Abstract
The mycobacteriophage Ms6 is a temperate double-stranded DNA (dsDNA) bacteriophage which, in addition to the predicted endolysin (LysA)-holin (Gp4) lysis system, encodes three additional proteins within its lysis module: Gp1, LysB, and Gp5. Ms6 Gp4 was previously described as a class II holin-like protein. By analysis of the amino acid sequence of Gp4, an N-terminal signal-arrest-release (SAR) domain was identified, followed by a typical transmembrane domain (TMD), features which have previously been observed for pinholins. A second putative holin gene (gp5) encoding a protein with a predicted single TMD at the N-terminal region was identified at the end of the Ms6 lytic operon. Neither the putative class II holin nor the single TMD polypeptide could trigger lysis in pairwise combinations with the endolysin LysA in Escherichia coli. One-step growth curves and single-burst-size experiments of different Ms6 derivatives with deletions in different regions of the lysis operon demonstrated that the gene products of gp4 and gp5, although nonessential for phage viability, appear to play a role in controlling the timing of lysis: an Ms6 mutant with a deletion of gp4 (Ms6Δgp4) caused slightly accelerated lysis, whereas an Ms6Δgp5 deletion mutant delayed lysis, which is consistent with holin function. Additionally, cross-linking experiments showed that Ms6 Gp4 and Gp5 oligomerize and that both proteins interact. Our results suggest that in Ms6 infection, the correct and programmed timing of lysis is achieved by the combined action of Gp4 and Gp5.
INTRODUCTION
The majority of double-stranded DNA (dsDNA) bacteriophages described to date terminate each infection cycle through the programmed and regulated activity of two phage-encoded proteins, the endolysin and the holin, a small membrane protein that controls the endolysin function and the access to the peptidoglycan (39, 40). Endolysins are characterized by their ability to directly target covalent bonds in the peptidoglycan layer of the bacterial cell wall; the result of this activity is disruption of the rigid murein layer and release of newly synthesized virions (17, 42). During phage assembly, holin molecules accumulate in the cytoplasmic membrane without a detectable effect on the host (11, 37). Then, at an allele-specific time programmed into their primary structure, holins trigger to disrupt the cytoplasmic membrane (8, 11). Holins are extremely diverse and are found with at least three membrane topologies in many unrelated sequence families, suggesting that they may have evolved from multiple distinct origins to allow precisely scheduled efficient lysis and rapid adjustment of the lysis time, either on the basis of genetic selection or, in some cases, in real time in response to environmental changes (37, 41). The canonical holins, such as those of phages λ and T4, form very large holes that allow fully folded and active endolysins accumulated in the cytosol to pass through the cytoplasmic membrane and attack the peptidoglycan. These holes are nonspecific and allow the passage of unrelated endolysins (41) and proteins larger than 480 kDa (38). In addition, hole formation is absolutely required for lysis. Many phages also encode an antiholin, which contributes to control the timing of host lysis by inhibiting the holin. In some cases, the antiholin is encoded by the holin gene, with an additional N-terminal extension of several amino acids—a dual-start motif (1)—or is translated from an alternative intragenic start codon (7, 35). In other cases, the antiholin is encoded by an independent gene (27, 36, 44).
Recently, an alternative and remarkably different class of holin-endolysin systems became known (22, 23). This class, represented by the lambdoid bacteriophage 21, utilizes endolysins having N-terminal secretory signal-arrest-release (SAR) signals and pinholins. For phages encoding SAR endolysins, the holin protein needs only to produce lesions large enough to allow the passage of ions and depolarize the cytoplasmic membrane in order to fulfill its role in controlling the timing of lysis. Indeed, unlike lesions formed by the λ holin, lesions formed by the phage 21 holin do not allow the passage of λ endolysin (23). The term “pinholin” has been proposed to differentiate the small-hole (pinhole)-forming character of the phage 21 holin from the canonical holins that form large, nonspecific holes (23).
