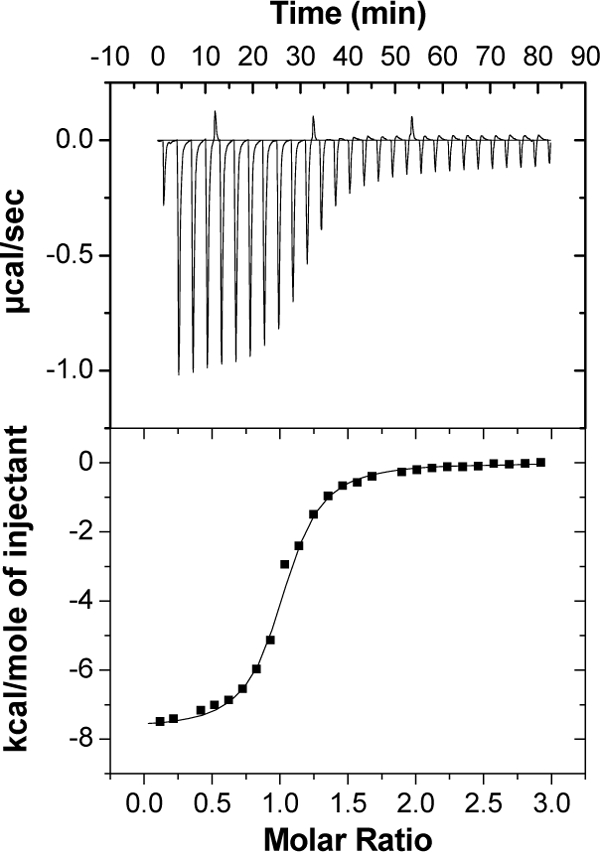
Representative isothermal calorimetric titration curve of AraP with arabinose. Titration calorimetry measurements were performed with a MicroCal VP-ITC titration calorimeter at 30°C. The top panel shows the calorimetric titration of AraP with arabinose, and the bottom panel displays the integrated injection heats from the upper panel, corrected for control dilution heats. The solid line is the curve of best fit that was used to derive the binding parameters.
Mapping of the 5′ end of the araEGH transcript.
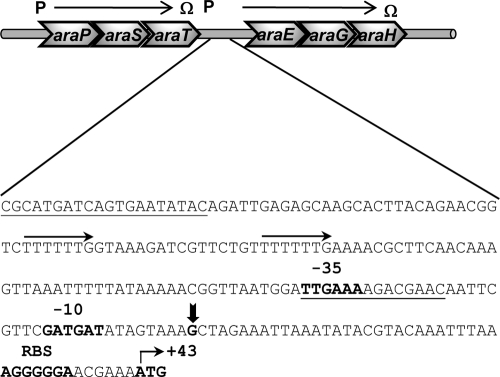
The genetic context of the two operons araPST and araEGH suggests that AraT is a response regulator that regulates the expression of the AraEGH ABC sugar transporter. To identify the 5′ end of the araEGH transcript, we utilized 5′ RACE analysis. The RACE products were cloned into pGEM, introduced into E. coli XL1-Blue, and subsequently sequenced. Eight independent clones were sequenced, and the apparent transcriptional start point, corresponding to a G residue, was the same for all clones (Fig. 3). The −35 sequence (TTGAAA) is a near match to the σA consensus sequence TTGACA and is separated by 17 bp from the potential −10 region (GATGAT), which differs from the B. subtilis consensus, TATAAT, by two nucleotides (48) (Fig. 3). The araE promoter region contains two direct repeats (TTTTTTG) separated by 16 bp, which may function as the recognition sequences for the response regulator AraT (Fig. 3).
Fig. 3.
Mapping of the 5′ end of the araEGH transcript by 5′ RACE analysis. Total RNA was isolated from mid-exponential-phase cultures of G. stearothermophilus T-6 grown on minimal media supplemented with 0.5% arabinose as the sole carbon source. Vertical arrow indicates the transcriptional start point (+1). The potential directed repeat binding sites for the response regulator AraT are indicated by horizontal arrows. The −35 and −10 regions, the ribosome-binding site (RBS), and the ATG initiating codon are in bold. The sequences of the primers used to synthesize the PCR product for the gel mobility shift assay (Fig. 4) with AraT are underlined.
AraT binds to the araE promoter region. To test whether AraT, the putative response regulator, can bind to the araE promoter region, gel mobility shift assays were performed. The araT gene was fused to a His6 sequence, and the gene product was purified from E. coli by Ni+-nitrilotriacetic acid chromatography. Based on SDS-PAGE analysis, the purity of the protein was over 95%. A 139-bp DNA fragment corresponding to positions −163 to −24 with respect to the araE transcriptional start site (+1) was used as a probe for testing of AraT-DNA interaction (Fig. 3). Gel mobility shift assays indicated that the purified His6-AraT protein binds the araE promoter region; a complete shift was seen in the presence of 0.6 μM AraT (Fig. 4). This relatively high concentration of AraT required for the shift may reflect the fact that efficient binding of AraT requires its phosphorylation mediated by AraS (the histidine kinase) (1). The binding appears to be specific, since AraT did not shift an unrelated 220-bp fragment containing the xynE promoter region (Fig. 4, lanes 6 to 8).
Fig. 4.

Gel mobility shift assay for binding of AraT to the araE promoter region. His6-AraT and a radioactively labeled 139-bp DNA fragment containing the araE promoter were incubated as described in Materials and Methods. Lane 1 contains no protein. Lanes 2 to 5 contain increasing concentrations of purified AraT. Lanes 6 to 8 contain the irrelevant promoter of xynE.
Sequence analysis of the l-arabinose utilization genes (araDBA-abp-abnB).
