Abstract
Synechocystis sp. strain PCC 6803 cultured at 30°C losses viability quickly under chill (5°C)-light stress but becomes highly tolerant to the stress after conditioning at 15°C (Y. Yang, C. Yin, W. Li, and X. Xu, J. Bacteriol. 190:1554–1560, 2008). Hypothetically, certain factors induced during preconditioning are involved in acquisition of chill-light tolerance. In this study, Rbp1 (RNA-binding protein 1) rather than Rbp2 was found to be accumulated during preconditioning, and the accumulation of Rbp1 was correlated with the increase of chill-light tolerance. Inactivation of its encoding gene rbp1 led to a great reduction in the acquired chill-light tolerance, while ectopic expression of rbp1 enabled the cyanobacterium to survive the chill-light stress without preconditioning. Microarray analyses suggested that the Rbp1-dependent chill-light tolerance may not be based on its influence on mRNA abundance of certain genes. Similarly to that in Synechocystis, the Rbp1 homologue(s) can be accumulated in Microcystis cells collected from a subtropic lake in low-temperature seasons. Rbp1 is the first factor shown to be both accumulated early during preconditioning and directly involved in development of chill-light tolerance in Synechocystis. Its accumulation may greatly enhance the overwintering capability in certain groups of cyanobacteria.
INTRODUCTION
Some cyanobacteria form water blooms in eutrophic freshwater lakes, causing serious environmental problems. The bloom-forming cyanobacteria, such as Microcystis sp., overwinter on sediment surface and reinitiate growth in spring in shallow areas with sufficient light (3, 13, 19, 23). In such areas, overwintering cyanobacteria are stressed by chill and light rather than by chill alone. Studies with Synechocystis sp. strain PCC 6803 (here referred to as Synechocystis) showed that a unicellular cyanobacterium could acquire chill (5°C)-light (100 μmol photons m−2 s−1) tolerance after preconditioning at a suboptimal low temperature, such as 15°C (26). A similar phenomenon was also found in the bloom-forming species Microcystis sp. (26). These findings suggest that cyanobacteria may develop the capability to overwinter in early winter, or before, when water temperatures decrease.
The acquired chill-light tolerance (ACLT) in cyanobacteria could be based on gene regulation or accumulation of certain metabolites during preconditioning. In Synechocystis, chill-light tolerance is rapidly increased within 24 h of preconditioning (26). ccr1 (sll1242, previously called ccr-1) is a gene required for growth at 15°C (27). When Synechocystis cells are directly transferred from 30°C to a chill (5°C)-light stress, the gene is also required for the ability to reinitiate growth. However, because it is almost not induced within 24 h at 15°C, ccr1 is probably not the key factor for the development of chill-light tolerance during preconditioning. In the same cyanobacterium, α-tocopherol was shown to be essential to the ACLT, but its level was only slightly increased within 48 h of preconditioning (26). The slight increase of α-tocopherol is not sufficient to explain the great increase of chill-light tolerance.
Many genes regulated in response to cold or cold-light stress had been identified in different species of cyanobacteria before. For examples, the expression levels of some RNA-binding protein genes (rbp) (4, 12) are upregulated upon downshift of temperature. In addition, fatty acid desaturase genes desA, desB, and desD and those encoding ribosomal protein S21, Clp family proteases, and RNA helicases are also induced in cyanobacteria by cold or cold/light stress (10). The roles of cold- or cold/light-induced genes in ACLT remain to be experimentally investigated.
All cyanobacterial RNA-binding proteins contain a single RNA recognition motif (RRM) and are divided into two classes: RbpG and its homologues possess a long conserved C-terminal domain (class II), while most others do not (class I) (7). Class I Rbp proteins are further divided into two types: those that possess a short C-terminal glycine-rich domain and are cold inducible (referred to as type I) and those that possess no C-terminal glycine-rich domain and show no or only a slight response to cold induction (referred to as type II) (12, 16). RNA-binding proteins could be involved in many posttranscriptional regulation processes (1). In the filamentous species Anabaena variabilis, a type I RNA-binding protein, RbpA1, affects the maintenance of normal gene expression (17). In Synechocystis, a type II RNA-binding protein, Rbp3, is specifically required for maintaining the mRNA levels of desA, desB, desD, and ccr1 (20). On the other hand, RNA-binding proteins may affect the physiological processes in cyanobacteria at low temperature. The rbpA1 mutant of A. variabilis showed abnormal regulation of heterocyst differentiation at a low temperature (17). In a unicellular species, Synechococcus sp. strain PCC 7942, an rbp1 mutant showed greatly reduced growth at 20°C (18). An rbp3 mutant of Synechocystis remained unchanged in growth at 15°C but showed reduced fatty acid desaturation of membrane lipids at both 15°C and 30°C (20). Probably, the reduction of total polyunsaturated fatty acids in the mutant has not attained the extent that would significantly affect growth at the low temperature.
Although no Rbp protein has been shown to be required for survival at a temperature as low as 4°C in cyanobacteria, it has been found to be involved in adaptation to cold stress at such a low temperature in a higher plant (8). We wondered if the accumulation of any Rbp protein during preconditioning was involved in acquisition of chill-light tolerance in cyanobacteria. In Synechocystis, there are 3 predicted rbp genes. We found that Rbp1 in this cyanobacterium was accumulated during preconditioning and played a key role in the development of chill-light tolerance. In addition, using samples collected from a lake, we found that Rbp1 was indeed accumulated in the bloom-forming cyanobacterium Microcystis in low-temperature seasons.
MATERIALS AND METHODS
Measurements of ARG, RACLT, and fatty acid composition.
Synechocystis and its derivatives are listed in Table 1. The cyanobacterium was cultured in BG11 with (mixotrophic) or without (autotrophic) glucose (5 mM) in flasks on a shaker at 30°C under continuous illumination of 30 μmol photons m−2 s−1. Measurements of the ability to reinitiate growth (ARG), the relative acquired chill-light tolerance (RACLT), and fatty acid composition were performed as previously described (26, 27). For growth of transformants, kanamycin (Km), erythromycin (Em), or spectinomycin (Sp) was added to the medium at 30 μg ml−1, 10 μg ml−1, or 5 μg ml−1. All values are means of 3 independent experiments with standard deviations.
