Abstract
Moraxella catarrhalis is a Gram-negative obligate aerobe that is an important cause of human respiratory tract infections. The M. catarrhalis genome encodes a predicted truncated denitrification pathway that reduces nitrate to nitrous oxide. We have previously shown that expression of both the M. catarrhalis aniA (encoding a nitrite reductase) and norB (encoding a putative nitric oxide reductase) genes is repressed by the transcriptional regulator NsrR under aerobic conditions and that M. catarrhalis O35E nsrR mutants are unable to grow in the presence of low concentrations of nitrite (W. Wang, et al., J. Bacteriol. 190:7762–7772, 2008). In this study, we constructed an M. catarrhalis norB mutant and showed that planktonic growth of this mutant is inhibited by low levels of nitrite, whether or not an nsrR mutation is present. To determine the importance of NorB in this truncated denitrification pathway, we analyzed the metabolism of nitrogen oxides by norB, aniA norB, and nsrR norB mutants. We found that norB mutants are unable to reduce nitric oxide and produce little or no nitrous oxide from nitrite. Furthermore, nitric oxide produced from nitrite by the AniA protein is bactericidal for a Moraxella catarrhalis O35E norB mutant but not for wild-type O35E bacteria under aerobic growth conditions in vitro, suggesting that nitric oxide catabolism in M. catarrhalis is accomplished primarily by the norB gene product. Measurement of bacterial protein S-nitrosylation directly implicates nitrosative stress resulting from AniA-dependent nitric oxide formation as a cause of the growth inhibition of norB and nsrR mutants by nitrite.
INTRODUCTION
Moraxella catarrhalis is an obligately aerobic Gram-negative bacterium that colonizes the human upper respiratory tract. For many decades, Moraxella catarrhalis was considered to be a harmless member of the normal flora and was known as Neisseria catarrhalis due to its morphological similarities to commensal Neisseria species (47). Recently, M. catarrhalis has been recognized as an important pathogen in both the upper and lower respiratory tracts (45). M. catarrhalis is the third leading bacterial cause of acute otitis media (32, 44, 67) in infants and very young children and the second most common bacterial cause of exacerbations of chronic obstructive pulmonary disease (COPD) in adults (43, 46, 58). It is estimated that 2 to 4 million exacerbations of COPD in the United States are attributable to M. catarrhalis infection each year (46). M. catarrhalis has been implicated in other infections, including community-acquired pneumonia (64), and extremely rarely may cause fatal bacteremia or pneumonia in patients with preexisting health conditions, such as immunodeficiency or impaired airway defenses (57).
Studies show that nasopharyngeal colonization with M. catarrhalis is common in infants and young children, and a high rate of colonization is associated with an increased risk of otitis media (16, 31). Recent surveys of nasopharyngeal colonization of Streptococcus pneumoniae, nontypeable Haemophilus influenzae, and M. catarrhalis showed that colonization with M. catarrhalis is highest among these pathogens surveyed in children between 2 and 12 years of age (42). M. catarrhalis frequently cocolonizes human nasopharyngeal mucosal surfaces with other bacteria, including Streptococcus pneumoniae (31, 42), Staphylococcus aureus (31), and H. influenzae (31, 68). Efforts to identify M. catarrhalis adhesins have uncovered several bacterial surface proteins that facilitate M. catarrhalis attachment to human epithelial cells in vitro (20, 27, 28, 36, 40, 41, 52, 54). Researchers have also identified several M. catarrhalis gene products that are important for growth under various in vitro conditions (1, 5, 10, 21, 41, 50). However, mechanisms of M. catarrhalis colonization of the nasopharyngeal mucosa remain to be fully elucidated. It was recently reported that M. catarrhalis forms biofilms on the middle ear mucosa in children with otitis media (24). It is likely that M. catarrhalis exists in biofilms together with other commensal bacteria in the human nasopharynx. In a chinchilla infection model, the persistence of M. catarrhalis within polymicrobial biofilms was shown to be facilitated by H. influenzae (4).
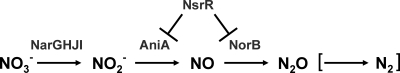
Gene expression during biofilm formation by M. catarrhalis in vitro has recently been examined (11, 41, 50, 71). Genes belonging to the M. catarrhalis truncated denitrification pathway (including the narGHJI cluster, aniA, and norB) (Fig. 1) were among the most highly upregulated genes in biofilm-grown cells (71). The M. catarrhalis transcriptional regulator NsrR represses the expression of both aniA and norB (Fig. 1) during aerobic growth. M. catarrhalis nsrR mutants are unable to grow in the presence of low concentrations of nitrite (72), but growth of an M. catarrhalis nsrR mutant in the presence of nitrite can be completely restored by disrupting the aniA gene to prevent the generation of nitric oxide (NO·) (72). These observations suggest that the reduction of nitrite to nitric oxide can be toxic for M. catarrhalis.
Fig. 1.
Truncated denitrification pathway in M. catarrhalis. The truncated denitrification pathway in M. catarrhalis involves three enzymatic steps: reduction of nitrate (NO3−) to nitrite (NO2−) by the nitrate reductase complex NarGHJI, reduction of NO2− to nitric oxide (NO·) by the nitrite reductase AniA, and reduction of NO· to nitrous oxide (N2O) by the nitric oxide reductase NorB. M. catarrhalis apparently lacks the ability to reduce N2O to N2 (indicated by brackets). The M. catarrhalis transcriptional regulator NsrR represses the expression of both AniA and NorB under aerobic growth conditions.
This study was undertaken to determine whether NorB metabolizes NO· to prevent its toxic effects. Here we show that the M. catarrhalis norB gene product reduces NO· to nitrous oxide (N2O) and that norB is required for M. catarrhalis growth in the presence of low levels of nitrite, in either the presence or absence of an nsrR mutation. We also show that NO· generated by AniA from the reduction of nitrite is bactericidal for an M. catarrhalis O35E norB mutant but not for an isogenic wild-type strain, suggesting that M. catarrhalis relies primarily on NorB for NO· detoxification. The AniA-dependent generation of NO· from nitrite increases bacterial protein S nitrosylation levels in M. catarrhalis strains expressing AniA, and increased levels of AniA expression in norB and nsrR mutants correlate with elevated levels of protein S nitrosylation. This demonstrates that NO· generated by the truncated denitrification pathway can cause nitrosative stress for M. catarrhalis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. catarrhalis strains used in this study are listed in Table 1. Bacterial culture conditions are as described previously (72). To measure the effect of nitrite on bacterial growth, a final concentration of 5 mM NaNO2 was added to brain heart infusion (BHI) broth. Bacterial growth was monitored turbidimetrically every hour or by testing bacterial viability at the beginning (0 h) and end (6 h) of aerobic growth in vitro.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description or genotype | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 43617 | Wild-type strain | ATCC |
| O35E | Wild-type strain | 26 |
| ETSU-9 | Wild-type strain | Steven Berk |
| O35E norB | norB::Kanr | This study |
| O35E ΔnsrR | nsrR deletion mutant | 72 |
| O35E ΔnsrR norB | nsrR norB double mutant | This study |
| O35E ΔaniA | aniA deletion mutant | This study |
| O35E ΔaniA norB | aniA norB double mutant | This study |
| 7169 | Wild-type strain | Anthony Campagnari |
| Plasmids | ||
| pWW115 | Specr, cloning vector for M. catarrhalis | 70 |
| pWW149 | pWW115 containing the wild-type ATCC 43617 norB gene | This study |
Whole-cell lysate preparation and Western blot analysis.
