Abstract
A gene, designated atlS, encoding a major autolysin from Streptococcus gordonii, was identified and characterized. The predicted AtlS protein is 1,160 amino acids and 127 kDa and has a conserved β1,4-N-acetylmuramidase domain. Zymographic analysis of wild-type S. gordonii revealed peptidoglycan hydrolase activities with molecular masses of 130 and 90 kDa that were absent in an atlS deletion mutant. Western blotting revealed that the 90-kDa band was derived from the 130-kDa protein. Inactivation of atlS resulted in formation of long chains by the cells, markedly decreased autolytic capacity, poor biofilm formation, diminished tolerance of acid and oxidative stress, and decreased production of extracellular DNA (eDNA). The biofilm-forming capacity of the atlS mutant could be almost completely restored to that of the wild-type strain by adding purified recombinant AtlA autolysin of S. mutans but was only partially restored by addition of eDNA. Autolysis, eDNA release, and atlS expression increased sharply when cells entered stationary phase and were greatly enhanced in cells growing with aeration. The LytST and VicRK two-component systems were both required for the induction of atlS by aeration, and purified LytT was able to bind to the promoter region of atlS in vitro. Thus, AtlS and its associated regulatory cascade dominantly control phenotypes of S. gordonii that are critical to colonization, persistence, and competition with other commensal and pathogenic oral bacteria in response to the redox environment and growth domain.
INTRODUCTION
Autolysis, which is triggered by a self-digestion of the cell wall by peptidoglycan hydrolases, can be a mechanism for programmed cell death in bacteria but also plays critical roles in cell wall turnover, cell separation, antibiotic resistance, adherence, genetic competence, and protein secretion (10, 23, 25, 41, 61). In recent years, autolysis has been shown to play important roles in biofilm development and dispersal (4, 8, 16, 54), although the underlying mechanisms are not fully understood. One of the benefits of autolysis to biofilm formation and persistence is believed to be the removal of old or damaged cells in a way that can promote the survival of the population during stresses (53, 68). In addition, extracellular DNA (eDNA) released by autolysis could promote intercellular adherence and thus stabilize biofilms (6, 17, 49, 51, 62, 70, 72).
The importance of autolysins of oral streptococci has been highlighted in recent years. Initial studies using functional genomic analyses demonstrated that the AtlA protein of Streptococcus mutans was crucial for biofilm development (3, 4, 11, 59). A role for this protein in a variety of virulence-related phenotypes, including surface protein biogenesis, was also demonstrated (3, 4, 11, 59). Subsequently, characterization of apparent atlA homologues in Streptococcus sobrinus (73) and Streptococcus downei (67) provided further evidence of the importance of these proteins to cellular physiology, homeostasis, and properties related to colonization and persistence. More recently, Ahn and coworkers showed that a complex posttranscriptional network affected by oxygen and glucose concentration modulates autolysin gene expression, as well as localization, maturation, and activity of AtlA of S. mutans (5).
Streptococcus gordonii, an early colonizer of the oral cavity of infants and of the teeth, can comprise a substantial proportion of the biofilms on healthy dental surfaces (1, 9, 47, 65). Colonization by S. gordonii is believed to be beneficial to the host because of its contribution to pH homeostasis in oral biofilms through the hydrolysis of arginine in saliva and the diet (13, 44). S. gordonii also promotes biofilm development and diversity and is believed to enhance the colonization and growth of some oral commensals through metabolic interdependence, surface protein interactions, and quorum sensing (21, 29, 45, 46, 66), Another important function of S. gordonii in oral biofilms is associated with its demonstrated abilities to antagonize the growth of the caries pathogen Streptococcus mutans (26, 31, 71).
The relatively recent discovery of the importance of eDNA release by bacteria in biofilm formation and stability (6, 17, 49, 51, 62, 70, 72) has stimulated interest in the mechanisms regulating externalization of DNA. The release of eDNA is typically a consequence of cell lysis, as reported for Streptococcus pneumoniae, in which a subpopulation of cells undergoes lysis during competence development (43, 63). In contrast, eDNA release from S. gordonii was reported to be inducible by exogenous H2O2 but independent of cell lysis (30). By computer analysis of the genome of S. gordonii strain Challis at http://www.oralgen.lanl.gov/, we identified a gene, SGO.2013, that encodes a protein with 32% identity to the AtlA autolysin of S. mutans. In this report, we present data on the characterization and regulation of SGO.2013 and the involvement of its gene product in eDNA release, biofilm formation, and stress tolerance.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
Escherichia coli DH10B was grown in Luria broth, and S. gordonii DL1 and its derivatives were cultured in brain heart infusion (BHI) broth (Difco). For selection of antibiotic-resistant colonies after genetic transformation, ampicillin (100 μg ml−1 for E. coli), erythromycin (300 μg ml−1 for E. coli or 10 μg ml−1 for S. gordonii), or kanamycin (50 μg ml−1) for E. coli or 1 mg ml−1 for S. gordonii) were added to the media. For biofilm formation assays, S. gordonii strains were grown in 1/4-strength BHI medium (BHI medium diluted 1:3 with distilled water [dH2O]) supplemented with 10 mM sucrose. Chemical reagents and antibiotics were obtained from Sigma (St. Louis, MO).
Construction of mutant strains.
Strains used in this study are listed in Table 1, and primers used for deletion mutagenesis are listed in Table 2. To construct a reporter gene fusion for measuring transcription from the atlS promoter (SGO.2013), a 624-bp fragment immediately 5′ to the start codon of atlS was amplified by PCR with primers PSG2013-5′-1 and PSG2013-3′ (3). To construct a reporter gene fusion for measuring transcription from the lytT gene promoter, a 300-bp fragment immediately 5′ to the start codon of lytT was amplified by PCR with primers PlytT-5′ and PlytT-3′ (3) that included BamHI recognition sequences. The products harboring PSG2013 and PlytT were fused with the promoterless cat gene derived from pC194 (14, 27). After the correct sequence of the promoter fusion was confirmed, the PSG2013-cat and PlytT-cat constructs were cloned onto plasmid pYQ1 (20), which allows for stable integration of the gene fusion into the gtfG gene of S. gordonii. The constructs were transformed into competent S. gordonii DL1 to construct strains SgWT and SgWT2, which carry the PSG2013-cat and PlytT-cat gene fusions, respectively (3). All strains were then verified by PCR followed by DNA sequence analysis.
Table 1.
Strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| SgWT | S. gordonii DL1/PatlS-cat | This study |
| UA159 | Wild type | University of Alabama, Birmingham |
| AtlS | ΔatlS S. gordonii/PatlS-cat | This study |
| SglytT | ΔlytT S. gordonii/PatlS-cat | This study |
| SglytS | ΔlytS S. gordonii/PatlS-cat | This study |
| SglytST | ΔlytST S. gordonii/PatlS-cat | This study |
| SgvicK | ΔvicK S. gordonii/PatlS-cat | This study |
| SgvicR | ΔvicR S. gordonii/PatlS-cat | This study |
| SgvicRK | ΔvicRK S. gordonii/PatlS-cat | This study |
| SgWT2 | S. gordonii DL1/PlytT-cat | This study |
| SgvicR2 | ΔvicR S. gordonii/PlytT-cat | This study |
| SgvicK2 | ΔvicK S. gordonii/PlytT-cat | This study |
| SgvicRK2 | ΔvicRK S. gordonii/PlytT-cat | This study |
| SgWT-pDL278 | SgWT/pDL278 | This study |
| SgWT-clytST | SgWT/pDL-lytST | This study |
| SgWT-cvicRK | SgWT/pDL-vicRK | This study |
| SglytST-clytST | SglytST/pDL-lytST | This study |
| SgvicRK-cvicRK | SgvicRK/pDL-vicRK | This study |
Table 2.