The genetic organization of the mycobacteriophage Ms6 lysis functions was previously described (4). In addition to the endolysin (lysA) and a holin-like gene (gp4), three accessory lysis genes restricted to mycobacteriophages were also identified: gp1, lysB, and gp5. The gp1 gene was recently identified as encoding a chaperone-like protein that specifically interacts with the N-terminal region of LysA and is involved in its delivery to the peptidoglycan in a holin-independent manner (2). The Ms6 holin-like protein, encoded by gp4 (previously named hol), shares some structural characteristics with class II holins, which are usually hydrophobic in nature and small, with a hydrophilic carboxy-terminal domain and two potential transmembrane domains (TMD1 and TMD2). Gp4 holin function was also supported by its ability to complement a λ phage S mutant (suggesting that the Ms6 holin allows the nonspecific release of the λR endolysin to the periplasm) and by the lethal phenotype observed when Gp4 is overexpressed in Escherichia coli, which is explained by the introduction of nonspecific lesions in the cytoplasmic membrane. However, unlike some holins, such as λ S, Gp4 lacks a dual-start motif (4). In addition, a second putative holin-like gene (gp5) encoding a protein with a predicted single TMD at the N-terminal region was identified at the end of the Ms6 lytic operon. In this work we studied the function of gp4 and gp5 gene products and demonstrated that, although nonessential for the Ms6 infective cycle, both appear to play a role in controlling the timing of lysis in mycobacteria. We also present evidence that the Ms6 holin-like protein encoded by gp4 (here, designated Gp4 to avoid confusion) has characteristic features of a pinholin and that Gp5 encodes a holin-like protein, and we suggest that the combined action of these two proteins is essential to effect host cell lysis at the correctly timed programmed lysis.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and culture conditions.
Bacterial strains, phages, and plasmids used throughout this study are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani (LB) broth or agar supplemented with 100 μg ml−1 ampicillin or 30 μg ml−1 kanamycin, when appropriate. Mycobacterium smegmatis recombinant strains were grown at 37°C in 7H9 medium (Difco) supplemented with 0.05% Tween 80 and 0.5% glucose, with shaking, or in Middlebrook 7H10 (Difco) medium containing 15 μg ml−1 kanamycin. For induced conditions, 0.2% succinate and 0.2% acetamide were also added to media.
Table 1.
Strains, bacteriophages, and plasmids used in this studya
| Strain, bacteriophage, or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Stratagene |
| BL21 | F−ompT hsdSB(rB− mB−) gal dcm | Novagen |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| M. smegmatis | 31 | |
| mc2155 | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | |
| Bacteriophages | ||
| D29 | Lytic phage that infects both fast- and slow-growing mycobacterial species | Institute Pasteur collection |
| λgt11 | cIts857 Sam100 | Stratagene |
| Ms6wt | Temperate bacteriophage from M. smegmatis | 26 |
| Ms6Δgp1 | 213-bp in-frame deletion of the Ms6 gp1 gene | 2 |
| Ms6Δgp4 | 210-bp in-frame deletion of the Ms6 gp4 gene | This study |
| Ms6Δgp5 | 366-bp in-frame deletion of the Ms6 gp5 gene | This study |
| Ms6Δgp1 gp4 | 213-bp and 210-bp in-frame deletions of the Ms6 gp1 and gp4 genes, respectively | This study |
| Ms6Δgp1 gp5 | 213-bp and 366-bp in-frame deletions of the Ms6 gp1 and gp5 genes, respectively | This study |
| Plasmids | ||
| pQE30 | Expression vector, T5 promoter; AmprlacIq | Qiagen |
| pET29a(+) | Expression vector, T7 promoter; Kanr | Novagen |
| pJV53 | Derivative of pLAM12 with Che9c 60 and 61 under control of the acetamidase promoter; Kanr | 34 |
| pMG231A | lysA cloned into pQE30 | 4 |
| pMP300 | Ms6 lysA and gp4 cloned in pQE30 | 4 |
| pMP310 | Ms6 gp4 cloned in pQE30 | 4 |
| pMJC21 | Ms6 gp5 cloned in pQE30 | This study |
| pMJC22 | Ms6 gp4 and gp5 cloned in pQE30 | This study |
| pMJC23 | D29 gp11 cloned in pQE30 | This study |
| pMJC24 | λR and Ms6 gp4 cloned in pQE30 | This study |
| pMJC25 | λR and Ms6 gp5 cloned in pQE30 | This study |
| pMJC27 | Ms6 gp5 cloned in pMG231A | This study |
| pMJC28 | Ms6 gp4 and gp5 cloned in pMG231A | This study |
| pMJC29 | D29 gp11 cloned in pMG231A | This study |
| pMJC30 | Ms6 gp4 cloned in pET29a(+) | This study |
| pMJC31 | Ms6 gp5 cloned in pET29a(+) | This study |
| pMJC32 | Ms6 gp4 and gp5 cloned in pET29a(+) | This study |
The accession number for the Ms6 lysis genes is AF319619.