Table 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Derivation, relevant characteristics, or sequences (5′→3′)a | Source, reference(s), or gene |
|---|---|---|
| Synechocystis sp. strains | ||
| PCC 6803 | Wild type, a glucose-tolerant strain | J. Zhao, Beijing University/Institute of Hydrobiology |
| DRHB818b | Kmr, ggpS::C.K, Synechocystis 6803 transformed with pHB818 | This study |
| DRHB2548 | Cmr Emr, rbp1::C.CE2, Synechocystis 6803 transformed with pHB2548 | This study |
| DRHB2548/DRHB2788 | Cmr Emr Kmr, rbp1::C.CE2 complemented with the wild-type rbp1, mutant DRHB2548 transformed with pHB2788 | This study |
| DRHB2549 | Kmr, rbp2::C.K, Synechocystis 6803 transformed with pHB2549 | This study |
| DRHB2791 | Smr Spr, PrbcL-ccr2 in addition to the indigenous ccr2; Synechocystis 6803 transformed with pHB2791 to introduce PrbcL-ccr2 into a neutral platform (6, 24) of the genome | This study |
| DRHB3288 | Smr Spr, PrbcL-rbp1 in addition to the indigenous rbp1; Synechocystis 6803 transformed with pHB3288 to introduce PrbcL-rbp1 into a neutral platform (6, 24) of the genome | This study |
| Plasmidsc | ||
| pHB796 | Apr; the PCR fragment containing ggpS, amplified with primers sll1566-1 and sll1566-2, cloned into pMD18-T | This study |
| pHB818 | Apr Kmr; the C.K cassette excised with PvuII from pRL446, blunted with T4 DNA polymerase, cloned into the BalI site of ggpS within pHB796 | This study |
| pHB2489 | Apr; the PCR fragment containing rbp1 coding region, amplified with primers sll0517-a1 and sll0517-a2, cloned into pMD18-T | This study |
| pHB2502 | Apr; the PCR fragment overlapping the 5′ end and upstream sequence of rbp1, amplified with primers sll0517-k1 and sll0517-k2, cloned into pMD18-T | This study |
| pHB2503 | Apr; the PCR fragment overlapping the 3′ end and downstream sequence of rbp1, amplified with primers sll0517-k3 and sll0517-k4, cloned into pMD18-T | This study |
| pHB2504 | Apr; the PCR fragment overlapping the 5′ end and upstream sequence of rbp2, amplified with primers ssr1480-k1 and ssr1480-k2, cloned into pMD18-T | This study |
| pHB2505 | Apr; the PCR fragment overlapping the 3′ end and downstream sequence of rbp2, amplified with primers ssr1480-k3 and ssr1480-k4, cloned into pMD18-T | This study |
| pHB2535 | Apr Cmr Emr; the C.CE2 cassette excised with BamHI from pRL598, blunted with T4 DNA polymerase, cloned into the blunted SalI site of pHB2503 | This study |
| pHB2536 | Apr Kmr; the C.K cassette excised with BamHI from pRL446, blunted with T4 DNA polymerase, cloned into the blunted SalI site of pHB2505 | This study |
| pHB2548 | Apr Cmr Emr; the DNA fragment containing C.CE2 and the downstream sequence of rbp1 excised with PstI and SmaI from pHB2535, blunted with T4 DNA polymerase, cloned into the SmaI site of pHB2502, with C.CE2 positioned between the flanking sequences of rbp1 | This study |
| pHB2549 | Apr Kmr; the DNA fragment containing C.K and the downstream sequence of rbp2 was excised with SmaI and PstI from pHB2536, blunted with T4 DNA polymerase, cloned into the SmaI site of pHB2504, with C.K positioned between the flanking sequences of rbp2 | This study |
| pHB2560 | Kmr; the DNA fragment excised with NcoI and XhoI from pHB2489, cloned into pET41a, for production of Synechocystis Rbp1 with six-His tag in Escherichia coli | This study |
| pHB2706 | Apr; the PCR fragment containing ccr2, amplified with primers ccr2-e1 and ccr2-e2, cloned into pMD18-T | This study |
| pHB2739 | Apr; the PCR fragment containing the rbcL promoter amplified with primers PrbcL-1 and PrbcL-6, cloned into pMD18-T | This study |
| pHB2759 | Apr Spr; Ω cassette excised with DraI from pRL57, cloned into SalI-cut and T4 DNA polymerase-blunted pHB2739 | This study |
| pHB2761 | Apr; the PCR fragment containing rbp1, amplified with primers sll0517-k1 and sll0517-a4, cloned into pMD18-T | This study |
| pHB2768 | Apr Kmr; the C.K cassette excised with BamHI from pRL446, cloned into the BamHI site of pHB2761 | This study |
| pHB2770 | Apr Spr; Ω-PrbcL excised with XbaI/PstI from pHB2759, blunted with T4 DNA polymerase, cloned into XbaI-cut and T4 DNA polymerase-blunted pHB2706 | This study |
| pHB2788 | Apr Kmr; the fragment containing the C.K cassette and rbp1 excised with PvuII from pHB2768, cloned between the blunted EcoRI sites of pKW1188 | This study |
| pHB2791 | Apr Spr; Ω-PrbcL-ccr2 excised with SmaI and HincII from pHB2770, cloned between the blunted EcoRI sites of pKW1188 | This study |
| pHB3208 | Apr; the PCR fragment containing the open reading frame of rbp1, amplified with primers sll0517-oe1 and sll0517-a4, cloned into pMD18-T | This study |
| pHB3284 | Apr Smr Spr; the Ω cassette excised with DraI from pRL57, cloned into the blunted SacI site of pHB2739, downstream of the cloned PrbcL | This study |
| pHB3287 | Apr Smr Spr; the DNA fragment containing PrbcL and Ω cassette, excised with PvuII and SalI from pHB3284 and blunted with T4 DNA polymerase, cloned between the blunted EcoRI sites of pKW1188, replacing the kanamycin resistance gene | This study |
| pHB3288 | Apr Smr Spr; the DNA fragment containing the rbp1 coding region, excised with SalI and XbaI from pHB3208 and blunted with T4 DNA polymerase, cloned into SmaI-cut and dephosphorylated pHB3287, located between PrbcL and Ω cassette, oriented as PrbcL | This study |
| pET21b | Apr; overexpression vector | Novagen, EMD Chemicals Inc. |
| pET41a | Kmr; overexpression vector | Novagen |
| pKW1188 | Apr Kmr; a plasmid bearing a neutral integrative platform for Synechocystis 6803 | 6, 24 |
| pMD18-T | Apr; cloning vector | Takara, Japan |
| pRL57 | Kmr Smr Spr; a pDU1-based plasmid containing the spectinomycin resistance cassette Ω | 5 |
| pRL446 | Apr Kmr; a plasmid containing the kanamycin resistance cassette C.K | NCBI GenBank accession no. EU346690 |
| pRL598 | Apr Cmr Emr; a plasmid containing the chloramphenicol and erythromycin resistance cassette C.CE2 | 5 |