Primers used in this study
| Primers | Sequencea | Application | Source |
|---|---|---|---|
| AtlS-5′ | 5′-CCAACGAGAGGGTCTTAG-3′ | Deletion of atlS | This study |
| AtlS-BamHI-3′ | 5′-CTGTCTGGATCCAGACATTATAGCATG-3′ | Deletion of atlS | This study |
| AtlS-BamHI-5′ | 5′-CAGCCTGGATCCAGCTTAGTAAAG-3′ | Deletion of atlS | This study |
| AtlS-3′ | 5′-CGGTCGTGACCGCCATGATCGAAG-3′ | Deletion of atlS | This study |
| PatlS-5′-biotin | 5′-/5Biosg/CGATGGCAGTTATAGGGAC-3′ | Amplification of atlS promoter probe with biotine for EMSA | This study |
| LytT-5′ | 5′-CAAGCCGCCTACCCTGTTATTGATGC-3′ | Deletion of lytT | This study |
| LytT-BamHI-3′ | 5′-CCCTGTCTTACCGGATCCTCATCCTCTAC-3′ | Deletion of lytT | This study |
| LytT-BamHI-5′ | 5′-GTGGTAGGGATCCCGCCTAGACAG-3′ | Deletion of lytT | This study |
| LytT-3′ | 5′-GGCTTTGAGACTATTGTAGCCGATATCCA-3′ | Deletion of lytT | This study |
| LytS-5′ | 5′-GGCTTTTCAACTAGGATTTAGTCCATC-3′ | Deletion of lytS | This study |
| LytS-BamHI −3′ | 5′-CGATCAACTGGATCCGAAGCGGATAGCG-3′ | Deletion of lytS | This study |
| LytS-BamHI-5′ | 5′-TCTGCCAGGATCCAATGATAAATAGTCG-3′ | Deletion of lytS | This study |
| LytS-3′ | 5′-TCCCCATAACATAGCGGAACCAAG-3′ | Deletion of lytS | This study |
| ClytST-BamHI-5′ | 5′-GCCAAGGATCCCCCGTCCTATCAGTCCTGAT-3′ | Construction of lytST complementation | This study |
| ClytST-EcoRI-3′ | 5′-CTTTCCGAATTCATGTCCCGACTATATGTC-3′ | Construction of lytST complementation | This study |
| VicR-5′-1 | 5′-GATGGTCGTGAAGCTCTTGA-3′ | Deletion of vicK | This study |
| VicR-BamHI-3′ | 5′-GGCGAGGATCCCTTCGATTCTC-3′ | Deletion of vicK | This study |
| VicK-BamHI-5′ | 5′-GGAGGATCCCTGGGAAAGTGAAG-3′ | Deletion of vicK | This study |
| VicK-3′ | 5′-CCGATAAAATTGTGGTGCCGCCGC-3′ | Deletion of vicK | 35 |
| VicR-5′-2 | 5′-CAAGGGTGCCTTCCCAACATGGC-3′ | Deletion of vicR | 35 |
| VicR-BamHI-3′-2 | 5′-GCCGGATCCACTTCATAGCCCTC-3′ | Deletion of vicR | 35 |
| VicR-BamHI-5′ | 5′-CTCGTCGTGGATCCGGCTAC-3′ | Deletion of vicR | 35 |
| VicR-3′ | 5′-GAACAGCTACCAAACCAGAG-3′ | Deletion of vicR | 35 |
| CvicRK-BamHI-5′ | 5′-GCTACGGATCCAAAGAAACACATACGGTAGC-3′ | Construction of vicRK complementation | This study |
| CvicRK-SphI-5′ | 5′-CCACTAGCATGCGATGCTAAAATGCTATAC-3′ | Construction of vicRK complementation | This study |
| LytT-MBP-5′ | 5′-TCCAGTATAAATAAGGGATCCAATCTTGTG-3′ | Construction of LytT-BMP protein | This study |
| LytT-MBP-3′ | 5′-GGATAAGAAGTCGACAGCGTTTCATTTCTCC-3′ | Construction of LytT-BMP protein | This study |
| PlytT-5′ | 5′-GCCAAGGATCCCCCGGGCTA-3′ | Construction of lytT promoter fusion | This study |
| PlytT-3′ | 5′-GAATACACGGATCCCAACTCC-3′ | Construction of lytT promoter fusion | This study |
| 16S rRNA-S | 5′-CACACCGCCCGTCACACC-3′ | eDNA quantification | This study |
| 16S rRNA-AS | 5′-CAGCCGCACCTTCCGATACG-3′ | eDNA quantification | This study |
Boldface indicates engineered restriction sites.
To make deletions of the genes of interest, 5′- and 3′-flanking regions of each gene were amplified from chromosomal DNA from S. gordonii DL1, ligated together using BamHI sites designed into each primer set, and cloned into the pGEM-T Easy vector (Promega, Madison, WI). These plasmids were digested with BamHI, and a nonpolar kanamycin (NPKm) cassette from pALH124 (3), which lacks its own promoter, was inserted (3). The desired mutagenic plasmids were identified by PCR amplification using vector-originated M13 primers. The plasmids were isolated and used to transform competent SgWT or SgWT2, selecting on BHI agar containing Km. In all cases, double-crossover mutants of each gene were confirmed by PCR and DNA sequencing, including sequencing of the flanking regions to ensure that no undesired mutations were inadvertently introduced. To construct lyt- and vic-complemented strains, DNA fragments containing lytST and vicRK, along with their promoter regions, were amplified with primers (Table 1) and inserted into the E. coli-Streptococcus shuttle vector pDL278 (32), to yield plasmids pDL-lytST and pDL-vicRK, respectively. These recombinant plasmids were introduced into strains carrying PatlS-cat gene fusions, including the wild-type background and strains lacking LytST or VicRK. All strains were then verified by PCR followed by DNA sequence analysis.
Growth kinetics.
Growth of strains of S. gordonii in BHI (pH 7.0) or BHI that was acidified to pH 5.5 with HCl (BHI/HCl), under aerobic or anaerobic conditions (4), was monitored using a Bioscreen C (Growth Curves USA, NJ) with multiwell disposable microtiter plates. An aliquot (3 μl) from an overnight culture was inoculated in at least triplicate into wells containing 300 μl of BHI or BHI/HCl. All inocula were adjusted to the same optical density at 600 nm (OD600) before dilution. To assess the ability of cells to grow in the presence of oxidative stressors, cells from overnight cultures were transferred to prewarmed BHI and grown at 37°C in a 5% CO2 aerobic atmosphere to an OD600 of 0.5. The cells were then diluted into fresh BHI containing 0.003% H2O2 (Fisher Scientific) or 25 mM paraquat (methyl viologen; Sigma), and the impact of the agents on bacterial growth was monitored in a Bioscreen C at 37°C under aerobic conditions or with an overlay of two drops of sterile mineral oil to create relatively anaerobic conditions.
Autolysis, biofilm, zymographic, CAT, and eDNA assays.