Within the 38-kb chromosomal segment, we identified a cluster of five genes that are likely to encode the l-arabinose catabolic genes (araD, araB, and araA), a β-l-arabinopyranosidase gene (abp), and an intracellular arabinanase gene (abnB). The genes araA, araB, and araD encode the first three enzymes of l-arabinose catabolism: l-arabinose isomerase, l-ribulose kinase, and l-ribulose epimerase, respectively. The gene products convert arabinose to xylulose 5-phosphate, which can enter the pentose phosphate cycle. The abp gene product belongs to glycoside hydrolase family 27 (GH27). In strain T-6 the abp gene is interrupted by the insertion sequence (IS) IS5377. We have cloned the gene without the insertion sequence, and preliminary results revealed that Abp is active toward p-nitrophenyl-β-l-arabinopyranoside, suggesting that the enzyme is an arabinopyranosidase. A homologous enzyme from Streptomyces avermitilis was recently characterized (34). The abnB gene encodes a 315-amino-acid protein that is a member of the intracellular inverting GH family 43 arabinanases. Previous viscosity and reducing power measurements, together with product analysis for the hydrolysis of linear arabinan, indicated that the enzyme works in an endo mode of action (3). ITC studies of a catalytic mutant with various arabino-oligosaccharides suggested that the enzyme active site can accommodate at least five arabinose units (3).
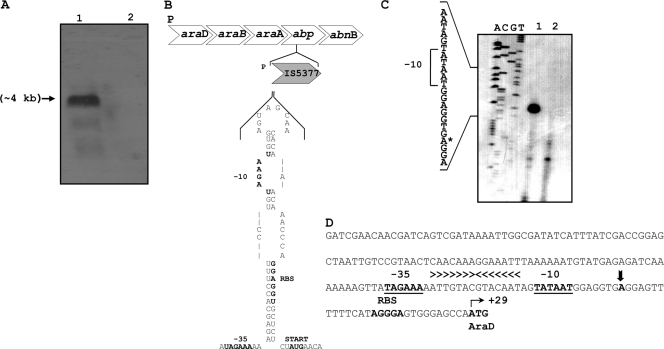
Transcriptional analyses of the araDBA-abp-abnB operon.
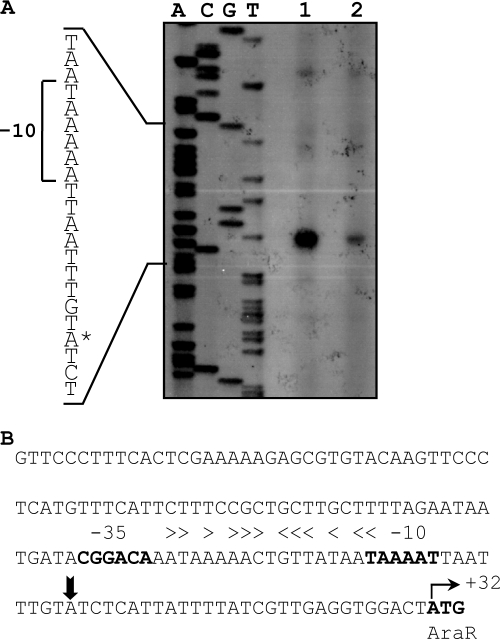
The genetic organization of araDBA-abp-abnB suggests that the five genes constitute an operon. To determine the transcript size, Northern blot analysis was performed. Total RNA was isolated from T-6 cultures grown in the presence or absence of arabinose (a potential inducer) and was hybridized to an araD DNA probe. Northern blot analysis indicated that both probes hybridized to an RNA 4 kb in length (Fig. 5A). The obtained mRNA size matched the expected size up to the transcriptional termination signal, which is formed by a stem-loop structure within the −10 region and the Shine-Dalgarno site in IS5377 (Fig. 5B). This secondary structure is formed only if the transcript begins outside the native promoter of IS5377, thus allowing control of the frequency of transposition (71). The araDBA transcript was not detected in cultures grown without arabinose (Fig. 5A), suggesting that arabinose is the molecular inducer of the araDBA-abp-abnB operon. Using primer extension analysis, the apparent transcriptional start point of the araD operon was assigned to an A nucleotide 28 bases upstream from the initiation ATG codon of araD (Fig. 5C). The potential −35 region (TAGAAA), with four of six bases matching the σA consensus, is separated by 17 bp from the potential −10 region (TATAAT), which matches the σA consensus perfectly (37, 48). A perfect 14-bp inverted repeat (5′-ATTGTACGTACAAT-3′) resembling the araR operator sites from Bacillus subtilis (49) is located between the −35 and −10 regions of the promoter.
Fig. 5.
Transcriptional analyses of the araDBA-abp-abnB transcript. (A) Northern blot analysis of the araDBA-abp-abnB transcript. Total RNA was extracted from mid-exponential-phase cultures of G. stearothermophilus T-6 grown in BSM supplemented with 0.5% arabinose (lane 1) or with 0.5% glucose (lane 2) as the sole carbon source. The RNA was subjected to electrophoresis, transferred to nitrocellulose, and annealed to a 687-bp 32P-labeled DNA probe for araD. (B) Schematic description of the possible stem-loop structure found in the untranslated region of IS5377. The predicted −35 and −10 sequences and the ribosome-binding site of IS5377 are in bold. (C) Mapping of the 5′ termini of the araDBA-abp-abnB transcript by primer extension analysis. Extension products resulting from RNA obtained from cultures grown on 0.5% arabinose (lane 1) or with 0.5% glucose (lane 2) as the carbon source are shown. Dideoxynucleotide sequencing reactions were carried out with the same primer used for the RT reactions. The position of the transcriptional start point is indicated by an asterisk on the inferred nontemplate strand sequence. (D) Sequence of the araD regulatory region. The transcriptional start point (+1) is indicated by a vertical arrowhead. The −35 and −10 regions for σA binding, the ribosome-binding site (RBS), and the ATG initiating codon are in boldface letters. The araR operator is indicated by horizontal arrowheads above the inverted repeat sequence.
Sequence analysis of the l-arabinan utilization genes (abnEFJ-abnA-abfBA-araJKLMN).