Whole-cell lysates were prepared from BHI agar-grown cells as described previously (49). Western blot analysis was performed as described previously (69), except that the mouse polyclonal AniA antibody (72) and monoclonal antibody (MAb) 10F3 were used as primary antibodies to detect the M. catarrhalis AniA and CopB proteins, respectively.
Construction of M. catarrhalis norB mutants.
The kanamycin-sensitive O35E ΔnsrR mutant and the kanamycin-resistant O35E aniA mutant were described previously (72). The kanamycin-sensitive O35E aniA deletion mutant, designated O35E ΔaniA, was constructed by transforming O35E aniA using the ΔANIA DNA fragment (72). One of the resulting kanamycin-sensitive transformants was confirmed to be a ΔaniA mutant by anchored PCR and sequence analysis (data not shown).
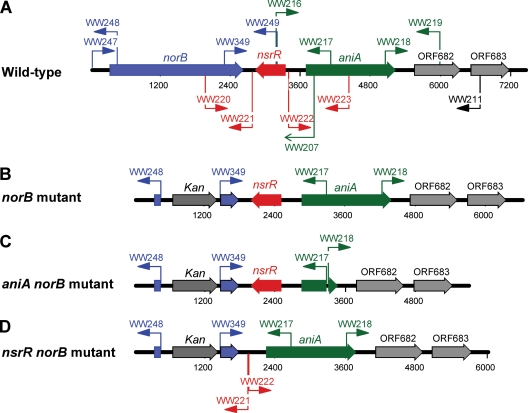
To construct M. catarrhalis O35E norB mutants, the oligonucleotide primer pairs WW247-WW248 and WW349-WW249 (Fig. 2A and Table 2) were used for PCR amplification using genomic DNA of M. catarrhalis ATCC 43617 as the template. The oligonucleotide primers WW248 and WW349 contain nucleotides (Table 2, underlined sequences) that are identical to the 5′ and the 3′ nucleotide residues of the kan cassette (71) from plasmid pAC7 (74). PCR amplification products were purified using a gel extraction kit (Qiagen) and, together with the kan cassette (71), were used as DNA templates for sequential overlapping extension PCR amplifications (29) (Table 3). The final amplicon, designated ΔNORB-KAN, was confirmed by DNA sequence analysis (data not shown) and used to transform wild-type O35E, the O35E ΔaniA mutant, the O35E ΔnsrR mutant, wild-type M. catarrhalis 7169, and ETSU-9. Kanamycin-resistant transformants were confirmed as the O35E norB mutant (Fig. 2B), O35E aniA norB mutant (Fig. 2C), O35E nsrR norB mutant (Fig. 2D), and ETSU-9 norB mutant (Fig. 2B) strains, respectively, by anchored PCR using the oligonucleotide primer pair WW247-WW217 (Fig. 2A) followed by sequence analysis (data not shown).
Fig. 2.
Schematic representation of M. catarrhalis gene products involved in the truncated denitrification pathway and construction of relevant mutants. Schematic diagram of the M. catarrhalis chromosomal locus containing the norB, nsrR, and aniA genes and flanking regions in the wild-type O35E strain (A), the O35E norB mutant (B), the O35E aniA norB mutant (C), and the O35E nsrR norB mutant (D). The relative positions of the different primers used for PCR are indicated by the arrows.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| WW247 | TAGGATCCAATCACACTTAGGATTATCA |
| WW248 | CGGAGCCTGCAGCCCGGGTGGTACGGTAAATCTCAAAa |
| WW349 | CTAGATTTAGATGTCGGGCTTTGTGCTGTTGATTGTb |
| WW249 | AGTTGAGCTCGCTTAAAGTCGTTGACAGTGC |
Nucleotides of WW248 that overlap primer WW195 are underlined.
Nucleotides of WW349 that overlap primer WW196 are underlined.
Table 3.
PCR extension
| Primer pair | DNA template(s) | DNA polymerase | PCR-amplified DNA fragments |
|---|---|---|---|
| WW247-WW248 | ATCC 43617 genomic DNA | Pfu | up |
| WW349-WW249 | ATCC 43617 genomic DNA | Pfu | down |
| WW195-WW196 | pAC7 plasmid DNA | Pfu | kan |
| WW247-WW196 | up and kan PCR DNAs | ExTaq | up-kan |
| WW195-WW249 | kan and down PCR DNAs | ExTaq | kan-down |
| WW247-WW249 | up-kan and kan-down PCR DNAs | ExTaq | ΔNORB-KAN |
Repair of the M. catarrhalis O35E norB mutant.
A DNA fragment containing the wild-type norB gene was amplified by PCR using primer pair WW247-WW207 (Fig. 2A) with genomic DNA of M. catarrhalis ATCC 43617 as the DNA template. This DNA fragment was used to transform the kanamycin-resistant O35E norB mutant. One of the kanamycin-sensitive transformants, designated O35E norB (norB), was confirmed to contain a wild-type ATCC 43617 norB gene by anchored PCR with primer pair WW220-WW217 (Fig. 2A), which was followed by nucleotide sequence analysis (data not shown).
Measurement of NO. consumption.
Consumption of chemically generated NO· by M. catarrhalis cells was measured as described previously (72). Briefly, wild-type O35E, norB mutant, nsrR norB mutant, and aniA norB mutant M. catarrhalis cells were grown in BHI medium to an optical density at 600 nm (OD600) of 2.0. Cells were washed and resuspended in freshly prepared BHI to an OD600 of 1.0. Approximately 3 ml of cells was assayed for NO· consumption in a sealed vessel. At approximately 0.5 min, the NO· -releasing reagent Proli-NO (half-life of 1.8 s) was added to the cell suspension to a final concentration of 10 μM, which releases a total of 20 μM NO· The concentration of dissolved NO· remaining over time was monitored using an ISO-NOPMC Mark II electrode (WPI Instruments).