Autolysis and the crystal violet/microtiter biofilm assays were performed as previously described (4). A zymographic analysis of cell wall-associated murein hydrolases was conducted as described by Qoronfleh and Wilkinson (52). To collect extracellular murein hydrolases of mutants of S. gordonii, cells were cultured in 50 ml BHI medium and collected at an OD600 of 0.9. Cell pellets were resuspended in 500 μl of 4% SDS and incubated for 60 min at room temperature with agitation, followed by centrifugation at 13,000 × g for 5 min. The supernatant fluids were collected and combined with an equal volume of 50 mM Tris (pH 6.5)-10% glycerol. To prepare substrates for the zymogram, an overnight culture (800 ml) of S. gordonii DL1 grown in BHI medium was collected and the pellet was washed four times with distilled H2O (dH2O). The pellet was suspended in 60 ml of 4% SDS and boiled for 30 min. The heat-killed cells were washed five times in dH2O, and the pellet was saved. The pellet was resuspended in dH2O, and the suspension was added to a polyacrylamide gel at a final concentration of 1% (vol/vol) (48), prior to casting and polymerization. CAT activity was measured by the spectrophotometric method of Shaw (58) as previously described (37). eDNA was collected and quantified as described by Kreth et al. (30).
Cloning, expression, and purification of recombinant proteins.
Recombinant plasmids expressing LytT lacking its predicted signal sequence was constructed by PCR cloning of the relevant DNA fragments into the vector pMAL-p2X (New England BioLabs) using primers listed in Table 1. The coding sequence for LytT was cloned in-frame behind the malE gene to create a maltose binding protein (MBP) fusion protein. The proteins were overproduced in E. coli DH10B by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) and purified as soluble proteins on affinity columns as recommended by the supplier (Fisher Scientific).
Western blot assays.
Exponentially growing cells (OD600, 0.5 to 0.6) were centrifuged and washed twice with Tris-buffered saline (10 mM Tris, 0.9% NaCl, pH 7.4). Whole-cell lysates were obtained by homogenization in SDS boiling buffer (60 mM Tris-HCl, pH 6.8, 10% glycerol, 5% SDS) in the presence of glass beads, followed by centrifugation at 2,000 × g for 10 min (15). Protein samples were separated by SDS-PAGE, blotted onto polyvinylidene difluoride (PVDF) membranes, and incubated with affinity-purified (33) AtlA antibody (3) followed by peroxidase-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, MD). Signals were disclosed using the SuperSignal West Pico chemiluminescent kit (Thermo, Waltham, MA).
EMSA.
An electrophoretic mobility shift assay (EMSA) was carried out following a previously published protocol (69). Briefly, a DNA fragment containing the predicted promoter region of atlS was amplified by PCR with biotin-labeled primers. Approximately 10 fmol of biotin-labeled probe was used in combination with different concentrations of purified recombinant-LytT protein in a 10-μl reaction mixture containing 10 mM HEPES (pH 7.9), 50 mM KCl, 10 mM EDTA, 5 mM MgCl2, and 2 μg poly(dI-dC). After incubation at room temperature for 1 h, the DNA-protein samples were resolved in a nondenaturing, low-ionic-strength polyacrylamide gel and blotted onto hybridization transfer membranes (Perkin-Elmer). Signals were developed using the Chemiluminescent nucleic acid detection module (Thermo Scientific).
RESULTS
Characterization of SGO2013 (AtlS) activity.
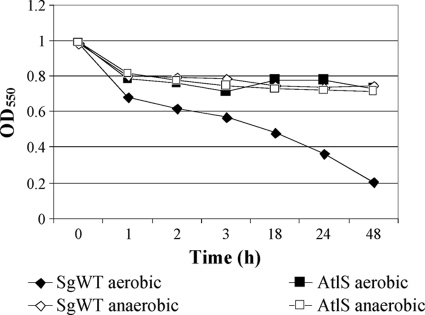
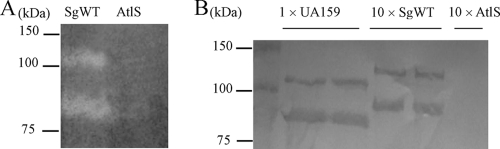
To determine whether SGO.2013 encoded an autolysin, the autolytic activity of an SGO.2013-deficient mutant was compared with that of the wild-type strain. The mutant strain exhibited a markedly lower autolytic rate and extent of autolysis than the parental strain (Fig. 1). The autolysin profiles of the mutant and wild-type strains were also confirmed by zymographic analysis (Fig. 2A). Two major cell wall hydrolase bands with apparent masses of 130 and 90 kDa were present in whole-cell extracts from the wild-type strain (Fig. 2A), but neither band was detected in the SGO.2013 mutant (Fig. 2A). Western blotting using antibodies that were raised against a purified, recombinant S. mutans AtlA autolysin (3), which were affinity purified using a 6-His-tagged AtlA protein (3), also disclosed proteins of 130 and 90 kDa (Fig. 2B). Thus, similar to atlA of S. mutans (Fig. 2B), SGO.2013 encodes a gene product, heretofore designated AtlS, which is present as a higher-molecular-mass species and as a lower-molecular-weight protein that likely arises from proteolysis of full-length AtlS.
Fig. 1.
Comparison of autolysis by the SgWT and atlS mutant strains. An autolysis assay was carried out as described in Materials and Methods.
Fig. 2.
(A) Zymographic assay: autolysin profiles of the SgWT and AtlS-deficient strains determined by renaturing SDS-PAGE using a 7.5% polyacrylamide gel containing 1% (wet weight) of cell walls prepared from S. gordonii. (B) Western blot analysis of proteins extracted from SgWT and atlS mutant strains after incubation in 4% SDS extracts. Of note, in order to detect similar signals for the AtlA and AtlS proteins with the anti-AtlA antibody, it was necessary to apply10-fold more surface protein extract from S. gordonii to the gel than is required for S. mutans (Fig. 2B). Thus, duplicate lanes labeled 1 × UA159 contain proteins extracted from S. mutans UA159. Duplicate lanes labeled 10 × SgWT and lanes labeled 10 × AtlS indicate that 10-fold more protein from the wild-type strain or atlS mutant of S. gordonii, respectively, was loaded on the gel than in the S. mutans lanes. Following SDS-PAGE, proteins were transferred onto a nitrocellulose membrane and subjected to Western blotting using an affinity-purified anti-AtlA polyclonal antiserum at a dilution of 1:500. See Materials and Methods for more details.
AtlS affects stress tolerance by S. gordonii.
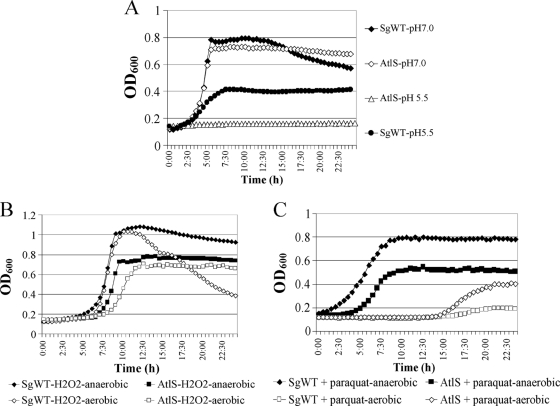
The atlS mutant strain formed longer chains of cells than the wild-type strain and clumped extensively in broth culture (data not shown). To examine whether AtlS of S. gordonii affected acid tolerance, SgWT and the atlS mutant were cultured in BHI (pH 7.0) or BHI/HCl broth (pH 5.5). In pH 7.0 medium, the growth curve of the atlS mutant was similar to that of SgWT (Fig. 3A) and the atlS mutant achieved modestly higher final optical densities, probably due to decreased autolysis of the mutant. In medium that had been adjusted to pH 5.5, the wild-type strain grew more slowly and achieved substantially lower final optical densities than were achieved at pH 7.0. In contrast to the parental strain, the atlS mutant was not able to grow in BHI/HCl broth (Fig. 3A).
Fig. 3.
Growth curves of SgWT and atlS mutant strains under aerobic or anaerobic conditions in BHI (pH 7.0) or BHI that was acidified to pH 5.5 with HCl (A); in BHI broth with or without 0.003% hydrogen perioxide (B); and in BHI broth with or without 25 mM paraquat (C). Optical density at 600 nm was determined every 30 min for 24 h using a Bioscreen C.