Following the araDBA-abp-abnB operon, there is a cluster of 11 genes, abnEFJ-abnA-abfBA-araJKLMN, that appear to be expressed as a single transcription unit, based on the absence of apparent transcription terminators between abnE and araN. AbnE shows homology to bacterial extracellular sugar-binding proteins and contains the typical characteristic features of signal peptides of bacterial lipoproteins. AbnF and AbnJ are likely to be the transmembrane proteins forming the transporter. Hydrophobic-moment analysis of AbnF and AbnJ predicted six transmembrane helices. The two proteins contain a conserved hydrophilic segment with the consensus sequence EAAX3GX9IXLP; this sequence is typical of integral membrane proteins from binding protein-dependent permeases (32). This subcluster contains proteins involved in the transport of short sugars and oligosaccharides. The gene for the ATP-binding protein is presumably located elsewhere on the chromosome. The abnA gene encodes an 848-residue protein with a calculated molecular mass of 93 kDa and shows high similarity to extracellular arabinanases of glycoside hydrolase family 43. AbnA hydrolyzes native substitute arabinan (sugar beet arabinan) and acts as an endoarabinanase (2). The cluster contains two adjacent genes for α-l-arabinofuranosidases, abfA and abfB, both of which belong to the retaining GH-51 family (33, 62). Sequence homology suggests that the last five genes of the cluster encode an NAD(P) sugar-dehydrogenase (araJ), aldose-1-epimerase (araK), a sugar-phosphatase (araL), an NAD(P)-dependent glycerol-1-dehydrogenase (araM), and a hypothetical protein (araN). It is likely that the araJKLMN cluster constitutes a novel, alternative pathway for the utilization of pentoses in G. stearothermophilus.
Transcriptional analysis of the abnEFJ-abnA-abfBA-araJKLMN operon.
The genes abnEFJ-abnA-abfBA-araJKLMN are all transcribed in the same direction and potentially constitute a polycistronic operon. To determine whether the abnEFJ-abnA-abfBA-araJKLMN cluster constitutes a single transcriptional unit, Northern blot analysis was performed. Total RNA was isolated from T-6 cultures grown in the presence or absence of arabinose (a potential inducer) and was annealed to a DNA probe for abfA. The probe hybridized to high-molecular-weight RNAs with a maximum length of over 15 kb (data not shown). No hybridization with mRNA from cultures grown without arabinose was detected, suggesting that arabinose can function as a molecular inducer. Using 5′ RACE analysis, the apparent transcriptional start point of the abnE promoter was identified and assigned to an A nucleotide 132 bases upstream from the ATG initiation codon of abnE (Fig. 6). The apparent −35 sequence, TTGACA, matches the σA consensus perfectly (37, 48) and is separated by 17 bp from the potential −10 region (TAATAT), which differs by two nucleotides from the σA consensus sequence, TATAAT (31, 37, 48). The promoter region contains an inverted repeat (5′-TGTACGAACA-3′) that can potentially function as the AraR repressor-binding site (Fig. 6). A potential operator site for catabolite-responsive regulation was identified and is located at positions +5 to +19 with respect to the transcription start point (Fig. 6).
Fig. 6.
Mapping of the 5′ end of the abnEFJ-abnA-abfBA-araJKLMN transcript by 5′ RACE analysis. Total RNA was isolated from mid-exponential-phase cultures of G. stearothermophilus T-6 grown on minimal medium supplemented with 0.5% arabinose as the sole carbon source. The transcriptional start point (+1) is indicated by a vertical arrowhead. The −35 and −10 regions for σA binding, the ribosome-binding site (RBS), the ATG initiating codon, and a putative catabolite-responsive element (cre) site for CcpA are in boldface letters. The consensus cre site is TGWAAANC|GNTNWTCA (W = A or T), where underlined letters represent the most critical bases, N is any base, and the vertical line denotes an axis of symmetry (45). The araR operator is indicated by horizontal arrowheads above the inverted repeat sequence.
AbnE is an arabino-oligosaccharide-binding protein.
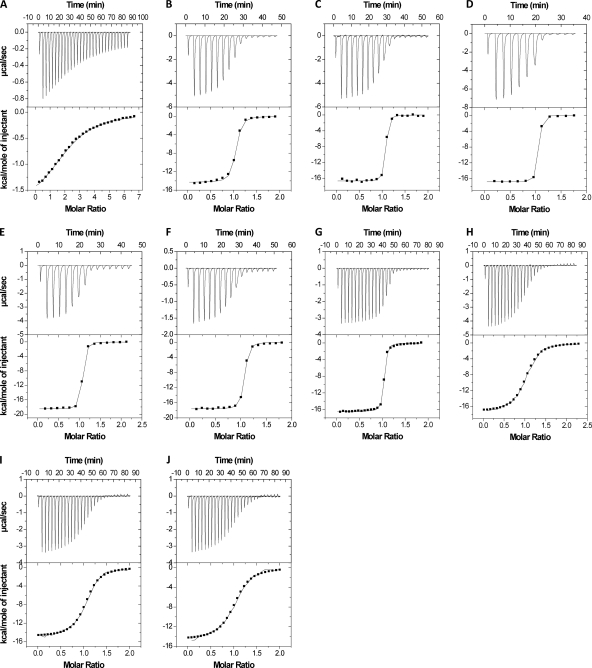
The abnEFJ genes are likely to code for a sugar ABC transport system. The ability of AbnE to bind linear and branched arabino-oligosaccharides was demonstrated using ITC (Fig. 7). The thermodynamic parameters of binding are summarized in Table 3. In general, most of the binding interactions were enthalpy driven and the titration curves fit very well into a single binding site model with a calculated n of 1. AbnE interacts with linear arabino-oligosaccharides (A3-8) with binding constants (KD) in the nanomolar range and with branched arabino-oligosaccharides in the micromolar range (Table 3).
Fig. 7.