Plasmid construction.
Unless indicated otherwise, the DNA fragments were obtained by PCR using Ms6 genomic DNA as a template. DNA amplification, plasmid isolation, and electrophoresis were carried out using standard techniques (28). E. coli and M. smegmatis mc2155 cells were transformed as described previously (28, 31). Restriction enzymes and T4 DNA ligase (New England BioLabs) were used according to the supplier's recommendations. All oligonucleotides were from Thermo Scientific and are listed in Table S1 in the supplemental material.
In order to construct plasmids pMJC21 and pMJC22, DNA fragments containing the gp5 or gp4 and gp5 genes were obtained by PCR amplification with primers Porf5a/Porf5-c2 or Porf4-1/Porf5-c2, respectively. Primers were designed in order to generate restriction sites, and the DNA fragments were inserted in the same sites of vector pQE30 (Qiagen), allowing fusion to a hexahistidine tag at the N terminus. To obtain plasmid pMJC23, the DNA fragment containing the gene gp11 was amplified by PCR using D29 genomic DNA as a template with primers PholD29fwd/PholD29rv and cloned into SacI/HindIII sites of pQE30. pMJC24 and pMJC25 were constructed in two steps: the λR gene was amplified using the genomic DNA of bacteriophage λgt11 as a template with primers PλRfwd/PλRrv and cloned into BamHI/SacI sites of pQE30. The gp4 or gp5 gene was amplified by PCR using Ms6 genomic DNA as a template with primers Porf4-1/Porf4-c1 or gp5RBSfwd/Porf5-c2 and cloned into SacI/HindIII sites of pQE30:λR, generating plasmids pMJC24 and pMJC25, respectively. pMJC27, pMJC28, and pMJC29 were obtained by amplifying gp5 or gp4 and gp5 with primers gp5RBSfwd/Porf5-c2 or Porf4-1/Porf5-c2 or D29 gp11 with primers PholD29fwd/PholD29rv using the genomic DNA of mycobacteriophage D29 as a template. The DNA fragments were introduced into the SacI/HindIII sites of pMG231A. To obtain plasmids pMJC30, pMJC31, and pMJC32, the gp4, gp5, or gp4 and gp5 genes of Ms6 were amplified with primers Prgp4Ms6fwd/gp4Ms6rv, gp5Ms6fwd/gp5Ms6rv, or Prgp4Ms6fwd/gp5Ms6rv, respectively, and cloned into BamHI/HindIII sites of pET29a(+). All constructs were validated by verifying the nucleotide sequence of the insert.
Protein interaction experiments: cross-linking.
Bis (sulfosuccinimidyl) suberate (BS3) cross-linker stock solution at a 10 mM final concentration was prepared immediately before use to decrease the extent of hydrolysis in 20 mM Na-HEPES–200 mM NaCl, pH 7.0. For in vitro cross-linking experiments, E. coli BL21(DE3) carrying plasmid pMJC30, pMJC31, or pMJC32 was induced at the logarithmic growth phase with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and 10-ml samples were withdrawn and pelleted after 1 h. Cells were resuspended in phosphate-buffered saline (PBS), broken by sonication, and centrifuged at 4°C. The proteins of the membrane fraction were extracted with 1% Triton X-100 for 2 h at 37°C. The detergent fraction was treated with BS3 solution to a final concentration between 1 and 5 mM at room temperature for 30 min. In the control samples, the cross-linker was omitted. After incubation at room temperature, samples were resuspended in SDS-PAGE sample buffer that quenches the reaction. Aliquots were subjected to SDS-PAGE, and Gp4 or Gp5 proteins were detected by Western blotting using horseradish peroxidase (HRP)-conjugated anti-His6 monoclonal antibody (Roche).