To examine whether AtlS of S. gordonii was required for oxidative stress tolerance, the SgWT and atlS mutant strains were cultured in BHI medium with 0.003% hydrogen peroxide or 25 mM paraquat under aerobic conditions, or with a mineral oil overlay to create a more anaerobic environment (4). In the presence of hydrogen peroxide under aerobic conditions, the SgWT strain displayed a slightly extended lag phase and a modestly lower growth rate and attained a maximum optical density similar to that obtained when the strain is cultured in anaerobic conditions (Fig. 3B). However, lysis of the cells was much more rapid and extensive in cells grown in aerobic conditions than in those growing under anaerobic conditions (Fig. 3B). The atlS mutant consistently displayed a longer lag phase and slower growth and achieved lower final optical densities in the presence of hydrogen peroxide under anaerobic and aerobic conditions (Fig. 3B) than SgWT, but no obvious lysis was detected in the atlS mutant. Similarly, in the presence of 25 mM paraquat, the atlS mutant grew more slowly and achieved lower final optical densities than the SgWT strain (Fig. 3C). Collectively, the results indicate that a compromised ability to cope with oxidative stress is associated with AtlS deficiency and provide more evidence that the cells lacking AtlS are hyperresistant to autolysis.
AtlS-dependent eDNA release.
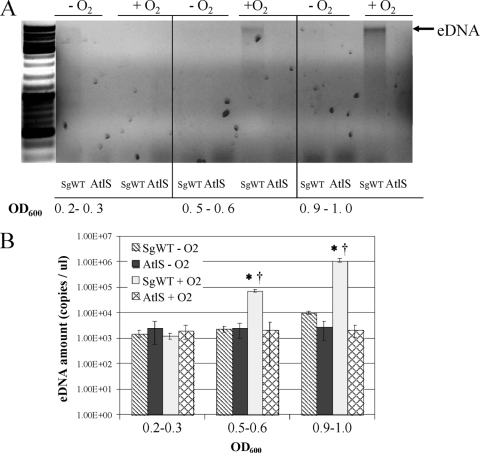
DNA release is typically a consequence of cell lysis. To examine the characteristics of extracellular DNA (eDNA) release by S. gordonii, the wild-type strain was cultured in BHI medium under aerobic and anaerobic conditions, and the cells were collected at different growth stages. The eDNA produced by a 1-ml culture was extracted and analyzed in a 0.8% agarose gel or quantified by real-time PCR with 16S rRNA primers (Table 2). When cells were cultured under aerobic conditions, eDNA released by the wild-type strain increased gradually from the early exponential phase (OD600, 0.2∼0.3) to stationary phase (OD600, 0.9∼1.0), with DNA bands characteristic of chromosomal DNA clearly visible (Fig. 4A). Notably, no eDNA production from anaerobically cultured cells was detectable by gel electrophoresis (Fig. 4). Consistent with these observations, quantitative real-time PCR revealed that aerobic cultures that had reached stationary phase produced 100-fold more eDNA than anaerobically grown cells in the same growth phase (Fig. 4B), indicating that oxygen or one of its metabolites plays an important role in triggering DNA release. In contrast to the wild-type strain, very low levels of eDNA were detected in the atlS mutant in all growth phases under aerobic or anaerobic conditions, providing strong evidence that AtlS and cell lysis contribute in major ways to eDNA production by S. gordonii.
Fig. 4.
eDNA release by SgWT and AtlS-deficient S. gordonii as a function of growth domain. Cells were cultured in BHI broth under aerobic or anaerobic conditions. (A) Agarose gel electrophoresis (0.8%) of eDNA stained with ethidium bromide as described in the Materials and Methods section. The optical density (OD600) of the culture when harvested is presented below each panel, and the photographs are representative of three independent experiments that yielded similar results. (B) Quantitative real-time PCR of eDNA using the 16S rRNA gene as the amplification target. Data were normalized as detailed in Materials and Methods and represent the means and standard deviations of results of two independent experiments done in duplicate on different days. *, statistically significant differences among the same strain collected at different growth phases (P < 0.05 [Student t test]). †, statistically significant differences between the same strain cultured anaerobically and aerobically (P < 0.05 [Student t test]).
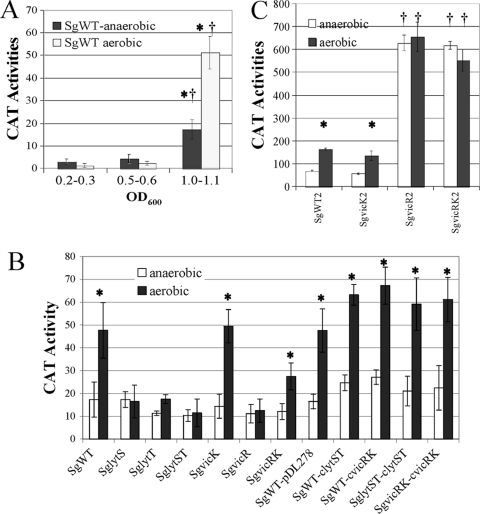
Regulation of atlS expression.
The SgWT strain, which carries a cat gene fused to the atlS promoter, was grown in TY medium containing 25 mM galactose under aerobic or anaerobic conditions. CAT activity arising from PatlS expression was examined at different growth phases. From the early exponential phase (OD600, 0.2∼0.3) to midexponential phase (OD600, 0.5∼0.6), atlS expression of cells was at a basal level, under both aerobic and anaerobic conditions (see Fig. 6A). However, PatlS expression was increased more than 10-fold after cells entered stationary phase (OD600, 0.9∼1.0), with aerobically grown cells showing 2.5-fold-higher atlS expression than anaerobically grown cells in stationary phase (see Fig. 6A). Thus, atlS expression is both growth phase and oxygen dependent in S. gordonii.
Fig. 6.
CAT activities of SgWT carrying PatlS-cat cultured in TY medium containing 25 mM galactose at different growth phases, under aerobic or anaerobic conditions (A), SgWT and its derivatives cultured in TY medium containing 25 mM galactose and collected at stationary phase, under aerobic or anaerobic conditions (B), and SgWT2 carrying PlytT-cat and its derivatives cultured in BHI broth, under aerobic or anaerobic conditions (C). Cells were collected at the midexponential phase of growth. The values of the columns are the average of a minimum of nine separate cultures for each strain and condition. *, statistically significant differences between the same strain cultured anaerobically or aerobically (P < 0.05 [Student t test]). †, statistically significant differences between the wild-type and mutant strains cultured under the same conditions (P < 0.05 [Student t test]).
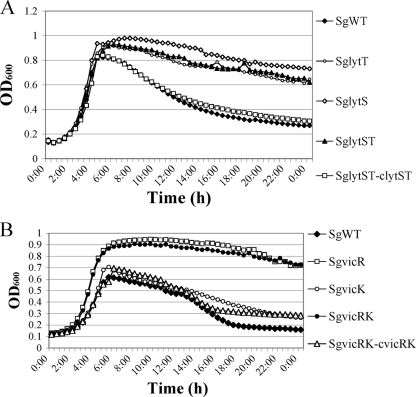
Previous studies demonstrated that the LytST and VicRK two-component systems (TCS) were involved in regulating autolysis in some Gram-positive bacteria, including S. pneumoniae, S. aureus, and S. mutans (4, 24, 50, 57). To determine whether these signal transduction systems also affected autolysis in S. gordonii, the entire lytST or vicRK operon was replaced by a nonpolar kanamycin cassette (10). To evaluate the function of individual components of the TCS, the lytS, lytT, vicR, and vicK genes were disrupted individually by nonpolar insertions (Table 1). All vic and lyt mutants formed long chains and clumped when growing in BHI (data not shown). With the exception of the strain carrying only the vicK mutation, SgvicK, all vic and lyt mutants were substantially more resistant to autolysis than the SgWT strain (Fig. 5). When the vicRK and lytST mutants were complemented with plasmid-borne copies of these genes, growth curves similar to the wild-type genetic background were observed (Fig. 5).