Representative ITC curves of the interactions of various linear and branched arabino-oligosaccharides with AbnE. All ITC experiments were performed at 30°C. Shown are the ITC curves of AbnE with the following sugars: arabinobiose (A), arabinotriose (B), arabinotetraose (C), arabinopentaose (D), arabinohexaose (E), arabinoheptaose (F), arabinooctaose (G), and branched arabino-oligosaccharides A5, A7, and A8 (H, I, and J, respectively). The top half of each panel shows the calorimetric titration of AbnE with ligand, and the lower half displays the integrated injection heats from the upper half.
Table 3.
Binding of AbnE to linear and branched arabino-oligosaccharides: thermodynamic parameters, and dissociation constantsa
| Sugar | KB × 106 (M−1) | KD (1/KB, μM) | ΔHB (kcal/mol) | TΔSB (kcal/mol) | ΔGB (kcal/mol) |
|---|---|---|---|---|---|
| A2 | 0.021 ± 0.001 | 48 | −1.9 ± 0.0 | 4.1 | −6.0 |
| A3 | 4.5 ± 0.5 | 0.22 | −14.3 ± 0.1 | −5.1 | −9.2 |
| A4 | 23.6 ± 5.6 | 0.04 | −16.7 ± 0.1 | −6.5 | −10.2 |
| A5 | 43.4 ± 9.2 | 0.02 | −16.8 ± 0.1 | −6.2 | −10.6 |
| A6 | 28.3 ± 4.3 | 0.04 | −18.5 ± 0.1 | −8.2 | −10.3 |
| A7 | 44.5 ± 6.2 | 0.02 | −17.6 ± 0.1 | −7.0 | −10.6 |
| A8 | 18.3 ± 2.2 | 0.05 | −16.5 ± 0.1 | −6.4 | −10.1 |
| A5b | 0.53 ± 0.03 | 1.9 | −17.3 ± 0.1 | −9.4 | −7.9 |
| A7b | 0.63 ± 0.04 | 1.6 | −15.0 ± 0.1 | −6.9 | −8.1 |
| A8b | 0.44 ± 0.04 | 2.2 | −14.7 ± 0.1 | −6.9 | −7.8 |
Binding experiments were performed at 30°C. ΔHB, binding enthalpy; ΔSB, entropy of binding; ΔGB, free energy for binding. A2 to A8 are arabino-oligosaccharides.
Average sizes of branched arabino-oligosaccharides obtained by enzymatic digestion of sugar beet arabinan (see Materials and Methods).
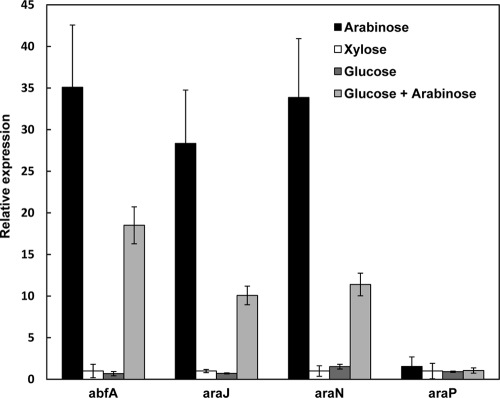
mRNA expression of abfA, araJ, araN, and araP genes.
To study the mRNA expression profiles of specific genes of the arabinan utilization system, T-6 cultures were grown in the presence of different sugars and total RNA was extracted from logarithmic-phase batch cultures. The cDNA was amplified with primers specific to abfA, araJ, araN, and araP, as well as for a 16S rRNA gene that was used for normalization. Relative expression was measured by real-time PCR and is presented in Fig. 8. The expression level of abfA, araJ, and araN in an arabinose-grown culture was about 28- to 35-fold higher than in a culture grown on xylose or glucose. In addition, the level of expression was reduced when the culture was grown in the presence of glucose and arabinose (Fig. 8). These results are also in line with the transcript analysis showing that abfA is cotranscribed with the araJKLMN genes. Interestingly, the gene for the arabinose-binding protein, araP, which may be part of the arabinose sensing system, appears to be expressed constitutively at low levels.
Fig. 8.
Quantitative real-time RT-PCR analysis of abfA, araJ, araN, and araP gene expression. Total RNA was extracted from mid-exponential-phase cultures of G. stearothermophilus T-6 grown in BSM supplemented with 0.5% arabinose, 0.5% xylose, or 0.5% glucose as the sole carbon source or with 0.5% arabinose and 0.5% glucose. Normalization was performed using the 16S rRNA gene.
AraR binds the araD and abnE promoters in vitro.
The araR gene is a monocistronic gene and encodes a protein with high similarity to repressors of the GalR-LacI family. Primer extension analysis indicated that the araR gene is expressed at low levels in the presence of glucose but its expression is stimulated in the presence of arabinose (Fig. 9A). In addition, the promoter region of araR contains an inverted repeat sequence resembling that of the araR operator (Fig. 9B). These results indicate that the repressor AraR is a negative autoregulator. The AraR protein appears to be highly toxic to E. coli, perhaps due to binding to essential elements on host DNA. Only by applying stringent control over basal expression levels of T7 RNA polymerase [using JM109(DE3)(pLysS) cells and the pET11d vector] was it possible to produce the araR gene product successfully in E. coli. The mobility of DNA fragments containing either the araD or the abnE promoter region was retarded when the fragments were incubated with a cell extract of E. coli expressing AraR (Fig. 10A and B). The binding specificity was confirmed by showing that AraR did not shift an unrelated 32-bp fragment containing a 14-bp inverted repeat (the GlcUA operator) (Fig. 10B, lane 7). To test whether arabinose can act as the inducer of the abnEFJ-abnA-abfBA-araJKLMN operon, the binding of AraR to the abnE promoter was assayed in the presence of various sugars. Binding was completely prevented in the presence of 0.2 mM arabinose, while arabino-oligosaccharides and glucose had no effect (Fig. 10C). These results are consistent with a previously shown Northern blot analysis (Fig. 5) indicating that arabinose can act as the molecular inducer of the arabinose and arabinan operons.
Fig. 9.