Measurement of NO2− consumption.
NO2− consumption by M. catarrhalis cells was measured as described previously (72). Briefly, after M. catarrhalis strains were resuspended in BHI to an OD600 of 1.0 as described immediately above, NaNO2 was added to a final concentration of 5 mM. The concentration of remaining NO2− was determined using the Griess reaction as described previously (72).
Measurement of NO· and N2O production.
Production of NO· and N2O by M. catarrhalis cells from the reduction of NO2− was measured as described previously (72), except that a lower concentration of NO2− (500 μM) was used to allow NO· and N2O to be measured simultaneously. The NO· level was monitored as described above, and the N2O level was monitored using an oxygen-insensitive, N2O-specific probe (N2O-50-3112 [Unisense AS, Aarhus, Denmark]) connected to a PA2000 picoammeter (Unisense AS) and an analog-to-digital converter (A/D converter), ADC 216 (Unisense AS).
Detection of bacterial protein S-nitrosylation.
A patented (Glythera Limited, United Kingdom) S-nitrosothiol (SNO) group binding reagent, designated SNOB, was used to detect protein S-nitrosylation. The SNOB reagents specifically bind SNO protein groups in a single chemical step. The biotin tag of the SNOB reagent allows the visualization of S-nitrosylated proteins using a streptavidin-horseradish peroxidase (HRP) conjugate in a Western blot assay, in which the intensities of protein bands reflect the relative levels of S nitrosylation. Briefly, an M. catarrhalis cell suspension (at a cell density of 260 Klett units) was added to 1 ml BHI containing SNOB reagent, with or without nitrite. The final concentrations of SNOB and nitrite were 1 mM and 3 mM (if added), respectively, and the final cell density was approximately 1 OD600 (5 × 108 CFU/ml). The mixtures were incubated at 30°C for 30 min, bacterial cells harvested by centrifugation, and cell pellets washed twice with ice-cold 1× phosphate-buffered saline (PBS) to remove unbound SNOB reagent. Whole-cell lysates (0.2 ml each) were prepared for Western blotting as described previously (14), except that the streptavidin–β-peroxidase (POD) conjugate (Roche) was used to detect the biotin tag of SNOB bound to S-nitrosylated proteins.
RESULTS
Construction of M. catarrhalis norB mutants.
The wild-type M. catarrhalis O35E genetic locus containing the norB, nsrR, and aniA genes is shown in a schematic diagram (Fig. 2A). Mutations in norB (Fig. 2B) were introduced using a PCR amplicon, ΔNORB-KAN, into the wild-type M. catarrhalis O35E, ETSU-9, and 7169 strain backgrounds by allelic exchange to replace the DNA sequence between the oligonucleotide primers WW248 and WW349 with a kan resistance cassette from plasmid pAC7, as described in Materials and Methods. Additional O35E norB mutations were constructed in both the O35E ΔaniA mutant (described in Materials and Methods) and the O35E ΔnsrR mutant (72) backgrounds, resulting in an M. catarrhalis O35E aniA norB mutant (Fig. 2C) and an O35E nsrR norB mutant (Fig. 2D), respectively. All norB mutants were confirmed by anchored PCR and DNA sequence analysis.
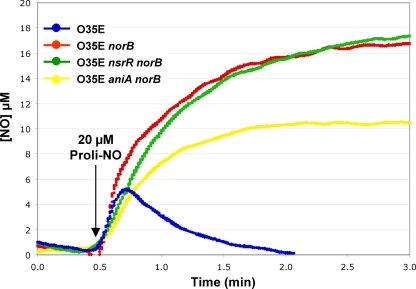
M. catarrhalis NorB is required for NO consumption.
The levels of consumption of chemically generated NO· by wild-type M. catarrhalis O35E and three norB mutant strains were compared as described in Materials and Methods. Wild-type O35E cells (Fig. 3, blue line) consumed NO·, as observed previously (72). The norB single mutant (Fig. 3, red line), the O35E nsrR norB mutant (Fig. 3, green line), and the O35E aniA norB mutant (Fig. 3, yellow line) strains failed to exhibit NO· consumption. After the addition of the NO· -releasing agent Proli-NO, NO· levels accumulated and were not consumed in the three O35E-derived strains carrying a norB mutation (Fig. 3). These assays were conducted in a sealed vessel, in which the remaining oxygen is consumed very rapidly, resulting in anaerobic conditions. These results indicate that the M. catarrhalis norB gene product is required for NO· consumption.
Fig. 3.
Consumption of chemically generated NO· by the wild-type and mutant strains of M. catarrhalis O35E. Cell suspensions of the wild-type strain, the norB mutant, the nsrR norB mutant, and the aniA norB mutant were exposed to 20 μM NO· produced by the addition of 10 μM proline nitric oxide (Proli-NO). The dissolved NO· concentration was monitored using an NO· -specific electrode. For reference, the NO· -consuming activity of BHI medium was determined (data not shown). This experiment was performed three times, and representative data are shown.
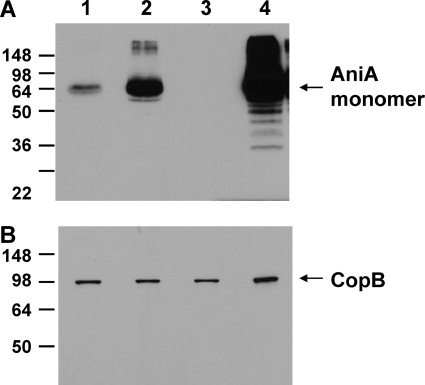
Analysis of AniA protein expression.