Fig. 5.
Growth and lysis of strains SgWT and lyt mutants (A) and SgWT and vic mutants (B) in BHI broth cultured under aerobic conditions. Optical density at 600 nm was determined every 30 min after shaking using a Bioscreen C.
To assess whether LytST and VicRK influenced autolysis of S. gordonii by regulating the expression of atlS, cells were grown to stationary phase in TY medium containing 25 mM galactose under aerobic or anaerobic conditions and CAT activities were measured. In the SgWT background, cells expressed 2.5-fold-higher CAT activity from PatlS under aerobic conditions than under anaerobic conditions (Fig. 6B), whereas the lyt mutants displayed a basal level of PatlS expression and no induction in aerobic conditions was evident (Fig. 6). A phenotype similar to that of the lyt mutants was observed in the SgvicR and SgvicRK mutants, whereas about a 2-fold induction of PatlS by oxygen still could be detected in the SgvicK mutant (Fig. 6B). The histidine kinase (LytS) and response regulators (LytT and VicR) were required for activation of the atlS gene expression by oxygen. Also of note, the complemented strains, SgvicRK-cvicRK and SglytST-clytST, and lytST or vicRK overexpressing strains (SgWT-cvicRK and SgWT-clytST) had higher levels of expression from the atlS promoter than the wild-type or mutant strains carrying the empty vector pDL278 (Fig. 6B). Collectively, these data demonstrate that the Lyt and Vic systems influence autolysis of S. gordonii by regulating the expression of atlS in response to oxygen and possibly other factors.
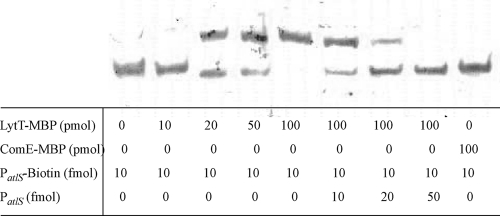
Purified LytT binds to the promoter region of atlS in vitro.
To explore whether the LytT response regulator could interact with the atlS promoter region, the entire lytT coding sequence was cloned onto plasmid pMAL-p2x to create a maltose-binding fusion protein to the N terminus of LytT. Soluble LytT-MBP protein was overproduced in E. coli and purified by affinity chromatography. The purified protein displayed the expected apparent molecular mass of 68 kDa following SDS-PAGE (data not shown). A DNA probe was constructed by PCR amplification of 300 bp of DNA immediately 5′ to the coding sequence of atlS with biotin-labeled primers. In the absence of LytT-MBP, the probe migrated to the bottom of the gel (Fig. 7). When recombinant LytT was added, the migration of PatlS was retarded and increasing concentrations of LytT-MBP caused more PatlS to shift (Fig. 7). Unlabeled PatlS DNA could effectively compete the interaction with LytT-MBP (Fig. 7). An irrelevant MBP-tagged protein, ComE-MBP, was used in mobility shift assays in place of LytT-MBP. ComE-MBP was constructed by cloning the entire comE coding sequence from S. mutans onto plasmid pMAL-p2x to create a translational fusion that added maltose binding protein to the N terminus of ComE. ComE-MBP was purified using the same protocol as for LytT-MBP. Previous experiments conducted in our lab confirmed that the ComE-MBP fusion protein was active in an EMSA with a known ComE-activated promoter (data not shown). No shift of the biotinylated PatlS was detected when ComE-MBP was used at same concentrations as LytT-MBP (Fig. 7), confirming that MBP was not responsible for the binding of recombinant LytT-MBP to its target DNA.
Fig. 7.
Electrophoretic mobility shift assays using LytT-MBP and a biotin-labeled fragment containing the atlS promoter region. Biotin-labeled PatlS probe was incubated with increasing concentrations of LytT-MBP at room temperature for 60 min and then analyzed on a low-ion-strength polyacrylamide gel as detailed in Materials and Methods.
Expression of lytST is affected by VicR.
Given the contrast in the behavior of the vicK mutant compared to that of the vicR or vicRK mutant strains in terms of atlS responsiveness to oxygen, we investigated the possibility that expression of the vicRK or lytST TCS might be influenced in response to oxygen. Thus, the SgWT2 and the vic mutant carrying the lytT promoter (PlytT) fused to cat were cultured in BHI under aerobic or anaerobic conditions. Cells were grown to an OD600 of 0.5∼0.6, and CAT activities were measured. Approximately 2-fold-higher expression of PlytT was detected under aerobic conditions than under anaerobic conditions in the SgWT2 and the SgvicK2 mutant (Fig. 6C). However, both the SgvicR2 and SgvicRK2 mutants displayed a similar high level of expression of lytT under aerobic and anaerobic conditions (Fig. 6C), suggesting a possible role of the VicRK system in repression of lytT in response to redox.
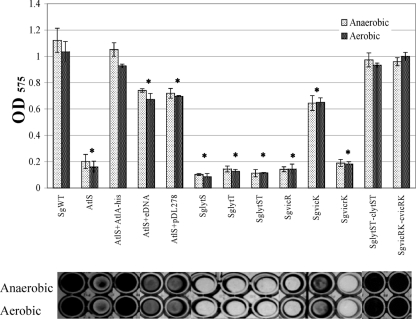
AtlS-dependent biofilm formation in response to oxygen.
AtlA of S. mutans was originally identified by virtue of its profound effect on biofilm formation (3), so we assessed the capacity of the SgWT and atlS mutant strains to form biofilms in 96-well microtiter plates (Fig. 8). The strains were cultured in 200 μl of 1/4-strength BHI broth with 10 mM sucrose for 48 h, with or without an overlay of three drops of sterile mineral oil to create relatively anaerobic or aerobic culture conditions, respectively. SgWT formed biofilms very efficiently, but the atlS mutant formed nearly 80% less biofilm (Fig. 8). Notably, the vic and lyt mutants were also defective in biofilm formation at a level comparable to that for the atlS mutant, whereas the vic- and lyt-complemented strains formed biofilms as efficiently as the wild-type strain (Fig. 8). To further confirm the requirement for AtlS for efficient in vitro biofilm formation by S. gordonii, purified 6-His-tagged, full-length AtlA protein from S. mutans (4) was added into the biofilm culture medium at a final concentration of 2.5 μM. Similar to what was reported for AtlA-deficient S. mutans (4), purified AtlA could restore biofilm formation by the atlS mutant to near wild-type levels (Fig. 8).
Fig. 8.
Biofilm formation assay. Cultures were grown in 1/4-strength BHI medium supplemented with 10 mM sucrose under aerobic or anaerobic conditions for 48 h. The top panel is quantification of the crystal violet-stained biofilms as detailed in the text. Data are representatives of the means of results of at least two separate experiments that were performed in triplicate, with error bars delineating standard deviations. *, statistically significant differences between the wild-type and mutant strains cultured under the same conditions (P < 0.05 [Student t test]). The bottom panel shows representative biofilms corresponding to the sample quantified in the bar graph above.