Mapping of the 5′ end of the araR gene. (A) Total RNA was extracted from mid-exponential-phase cultures of G. stearothermophilus T-6 grown in BSM supplemented with 0.5% arabinose or with 0.5% glucose as the carbon source. Shown are extension products that resulted from RNA extracted from cultures grown on 0.5% arabinose (lane 1) or with 0.5% glucose (lane 2). Dideoxynucleotide sequencing reactions were carried out with the same primer used for the RT reactions. (B) Sequence of the araR regulatory region. The −35 and −10 regions for σA binding and the ATG initiating codon are in boldface letters. The araR operator is indicated by horizontal arrowheads above the inverted repeat sequence.
Fig. 10.
Gel retardation analyses of AraR binding to the araD and abnE promoter regions. (A) All lanes contained about 0.12 ng of a radioactively labeled 42-bp DNA fragment containing the araD promoter. Lane 1 contained 100 μg of crude extract from E. coli cells carrying only the vector (pET11d). Lane 2 contained no extract. Lanes 3 to 8 contained different concentrations of crude extracts of E. coli cells producing AraR. (B) All lanes contained about 0.12 ng of a radioactively labeled DNA fragment containing the abnE promoter. Lane 1 contained 100 μg of crude extract from cells carrying only the vector (pET11d). Lane 2 contained no extract. Lanes 3 to 6 contained different concentrations of crude extracts of E. coli cells producing AraR. Lane 7 contained an unrelated 32-bp fragment containing a 14-bp inverted repeat (the GlcUA operator). (C) Binding of AraR to the abnE promoter in the presence of various sugars. Each lane contained 0.12 ng of labeled DNA, crude extract of E. coli cells producing AraR (100 ng/μl), and the following sugar at 0.2 mM: arabinoheptaose (A7) (lane 1), arabinohexaose (A6) (lane 2), arabinopentaose (A5) (lane 3), arabinotetraose (A4) (lane 4), arabinotriose (A3) (lane 5), arabinobiose (A2) (lane 6), arabinose (A1) (lane 7), or glucose (Glu) (lane 8).
DISCUSSION
The arabinan utilization system of G. stearothermophilus T-6 is encoded in a 38-kb segment (Fig. 11) that includes genes for two arabinanases (AbnA and AbnB), two arabinofuranosidases (AbfA and AbfB), a β-l-arabinopyranosidase (Abp), an ABC transporter for arabinose (AraEGH), an ABC transporter for arabino-oligosaccharides (AbnEFJ), a putative three-component sensing system (AraPST), an arabinose-inactivated transcriptional repressor (AraR), arabinose-metabolizing enzymes (AraDBA), and the AraJKLMN cluster that presumably constitutes an alternative pathway for arabinose utilization.
Fig. 11.
The l-arabinan utilization system in G. stearothermophilus T-6. Free arabinose at a very low concentration activates the arabinan utilization system via a three-component sensing system (araPST). Outside the cell, arabinose interacts with AraP, which presents the sugar to the sensor histidine kinase (AraS). AraS phosphorylates the response regulator (AraT), which in turn binds to the araE promoter and activates the expression of the ABC transport system for arabinose (AraEGH). In the presence of arabinose, the AraR repressor is inactivated, resulting in arabinan utilization gene expression. Arabinan comprises a backbone of α-1,5-linked l-arabinofuranosyl units, which are further decorated with α-1,2- and α-1,3-linked l-arabinofuranosides. The key enzyme in arabinan degradation is an extracellular endo-1,5-α-arabinanase (AbnA). AbnA cleaves the main backbone of arabinan and generates short branched arabino-oligosaccharides. These units enter the cell via a specific ABC transport system (AbnEFJ). Intracellular α-l-arabinofuranosidases (AbfA and AbfB) and β-l-arabinopyranosidase (Abp) remove the arabinose substitutions to generate short linear arabino-oligosaccharides that are then further degraded by an intracellular arabinanase (AbnB) into arabinose monomers.
A potential three-component sensing system for regulating the expression of the arabinose transporter.
Based on their primary sequences, the araS and araT products constitute a two-component sensing system. AraS is a class I histidine kinase (24) and has two putative transmembrane (TM) segments and a conserved C-terminal cytoplasmic region containing the ATP-binding kinase domain. AraT is the response regulator which is most likely phosphorylated by AraS. Unexpectedly, the gene cluster encodes an additional protein, AraP, an arabinose-binding lipoprotein, suggesting that AraP, AraS, and AraT might constitute a unique three-component system in which the arabinose receptor protein mediates signal transduction. The araP mRNA level appeared to be low and constant under different growth conditions, suggesting that the operon is expressed constitutively, thus allowing the cell to sense arabinose when it is available. We hypothesize that AraP bound to arabinose activates the sensor histidine kinase (AraS) either by direct protein-protein interaction or by delivering arabinose to AraS, which in turn phosphorylates the response regulator (AraT). Phosphorylated AraT binds to the araE promoter and activates the expression of the ABC transport system for arabinose (AraEGH) (Fig. 11). The potential binding sites for AraT are most likely located upstream of the −35 region, thus allowing direct interaction of the activator with the carboxy-terminal domain of the RNA polymerase α subunit (8, 57). Indeed, the region upstream of the −35 site contains two direct repeat sequences of TTTTTTG separated by 16 nucleotides. Direct repeats are common binding sites for activators (10, 11) and are also found in the upstream region of xynE (part of the xylotriose ABC transporter) in G. stearothermophilus (68).
This unique three-component sensing system for arabinose may provide the bacterium a highly sensitive and rapidly reacting mechanism for detecting and utilizing extracellular arabinose. A major challenge for soil bacteria is the identification of high-molecular-weight polymers that cannot enter the cell. G. stearothermophilus is geared to identify small amounts of arabinose (as a sign for arabinan) in order to activate the arabinan utilization system. Interestingly, we have recently characterized a different sensing mechanism in C. thermocellum by which alternate sigma factors are expressed in response to the presence of extracellular polysaccharides (51). Three-component regulatory systems were characterized in other cases, including the transport of trimethylamine N-oxide for respiration, and in antimicrobial resistance (7, 42). To our knowledge, this is the first example of a three-component sugar transport-regulating system.