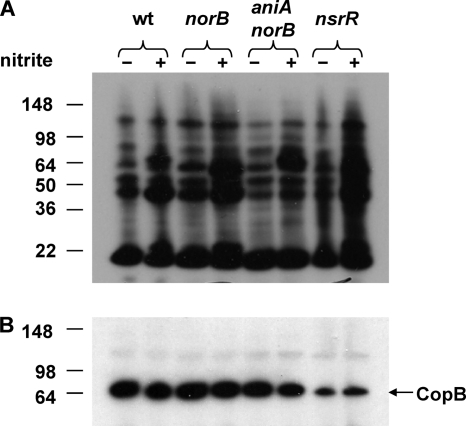
In this study, nitrite was used as a source of NO· biologically generated by the action of the M. catarrhalis AniA protein. The expression of AniA protein by M. catarrhalis O35E strains was determined by Western blotting using a mouse polyclonal AniA antibody (72). Interestingly, expression of AniA protein in the M. catarrhalis O35E norB mutant (Fig. 4A, lane 2) was higher than in wild-type O35E (Fig. 4A, lane 1). As expected, the M. catarrhalis O35E aniA norB mutant (Fig. 4A, lane 3) did not express AniA, and the M. catarrhalis O35E nsrR norB mutant (Fig. 4A, lane 4) exhibited the highest expression of AniA, similar to that of an O35E nsrR mutant (72). Expression of the M. catarrhalis CopB protein was measured as a loading control (Fig. 4B).
Fig. 4.
Expression of AniA protein in wild-type and mutant strains of M. catarrhalis O35E. Whole-cell lysates were probed by Western blot analysis with polyclonal AniA antiserum (A) or with the CopB-specific MAb 10F3 (26) (B) as the primary antibody. Lane 1, wild-type O35E; lane 2, O35E norB; lane 3, O35E norB aniA; lane 4, O35E norB nsrR. The position of the putative AniA monomers is indicated by an arrow on the right side of panel A. The CopB outer membrane protein was used as a loading control, and its position is indicated by an arrow on the right side of panel B. Molecular weight position markers (in thousands) are present on the left side of each panel.
AniA is required for NO2− consumption.
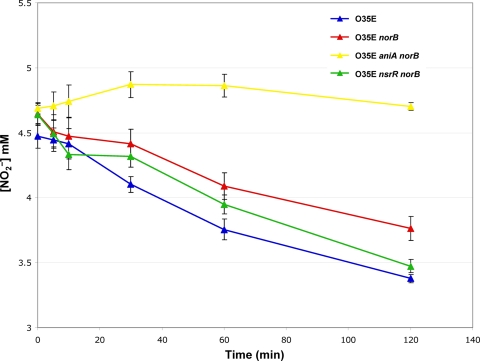
To confirm that the AniA protein expressed by M. catarrhalis norB mutants is functional, NO2− consumption by mutant M. catarrhalis strains was determined as described in Materials and Methods. The three AniA-expressing M. catarrhalis strains, including wild-type O35E (Fig. 5, blue line), the norB mutant (Fig. 5, red line), and the nsrR norB double mutant (Fig. 5, green line) were able to consume NO2−. In contrast, the O35E aniA norB mutant (Fig. 5, yellow line) did not consume NO2−. This result confirmed that the AniA proteins expressed by wild-type O35E, the O35E norB mutant, and the O35E nsrR norB mutant are functional. Although both the O35E norB and O35E nsrR norB mutants expressed higher levels of AniA than the parental wild-type strain O35E (Fig. 4A), the two norB mutants did not consume NO2− significantly faster than wild-type O35E (Fig. 5).
Fig. 5.
NO2− consumption by wild-type and mutant strains of M. catarrhalis O35E. Cell suspensions of the wild-type strain, the norB mutant, the nsrR norB mutant, and the aniA norB mutant in BHI medium supplemented with 5 mM NaNO2 were monitored for the presence of NO2− using the Griess reaction. The data shown are the averages of results from three independent experiments, with standard errors indicated.
Production of NO· and N2O by wild-type and mutant M. catarrhalis O35E strains.
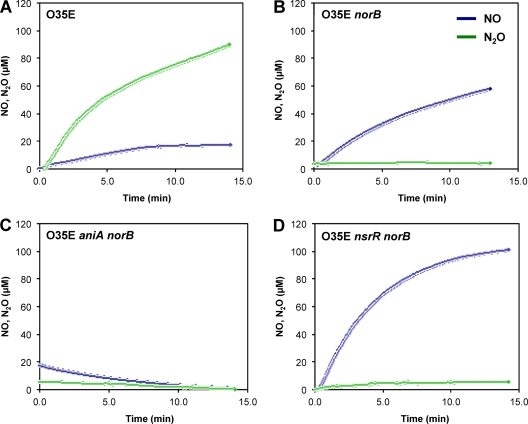
Following the addition of 500 μM NaNO2, wild-type M. catarrhalis simultaneously produced and consumed NO· to form the final product, N2O (Fig. 6A). The nsrR norB and norB mutants consumed nitrite to form NO·, which accumulated to high levels in both cultures (Fig. 6B and D). A low level of erratic signal of the N2O sensor observed in the experiments with the O35E norB and O35E nsrR norB mutants was due to chemical reduction of NO· to N2O within the sensor compartment occurring at high concentrations of NO· As expected, the O35E aniA norB mutant failed to generate either NO· or N2O from nitrite (Fig. 6C). This study confirms that AniA is required for nitrite reduction to NO· and that NorB reduces NO· to N2O in the truncated M. catarrhalis denitrification pathway.
Fig. 6.
Production of NO and N2O from NO2− by wild-type and mutant strains of M. catarrhalis O35E. Cell suspensions of the wild-type strain (A), the norB mutant (B), the aniA norB mutant (C), and the aniA norB mutant (D) in BHI medium supplemented with 500 μM NaNO2 were monitored for 15 min for the presence of NO· (blue lines) using an NO· -specific electrode. Probe specificity was affirmed by the ability of an NO· scavenger, Carboxy-PTIO, to quench measurable signal (not shown). The presence of N2O (green lines) was measured simultaneously using an oxygen-insensitive, N2O-specific probe as described in Materials and Methods. The data shown are a representative set of results from two independent experiments.
Effect of NO2− on growth of the wild-type, mutant, and repaired mutant strains of M. catarrhalis.
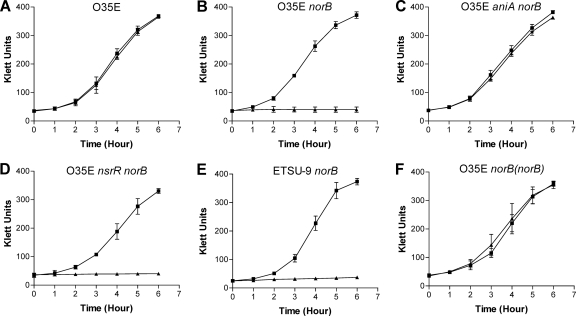
To investigate the biological relevance of the M. catarrhalis norB gene product, the effect of nitrite at low concentrations on the growth of M. catarrhalis O35E strains was examined. The presence of 5 mM NaNO2 had no effect on the aerobic growth of wild-type O35E (Fig. 7A), as reported previously (72). In contrast, the growth of the O35E norB mutant and the O35E nsrR norB mutant was completely inhibited by 5 mM NO2− (Fig. 7B and D). This NO2−-dependent growth inhibition was completely relieved by disrupting the aniA gene in an O35E norB mutant background (Fig. 7C). The growth of the M. catarrhalis 7169 norB mutant (data not shown) and the ETSU-9 norB mutant (Fig. 7E) was also completely inhibited by NO2−, suggesting that the growth inhibition of a norB mutant by nitrite is not strain specific.