The release of bacterial DNA is believed to stabilize cell-to-cell adherence and biofilm architecture (6, 17, 49, 51, 62, 70, 72). To test if eDNA release from AtlS-dependent lysis played a role in biofilm formation by S. gordonii, atlS mutants were cultured in biofilm medium with 50 ng/μl of eDNA extracted from aerobically grown wild-type S. gordonii (Fig. 4). Under these conditions, the atlS mutant was able to form about 70% of the biofilm mass of the SgWT strain (Fig. 8), but doubling of the concentration of eDNA in the culture medium did not restore biofilm biomass to wild-type levels (data not shown). Thus, a compromised ability of the atlS mutant to form biofilms may be only partially attributable to decreased release of eDNA. Plasmid DNA (pDL278, 50 ng/μl) (32) was also used as the eDNA source with the atlS mutant, and biofilm biomass was formed at a level similar to that formed with added chromosomal DNA (Fig. 8), indicating that there is no specificity for eDNA to promote biofilm formation and that restoration of the biofilm phenotype did not require the presence of wild-type chromosomal DNA.
DISCUSSION
Dental caries remains a major health problem, and this diet-related infectious disease has a strong ecological basis. Specifically, development of the disease is associated with increases in the proportions of acid-tolerant species, including S. mutans and lactobacilli, usually at the expense of beneficial commensal organisms (7, 18, 22, 60). Evidence continues to emerge that there is intense interspecies competition between oral commensals and caries pathogens. In addition to having similar nutritional requirements and thus competing for common nutrients, these organisms can antagonize the growth of one another through metabolic pathways and production of antimicrobial peptides. The AtlA autolysin of S. mutans is tremendously important to cellular homeostasis and is highly regulated at the transcriptional and posttranscriptional levels by environmental factors that are relevant to oral biofilm ecology and composition, e.g., oxygen and carbohydrate availability (3, 4, 11, 59). Thus, the autolytic pathway represents a potential target for anti-caries therapies (3, 4, 11, 59). Importantly, though, an effective anti-caries therapy should not have a detrimental influence on beneficial commensal microorganisms. This report is the first description of an autolysin of the abundant oral commensal S. gordonii and its role in biofilm formation, stress tolerance, autolysis, and eDNA production. Collectively, the data highlight similarities and some notable differences in the regulation and the roles in cellular processes of the apparent autolysin homologues of a health-associated commensal and a known caries pathogen (these data are summarized in part in Table 3).
Table 3.
Comparison of AtlA and AtlSa
| Parameter | Result for: |
|
|---|---|---|
| AtlA | AtlS | |
| Origins | S. mutans | S. gordonii |
| Encoding genes | SMu0630 | SGO2013 |
| Gene locus | Multigene operon | Single-gene operon |
| Protein characteristics | 977 amino acids, 107 kDa, and has a conserved β1,4-N-acetylmuramidase domain located at C-terminal (3) | 1,160 amino acids, 127 kDa, and has a conserved β1,4-N-acetylmuramidase domain located at N-terminal |
| Involvement in the cell physiology | Normal cell wall biosynthesis, stress tolerance, autolysis, competence, and biofilm formation (3, 4) | Normal cell wall biosynthesis, stress tolerance, autolysis, and biofilm formation |
| Contribution to eDNA release | NR | Major |
| Processing for protein maturation | Cleavage (3) | Cleavage |
| Inducible conditions | Oxidative stress (4) | Oxidative stress and CSP |
| Transcriptional regulation | ||
| Promoter | At least three promoters (3) | One proximal promoter |
| Regulator | NR | VicRK and LytST |
| Direct regulator | NR | LytT |
| Posttranscription regulation | ThmA, Smu0629, and VicRK (4) | ND |
NR, not reported; ND, not determined.
Studies with various Gram-positive bacteria, including S. pneumoniae, S. aureus, and S. mutans, demonstrate that induction of autolysis may be a response by cells to environmental stresses, often involving quorum sensing, that could enhance the survival of the population as a whole (53, 68). For example, LytA-mediated autolysis of S. pneumoniae can be triggered by competence development (24), acid stress (50), or antibiotics (34). In S. aureus, metabolism of carbohydrates regulates cid- and lrg-dependent autolysis, with both glucose and acetate affecting autolytic behavior (56). Regulation of the gene for, and the activity of, the AtlA autolysin of S. mutans (59), which dominantly controls cell separation, biofilm formation, and autolysis (3, 11, 59), is very complex. For example, the atlA gene is in an operon with at least four additional genes, and transcription of atlA is influenced by at least three promoters (3, 11, 59). Aeration is a major trigger for AtlA-dependent autolysis in S. mutans, and the response to aerobiosis is controlled by the VicRK two-component system (4). While regulation of autolysis by S. gordonii has some properties in common with that by these organisms, important differences exist.
The cross-reaction of anti-AtlA antibody with the AtlS protein by Western blot analysis (Fig. 2B) confirmed the prediction from computer algorithms that AtlS and AtlA share substantial amino acid sequence identity (32%) and that these autolysins are produced as two active isoforms, presumably as a result of proteolytic cleavage of the full-length, secreted proteins (3). Also, as shown, AtlA could function to restore biofilm formation by an atlS mutant of S. gordonii. In spite of these sequence and functional similarities, there are some important differences in the contribution of these enzymes to cellular physiology and homeostasis. For example, loss of AtlA had a profoundly negative impact on the development of genetic competence in S. mutans (4), whereas inactivation of atlS did not adversely affect the transformation efficiency of S. gordonii (data not shown). In addition, compromised tolerance of acid stress was strongly associated with a deficiency of AtlS in S. gordonii (Fig. 3A), but strains of S. mutans lacking AtlA did not show a similar phenotype (4).
Substantial variations in transcriptional and posttranscriptional control mechanisms for atlA and atlS are also apparent. Transcriptional studies revealed that atlA is part of a multigene operon under the control of at least three promoters (3), but AtlS appears to be encoded as a single cistron (Fig. 6A) (10) driven from a proximal promoter. Proper maturation of AtlA (reference 3 and Fig. 2) of S. mutans requires the ThmA and SMu0629 proteins encoded in the atlA operon (3, 4). In contrast, the genes encoding proteins with the highest degree of similarity to ThmA and Smu0629 are not genetically linked to atlS or to one another. Thus, basic aspects of the organization of transcription and of the regulatory machinery for autolysin maturation differ in these two organisms.
In both S. mutans and S. gordonii, oxygen was a major trigger for AtlS- and AtlA-dependent autolysis (4), but clear differences in the signal transduction pathways for this activation exist. In S. mutans, the VicRK TCS affects AtlA levels and maturation by regulating the transcription of the SMu0629 gene, which is required for efficient maturation of AtlA (4). In contrast, the expression of atlS of S. gordonii is regulated by oxygen primarily at the transcriptional level (Fig. 6A), and both the VicRK and LytST TCS were required for the induction of atlS transcription in response to oxygen (Fig. 6B). The effect of LytT on atlS transcription is apparently direct, as LytT could bind to the promoter region of atlS in vitro (Fig. 7). In contrast, the effects of Vic appear to be exerted through effects on the expression of the lytST operon. As evidenced by the different behaviors of strains lacking VicK or VicR (Fig. 6C), it could be proposed that the VicR response regulator may repress lytT expression in the absence of VicK kinase activity. In this case, VicR phosphorylation by VicK in response to redox may alleviate lytT repression, leading to induction of lytT by oxygen. However, cross-regulation of VicR by the LytT histidine kinase or other kinases in response to oxygen or other signals may contribute to differential expression of atlS. Collectively, then, it appears that S. gordonii modulates atlS expression in response to the redox environment, and possibly other signals, through a VicRK-LytST cascade, which would allow the organism to alter envelope biogenesis, autolytic activity, and eDNA release to adapt to microenvironments. It is also notable that the Vic, Com, and Cia TCS have different impacts on expression of the genes for the agmatine deiminase and arginine deiminase pathways, which are highly similar alkali-generating systems of S. mutans and S. gordonii, respectively (35, 36). Thus, this study provides further support for the idea that ecological and physiologic pressures may have led to evolutionary divergence in the roles of the Vic, Com, and Cia TCS of these two organisms.