The arabinan utilization system in G. stearothermophilus.
The arabinan utilization strategy in G. stearothermophilus is based on a unique three-component sensing system for extracellular arabinose (a sign for arabinan). The three-component sensing system activates the arabinan utilization system, including a single extracellular endoarabinanase (AbnA) that hydrolyzes the main high-molecular-weight arabinan polymer to produce short branched arabino-oligosaccharides and arabinose (Fig. 11).
The arabino-oligosaccharides products enter the cell via a specific ABC sugar transporter, AbnEFJ, and the final hydrolysis of the modified arabino-oligosaccharides is accomplished by the action of intracellular enzymes. These enzymes are two intracellular α-l-arabinofuranosidases (AbfA and AbfB) that remove α-1,2- and α-1,3-linked arabinofuranosides substitutions and a β-l-arabinopyranosidase that hydrolyzes β-l-arabinopyranoses from arabino-oligomers. Intracellular arabinanase (AbnB) hydrolyzes the short unmodified arabino-oligomers into arabinose monomers, which are converted into xylulose-5-phosphate by l-arabinose isomerase, l-ribulose kinase, and l-ribulose epimerase (araDBA) and subsequently enter the pentose cycle.
In B. subtilis, the arabinan utilization system resembles that of G. stearothermophilus but is based on two extracellular endo-α-1,5-l-arabinanases (36). The resulting products, arabinose and arabinose oligomers, are transported by specific transport systems, AraE, a proton symporter, and AraNPQ, an ABC-type transporter for arabino-oligosaccharides, and presumably an additional, unidentified transporter (22). Inside the cell, arabino oligomers are further hydrolyzed by the concerted action of the two α-l-arabinofuranosidases (35).
Regulation of the l-arabinan and l-arabinose utilization system.
The expression of l-arabinan/arabinose-related genes is negatively regulated by AraR and probably by the catabolite control protein A (CcpA). The system is induced by arabinose, and gel retardation analyses demonstrated that the binding of AraR to the araD and abnE promoters is completely prevented in the presence of l-arabinose (and not by arabino-oligosaccharides), suggesting that arabinose is the true molecular inducer. A potential catabolite-responsive element site for CcpA binding is found in the promoter region of the abnE gene (Fig. 6). Indeed, when strain T-6 is grown in minimal medium containing arabinose and glucose, the expression of abfA is repressed by 48% compared to growth without glucose. The AraR protein appears to negatively control the expression of the abnEFJ-abnA-abfBA-araJKLMN and araDBA-abp-abnB operons. The promoter region of the monocistronic araR gene contains the putative inverted repeat binding site of AraR. Thus, AraR probably regulates its own expression. This type of repressor autoregulation allows the cells to respond more gradually to increasing concentrations of arabinose and was also observed with XylR (41) and UxuR (67), which negatively regulate the xylan and glucuronic acid utilization systems in G. stearothermophilus, respectively.
Strain T-6 possesses dedicated ABC transporters for arabino-oligosaccharides and for arabinose.
The abnEFJ genes encode an ABC transport system that belongs to the carbohydrate uptake transporter 1 (CUT1) family. This family contains mainly transporters for di- and oligosaccharides, in addition to glycerol phosphate and polyols (http://www.tcdb.org) (15, 61), and consists of two integral membrane domains/proteins, an ATPase subunit, and an extracytoplasmic solute-binding protein. Since G. stearothermophilus T-6 encodes an extracellular arabinanase, AbnA, it requires a specific transporter for arabino-oligosaccharides. Using ITC measurements, we demonstrated that AbnE specifically binds linear and branched arabino-oligosaccharides. The dissociation constants, KD, of AbnE was 0.02 μM for linear arabino-oligosaccharide (A5) and 2.2 μM for branched arabino-oligosaccharide (A8). Similar KD values have been reported for other bacterial sugar-binding proteins; for example, the KD of XynE from G. stearothermophilus is 0.08 μM for xylortiose and 1.4 μM for xylohexaose (68). The xylobiose-binding lipoprotein (BxlE) from Streptomyces thermoviolaceus has a KD of 0.008 μM (72), whereas the cellodextrin lipid-anchored binding protein from Streptomyces reticuli showed a KD of 1.5 μM (63). Dissociation constants in the micromolar range were also measured for cellodextrins with the sugar-binding lipoproteins (CbpB to -D) from C. thermocellum (52). Interestingly, the dissociation constants of AbnE for the linear arabino-oligosaccharides (A4 to A8) are in the nanomolar range, which is 2 orders of magnitude smaller than the values obtained for the natural branched arabino-oligosaccharides, the main products of the extracellular arabinanase, AbnA.
It appears that the gene for the ATP-binding protein is not a part of the ABC transporter AbnEFJ. Perhaps this system uses the ATP-binding protein AraG, which is part of the arabinose ABC transporter. The expression of araEGH is activated in the presence of arabinose, providing sufficient amounts of AraG.
The araJKLMN genes may constitute an alternative pathway for arabinose utilization.
It is tempting to speculate that the araJKLMN genes constitute a new, alternative pathway for l-arabinose utilization. This conclusion is based on the following observations. The cluster is induced by arabinose and cotranscribed with an ABC transport system for arabino-oligosaccharides, extracellular arabinanase, and two arabinofuranosidases. In addition, AraJ, AraK, AraL, and AraM show similarity to enzymes involved in sugar metabolism. Indeed, our preliminary results show that the NAD(P) sugar dehydrogenase (AraJ) is capable of oxidizing arabinose. Additionally, AraJ has 20% identity to an arabinose dehydrogenase from Caulobacter crescentus, which is the first enzyme for an alternative arabinose utilization pathway in this bacterium (77).