Beta-galactosidase activity assay.
β-Galactosidase activity (20) was measured in the supernatants of induced E. coli BL21 cells carrying plasmids pQE30, pMP310, pMJC21, pMJC22, and pMJC23. Following a 1-h induction with 1 mM IPTG, 1-ml aliquots of exponential growing cultures were centrifuged, and 30 μl of supernatant was added to 66 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) solution (Sigma) (4 mg/ml in 0.1 M sodium phosphate buffer, pH 7.5), 3 μl of 4.5 M β-mercaptoethanol, 0.1 M MgCl2 solution, and 200 μl of 0.1 M sodium phosphate buffer, pH 7.5. The reaction was performed at 37°C for 30 min and then stopped by the addition of 500 μl of Na2CO3. The amount of o-nitrophenol released was measured at 405 nm. Enzyme activity was expressed in arbitrary units of the optical density at 405 nm (OD405) ml−1 of culture min−1.
Construction of Ms6 mutants.
Construction of Ms6 mutants was performed using bacteriophage recombineering of electroporated DNA (BRED) in M. smegmatis. Recombineering substrates and BRED strategy were done as described previously (2, 19). Briefly, for deletion of the Ms6 gp4, gp5, or gp4 and gp5 genes (yielding Ms6Δgp4, Ms6Δgp5, or Ms6Δgp4 gp5, respectively), 100-bp oligonucleotides, PrΔgp4, PrΔgp5, or PrΔgp4gp5, that have 50 bp of homology upstream and downstream of the region to be deleted, were extended by PCR using two 75-bp extender primers, PrExtΔgp4fwd/PrExtΔgp4rv, PrExtΔgp5fwd/PrExtΔgp5rv, or PrExtΔgp4fwd/PrExtΔgp5rv, respectively, which have 25 bp of homology to the ends of the 100-mer and add an additional 50 bp of homology on either end. For deletion of the Ms6 gp1 gene, a 100-bp oligonucleotide (PrΔgp1) was extended with the primers PrExtΔgp1fwd/PrExtΔgp1rv. The final 200-bp dsDNA products were purified using a MinElute PCR purification kit (Qiagen) and coelectroporated with Ms6 genomic DNA (for gp4, gp5, or gp4 and gp5 deletion) or with Ms6Δgp4 or Ms6Δgp5 genomic DNA (for gp1 deletion) into electrocompetent recombineering cells of M. smegmatis mc2155/pJV53. Cells were resuspended in 7H9 medium supplemented with 0.5% glucose and 1 mM CaCl2, incubated at 37°C for 2 h, and plated as top agar lawns with M. smegmatis mc2155. Phage plaques were picked into 100 μl of phage buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgSO4, 68.5 mM NaCl, 1 mM CaCl2), eluted for 2 h at room temperature, and analyzed by PCR with primers PrP1Fwd/PrlysA180bprv flanking the gp1 deletion or with primers lysBfwd/Ms6rv to detect the gp4, gp5, or gp4 and gp5 deletions. Mixed primary plaques containing both the deletion mutant and the wild-type (wt) DNA were eluted as described above, and serial dilutions were plated with M. smegmatis. Individual secondary plaques or lysates were screened by PCR with primers flanking the deletions for the presence of pure mutant phage.
One-step growth curves and burst size determination.