It is also of interest that apparent LytST homologues in S. mutans affected the regulation of autolysis of S. mutans not by influencing atlA expression but by regulating the transcription of lrgAB and cidAB, which encode predicted holin:antiholin complexes (5). Similar to that of S. pneumoniae and S. aureus, the lrgAB operon of S. mutans is located adjacent to the lytST operon (12, 55, 57). In contrast, the S. gordonii lrgAB genes are distant from the lytST operon and we could find no evidence for transcriptional regulation of lrg by LytST in this organism (data not shown). In addition, we were unable to identify cidAB genes or other genes that encoded LrgAB-like proteins in the S. gordonii genome. In addition to availability of oxygen, glucose levels were revealed to affect autolysis of S. mutans through the CcpA protein (2), which was able to bind directly to the promoter region of lrgA (7). However, we have no evidence to support that autolysis of S. gordonii is regulated by glucose availability, nor have we been able to observe binding of a purified, recombinant CcpA protein (2, 18) to the promoter regions of the atlS gene of S. gordonii (data not shown). These findings highlight additional fundamental differences between S. gordonii and S. mutans in control of autolysin expression, maturation, and localization.
S. gordonii and S. pneumoniae are both naturally competent for genetic transformation and have evolutionarily similar competence regulatory pathways, whereas the CSP system of S. mutans appears to have evolved from a bacteriocin regulatory cascade (26, 28) and comX expression is governed primarily by the ComRS system (40). Notably, the expression of cbpD and lytA, which encode autolysins of S. pneumoniae, increases sharply when cells enter stationary phase and can be induced by the addition of CSP (63), but cells can be protected from self-lysis by induction of an immunity protein, ComM (63). S. mutans can also be induced to lyse by high concentrations (2 μM) of its cognate CSP through pathways distinct from those in S. pneumoniae (24), although we found that atlA expression can be stimulated by high concentrations of S. mutans CSP (data not shown). Notably, pure CSP that was synthesized based on the S. gordonii ComC sequence could activate atlS transcription and induce cell lysis (data not shown), but S. gordonii lacks apparent cbpD and comM homologues. Therefore, there is considerable evolutionary divergence in the interconnection of the competence and lytic control pathways among the oral streptococci, as well as between the viridans group of streptococci and S. pneumoniae.
In this report, we provide direct evidence that AtlS is required for efficient eDNA release by S. gordonii (Fig. 4) and that aeration is a major environmental trigger for eDNA release. Previously, it was reported that eDNA release of S. gordonii could be stimulated by 1 mM H2O2 and occurred without detectable lysis of the cells (30). More recently, Zheng and coworkers showed that H2O2 production by pyruvate oxidase is a contributing factor to lysis of S. sanguinis, a close relative of S. gordonii (74). In this study, we observed that 0.003% H2O2 (4) was not sufficient to trigger cell lysis of S. gordonii (Fig. 3B) or to enhance atlS expression (data not shown) when cells were cultured anaerobically, suggesting that oxygen itself or a metabolite of oxygen other than H2O2 serves as a stimulus for cell lysis. While it was noted that there was a basal level of eDNA released from cells that were cultured under relatively anaerobic conditions, the production of the overwhelming majority of eDNA by S. gordonii in aerobic conditions and in stationary phase under the conditions utilized in this study is dependent on the presence of the AtlS autolysin and associated with cell lysis.
In many species, eDNA contributes to biofilm formation as a component of the extracellular biofilm matrix, impacting cell-to-cell adherence and biofilm architecture (6, 17, 49, 51, 62, 70, 72). Supporting a role for eDNA in the development of S. gordonii biofilms is the observation that biofilm formation by strains lacking AtlA could be partially restored by the addition of either chromosomal or plasmid DNA (Fig. 8). The fact that complete restoration of biofilm formation by eDNA could not be achieved could simply indicate that the production of eDNA must be spatially or temporally regulated for optimal development of stable biofilms. As likely, the impact of loss of AtlS on the biogenesis of a normal cell surface, which was a striking phenotype of the atlA mutant of S. mutans (3), could account for a failure of the mutant strain to produce wild-type levels of biofilm. From a mechanistic perspective, altered peptidoglycan structure in the atlS mutant could impact osmo-adaptation or adaptation to other stresses associated with the mass transport limitations in three-dimensional biofilms (38). Likewise, long cell chains and the associated decrease in available cell surface area for biofilm-promoting interactions may contribute to poor cell coaggregation and biofilm formation (64).
It was of significant interest that the biofilm-forming capacity of the atlS mutant could be restored to a level comparable to that of the wild-type strain by the addition of 2.5 μM AtlA protein to the cultures (Fig. 8), which implies that AtlS is able to recognize S. mutans cell walls as a suitable substrate. Interestingly, addition of relatively high concentrations of S. mutans AtlA protein caused obvious cell lysis of S. gordonii (data not shown). We are presently examining whether a similar antagonistic impact of AtlS on S. mutans can be observed in vitro, but these preliminary results point to the possibility that autolysin release in dental biofilms could modulate the composition and activity of oral microbial populations. Such antagonistic potential would add to the growing body of evidence that there are multiple strategies used by oral streptococci to establish and persist in healthy and cariogenic biofilms.
S. gordonii is an earlier colonizer on dental surface and is also thought to be important for the cosubsequent colonization of other oral commensals (21, 29, 47). It is known that the oxygen tension and redox potential in early biofilms are very high compared to that of immature biofilms (39, 42). Whereas biofilm formation by S. gordonii does not appear to be adversely affected by aerobic conditions, cultivation of S. mutans under similar conditions causes potent inhibition of biofilm development by this caries pathogen. Given the differences in the signal transduction pathways and regulation mechanisms controlling the expression of the genes for, and the maturation of, the AtlA and AtlS autolysins, development of therapeutic strategies that selectively disrupt the autolytic pathways of a prominent caries pathogen without disrupting normal functions of the commensal streptococci may be an effective means of modulating oral biofilm composition and, thus, oral biofilm pathogenic potential. Efforts are under way to identify the signals regulating autolysin gene expression, processing, and activity.
ACKNOWLEDGMENTS
We thank Sang-Joon Ahn and Lin Zeng for helpful suggestions.