| Primers (5′→3′) | ||
| ccr2-e1 | GGCTGTTACTCCAGACCCA | ccr2 |
| ccr2-e2 | AGCAAGACAACAATGGACAGGA | |
| sll1566-1 | CCTGGTCAATGGATTCGTCC | ggpS |
| sll1566-2 | GTGAGCCCTACGACGAAGT | |
| gvpAC-1 | C(C/T)TACCTCAAATATGCTGAAGC | gvpA-gvpC intergenic sequence |
| gvpAC-2 | TGCCTGTTCTTGCGCTTGT | |
| PrbcL-1 | CCGATGAAGTGGTGGAGCA | rbcL |
| PrbcL-6 | GGTCAGTCCTCCATAAACATTG | |
| sll0517-a1 | ACCATGGTGTCAATTTATGTAGGCAACCTGTCC | rbp1 |
| sll0517-a2 | TTCTCGAGGTAGCGGCTACCACCATAGCT | |
| sll0517-a3d | TCTCATATGTCAATTTATGTAGGCAACCTGTCC | |
| sll0517-a4d | TTTCTCGAGTGGTGGAACGACGGCGAA | |
| sll0517-k1 | GTAGAAACGGGTACTGGTCATG | |
| sll0517-k2 | GTTGCCTACATAAATTGACATGGATT | |
| sll0517-k3 | TTCCTTTGGTGGCGGTCGT | |
| sll0517-k4 | CTCCTCCGAATCCTTGCGAA | |
| sll0517-oe1 | GTTTTTGGAGAAAATCCATGTCAA | |
| ssr1480-a3d | TTTCATATGTCCATTTATGTCGGGAACCTTTCTT | rbp2 |
| ssr1480-a4d | TTTCTCGAGGACTCAAACACCTTCCCTTCTACAA | |
| ssr1480-k1 | CGGCTACTGTGAATCTTTGGA | |
| ssr1480-k2 | GGTTCCCGACATAAATGGACA | |
| ssr1480-k3 | AAAGCAAGACCGAGAACCCCT | |
| ssr1480-k4 | ACTCCCTTCAAATCTGGCTTCA | |
| rnpB-1d | GTTAGGGAGGGAGTTGCGG | rnpB |
| rnpB-2d | AAGAGAGTTAGTCGTAAGCCG |
Abbreviations: Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin.
DRHBxxxx refers to a product of double homologous recombination between plasmid pHBxxxx and the Synechocystis sp. genome.
Unless stated otherwise, the template for PCRs was Synechocystis sp. genomic DNA.
These primers were used to generate probes for Northern blot hybridizations.
For assays of ARG, cells diluted to an optical density at 730 nm (OD730) of 0.05 were exposed to chill (5°C)-light (100 μmol photons m−2 s−1) stress and allowed to grow mixotrophically in test tubes at 30°C for 4 days, and then OD730 (treated) and OD730 (control) were measured. The rbp2 mutant, as an exception, was grown only in flasks on a shaker. The control was Synechocystis diluted to an OD730 of 0.05 but not chill light stressed before growth at 30°C. The ARG was calculated as OD730 (treated)/OD730 (control) × 100%.
To evaluate the effect of preconditioning on chill-light tolerance, cells preconditioned or not were exposed to chill-light stress for 8 days and transferred to 30°C for growth, and the increase of ARG due to preconditioning was calculated. The RACLT of a mutant was then calculated as the percentage of the preconditioning-induced increase of ARG of the mutant relative to that of the wild type (26). Unlike mutants defective in tocopherol synthesis (15), the rbp1 mutant was not sensitive to glucose. Therefore, it was not necessary to avoid glucose in the test of its RACLT. Due to the poor growth of the rbp2 mutant in test tubes, its exposure to the chill-light stress and reinitiated growth at 30°C were carried out in 250-ml flasks instead of test tubes.
Molecular cloning and mutant construction.
Molecular cloning was performed using standard methods. Tool enzymes or kits were used per manufacturers' instructions. PCRs were performed using primers listed in Table 1. Clones of PCR products were confirmed by sequencing.
Table 1 describes the details of plasmid construction. In brief, pHB818 is the plasmid used to inactivate ggpS with C.K, pHB2548 is the plasmid used to inactivate rbp1 with C.CE2, pHB2549 is the plasmid used to inactivate rbp2 with C.K, pHB2788 is the plasmid carrying C.K-rbp1 within an integrative platform and used to complement the rbp1::C.CE2 mutant of Synechocystis, pHB2791 is the plasmid carrying Ω-PrbcL-ccr2 within an integrative platform and used to overexpress ccr2, and pHB3288 is the plasmid carrying Ω-PrbcL-rbp1 within an integrative platform and used to overexpress rbp1. C.CE2, C.K, and Ω are chloramphenicol/erythromycin, kanamycin, and spectinomycin resistance cassettes excised from pRL598 (5), pRL446 (NCBI GenBank accession no. EU346690), and pRL57 (5), respectively.
For targeted insertion of a gene, Synechocystis was transformed with the corresponding plasmids according to the work of Williams (24) and the resultant transformants were streaked on plates and cultured in liquid medium with appropriate antibiotics. The complete segregation of mutants was confirmed by PCR. Constructed strains and primers used are also listed in Table 1.
Analyses of gene expression in Synechocystis.
Synechocystis cells used in gene expression analyses were cultured mixotrophically at 30°C. The cultures were quickly cooled to 15°C or 5°C in a water bath and then transferred to an illuminating incubator or refrigerator set at the corresponding temperatures. Cells were collected at different stages as indicated. Total RNA was extracted from Synechocystis using TRIzol reagent (Invitrogen), treated with RNase-free DNase I (Takara, Japan) to eliminate contaminating chromosomal DNA, and examined by agarose gel electrophoresis. Soluble proteins were prepared from 300 ml of Synechocystis cells by sonication and ultracentrifugation in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), a protease inhibitor, and precipitated by being mixed with equal volumes of ice-cold acetone containing 10% trichloroacetic acid and 0.07% β-mercaptoethanol at −20°C. To test protein stability in Synechocystis under chill-light conditions, chloramphenicol (50 μg ml−1) was added to the culture after cooling to 5°C.
For Northern blot analysis, DNA probes were prepared by PCR using primers listed in Table 1. PCR products were labeled by incorporation of digoxigenin-dUTP. Total RNA was separated by electrophoresis on an agarose-formaldehyde gel and blotted onto an Immobilon-Ny+ membrane (Millipore) by capillary transfer. Hybridization and immunological detection were performed with DIG High Prime DNA Labeling and Detection Starter Kit I (Roche) according to the manufacturer's recommendations. The transcription of rnpB (RNase P subunit B) (22) was used as the internal control.