One-step growth curve and burst size determination were described previously (2). The one-step assays were carried out in cells in exponential growth using a multiplicity of infection (MOI) of 1. M. smegmatis cells were pelleted and resuspended in 1 ml of a phage suspension (wt Ms6 [Ms6wt], Ms6Δgp1, Ms6Δgp4, Ms6Δgp5, Ms6Δgp1 gp4 or Ms6Δgp1 gp5) supplemented with 1 mM CaCl2. The mixture was incubated for 50 min at 37°C to allow adsorption of the phage. One hundred microliters of 0.4% H2SO4 was added to inactivate the nonadsorbed phage, and the incubation was continued for five min. The suspension was neutralized with 100 μl of 0.4% NaOH and diluted 1:100 in 7H9 medium supplemented with 0.5% glucose and 1 mM CaCl2. One-milliliter samples were withdrawn every 30 min up to 300 min. One hundred microliters of serial dilutions of each sample was plated with 200 μl of M. smegmatis cells on 7H10 medium as top agar lawns, and the phage titer for each sample was determined after 24 h of incubation at 37°C. The same experimental procedure was used for burst size determination except that 10 μl of infected cells was diluted in order to obtain one infected cell ml−1 in 7H9 medium. Samples of 1 ml of infected cultures were distributed in 50 tubes and incubated for 180 min at 37°C. A total of 200 μl of M. smegmatis cells and top agar (4 ml) were added to each tube and plated on 7H10 medium. After 24 h at 37°C, the phage plaques were counted, and the Poisson distribution of [P(n)] was applied to determine the burst size (BS): P(n) = (e−c · cn)/n! (e < 1), where P(n) is the probability of samples having n infected cells, c is the average number of infected cells per tube, and BS = (total plaque count in the 50 plates)/(total number of infected cells).
RESULTS
Sequence analysis of the Ms6 holin-like genes.
The mycobacteriophage Ms6 gp4 encodes a 77-amino-acid polypeptide with a predicted molecular mass of 7.8 kDa that was previously described as a holin-like protein (4). This assumption was based on several features of Gp4: its high similarity with the Lactococcus lactis bacteriophage r1t holin, a deduced amino acid sequence sharing characteristics with the class II holins, its high toxicity when overexpressed in E. coli leading to a lethal phenotype, and, finally, the ability to complement a lambda S mutant. However, unlike other class II holins, such as the S holin of bacteriophage 21, Ms6 Gp4 lacks a dual-start motif (4). The availability of more than 60 mycobacteriophage sequenced genomes has placed the Ms6 holin in the gene phamily Pham 95, according to sequence similarity to putative holin genes from mycobacteriophages of subcluster F1 (Fig. 1A) (12). Recently, we have reported that export of the Ms6 endolysin (LysA) is holin independent and that LysA translocation across the cytoplasmic membrane is assisted by Gp1, a chaperone-like protein, encoded by the first gene of the Ms6 lysis cassette (2). These data, together with the absence of a lysis phenotype when Gp4 was coexpressed in E. coli with Ms6 endolysin (4), even when both proteins were shown to be expressed at detectable levels (M. J. Catalão, unpublished data), led us to reanalyze the amino acid sequence of Ms6 Gp4.
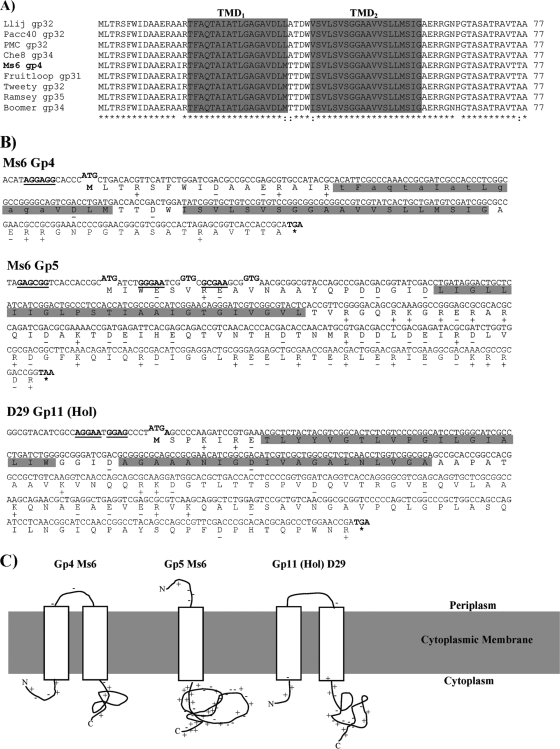
Fig. 1.