ACKNOWLEDGMENTS
This research was supported by the Israel Science Foundation (grant 500/10 to Y.S.) and the United States-Israel Binational Science Foundation, Jerusalem, Israel (grant 96-178 to Y.S.). Additional support was provided by the Otto Meyerhof Center for Biotechnology, Technion, established by the Minerva Foundation (Munich, Germany). Y.S. holds the Erwin and Rosl Pollak Chair in Biotechnology at the Technion.
We thank A. L. Sonenshein for the useful suggestions and critical reading of the manuscript.
Footnotes
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Abo-Amer A. E., et al. 2004. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J. Bacteriol. 186:1879–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abramovich-Naveh E. 2007. Structure-function studies on arabinases from Geobacillus stearothermophilus. Ph.D. thesis Technion-Israel Institute of Technology, Haifa, Israel [Google Scholar]
- 3. Alhassid A., et al. 2009. Crystal structure of an inverting GH 43 1,5-alpha-l-arabinanase from Geobacillus stearothermophilus complexed with its substrate. Biochem. J. 422:73–82 [DOI] [PubMed] [Google Scholar]
- 4. Alper H., Stephanopoulos G. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential. Nat. Rev. Microbiol. 7:715–723 [DOI] [PubMed] [Google Scholar]
- 5. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 6. Anantharaman V., Aravind L. 2000. CaChe—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535–537 [DOI] [PubMed] [Google Scholar]
- 7. Baraquet C., et al. 2006. TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281:38189–38199 [DOI] [PubMed] [Google Scholar]
- 8. Barnard A., Wolfe A., Busby S. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102–108 [DOI] [PubMed] [Google Scholar]
- 9. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521–554 [DOI] [PubMed] [Google Scholar]
- 10. Blanco A. G., Sola M., Gomis-Ruth F. X., Coll M. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701–713 [DOI] [PubMed] [Google Scholar]
- 11. Bordi C., et al. 2004. Genes regulated by TorR, the trimethylamine oxide response regulator of Shewanella oneidensis. J. Bacteriol. 186:4502–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bravman T., et al. 2003. Detailed kinetic analysis of a family 52 glycoside hydrolase: a beta-xylosidase from Geobacillus stearothermophilus. Biochemistry 42:10528–10536 [DOI] [PubMed] [Google Scholar]
- 13. Brüx C., et al. 2006. The structure of an inverting GH43 beta-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J. Mol. Biol. 359:97–109 [DOI] [PubMed] [Google Scholar]
- 14. Czjzek M., et al. 2005. Enzyme-substrate complex structures of a GH39 beta-xylosidase from Geobacillus stearothermophilus. J. Mol. Biol. 353:838–846 [DOI] [PubMed] [Google Scholar]
- 15. Dassa E., Bouige P. 2001. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152:211–229 [DOI] [PubMed] [Google Scholar]
- 16. Demain A. L., Newcomb M., Wu J. H. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Vries R. P., Visser J. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65:497–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doner L. W., Irwin P. L. 1992. Assay of reducing end-groups in oligosaccharide homologues with 2,2′-bicinchoninate. Anal. Biochem. 202:50–53 [DOI] [PubMed] [Google Scholar]
- 19. Eitinger T., Rodionov D. A., Grote M., Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 35:3–67 [DOI] [PubMed] [Google Scholar]
- 20. Faergeman N. J., Sigurskjold B. W., Kragelund B. B., Andersen K. V., Knudsen J. 1996. Thermodynamics of ligand binding to acyl-coenzyme A binding protein studied by titration calorimetry. Biochemistry 35:14118–14126 [DOI] [PubMed] [Google Scholar]
- 21. Farrell A. E., et al. 2006. Ethanol can contribute to energy and environmental goals. Science 311:506–508 [DOI] [PubMed] [Google Scholar]
- 22. Ferreira M. J., de Sa-Nogueíra I. 2010. A multitask ATPase serving different ABC-type sugar importers in Bacillus subtilis. J. Bacteriol. 192:5312–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fontes C. M., Gilbert H. J. 2010. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79:655–681 [DOI] [PubMed] [Google Scholar]
- 24. Foussard M., et al. 2001. The molecular puzzle of two-component signaling cascades. Microbes Infect. 3:417–424 [DOI] [PubMed] [Google Scholar]
- 25. Galazka J. M., et al. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86 [DOI] [PubMed] [Google Scholar]
- 26. Gallegos M. T., Schleif R., Bairoch A., Hofmann K., Ramos J. L. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gat O., Lapidot A., Alchanati I., Regueros C., Shoham Y. 1994. Cloning and DNA sequence of the gene coding for Bacillus stearothermophilus T-6 xylanase. Appl. Environ. Microbiol. 60:1889–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilbert H. J. 2010. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153:444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilson E., et al. 1988. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 7:3971–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gírio F. M., et al. 2010. Hemicelluloses for fuel ethanol: a review. Bioresour. Technol. 101:4775–4800 [DOI] [PubMed] [Google Scholar]
- 31. Helmann J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higgins C. F., et al. 1990. Binding protein-dependent transport systems. J. Bioenerg. Biomembr. 22:571–592 [DOI] [PubMed] [Google Scholar]
- 33. Hövel K., et al. 2003. Crystal structure and snapshots along the reaction pathway of a family 51 alpha-l-arabinofuranosidase. EMBO J. 22:4922–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ichinose H., et al. 2009. A beta-l-arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27. J. Biol. Chem. 284:25097–25106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inácio J. M., Correia I. L., de Sa-Nogueíra I. 2008. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154:2719–2729 [DOI] [PubMed] [Google Scholar]
- 36. Inácio J. M., de Sa-Nogueíra I. 2008. Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. J. Bacteriol. 190:4272–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jarmer H., et al. 2001. Sigma A recognition sites in the Bacillus subtilis genome. Microbiology 147:2417–2424 [DOI] [PubMed] [Google Scholar]