CyanoCHIP v.2.0 (Takara) was used for analyses of transcriptional profiles, and cDNA was labeled using an RNA fluorescence labeling core kit (Moloney murine leukemia virus [MMLV] version; Takara, Japan). Microarray slides were hybridized with labeled cDNA, washed, and then scanned with an Affymetrix 428 array scanner (Affymetrix). The spot intensities were determined using ImaGene v.3.0 software (BioDiscovery). Labeling of cDNA, hybridization, rinsing and scanning of microarrays, and data analyses were performed by Takara Company. Data were generated from 3 independent experiments, each with 2 repeats (2 × 3).
For Western blot analyses, proteins (18 μg in each sample) dissolved in the loading buffer were boiled and subjected to 15% SDS-PAGE and electroblotted onto nitrocellulose membranes (Millipore). In addition to quantification by Bradford's method (9), protein samples were examined by SDS-PAGE and Coomassie brilliant blue (CBB) staining to make sure that equal amounts of soluble proteins were loaded. Synechocystis Rbp1 was detected with the rabbit antiserum raised against recombinant Rbp1 and visualized using alkaline phosphatase-conjugated secondary antibody specific for rabbit IgG (Pierce) with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Amresco) as substrates. The recombinant Rbp1 was purified from Escherichia coli BL21(DE3) harboring pHB2560 or pHB2225 (Table 1) using the His-Bind purification kit (Novagen) according to the manufacturer's instructions.
Detection of Rbp1 in Microcystis cells collected from a lake.
Microcystis colonies were collected from the upper layer of Meiliang Bay of Lake Taihu (31°24′39.18″N, 120°11′14.34″E) monthly from November 2008 to October 2009 using a phytoplankton net (64 μm in mesh size). The collected cells were further concentrated using the same phytoplankton net, quickly frozen in liquid nitrogen, and stored in a −70°C freezer.
The frozen cyanobacterial cells were suspended in 40 mM Tris Cl (pH 8.0) with 1 mM PMSF. Cells were broken by sonication on ice for 2 min and centrifuged (6,000 × g) at 4°C for 20 min to remove cell debris. The supernatant was ultracentrifuged (100,000 × g) at 4°C for 1 h to remove cell membranes. The resulting soluble proteins were used for Western blot detection of Rbp1, and 50 μg of proteins was loaded onto each lane of an SDS-PAGE gel.
Typing of Microcystis based on gvpA-gvpC intergenic sequences.
About 0.1 ml of Microcystis colonies was washed once with 1.5 ml of TE buffer (50 mM Tris Cl, 100 mM Na2-EDTA, pH 8.0), resuspended in 1 ml of TE buffer with 1% SDS, and incubated at 37°C for 1 to 2 h until cells were lysed. After removal of proteins with proteinase K and removal of RNA with RNase A, the samples were extracted with phenol and chloroform. Total DNA was precipitated with ethanol at −20°C and dissolved in sterilized double-distilled water (ddH2O).
Using gvpAC-1 and gvpAC-2 (Table 1) as the primers and total Microcystis DNA as the template, PCR was performed to generate DNA fragments containing the gvpA-gvpC intergenic region (25). A mixture of Taq and Pfu DNA polymerases (1:1) was used to reduce rates of error in PCR. After addition of dA to their ends with Taq DNA polymerase, the PCR products were purified and cloned into pMD18-T (Takara, Japan). For Microcystis cells collected in each month, 50 clones of the PCR products were sequenced, and the percentages of different types of gvpA-gvpC intergenic sequences were calculated. Microcystis cells were classified into different types according to the gvpA-gvpC intergenic sequences. Sequences repeatedly found in PCR products for different months (like types 1 to 10) were considered to be without error. Based on sequences of 200 clones for samples from December 2008 to March 2009, more than 98% of clones should carry no PCR error.
RESULTS AND DISCUSSION
Rbp1 is accumulated in Synechocystis during preconditioning.
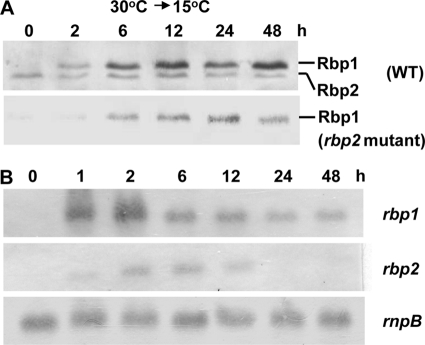
In Synechocystis, sll0517 (rbp1) and ssr1480 (rbp2) are predicted to encode two highly similar RNA-binding proteins (79% identity to each other) with the C-terminal glycine-rich domain (type I), while slr0193 (rbp3) is predicted to encode an RNA-binding protein without the glycine-rich domain (type II). In a previous report, rbp3 was shown to be slightly induced by exposure to cold (20). In this study, we generated the antiserum against Rbp1 and detected Rbp1 and Rbp2 in Synechocystis transferred from 30°C to 15°C by Western blot analysis. Detection with Rbp1 antiserum resulted in two bands close to each other (Fig. 1A). Based on the molecular mass, the upper band showing cold induction should correspond to Rbp1 (predicted molecular mass, 10.96 kDa), while the lower band showing no or transient slight induction should be Rbp2 (predicted molecular mass, 9.39 kDa). To confirm the assignment, we generated the rbp2::C.K mutant DRHB2549 (Table 1). With the inactivation of rbp2, the lower noninducible band disappeared. In other words, the upper inducible band must be Rbp1. These results indicated that Rbp1 rather than Rbp2 was accumulated in Synechocystis during preconditioning. We also detected these proteins' expression at mRNA level by Northern blot analyses. The rbp1 transcript was rapidly accumulated within 1 h of cold induction, reached the maximal level at 2 h, and thereafter slowly decreased to a relatively stable level within 24 h (Fig. 1B). The rbp2 transcript was also rapidly accumulated under the same conditions but decreased to an undetectable level after 12 h (Fig. 1B).
Fig. 1.
Western blot (A) and Northern blot (B) analyses of the expression of rbp1 and rbp2 in Synechocystis during preconditioning. Cells cultured at 30°C were transferred to 15°C with illumination of 30 μmol photons m−2 s−1 for different periods of time. The RNase P RNA subunit gene rnpB (22) was used as the internal control in the Northern blot analysis.
Accumulation of Rbp1 during preconditioning is correlated with acquisition of chill-light tolerance.