Holin-like proteins of mycobacteriophages Ms6 and D29. (A) CLUSTALW alignment of Ms6 Gp4 (AAG48320) with similar sequences of Pham 95 members included in subcluster F1: Llij Gp32 (ABD58248), Pacc40 Gp32 (YP002241616), PMC Gp32 (ABE67533), Che8 Gp34 (NP817372), Fruitloop Gp31 (YP002241716), Tweety Gp32 (YP001469265), Ramsey Gp35 (YP002241822), and Boomer Gp34 (YP002014250) (the primary accession numbers of the UniProtKB/TrEmbl database are given in parentheses). Identical (*) and highly similar (:) amino acids are indicated. Numbers refer to the amino acid positions. The two TMDs are indicated in a gray box. (B) Sequences of genes coding for the class II holin (gp4) and class III holin (gp5) of Ms6 and class II holin of D29 (gp11). Charged residues are indicated by a plus or minus sign. TMDs are indicated in gray. Amino acid residues in the SAR domain of Gp4 that are predicted to be weakly hydrophobic are shown in lowercase. Potential translation start codons and corresponding Shine-Dalgarno sequences are in bold and underlined. (C) Topological model for Ms6 Gp4 (N-in, C-in), Gp5 (N-out, C-in), and D29 Gp11 (Hol) (N-in, C-in).
The Ms6 Gp4 possesses two TMDs, the most hydrophobic of which is TMD2 (residues 39 to 58) (Fig. 1B) and a predicted N-in, C-in topology according to the HMMTOP (http://www.enzim.hu/hmmtop/) program from the Expasy server. TMD1, not predicted by every TMD search algorithm, is present from residues 17 to 34 and has characteristics of a SAR domain, with a high percentage (11 out of 18) of weakly hydrophobic or polar uncharged residues (Fig. 1B), like Ala, Gly, Gln, and Thr (22). The presence of a SAR domain followed by a typical TMD suggests that Ms6 Gp4 is a pinholin, analogous to other pinholins already characterized, such as the holin of phage 21, S2168 (21, 23), or the holin of phage Xfas53 (32). As already mentioned, the Ms6 lysis module is organized into five genes, and, so far, no function has been assigned to the last gene within the lysis cassette. gp5 has the potential to encode a 124-amino-acid polypeptide with a predicted molecular mass of 14.1 kDa. The gp5 gene starts at an ATG codon that overlaps the gp4 TGA stop codon in a different reading frame (4). A BLASTp search using the Ms6 Gp5 deduced amino acid sequence identified a number of putative proteins with unknown functions, restricted to the mycobacteriophage group of phages with a high degree of sequence identity. Owing to their related amino acid sequences, they have been recently grouped in mycobacteriophage gene phamily, Pham 96 (12). Analysis of the amino acid sequence of Gp5 using HMMTOP showed the presence of a single TMD membrane-spanning α-helix domain from residues 22 to 45 in the N terminus with a highly charged and hydrophilic C-terminal domain (Fig. 1B), structural characteristics of class III holins, and a predicted N-out, C-in topology (Fig. 1C). In addition to the location of Gp5 in the lysis cassette in the vicinity of the endolysin gene, overexpression of Gp5 in E. coli results in a drastic inhibition of cell growth (Fig. 2A), suggesting that the gp5 gene product might function as a holin-like protein forming lesions in the cell membrane. However, in contrast to Ms6 Gp4, Gp5 was unable to complement a λS holin-defective mutant (data not shown).
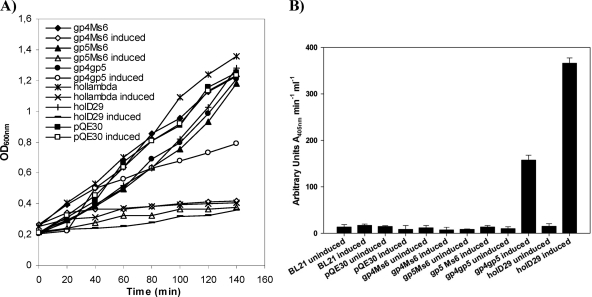
Fig. 2.