Fig. 7.
Effect of NO2− on growth of the wild-type, mutant, and repaired mutant strains of M. catarrhalis. Cells of the wild-type strain (A), the O35E norB mutant (B), the O35E aniA norB mutant (C), the O35E nsrR norB mutant (D), the ETSU-9 norB mutant (E), and the repaired strain O35E norB(norB) (F) were grown in BHI medium (■) or BHI containing 5 mM NaNO2 (▴). The data shown are the means of results from three independent growth experiments.
Gene repair was performed to restore a functional norB gene in the chromosome of the O35E norB mutant, resulting in the strain O35E norB(norB) as described in Materials and Methods. Growth of the repaired O35E norB(norB) strain was not affected by 5 mM NO2− (Fig. 7F). These experiments confirmed that a functional norB gene is essential for M. catarrhalis to grow in the presence of nitrite at low concentrations.
NO· produced from nitrite by the AniA protein is bactericidal for a Moraxella catarrhalis O35E norB mutant.
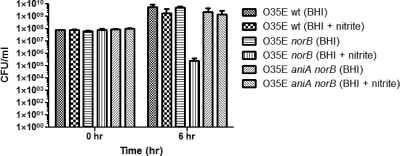
The viability of M. catarrhalis O35E strains under aerobic growth at 37°C in BHI with or without the addition of nitrite at low concentrations was examined. Wild-type M. catarrhalis O35E, norB mutant, and aniA norB mutant cells were used to inoculate BHI with or without the addition of 5 mM NaNO2 to a final concentration of approximately 108 CFU/ml (Fig. 8, 0 h). The growth of both the wild-type O35E and aniA norB mutant strains was not affected by nitrite, as viable counts for these strains increased to ∼9 × 109 CFU/ml in BHI either with or without NaNO2 (Fig. 8, 6 h). In contrast, viable counts of the O35E norB mutant increased to ∼9 × 109 CFU/ml in BHI but decreased to 105 CFU/ml in BHI containing nitrite. NO· produced from nitrite is bactericidal for an M. catarrhalis O35E norB mutant and the ETSU-9 norB mutant (data not shown) but not for wild-type M. catarrhalis, indicating that M. catarrhalis relies on NorB for NO· detoxification.
Fig. 8.
NO· produced from nitrite by the AniA protein is bactericidal for an M. catarrhalis O35E norB mutant. Cells of the wild-type strain (wt), the norB mutant (norB), and the aniA norB mutant (aniA norB) were grown in BHI without or with the addition of 5 mM nitrite (+ nitrite). Aliquots from each culture were taken at the beginning (0 h) and end (6 h) of aerobic growth for the determination of viable cell counts (CFU/ml). The data shown are the means of results from three independent growth experiments.
NO· produced from nitrite by the AniA protein increases bacterial protein S-nitrosylation.
To investigate whether NO· produced from nitrite by the AniA protein is responsible for the nitrite-dependent growth inhibition of both the M. catarrhalis O35E nsrR mutant and the O35E norB mutant, bacterial protein S-nitrosylation profiles were measured as described in Materials and Methods. Studies have shown that nitrite/nitrate is present in various animal tissues at levels between 0.5 and 50 μM (2, 48), which can support NO generation (38). We have briefly determined that BHI contains approximately 50 μM nitrate (data not shown), which can be reduced to a trace amount of NO· Protein S nitrosylation in M. catarrhalis cells grown in medium without the addition of nitrite was measured as the steady-state endogenous S-nitrosylation level (Fig. 9A, − lanes). Increased S-nitrosylation was observed in all three AniA-expressing O35E strains in the presence of nitrite, especially a band slightly above 22 kDa that was present only in all three AniA-expressing O35E strains in the presence of nitrite (Fig. 9A, lanes wt +, norB +, and nsrR +). Increased S-nitrosylation levels, indicative of nitrosative stress, correlated with AniA expression levels in the presence of nitrite: highest in the O35E nsrR mutant (72), modest in wild-type O35E (Fig. 4, lane 1), and intermediate in the O35E norB mutant (Fig. 4, lane 2). As expected, the O35E aniA norB mutant did not express AniA protein (Fig. 4, lane 3), and its overall level of protein S-nitrosylation was not substantially affected by the presence of nitrite (Fig. 9, lane aniA norB +), except in a protein migrating at ∼64 kDa. This might reflect the high sensitivity of the SNOB reagent for S-nitrosothiols and the nonenzymatic generation of NO· from the added nitrite. Fig. 9 shows representative data from two experiments. The identities of S-nitrosylated proteins are presently unknown. These observations suggest that high levels of nitrosative stress can inhibit the growth of M. catarrhalis.
Fig. 9.
Bacterial cellular protein S-nitrosylation profiles of wild-type and mutant strains of M. catarrhalis O35E. (A) Cell suspensions of the wild-type strain, the norB mutant, the aniA norB mutant, and the nsrR mutant were incubated with SNOB reagent in medium with (+) or without (−) added nitrite. S-nitrosylated proteins were visualized using streptavidin-HRP. (B) M. catarrhalis CopB was detected as a loading control.