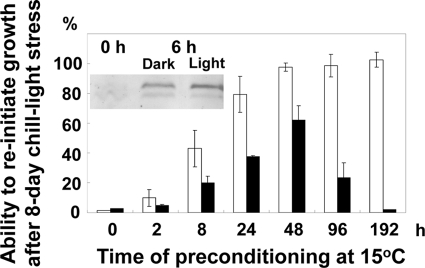
Upon preconditioning at 15°C, the chill (5°C)-light tolerance of Synechocystis is induced within 48 h to almost the maximal level (26). Parallel to the development of chill-light tolerance, Rbp1 started to be accumulated at a very early stage (within 2 h) of preconditioning and reached the maximal level within 12 to 48 h (Fig. 1A). In Anabaena sp. strain PCC 7120, microarray analysis suggested that the cold-induced expression of rbpA1/rbpA2 was independent of the light (4). Similarly, the accumulation of Rbp1 in Synechocystis transferred from 30°C to 15°C was independent of the light (Fig. 2). We then examined the role of light in the development of chill-light tolerance during preconditioning. The chill-light tolerance was evaluated based on the ability to reinitiate growth (ARG) after exposure to the chill-light stress for 8 days. Synechocystis can grow heterotrophically on glucose in the dark with a daily brief exposure to weak light (2). As shown in Fig. 2, removal of light during preconditioning in the presence of glucose reduced the maximal chill-light tolerance by ca. 40% at 48 h. Apparently, there should be both light-dependent and -independent processes involved in enhancement of chill-light tolerance. Accumulation of factors like Rbp1 may affect at least the light-independent preconditioning. However, we also noticed that the ARG decreased after incubation at 15°C for 48 h in the dark. Probably, utilization of glucose under such conditions could not provide sufficient energy to support cell activities after prolonged incubation.
Fig. 2.
Effects of light during preconditioning on the acquisition of chill-light tolerance. Cells were grown in BG11 with glucose at 30°C and pretreated at 15°C for different periods of time. The chill-light tolerance was evaluated as the ability to reinitiate growth (ARG). Empty bars, light of 30 μmol photons m−2 s−1; solid bars, no light. The inset shows Western blot detection of Rbp1 (the upper band) in Synechocystis treated at 15°C with or without light.
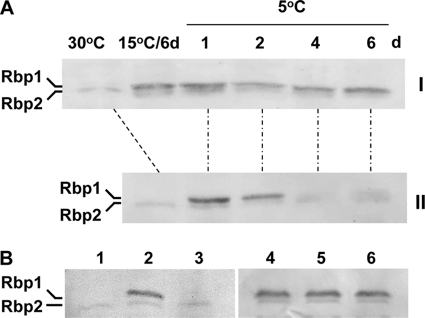
Previously, we reported that after pretreatment at 15°C, a significantly higher level of α-tocopherol could be maintained in Synechocystis under the chill-light stress (26). Similarly, we tested the effect of preconditioning on maintenance of Rbp1 level in the cyanobacterium. In cells grown at 30°C, Rbp1 was not detectable. When 30°C-grown cells were directly exposed to the chill-light stress, Rbp1 was synthesized rapidly on the first day and then decreased and almost disappeared after 4 days (Fig. 3A, panel II). In contrast, in cells preconditioned at 15°C, Rbp1 was maintained at a relatively high level after 4 days of chill-light stress (Fig. 3A, panel I).
Fig. 3.
Western blot analyses showing effects of preconditioning (A) and chloramphenicol (B) on the level of Rbp1 (the upper band) in Synechocystis under chill-light stress. (A) Synechocystis cells grown at 30°C were exposed to chill-light stress for different periods of time with (I) or without (II) preconditioning at 15°C for 6 days. (B) Synechocystis cells grown at 30°C (lane 1) were directly exposed to chill-light stress for 1 day without (lane 2) or with (lane 3) chloramphenicol or preconditioned for 6 days (lane 4) and exposed to chill-light stress for 4 days without (lane 5) or with (lane 6) chloramphenicol.
Protein levels can be affected by rates of synthesis and degradation. We examined the stability of Rbp1 under chill-light stress by using chloramphenicol as an inhibitor of protein synthesis. The antibiotics completely inhibited the accumulation of Rbp1 in cells directly exposed to chill-light stress but showed no effect on the level of Rbp1 in cells subjected to preconditioning (Fig. 3B). It appeared that preconditioning enhanced the stability of Rbp1 in the cyanobacterium under the chill-light stress. In contrast, Rbp2 seemed to be stable under the chill-light stress with or without pretreatment at 15°C.
Rbp1 plays an important role in ACLT.
To test the role of rbp1 in the ACLT, we constructed Synechocystis DRHB2548, the rbp1::C.CE2 mutant (Table 1). At 15°C, the rbp1 mutant grew very slowly (Table 2), but its viability remained essentially unchanged. When exposed to chill-light stress, however, the preconditioned rbp1 mutant showed greatly reduced viability relative to that of the wild type. The role of rbp1 in ACLT was evaluated with relative ACLT (% RACLT) (26). The rbp1 mutant showed greatly reduced ACLT relative to that of the wild type, and the phenotype was fully restored by complementation with the wild-type rbp1 (Table 2). Because rbp1 was induced by cold with or without the light, we measured its RACLT under the two conditions and found similar results. As a control, the ggpS::C.K mutant DRHB818 (Table 1) showed a very slight reduction in ACLT (89.9% ± 10.5%, preconditioned at 15°C in the light). ggpS is involved in synthesis of the osmolyte glucosylglycerol (11). ccr1 (sll1242) is also required for growth at 15°C, but unlike rbp1, it is induced at a late stage after transfer from 30°C to 15°C (27). The mutant sll1242::C.K2d reported before (27) showed an unstable RACLT varying from 41.8% to 90.0%. The rbp2 mutant showed pleiotropic phenotypes. It grew poorly in test tubes at 30°C and 15°C. In flasks with agitation, however, it grew as well as the wild type at both temperatures (Table 2). Inactivation of rbp2 also caused a significant reduction in RACLT. Based on the phenotypes of the mutant and the essentially unchanged level of Rbp2 in cold-induced cells, we think that this protein may not be specifically required for the ACLT. Any possible effect of Rbp2 in the ACLT will need further investigations.
Table 2.