(A) Expression of the holin-like proteins from mycobacteriophages Ms6 and D29 in E. coli. E. coli JM109 cells carrying plasmid pQE30 containing no insert or cloned genes were grown in LB broth at 37°C to an OD600 of 0.2. At time zero, transcription of cloned lysis genes was induced by addition of 1 mM IPTG. (B) Release of β-galactosidase from E. coli BL21 expressing Ms6 and D29 holin-like proteins. Activity was determined in the supernatants of induced cultures. Results are averages of triplicate experiments.
Interestingly, there are three possible start codons in Ms6 gp5 at positions 1, 6, and 10 that would produce 124 (Gp5124), 118 (Gp5118), and 114 (Gp5114) amino acid products (of 14.1, 13.5, and 13 kDa, respectively), all preceded by potential ribosome-binding sites (5′-GAGCGG-3′ for Gp5124, 5′-GGGAA-3′ for Gp5118, and 5′-GCGAAG-3′ for Gp5114). Gp5118 and Gp5124 have N-terminal extensions with one extra positively charged residue compared to Gp5114, which could retard hole formation and also confer a negative-dominant antiholin character. However, unlike 21 S21 and Xfas53 hol whose translation is regulated by the presence of RNA stem-loop structures overlapping the ribosome-binding sites that reduce holin translation in favor of antiholin synthesis (23, 32), no RNA stem-loops were identified upstream of gp5 mRNA.
Evaluation of holin lesion through β-galactosidase leakage.
The ability of cytoplasmic membrane hole formation by Ms6 Gp4 or Gp5 was investigated by β-galactosidase leakage from E. coli BL21 strains expressing Ms6 holin-like proteins Gp4, Gp5, or both (Fig. 2A). This assay has been used to search for proteins with canonical holin-like activity, as the damage caused to the cytoplasmic membrane by the holin protein is sufficient to allow the leakage of cytoplasmic contents, including large proteins such as the constitutively expressed β-galactosidase enzyme (3). Our results show that a β-galactosidase leakage phenomenon (measured by an increase in enzymatic activity) was not observed when Gp4 or Gp5 expression was induced, suggesting that the lesions formed by these proteins are not large enough to allow the passage through the cytoplasmic membrane of proteins as large as β-galactosidase (Fig. 2B). In contrast to what is observed in Ms6 and other related mycobacteriophages, the lysis cassette of mycobacteriophage D29, a phage grouped in subcluster A2 (12), does not possess homologues of the accessory lysis protein Gp1 or Gp5. D29 lytic genes are clustered together, with the holin-like gene (gp11) localized between the lysA and lysB genes (12). The D29 gp11 gene has the potential to code for a 141-amino-acid polypeptide with a predicted molecular mass of 14.6 kDa and possesses structural characteristics of class II holins, presenting two transmembrane domains from residues 8 to 26 (TMD1) and 31 to 50 (TMD2) (Fig. 1B), a highly charged and hydrophilic C-terminal domain, and N-in, C-in topology (Fig. 1C), as predicted by HMMTOP. In addition, D29 Gp11 (Hol) overexpression in E. coli inhibits cell growth as observed for holin-like proteins (Fig. 2A). As shown in Fig. 2A, induction of D29 Hol expression allowed the release of β-galactosidase to the culture medium, resulting in an increase of the enzymatic activity (377 arbitrary OD405 units ml−1 min−1) by comparison to the BL21 control cells (14 arbitrary OD405 units ml−1 min−1) (Fig. 2B). These results suggest that the D29 holin (Gp11) functions as a canonical holin, forming large lesions in the cytoplasmic membrane sufficient to allow the access of the cytoplasmic endolysin to the peptidoglycan and bring about effective lysis of the host. These data support a potential holin function for D29 Gp11 (Hol) as previously proposed (12, 25). Surprisingly, the concomitant expression of Ms6 Gp4 and Gp5 results in a moderate increase in the β-galactosidase activity (156 arbitrary OD405 units ml−1 min−1), suggesting that these two proteins together may form a larger hole lesion than the sole proteins.
Expression of holin/endolysin pairwise combinations in E. coli.