DISCUSSION
Bacterial denitrification pathways, consisting of four sequential enzymatic steps that reduce nitrate to gaseous nitrogen, have been identified in bacteria living in various environments (reviewed in reference 75). Anaerobic bacteria use denitrification pathways as an alternative means of energy production, with nitrogen oxides functioning as electron acceptors (75). Recently, denitrification by bacterial pathogens has attracted increasing interest (51). The nitrate reductase complex NarGHJI was shown to be involved in Mycobacterium bovis BCG virulence in an animal model (73) and was implicated in Pseudomonas aeruginosa biofilm formation and virulence (65, 66). The Brucella melitensis denitrification pathway is required for virulence in mice (6, 23). Denitrification promotes the growth of Neisseria meningitidis, a strictly aerobic human pathogen, under oxygen-limited conditions (3, 55). Barth and colleagues (8) recently showed that, among all Neisseria strains tested, a nitrate reductase complex could be identified only in Neisseria mucosa. However, a nitrite reductase (AniA or NirK) and nitric oxide reductase (NorB) were present in all Neisseria species tested. Interestingly, a nitrous oxide reductase (Nos) is present in some commensal Neisseria species but absent from the pathogenic species Neisseria gonorrhoeae and N. meningitidis. The fumarate and nitrate reductase (FNR), NarQP, and NsrR transcriptional regulators are highly conserved in Neisseria species. The transcriptional regulator NsrR is conserved in several M. catarrhalis clinical isolates; however, the ATCC 43617 genome does not encode an FNR protein regulator (72). Instead, the M. catarrhalis genome encodes a two-component system NarXL and a homologue of the DnrD protein which lacks the [4Fe-4S] center and belongs to a new subgroup of the FNR regulator family (72). The nitric oxide reductase NorB is responsible primarily for NO detoxification (61, 62) in Neisseria species, and a cycP gene product also functions in NO detoxification (3, 62). The M. catarrhalis ATCC 43617 genome has 18 open reading frames (ORFs) encoding cytochrome c proteins (71). Six of the M. catarrhalis cytochrome c proteins contain a heme-binding motif (CXXCH) (15). Four of these heme-binding cytochrome c proteins have a signal peptide sequence identified with SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/). Only ORF 192 (152 amino acids; also known as Msp22 [56]) has both a signal peptide sequence and a single CXXCH motif that is located at its C terminus, which are features specific for cytochrome c′ (cycP). However, a role for the ORF 192 protein in M. catarrhalis NO· detoxification has not been established.
It was recently reported that M. catarrhalis forms biofilms on the mucosal surface of the middle ear in children with chronic otitis media (24). Studies of M. catarrhalis gene expression have revealed that the expression of the enzymes comprising the truncated M. catarrhalis denitrification pathway was highly upregulated in biofilm-grown M. catarrhalis cells in vitro (71). Elevated expression of denitrification genes has been reported in other pathogens, including N. gonorrhoeae (17) and P. aeruginosa (65), during growth in biofilms in vitro. M. catarrhalis is known to reduce NO3− (reviewed in reference 12), although the genes that encode this activity (likely narGHJI) have not been fully described. An initial study showed that an M. catarrhalis narGH mutant grows as well in a continuous-flow biofilm system as its wild-type parent (71). The importance of denitrification for M. catarrhalis biofilm formation remains to be fully elucidated. Recent studies showed that a low level of NO· can promote biofilm dispersal in P. aeruginosa (65) and N. gonorrhoeae (18). In contrast, high chemically generated levels of NO· inhibit early-stage biofilm formation but enhance N. gonorrhoeae growth in established biofilms (18).
The present study shows that a functional norB gene is required for M. catarrhalis to reduce chemically generated NO· (Fig. 3, blue line), as NO· is not utilized by an O35E norB isogenic mutant (Fig. 3, red, green, and yellow lines). It is not apparent why M. catarrhalis norB mutant cells express higher levels of AniA (Fig. 4A, lane 2) than the wild type (Fig. 4A, lane 1), even when cells are grown on BHI agar without the addition of nitrite. The NsrR-repressed aerobic expression of M. catarrhalis AniA is insensitive to chemically generated NO· (from 50 μM spermine NONOate; half-life, about 39 min at 37°C) in wild-type O35E cells (72). Although both norB and nsrR norB mutants express significantly higher levels of AniA protein (Fig. 4, lanes 2 and 4), they do not reduce nitrite faster (Fig. 5, red and green lines) than the wild type (Fig. 5, blue line). In contrast, an M. catarrhalis nsrR mutant reduces nitrite more rapidly than the wild type (72). This is attributable to the accumulation of NO· in the absence of NorB, some of which undergoes auto-oxidation to nitrite (30), and the inability of the Griess reagent to distinguish NO· and nitrite.
Bacteria employ different mechanisms for NO· detoxification to withstand nitrosative stress. An NsrR-regulated and NO· -inducible flavohemoglobin protein (Hmp) is the major NO· detoxifier in Salmonella enterica serovar Typhimurium (7, 22) and Escherichia coli (19). The enteric pathogen S. Typhimurium requires Hmp for virulence in mice expressing inducible NO· synthase (7). In E. coli, Hmp is required for resistance to nitrosative stress (25) and for bacterial survival within macrophages (63). The M. catarrhalis ATCC 43617 genome does not contain a gene encoding a Hmp-like protein (72). Furthermore, a low level of nitrite completely inhibits the growth of an M. catarrhalis O35E norB mutant (Fig. 7B), strongly suggesting that this bacterium relies on NorB for NO· detoxification. It has not been immediately obvious why nitrite supplementation also completely inhibits the aerobic growth of an M. catarrhalis nsrR mutant, because this mutant rapidly reduces nitrite to nitrous oxide with little or no detectable steady-state NO· accumulation (72). To investigate whether an increased flux of NO· is responsible for nitrite-related inhibition of the aerobic growth of an M. catarrhalis nsrR mutant, we examined bacterial protein S-nitrosylation profiles.
In an enzyme-independent chemical reaction, NO· can bond covalently with the thiol groups of protein cysteine residues to form S-nitrosothiols (SNOs), a posttranslational modification of cellular proteins known as S-nitrosylation (59, 60). S-nitrosylation has been implicated in mammalian cell apoptosis (9, 33–35, 39, 53). A recent study reported that bacterial proteins involved in NO· detoxification, including NorB of N. meningitidis and the flavohemoglobins (Hmp) of S. enterica and E. coli, prevent host cell SNO formation (37).
S-nitrosylation of bacterial proteins was determined in wild-type O35E, norB mutant, aniA norB mutant, and nsrR mutant M. catarrhalis cells as described in Materials and Methods. The source of additional NO· was nitrite that was reduced by the activity of the M. catarrhalis AniA protein. The O35E aniA norB mutant does not express AniA (Fig. 4A, lane 3) and is unable to reduce nitrite (Fig. 5, yellow line). The O35E norB mutant and nsrR mutant strains express significantly higher levels of AniA (Fig. 4A, lane 2, and reference 72) than the parental strain, O35E (Fig. 4A, lane 1). By Western blot analysis, a band slightly above 22 kDa is present only in the three AniA-expressing O35E strains, and an overall increase in S-nitrosylation was observed in all AniA-expressing M. catarrhalis strains in the presence of nitrite (Fig. 9A, wt +, norB +, and nsrR + lanes). Increased S-nitrosylation correlated well with levels of AniA expression and was highest in an nsrR mutant (Fig. 9A, nsrR + lane) and lowest in wild-type O35E (Fig. 9A, wt + lane). These observations strongly suggest that NO· is the inhibitory factor responsible for the nitrite-dependent inhibition of the growth of the norB and nsrR mutant strains. Although the nsrR mutant exhibits low measurable steady-state NO· concentrations (72), the increased flux of NO· produced from nitrite reduction appears to cause nitrosative stress, which could account for growth inhibition by nitrite. The specific protein targets of S-nitrosylation in M. catarrhalis remain to be identified.