Growth rates and chill-light tolerance of Synechocystis strains
| Strain | Doublings day−1a |
% RACLT |
||
|---|---|---|---|---|
| 30°C | 15°C | Preconditioned in light | Preconditioned in dark | |
| Wild type | 2.0 ± 0.1 | 0.7 ± 0.02 | 100 | 62.0 ± 1.5 |
| rbp1::C.CE2b | 1.7 ± 0.3 | 0.1 ± 0.01 | 24.0 ± 2.8 | 9.3 ± 1.0 |
| Complemented rbp1::C.CE2c | 1.9 ± 0.2 | 0.6 ± 0.1 | 102 ± 4.6 | 72.9 ± 7.0 |
| rbp2::C.Kd | 2.0 ± 0.1 | 0.6 ± 0.01 | 36.6 ± 6.0 | Not tested |
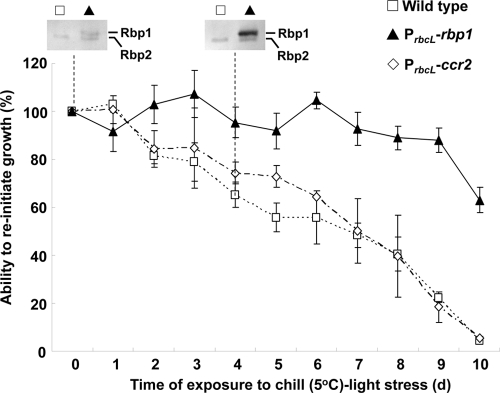
The role of Rbp1 in acquisition of chill-light tolerance was also tested by ectopic expression. We constructed Synechocystis strain DRHB3288, which expressed rbp1 from the promoter of rbcL in addition to the expression of the indigenous rbp1. In this strain, Ω-PrbcL-rbp1 was integrated into a neutral platform in the genome (Table 1), and the synthesis of Rbp1 at 30°C was detectable with Western blot analysis (Fig. 4). The upper band found in DRHB3288 (here referred to as the PrbcL-rbp1 strain) rather than in the wild-type strain was Rbp1, while the lower band found in both strains was Rbp2. Without preconditioning, the PrbcL-rbp1 strain showed significantly enhanced chill-light tolerance compared to that of the wild type (Fig. 4). As a control, we also overexpressed ccr2 (slr0815) from PrbcL in Synechocystis strain 6803 (Fig. 4). Unlike ccr1, ccr2 is a gene upregulated at an early stage of preconditioning and required for growth at 15°C (W. Li and X. Xu, unpublished data). Unlike rbp1, overexpression of ccr2 did not enhance the chill-light tolerance of the cyanobacterium (Fig. 4).
Fig. 4.
Enhanced chill-light tolerance by ectopic expression of rbp1 in Synechocystis. ccr2, a gene upregulated at an early stage of preconditioning and required for growth at 15°C (Li and Xu, unpublished), was used as a control. In addition to the single wild-type copy of each of rbp1 and ccr2, the overexpression strains DRHB3288 and DRHB2791 contain a second, PrbcL-promoted copy of rbp1 or ccr2 and are denoted as PrbcL-rbp1 or PrbcL-ccr2, respectively. Rbp1 was detected by Western blot analysis in cells exposed to the chill-light stress for 0 or 4 days. The lower band detected by Rbp1 antiserum was Rbp2 as shown in Fig. 1.
In the PrbcL-rbp1 strain grown at 30°C, the level of Rbp1 was further increased to a high level after 4 days of exposure to chill-light stress, while in the wild type, Rbp1 was not detectable at 30°C and after 4 days of chilling (Fig. 4). In the PrbcL-rbp1 strain, the indigenous copy of rbp1 should be induced to express within 24 h of chilling at 5°C as in the wild type (Fig. 3A, panel II). The accumulated Rbp1 on the 4th day should have resulted from a combination of preaccumulation (due to PrbcL-rbp1) and chilling induction (due to indigenous rbp1). Preaccumulated Rbp1 may directly or indirectly enhance the synthesis and/or stability of the same protein under the chill-light stress.
Rbp1-dependent chill-light tolerance may not be based on Rbp1 influence on mRNA abundance.
Unlike Rbp3 (20), Rbp1 showed no apparent effect on the desaturation degree of membrane lipids. The fatty acid compositions in the wild-type, rbp1::C.CE2, and PrbcL-rbp1 strains were similar to one another (see Table S1 in the supplemental material). Therefore, Rbp1 is probably not involved in the posttranscriptional regulation of fatty acid desaturase genes. Employing microarrays, we further analyzed changes of mRNA expression profile in Synechocystis 6803 in response to preconditioning at 15°C and inactivation or overexpression of rbp1. Table 3 is a list of genes whose mRNA levels were influenced by Rbp1. Most genes showed upregulation in the wild type during preconditioning and reduced expression in the rbp1::C.CE2 strain relative to the wild type or the opposite. However, such genes showed no or slight changes in the PrbcL-rbp1 strain at 30°C compared to the wild type. slr1764 (capA) was the only gene upregulated in response to the overexpression of rbp1 but showed almost no change in the rbp1::C.CE2 strain during preconditioning. Apparently, none of the genes was the direct target of the posttranscriptional regulation by Rbp1. The microarray analysis results suggested that the enhancement of chill-light tolerance by Rbp1 may not be due to its influence on mRNA abundance of certain genes.
Table 3.
List of genes that are up- or downregulated during preconditioning and affected by Rbp1 as shown in either the rbp1::C.CE2 or the PrbcL-rbp1 straina
| ORF | Product | Ratio |
||
|---|---|---|---|---|
| WT at 15°C/WT at 30°C | rbp1::C.CE2/WT at 15°C | PrbcL-rbp1/WT at 30°C | ||
| sll0219 | Potential FMN protein | 3.493 ± 1.489 | 0.498 ± 0.281 | 0.729 ± 0.123 |
| sll0517 | rbp1, RNA-binding protein | 5.659 ± 2.446 | 0.048 ± 0.015 | 2.475 ± 0.443 |
| sll0662 | 2.185 ± 0.418 | 0.337 ± 0.11 | 0.88 ± 0.221 | |
| sll0781 | 2.577 ± 0.576 | 0.299 ± 0.041 | 1.235 ± 0.191 | |
| sll1091 | 391-aa (43-kDa) bacteriochlorophyll synthase subunit | 2.827 ± 0.659 | 0.496 ± 0.115 | 1.1 ± 0.304 |
| sll1167 | Penicillin-binding protein 4 | 0.434 ± 0.063 | 2.221 ± 0.612 | 0.935 ± 0.123 |
| sll1476 | 2.433 ± 0.44 | 0.467 ± 0.122 | 0.977 ± 0.074 | |
| sll1926 | 2.465 ± 0.42 | 0.498 ± 0.108 | 0.93 ± 0.145 | |
| slr0447 | urtA, ABC-type urea transport system substrate-binding protein | 5.771 ± 0.303 | 0.189 ± 0.133 | 0.794 ± 0.394 |
| slr1136 | ctaC or coxB, cytochrome c oxidase subunit II | 3.091 ± 0.467 | 0.243 ± 0.044 | 0.869 ± 0.082 |
| slr1137 | ctaD, cytochrome c oxidase subunit I | 2.273 ± 0.346 | 0.219 ± 0.053 | 0.789 ± 0.09 |
| slr1138 | ctaE, cytochrome c oxidase subunit III | 2.95 ± 0.473 | 0.302 ± 0.075 | 0.834 ± 0.082 |
| slr1452 | Sulfate-binding protein SbpA | 0.164 ± 0.057 | 8.366 ± 5.581 | 1.000 ± 0.325 |
| slr1453 | Sulfate transport system permease protein | 0.437 ± 0.071 | 2.738 ± 0.914 | 1.043 ± 0.419 |
| slr1764 | capA, cAMP-binding protein, similar to tellurium resistance protein TerE | 2.309 ± 0.456 | 1.216 ± 0.309 | 3.187 ± 0.969 |
| ssl1263 | 2.763 ± 0.822 | 0.385 ± 0.158 | 0.957 ± 0.357 | |
| ssr1386 | ictA, inorganic carbon transport protein | 3.669 ± 0.62 | 0.495 ± 0.056 | 1.013 ± 0.265 |
Preconditioning at 15°C was performed in the light for 2 days. Data represent the means ± standard deviations calculated from 3 independent experiments, each with 2 repeats (2 × 3). Ratios of ≥2.0 indicate a significant increase in mRNA level; ratios of ≤0.5 indicate a significant decrease in mRNA level. Abbreviations: ORF, open reading frame; FMN, flavin mononucleotide; aa, amino acid; cAMP, cyclic AMP; WT, wild type.