This work was supported by Public Health Service grants DE013239 and DE019106 from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Aas J. A., et al. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abranches J., et al. 2008. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J. Bacteriol. 190:2340–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn S. J., Burne R. A. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J. Bacteriol. 188:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahn S. J., Burne R. A. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 189:6293–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahn S. J., Rice K. C., Oleas J., Bayles K. W., Burne R. A. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allesen-Holm M., et al. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 7. Avila M., Ojcius D. M., Yilmaz O. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bayles K. W. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5:721–726 [DOI] [PubMed] [Google Scholar]
- 9. Becker M. R., et al. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blackman S. A., Smith T. J., Foster S. J. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144(pt. 1):73–82 [DOI] [PubMed] [Google Scholar]
- 11. Brown T. A., Jr., et al. 2005. A hypothetical protein of Streptococcus mutans is critical for biofilm formation. Infect. Immun. 73:3147–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brunskill E. W., Bayles K. W. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burne R. A., Marquis R. E. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1–6 [DOI] [PubMed] [Google Scholar]
- 14. Chen Y. Y., Betzenhauser M. J., Burne R. A. 2002. cis-Acting elements that regulate the low-pH-inducible urease operon of Streptococcus salivarius. Microbiology 148:3599–3608 [DOI] [PubMed] [Google Scholar]
- 15. Chen Y. Y., Weaver C. A., Mendelsohn D. R., Burne R. A. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Claverys J. P., Havarstein L. S. 2007. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat. Rev. Microbiol. 5:219–229 [DOI] [PubMed] [Google Scholar]
- 17. Das T., Sharma P. K., Busscher H. J., van der Mei H. C., Krom B. P. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76:3405–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dewhirst F. E., et al. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Dong Y., Chen Y. Y., Burne R. A. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Egland P. G., Palmer R. J., Jr., Kolenbrander P. E. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. U. S. A. 101:16917–16922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Filoche S., Wong L., Sissons C. H. 2010. Oral biofilms: emerging concepts in microbial ecology. J. Dent. Res. 89:8–18 [DOI] [PubMed] [Google Scholar]
- 23. Groicher K. H., Firek B. A., Fujimoto D. F., Bayles K. W. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Havarstein L. S., Martin B., Johnsborg O., Granadel C., Claverys J. P. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297–1307 [DOI] [PubMed] [Google Scholar]
- 25. Heilmann C., Hussain M., Peters G., Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013–1024 [DOI] [PubMed] [Google Scholar]
- 26. Heng N. C., Tagg J. R., Tompkins G. R. 2007. Competence-dependent bacteriocin production by Streptococcus gordonii DL1 (Challis). J. Bacteriol. 189:1468–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horinouchi S., Weisblum B. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jenkinson H. F., Vickerman M. M. 2006. Genetics of sanguinis group streptococci, p. 347–355 In Fischetti V. A., Novick R. P., Ferretti J. J., Portnoy D. A., Rood J. I. (ed.), Gram-positive pathogens, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 29. Johnson B. P., et al. 2009. Interspecies signaling between Veillonella atypica and Streptococcus gordonii requires the transcription factor CcpA. J. Bacteriol. 191:5563–5565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kreth J., Vu H., Zhang Y., Herzberg M. C. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J. Bacteriol. 191:6281–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kreth J., Zhang Y., Herzberg M. C. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190:4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. LeBlanc D. J., Chen Y. Y., Lee L. N. 1993. Identification and characterization of a mobilization gene in the streptococcal plasmid, pVA380-1. Plasmid 30:296–302 [DOI] [PubMed] [Google Scholar]
- 33. Levin P. 2002. Light microscopy techniques for bacterial cell biology, p. 115–132 In Sansonetti P. J., Zychlinsky A. (ed.), Molecular cellular microbiology. Academic Press, Ltd., London, United Kingdom [Google Scholar]
- 34. Lewis K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y., Burne R. A. 2009. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J. Bacteriol. 191:7353–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Y., Burne R. A. 2009. Multiple two-component systems of Streptococcus mutans regulate agmatine deiminase gene expression and stress tolerance. J. Bacteriol. 191:7363–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y., Dong Y., Chen Y. Y., Burne R. A. 2008. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl. Environ. Microbiol. 74:5023–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loo C. Y., Corliss D. A., Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marquis R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Indust. Microbiol. 15:198–207 [DOI] [PubMed] [Google Scholar]
- 40. Mashburn-Warren L., Morrison D. A., Federle M. J. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mercier C., et al. 2002. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol. Microbiol. 46:235–243 [DOI] [PubMed] [Google Scholar]
- 42. Mettraux G. R., Gusberti F. A., Graf H. 1984. Oxygen tension (pO2) in untreated human periodontal pockets. J. Periodontol. 55:516–521 [DOI] [PubMed] [Google Scholar]
- 43. Moscoso M., Claverys J. P. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol. Microbiol. 54:783–794 [DOI] [PubMed] [Google Scholar]
- 44. Nascimento M. M., Gordan V. V., Garvan C. W., Browngardt C. M., Burne R. A. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol. Immunol. 24:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nicholas S. S. 2008. Oral vs IV corticosteroids for in-hospital treatment of COPD exacerbations. Chest 134:470. [DOI] [PubMed] [Google Scholar]
- 46. Nobbs A. H., Lamont R. J., Jenkinson H. F. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nyvad B., Kilian M. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267–272 [DOI] [PubMed] [Google Scholar]
- 48. Perry J. A., Cvitkovitch D. G., Levesque C. M. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol. Lett. 299:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petersen F. C., Tao L., Scheie A. A. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187:4392–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pinas G. E., Cortes P. R., Orio A. G., Echenique J. 2008. Acidic stress induces autolysis by a CSP-independent ComE pathway in Streptococcus pneumoniae. Microbiology 154:1300–1308 [DOI] [PubMed] [Google Scholar]
- 51. Qin Z., et al. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092 [DOI] [PubMed] [Google Scholar]
- 52. Qoronfleh M. W., Wilkinson B. J. 1986. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of beta-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob. Agents Chemother. 29:250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rice K. C., Bayles K. W. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729–738 [DOI] [PubMed] [Google Scholar]
- 54. Rice K. C., Bayles K. W. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72:85–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rice K. C., et al. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rice K. C., Nelson J. B., Patton T. G., Yang S. J., Bayles K. W. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sharma-Kuinkel B. K., et al. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 191:4767–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shaw W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737–755 [DOI] [PubMed] [Google Scholar]
- 59. Shibata Y., Kawada M., Nakano Y., Toyoshima K., Yamashita Y. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect. Immun. 73:3512–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Siqueira J. F., Jr., Rocas I. N. 2010. The oral microbiota: general overview, taxonomy, and nucleic acid techniques. Methods Mol. Biol. 666:55–69 [DOI] [PubMed] [Google Scholar]
- 61. Smith T. J., Blackman S. A., Foster S. J. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146(pt. 2):249–262 [DOI] [PubMed] [Google Scholar]
- 62. Steinberger R. E., Holden P. A. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71:5404–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Steinmoen H., Knutsen E., Havarstein L. S. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. U. S. A. 99:7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Suzuki N., Nakano Y., Kiyoura Y. 2006. Characterizing the specific coaggregation between Actinobacillus actinomycetemcomitans serotype c strains and Porphyromonas gingivalis ATCC 33277. Oral Microbiol. Immunol. 21:385–391 [DOI] [PubMed] [Google Scholar]
- 65. Suzuki N., Yoshida A., Nakano Y. 2005. Quantitative analysis of multi-species oral biofilms by TaqMan Real-Time PCR. Clin. Med. Res. 3:176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Takamatsu D., et al. 2005. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 58:380–392 [DOI] [PubMed] [Google Scholar]
- 67. Tamura H., Yamada A., Yoshida Y., Kato H. 2009. Identification and characterization of an autolysin gene, atlh, from Streptococcus downei. Curr. Microbiol. 58:432–437 [DOI] [PubMed] [Google Scholar]
- 68. Thomas V. C., Hancock L. E. 2009. Suicide and fratricide in bacterial biofilms. Int. J. Artif. Organs 32:537–544 [DOI] [PubMed] [Google Scholar]
- 69. Titgemeyer F., Hillen W. 2002. Global control of sugar metabolism: a Gram-positive solution. Antonie Van Leeuwenhoek 82:59–71 [PubMed] [Google Scholar]
- 70. Vilain S., Pretorius J. M., Theron J., Brozel V. S. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microbiol. 75:2861–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang B. Y., Kuramitsu H. K. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 71:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 73. Yamada A., Tamura H., Kato H. 2009. Identification and characterization of an autolysin gene, atlg, from Streptococcus sobrinus. FEMS Microbiol. Lett. 291:17–23 [DOI] [PubMed] [Google Scholar]
- 74. Zheng L., Chen Z., Itzek A., Ashby M., Kreth J. 2011. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J. Bacteriol. 193:516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]