The AniA protein of N. gonorrhoeae is expressed in vivo during infection in humans (13). The M. catarrhalis AniA protein, also known as Msp78 (56), has recently been shown to be present in patients during COPD exacerbations associated with M. catarrhalis infection (56). The transcriptional regulatory network that controls the expression of the truncated denitrification pathway in M. catarrhalis under various in vitro growth conditions is under active investigation. It is hoped that such work will help to elucidate the importance of denitrification in M. catarrhalis pathogenesis.
ACKNOWLEDGMENTS
This study was supported by FDA operating funds to W.W., PHS grant AI39557 to F.C.F., and PHS grant AI036344 to E.J.H.
We thank John Nelson, Anthony Campagnari, and Steven Berk for providing clinical isolates of M. catarrhalis used in this study, Flora Lichaa and Brian Mocca for assistance with NO· bactericidal experiments, and Willie F. Vann for information regarding the SNOB reagent.
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Aebi C., et al. 1996. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect. Immun. 64:2024–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akcay Y. D., Yalcin A., Sozmen E. Y. 2005. The effect of melatonin on lipid peroxidation and nitrite/nitrate levels, and on superoxide dismutase and catalase activities in kainic acid-induced injury. Cell. Mol. Biol. Lett. 10:321–329 [PubMed] [Google Scholar]
- 3. Anjum M. F., Stevanin T. M., Read R. C., Moir J. W. 2002. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184:2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Armbruster C. E., et al. 2010. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1:e00102–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Attia A. S., et al. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 73:2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baek S. H., Rajashekara G., Splitter G. A., Shapleigh J. P. 2004. Denitrification genes regulate Brucella virulence in mice. J. Bacteriol. 186:6025–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bang I. S., et al. 2006. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 281:28039–28047 [DOI] [PubMed] [Google Scholar]
- 8. Barth K. R., Isabella V. M., Clark V. L. 2009. Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology 155:4093–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benhar M., Stamler J. S. 2005. A central role for S-nitrosylation in apoptosis. Nat. Cell Biol. 7:645–646 [DOI] [PubMed] [Google Scholar]
- 10. Bonnah R. A., Wong H., Loosmore S. M., Schryvers A. B. 1999. Characterization of Moraxella (Branhamella) catarrhalis lbpB, lbpA, and lactoferrin receptor orf3 isogenic mutants. Infect. Immun. 67:1517–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budhani R. K., Struthers J. K. 1998. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 42:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Catlin B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark V. L., Knapp J. S., Thompson S., Klimpel K. W. 1988. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb. Pathog. 5:381–390 [DOI] [PubMed] [Google Scholar]
- 14. Cope L. D., et al. 1994. The 100 kDa heme:hemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13:863–873 [DOI] [PubMed] [Google Scholar]
- 15. Cross R., Aish J., Paston S. J., Poole R. K., Moir J. W. 2000. Cytochrome c′ from Rhodobacter capsulatus confers increased resistance to nitric oxide. J. Bacteriol. 182:1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faden H. S., Harabuchi Y., Hong J. J., and Tonawanda/Williamsburg Pediatrics 1994. Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media. J. Infect. Dis. 169:1312–1317 [DOI] [PubMed] [Google Scholar]
- 17. Falsetta M. L., et al. 2009. Transcriptional profiling identifies the metabolic phenotype of gonococcal biofilms. Infect. Immun. 77:3522–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Falsetta M. L., McEwan A. G., Jennings M. P., Apicella M. A. 2010. Anaerobic metabolism occurs in the substratum of gonococcal biofilms and may be sustained in part by nitric oxide. Infect. Immun. 78:2320–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Filenko N., et al. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 189:4410–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forsgren A., Brant M., Karamehmedovic M., Riesbeck K. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 71:3302–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furano K., Campagnari A. A. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 71:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilberthorpe N. J., Lee M. E., Stevanin T. M., Read R. C., Poole R. K. 2007. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-gamma-stimulated J774.2 macrophages. Microbiology 153:1756–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haine V., Dozot M., Dornand J., Letesson J. J., De B. X. 2006. NnrA is required for full virulence and regulates several Brucella melitensis denitrification genes. J. Bacteriol. 188:1615–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hall-Stoodley L., et al. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hausladen A., Gow A. J., Stamler J. S. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 95:14100–14105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helminen M. E., et al. 1993. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect. Immun. 61:2003–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holm M. M., Vanlerberg S. L., Foley I. M., Sledjeski D. D., Lafontaine E. R. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holm M. M., Vanlerberg S. L., Sledjeski D. D., Lafontaine E. R. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horton R. M., Cai Z., Ho S. N., Pease L. R. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 30. Ignarro L. J., Fukuto J. M., Griscavage J. M., Rogers N. E., Byrns R. E. 1993. Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from l-arginine. Proc. Natl. Acad. Sci. U. S. A. 90:8103–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jourdain S., et al. 14 December 2010. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2010.03410.x [DOI] [PubMed] [Google Scholar]
- 32. Karalus R., Campagnari A. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547–559 [DOI] [PubMed] [Google Scholar]
- 33. Kim Y. M., Bombeck C. A., Billiar T. R. 1999. Nitric oxide as a bifunctional regulator of apoptosis. Circ. Res. 84:253–256 [DOI] [PubMed] [Google Scholar]
- 34. Kim Y. M., Talanian R. V., Billiar T. R. 1997. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J. Biol. Chem. 272:31138–31148 [DOI] [PubMed] [Google Scholar]
- 35. Klassen S. S., Rabkin S. W. 2007. The role of p53 in nitric oxide-induced cardiomyocyte cell death. DNA Cell Biol. 26:465–475 [DOI] [PubMed] [Google Scholar]