Rbp1 is accumulated in overwintering Microcystis cells in a lake.
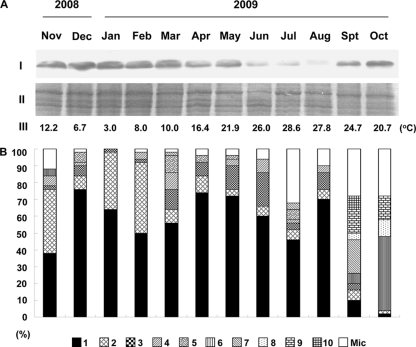
Our studies with Synechocystis showed that Rbp1 is at least one of the key factors in acquisition of chill-light tolerance. To find out if homologues of Rbp1 (for simplicity, also called Rbp1) are accumulated in bloom-forming cyanobacteria over the winter, we collected cyanobacterial cells from Meiliang Bay of Lake Taihu, a large shallow eutrophic lake in eastern China, in each month from November 2008 to October 2009. Microscopic examination showed that an overwhelming number of species in the samples were Microcystis (Fig. 5). According to the gvpA-gvpC intergenic sequence (25), Microcystis cells were classified into types 1 to 10 and miscellaneous (Fig. 6B). Type 1 remained predominant in the population from November 2008 to August 2009 and became minor in September and November 2009. Most other types showed great fluctuation in percentage from month to month.
Fig. 5.
Micrographs showing Microcystis colonies collected from Lake Taihu from November 2008 to October 2009.
Fig. 6.
Western blot detection of Rbp1 in Microcystis cells collected from Meiliang Bay of Lake Taihu in eastern China from November 2008 to October 2009. The monthly average water temperatures were based on the automatic monitoring records at the Taihu Ecosystem Research and Field Observation Station of the Nanjing Institute of Geography and Limnology. (A) Western blot analysis of Rbp1. I, Western blot detection; II, a part of the SDS-PAGE electrophoretogram showing that proteins were loaded at equal amounts; III, the average water temperature of the month. (B) The composition of the Microcystis population as shown with gvpA-gvpC intergenic sequences. The intergenic sequences designated types 1 to 10 in this figure are shown in Fig. S1 in the supplemental material.
According to the genome sequence data of two Microcystis aeruginosa strains, NIES-843 and PCC7806, two genes in NIES-843 and four genes in PCC7806 are highly similar to rbp1/rbp2 of Synechocystis at the amino acid level. It is possible to detect Rbp1 in Microcystis with the antiserum against Rbp1 of Synechocystis. Western blot analysis indeed showed a cold induction of Rbp1 in Microcystis PCC7806 (data not shown). Analyses of the monthly collected samples showed that Rbp1 was accumulated and maintained at high levels in Microcystis cells from November 2008 to March 2009 (Fig. 6A). During this period, the average water temperature in each month was below 13°C. With the increase of temperature, the abundance of Rbp1 was slightly reduced in April and May and greatly reduced in June, July, and August (Fig. 6A). As shown with types of gvpA-gvpC intergenic sequences, the Microcystis population remained relatively stable in the winter, spring, and summer (Fig. 6B), which suggests that these Microcystis species/strains indeed survived the long-term chill-light stress in the winter. In September and October of 2009, with the downshift of temperature, Rbp1 was accumulated again (Fig. 6A), and the Microcystis population underwent a great change in composition compared to that before (Fig. 6B). Apparently, Microcystis cells in the lake kept a high level of Rbp1 below 10°C over the winter until April and May, when the temperature allowed them to reinitiate growth (14, 21).
Rbp1 is proposed to be one of the key factors for the development of overwintering capability in cyanobacteria.
Previously, we reported that Synechocystis and Microcystis could rapidly lose viability under chill-light stress (27) and that preconditioning at 15°C greatly enhanced their chill-light tolerance (26). In this study, we showed that Rbp1 was accumulated during preconditioning and the accumulation of Rbp1 alone was sufficient to confer chill-light tolerance in Synechocystis. Western blot detection showed the accumulation of Rbp1 in Microcystis in low-temperature seasons. If the findings in Synechocystis can be extrapolated to Microcystis, the accumulation of Rbp1 should be an indication for its role in overwintering under natural conditions.
Before Rbp1, we had identified ccr1 (27), α-tocopherol (26), and Rbp3 (20) as factors involved in chill-light tolerance or regulation of gene expression required for chill-light tolerance. ccr1 was found to play an important role in the chill-light tolerance of 30°C-grown Synechocystis cells (without preconditioning at 15°C) (27). Because many genes are induced to express in cells during preconditioning, a gene with significant effects on the chill-light tolerance of 30°C-grown cells may not necessarily play an important role in the greatly enhanced chill-light tolerance of preconditioned cells. ccr1 is induced in Synechocystis at a late stage, namely, after 24 h, of exposure to 15°C in the light (27). Accordingly, it shows lesser or very slight effects on the acquisition of chill-light tolerance during preconditioning (RACLT, 41.8 to 90.0%). α-Tocopherol is essential for the acquired chill-light tolerance in Synechocystis, but it does not show a remarkable increase that is correlated with the development of chill-light tolerance during preconditioning (26). Probably, its level before preconditioning has been sufficient for supporting the increase of chill-light tolerance to the maximal level. Rbp3 affects the mRNA levels of fatty acid desaturase genes and ccr1 in Synechocystis. As a type II RNA-binding protein gene, rbp3 shows slight upregulation in cells transferred from 30°C to 15°C and affects fatty acid desaturation degree at both temperatures. Neither Rbp3 nor α-tocopherol is required for growth at 15°C.