- 38. Johnson J. L. 1981. Genetic characterization, p. 450–472 In Gerhardt P., Murray R. G. E., Costilow E. W., Nester W. A., Wood N. R., Krieg R., Philips G. B. (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 39. Khasin A., Alchanati I., Shoham Y. 1993. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl. Environ. Microbiol. 59:1725–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar R., Singh S., Singh O. V. 2008. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 35:377–391 [DOI] [PubMed] [Google Scholar]
- 41. Langut Y. 2006. Characterization of XylR, the repressor of the xylanolytic system in Geobacillus stearothermophilus. M.S. thesis Technion-Israel Institute of Technology, Haifa, Israel [Google Scholar]
- 42. Li M., et al. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104:9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Margulies M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208–218 [Google Scholar]
- 45. Miwa Y., et al. 1997. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol. Microbiol. 23:1203–1213 [DOI] [PubMed] [Google Scholar]
- 46. Mohnen D. 2008. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11:266–277 [DOI] [PubMed] [Google Scholar]
- 47. Moran C. P. 1990. Measuring gene expression in Bacillus, p. 267–293 In Harwood C. R., Cutting S. M. (ed.), Molecular biological methods for bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 48. Moran C. P., Jr., et al. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339–346 [DOI] [PubMed] [Google Scholar]
- 49. Mota L. J., Sarmento L. M., de Sa-Nogueíra I. 2001. Control of the arabinose regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity, and DNA looping. J. Bacteriol. 183:4190–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nagy T., et al. 2002. The membrane-bound alpha-glucuronidase from Pseudomonas cellulosa hydrolyzes 4-O-methyl-d-glucuronoxylooligosaccharides but not 4-O-methyl-d-glucuronoxylan. J. Bacteriol. 184:4925–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nataf Y., et al. 2010. Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc. Natl. Acad. Sci. U. S. A. 107:18646–18651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nataf Y., et al. 2009. Cellodextrin and laminaribiose ABC transporters in Clostridium thermocellum. J. Bacteriol. 191:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ouyang J., Yan M., Kong D., Xu L. 2006. A complete protein pattern of cellulase and hemicellulase genes in the filamentous fungus Trichoderma reesei. Biotechnol. J. 1:1266–1274 [DOI] [PubMed] [Google Scholar]
- 54. Pearson W. R., Lipman D. J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ragauskas A. J., et al. 2006. The path forward for biofuels and biomaterials. Science 311:484–489 [DOI] [PubMed] [Google Scholar]
- 56. Ren N., Wang A., Cao G., Xu J., Gao L. 2009. Bioconversion of lignocellulosic biomass to hydrogen: potential and challenges. Biotechnol. Adv. 27:1051–1060 [DOI] [PubMed] [Google Scholar]
- 57. Rhodius V. A., Busby S. J. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152–159 [DOI] [PubMed] [Google Scholar]
- 58. Robinson V. L., Buckler D. R., Stock A. M. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626–633 [DOI] [PubMed] [Google Scholar]
- 59. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 60. Scheller H. V., Ulvskov P. 2010. Hemicelluloses. Annu. Rev. Plant Biol. 61:263–289 [DOI] [PubMed] [Google Scholar]
- 61. Schneider E. 2001. ABC transporters catalyzing carbohydrate uptake. Res. Microbiol. 152:303–310 [DOI] [PubMed] [Google Scholar]
- 62. Shallom D., et al. 2002. Detailed kinetic analysis and identification of the nucleophile in alpha-l-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycoside hydrolase. J. Biol. Chem. 277:43667–43673 [DOI] [PubMed] [Google Scholar]
- 63. Shallom D., et al. 2005. Biochemical characterization and identification of the catalytic residues of a family 43 beta-d-xylosidase from Geobacillus stearothermophilus T-6. Biochemistry 44:387–397 [DOI] [PubMed] [Google Scholar]
- 64. Shallom D., Shoham Y. 2003. Microbial hemicellulases. Curr. Opin. Microbiol. 6:219–228 [DOI] [PubMed] [Google Scholar]
- 65. Shoham Y., Lamed R., Bayer E. A. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275–281 [DOI] [PubMed] [Google Scholar]
- 66. Shoham Y., et al. 1992. Delignification of wood pulp by a thermostable xylanase. Biodegradation 3:161–170 [Google Scholar]
- 67. Shulami S., Gat O., Sonenshein A. L., Shoham Y. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shulami S., et al. 2007. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl. Environ. Microbiol. 73:874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Taylor L. E., II, et al. 2006. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J. Bacteriol. 188:3849–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tian Y., et al. 2007. Structure-based design of robust glucose biosensors using a Thermotoga maritima periplasmic glucose-binding protein. Protein Sci. 16:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Timmerman K. P., Tu C. P. 1985. Complete sequence of IS3. Nucleic Acids Res. 13:2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tsujibo H., et al. 2004. Molecular characterization of a high-affinity xylobiose transporter of Streptomyces thermoviolaceus OPC-520 and its transcriptional regulation. J. Bacteriol. 186:1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van der Heide T., Poolman B. 2002. ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 3:938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Volz K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741–11753 [DOI] [PubMed] [Google Scholar]
- 75. von Heijne G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2:531–534 [DOI] [PubMed] [Google Scholar]
- 76. Warnecke F., et al. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565 [DOI] [PubMed] [Google Scholar]
- 77. Watanabe S., Kodaki T., Makino K. 2006. Cloning, expression, and characterization of bacterial l-arabinose 1-dehydrogenase involved in an alternative pathway of l-arabinose metabolism. J. Biol. Chem. 281:2612–2623 [DOI] [PubMed] [Google Scholar]
- 78. Wiseman T., Williston S., Brandts J. F., Lin L. N. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179:131–137 [DOI] [PubMed] [Google Scholar]
- 79. Yu H., et al. 2007. Microbial community succession and lignocellulose degradation during agricultural waste composting. Biodegradation 18:793–802 [DOI] [PubMed] [Google Scholar]
- 80. Zaide G., et al. 2001. Biochemical characterization and identification of catalytic residues in alpha-glucuronidase from Bacillus stearothermophilus T-6. Eur. J. Biochem. 268:3006–3016 [DOI] [PubMed] [Google Scholar]