- 36. Lafontaine E. R., et al. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laver J. R., et al. 2010. Bacterial nitric oxide detoxification prevents host cell S-nitrosothiol formation: a novel mechanism of bacterial pathogenesis. FASEB J. 24:286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Cui H., Kundu T. K., Alzawahra W., Zweier J. L. 2008. Nitric oxide production from nitrite occurs primarily in tissues not in the blood: critical role of xanthine oxidase and aldehyde oxidase. J. Biol. Chem. 283:17855–17863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li J., Bombeck C. A., Yang S., Kim Y. M., Billiar T. R. 1999. Nitric oxide suppresses apoptosis via interrupting caspase activation and mitochondrial dysfunction in cultured hepatocytes. J. Biol. Chem. 274:17325–17333 [DOI] [PubMed] [Google Scholar]
- 40. Lipski S. L., Akimana C., Timpe J. M., Wooten R. M., Lafontaine E. R. 2007. The Moraxella catarrhalis autotransporter McaP is a conserved surface protein that mediates adherence to human epithelial cells through its N-terminal passenger domain. Infect. Immun. 75:314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luke N. R., Jurcisek J. A., Bakaletz L. O., Campagnari A. A. 2007. Contribution of Moraxella catarrhalis type IV pili to nasopharyngeal colonization and biofilm formation. Infect. Immun. 75:5559–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mackenzie G. A., Leach A. J., Carapetis J. R., Fisher J., Morris P. S. 2010. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect. Dis. 10:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murphy T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murphy T. F. 2000. Bacterial otitis media: pathogenetic considerations. Pediatr. Infect. Dis. J. 19:S9–S15 [DOI] [PubMed] [Google Scholar]
- 45. Murphy T. F. 2005. Moraxella (Branhamella) catarrhalis and other gram-negative cocci, p. 2529 In Mandell G. L., Bennett J. E., Dolin R. (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 6th ed Churchill Livingstone, Philadelphia, PA [Google Scholar]
- 46. Murphy T. F., Brauer A. L., Grant B. J., Sethi S. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murphy T. F., Parameswaran G. I. 2009. Moraxella catarrhalis, a human respiratory tract pathogen. Clin. Infect. Dis. 49:124–131 [DOI] [PubMed] [Google Scholar]
- 48. Oliveira A. A., et al. 2007. Effects of levetiracetam in lipid peroxidation level, nitrite-nitrate formation and antioxidant enzymatic activity in mice brain after pilocarpine-induced seizures. Cell. Mol. Neurobiol. 27:395–406 doi:10.1007/s10571-006-9132-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Patrick C. C., et al. 1987. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypeable Haemophilus influenzae. Infect. Immun. 55:2902–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pearson M. M., Laurence C. A., Guinn S. E., Hansen E. J. 2006. Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin. Infect. Immun. 74:1588–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Philippot L. 2005. Denitrification in pathogenic bacteria: for better or worst? Trends Microbiol. 13:191–192 [DOI] [PubMed] [Google Scholar]
- 52. Plamondon P., Luke N. R., Campagnari A. A. 2007. Identification of a novel two-partner secretion locus in Moraxella catarrhalis. Infect. Immun. 75:2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rabkin S. W., Klassen S. S. 2007. Nitric oxide differentially regulates the gene expression of caspase genes but not some autophagic genes. Nitric Oxide 16:339–347 [DOI] [PubMed] [Google Scholar]
- 54. Reddy M. S., Murphy T. F., Faden H. S., Bernstein J. M. 1997. Middle ear mucin glycoprotein; purification and interaction with nontypeable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116:175–180 [DOI] [PubMed] [Google Scholar]
- 55. Rock J. D., et al. 2005. The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite, nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol. Microbiol. 58:800–809 [DOI] [PubMed] [Google Scholar]
- 56. Ruckdeschel E. A., Kirkham C., Lesse A. J., Hu Z., Murphy T. F. 2008. Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect. Immun. 76:1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sano N., et al. 2010. Moraxella catarrhalis bacteraemia associated with prosthetic vascular graft infection. J. Med. Microbiol. 59:245–250 [DOI] [PubMed] [Google Scholar]
- 58. Sethi S., Evans N., Grant B. J., Murphy T. F. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465–471 [DOI] [PubMed] [Google Scholar]
- 59. Stamler J. S., et al. 1992. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc. Natl. Acad. Sci. U. S. A. 89:7674–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stamler J. S., et al. 1992. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U. S. A. 89:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stevanin T. M., Laver J. R., Poole R. K., Moir J. W., Read R. C. 2007. Metabolism of nitric oxide by Neisseria meningitidis modifies release of NO-regulated cytokines and chemokines by human macrophages. Microbes Infect. 9:981–987 [DOI] [PubMed] [Google Scholar]
- 62. Stevanin T. M., Moir J. W., Read R. C. 2005. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect. Immun. 73:3322–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stevanin T. M., Read R. C., Poole R. K. 2007. The hmp gene encoding the NO-inducible flavohaemoglobin in Escherichia coli confers a protective advantage in resisting killing within macrophages, but not in vitro: links with swarming motility. Gene 398:62–68 [DOI] [PubMed] [Google Scholar]
- 64. Sy M. G., Robinson J. L. 2010. Community-acquired Moraxella catarrhalis pneumonia in previously healthy children. Pediatr. Pulmonol. 45:674–678 [DOI] [PubMed] [Google Scholar]
- 65. Van Alst N. E., Picardo K. F., Iglewski B. H., Haidaris C. G. 2007. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect. Immun. 75:3780–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Alst N. E., Wellington M., Clark V. L., Haidaris C. G., Iglewski B. H. 2009. Nitrite reductase NirS is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa. Infect. Immun. 77:4446–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Verduin C. M., Hol C., Fleer A., van Dijk H., Van Belkum A. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Verhaegh S. J., et al. 2011. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae. Microbiology 157:169–178 [DOI] [PubMed] [Google Scholar]
- 69. Wang W., et al. 2006. Development of a shuttle vector for Moraxella catarrhalis. Plasmid 55:50–57 [DOI] [PubMed] [Google Scholar]
- 70. Wang W., Hansen E. J. 2006. Plasmid pWW115, a cloning vector for use with Moraxella catarrhalis. Plasmid 56:133–137 [DOI] [PubMed] [Google Scholar]
- 71. Wang W., et al. 2007. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect. Immun. 75:4959–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang W., et al. 2008. Identification of a repressor of a truncated denitrification pathway in Moraxella catarrhalis. J. Bacteriol. 190:7762–7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weber I., Fritz C., Ruttkowski S., Kreft A., Bange F. C. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017–1025 [DOI] [PubMed] [Google Scholar]
- 74. Weinrauch Y., Msadek T., Kunst F., Dubnau D. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 173:5685–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zumft W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]