The previously identified factors are more or less involved in ACLT and may indirectly cooperate with Rbp1 and each other to enhance the chill-light tolerance. However, Rbp1 is the only one showing early accumulation during preconditioning. We propose that the accumulation of Rbp1 is one of the key steps in preparation for overwintering in Synechocystis, Microcystis, and possibly other cyanobacterial species. With the accumulated Rbp1, certain requisite proteins may be maintained at relatively high levels under the chill-light stress in winter, and the activities of these proteins enable cyanobacteria to repair cell damage and restore cell activities when favorable conditions recur in the following spring.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (30825003) and the State Key Basic Research Development Program of China (2008CB418001).
We thank Qing Tang, Weizhi Li, and Xiangzhi Zhu for technical assistance and Fanxiang Kong of the Nanjing Institute of Geography and Limnology for providing the monthly average water temperatures of Lake Taihu.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Albà M. M., Pagès M. 1998. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 3:15–21 [Google Scholar]
- 2. Anderson S. L., McIntosh L. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 173:2761–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brunberg A. K., Blomqvist P. 2003. Recruitment of Microcystis (Cyanophyceae) from lake sediments: the importance of littoral inocula. J. Phycol. 39:59–63 [Google Scholar]
- 4. Ehira S., Ohmori M., Sato N. 2005. Identification of low-temperature-regulated ORFs in the cyanobacterium Anabaena sp. strain PCC 7120: distinguishing the effects of low temperature from the effects of photosystem II excitation pressure. Plant Cell Physiol. 46:1237–1245 [DOI] [PubMed] [Google Scholar]
- 5. Elhai J., Wolk C. P. 1988. A versatile class of positive-selection vectors basted on the nonviability of palindrome-containing plasmids that allows the cloning into long polylinkers. Gene 68:119–138 [DOI] [PubMed] [Google Scholar]
- 6. Gao H., Xu X. 2009. Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity prior to the loss of thylakoid membranes. FEMS Microbiol. Lett. 292:63–70 [DOI] [PubMed] [Google Scholar]
- 7. Hamano T., et al. 2004. Characterization of RNA-binding properties of three types of RNA-binding proteins in Anabaena sp. PCC 7120. Cell. Mol. Biol. (Noisy-le-Grand) 50:613–624 [PubMed] [Google Scholar]
- 8. Kim Y.-O., Kang H. 2006. The role of a zinc finger-containing glycine-rich RNA-binding protein during the cold adaptation process in Arabidopsis thaliana. Plant Cell Physiol. 47:793–798 [DOI] [PubMed] [Google Scholar]
- 9. Kruger N. J. 2002. The Bradford method for protein quantitation, p. 15–21 In Walker J. M. (ed.), The protein protocols handbook, 2nd ed Humana Press, Totowa, NJ [Google Scholar]
- 10. Los D. A., Murata N. 1999. Responses to cold shock in cyanobacteria. J. Mol. Microbiol. Biotechnol. 1:221–230 [PubMed] [Google Scholar]
- 11. Marin K., Huckauf J., Fulda S., Hagemann M. 2002. Salt-dependent expression of glucosylglycerol-phosphate synthase, involved in osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 184:2870–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maruyama K., Sato N., Ohta N. 1999. Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res. 27:2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Preston T., Stewart W. D. P., Reynolds C. S. 1980. Bloom-forming cyanobacterium Microcystis aeruginosa overwinters on sediment surface. Nature 288:365–367 [Google Scholar]
- 14. Reynolds C. S. 1973. Growth and buoyancy of Microcystis aeruginosa Kutz emend. Elenkin in a shallow eutrophic lake. Proc. R. Soc. Lond. 184:29–50 [Google Scholar]
- 15. Sakuragi Y., Maeda H., Dellapenna D., Bryant D. A. 2006. Alpha-tocopherol plays a role in photosynthesis and macronutrient homeostasis of the cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol. 141:508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato N. 1995. A family of cold-regulated RNA-binding protein genes in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 23:2161–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sato N., Wada A. 1996. Disruption analysis of the gene for a cold-regulated RNA-binding protein, rbpA1, in Anabaena: cold-induced initiation of the heterocyst differentiation pathway. Plant Cell Physiol. 37:1150–1160 [DOI] [PubMed] [Google Scholar]
- 18. Sugita C., Mutsuda M., Sugiura M., Sugita M. 1999. Targeted deletion of genes for eukaryotic RNA-binding proteins, Rbp1 and Rbp2, in the cyanobacterium Synechococcus sp. strain PCC 7942: Rbp1 is indispensable for cell growth at low temperatures. FEMS Microbiol. Lett. 176:155–161 [Google Scholar]
- 19. Takamura N., Yasuno M., Sugahara K. 1984. Overwintering of Microcystis aeruginosa Kutz. in a shallow lake. J. Plankton Res. 6:1019–1029 [Google Scholar]
- 20. Tang Q., Tan X., Xu X. 2010. Effects of a type-II RNA-binding protein on fatty acid composition in Synechocystis sp. PCC 6803. Chin. Sci. Bull. 55:2416–2421 [Google Scholar]
- 21. Tao Y., Kong F., Cao H., Zhang X. 2005. Simulative recruitment of Microcystis from the surface sediment in Taihu lake. J. Lake Sci. 17:231–236 [Google Scholar]
- 22. Vioque A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Visser P. M., Ibelings B. W., Mur L. R. 1995. Autumnal sedimentation of Microcystis spp. as result of an increase in carbohydrate ballast at reduced temperature. J. Plankton Res. 17:919–933 [Google Scholar]
- 24. Williams J. G. K. 1988. Construction of specific mutations in photosystem II photosynthetic reaction center by engineering methods in Synechocystis 6803. Methods Enzymol. 167:766–778 [Google Scholar]
- 25. Xu M., Xu X., Gao H., Kong R. 2007. The high variability of gvpA-gvpC regions in Microcystis. Prog. Nat. Sci. 17:1290–1295 [Google Scholar]
- 26. Yang Y., Yin C., Li W., Xu X. 2008. α-Tocopherol is essential for acquired chill-light tolerance in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 190:1554–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yin C., Li W., Du Y., Kong R., Xu X. 2007. Identification of a gene, ccr-1 (sll1242), required for chill-light tolerance and growth at 15°C in Synechocystis sp. PCC 6803. Microbiology 153:1261–1267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.