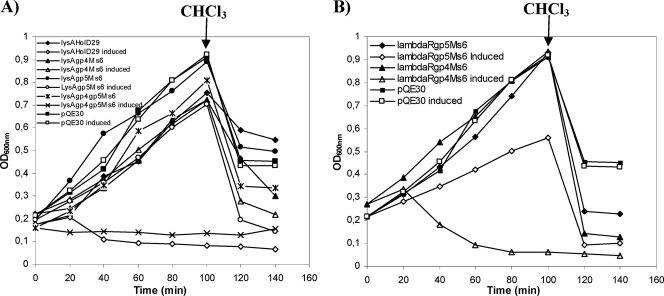
In contrast to what happens with phage endolysins that possess a narrow range of activity regarding the infected bacteria, holins are not species specific and do not specifically interact with the endolysins (29, 41). Since canonical holins, such as λS, form very large nonspecific holes that allow fully folded unrelated endolysins to pass through the membrane and attack the murein, we expressed in E. coli different pairwise combinations of endolysins/holins in an attempt to clarify the role of Ms6 Gp4 and Gp5 in bacterial lysis. We observed that the concomitant expression of Ms6 LysA with Gp4 or Gp5 was not sufficient to support a lysis phenotype in E. coli. However, coexpression of LysA with both Gp4 and Gp5 resulted in a slight decrease of the OD600 40 min after induction (Fig. 3A). The fact that the lack of a lysis phenotype was a consequence of the inability of Ms6 Gp4 or Gp5 to form lesions on the cytoplasmic membrane large enough to allow the passage of LysA to the periplasm was further confirmed by concomitantly expressing the Ms6 endolysin LysA with the D29 Gp11 holin. Lysis of E. coli was observed beginning 20 min after induction, which suggests that the D29 holin is functional in E. coli and allows the access of Ms6 LysA to the peptidoglycan (Fig. 3A). In addition, the Ms6 Gp4 but not Gp5 allows the access of the λ transglycosylase to the murein, as demonstrated by complementation of a λS mutant (4; M. J. Catalão, unpublished data), and unlike Gp4, Gp5 was unable to promote release of λR, the cytosolic endolysin of phage λ (Fig. 3B). We interpret this to mean that the Ms6 Gp4 or Gp5 alone makes holes too small to allow the passage of Ms6 LysA in E. coli.
Fig. 3.
(A) Effect of the expression of phage endolysin/holin pairwise combinations on E. coli growth. (B) Effect of the expression of pairwise combinations of the λR endolysin with the Ms6 holin-like proteins Gp4 and Gp5 in E. coli. E. coli JM109 cells carrying plasmid pQE30 containing no insert or cloned genes were grown in LB broth at 37°C to an OD600 of 0.2. At time zero, transcription of cloned lysis genes was induced by addition of 1 mM IPTG. At the time indicated by the arrow, 2% CHCl3 was added to the cultures.
Cross-linking of Gp4 and Gp5 in the E. coli cell membrane.
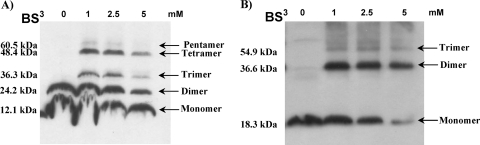
It is known that holins must oligomerize to achieve their lethal membrane effect (9). To identify the oligomeric states of Ms6 Gp4 and Gp5, the membrane fractions from E. coli expressing Gp4 or Gp5 (fused to an S tag at the N terminus and a His6 tag at the C terminus) from a derivative plasmid of pET29a were collected 60 min after induction; proteins were extracted from the membranes with Triton X-100 and subjected to cross-linking with the water-soluble membrane-impermeant, homobifunctional sulfo-N-hydroxy-succinimide ester, BS3. The presence of a band of 24.2 kDa in the absence of the cross-linker shows that Gp4 forms SDS-resistant dimers during membrane extraction with Triton X-100 (Fig. 4A). Furthermore, Gp4-specific bands corresponding to molecular masses of 12.1, 24.2, 36.3, 48.4, and 60.5 kDa, up to pentamers, could be detected by Western blot analysis (Fig. 4A). In contrast, Gp5, which does not complement a λS-defective mutant phage, formed only trimers but not higher oligomers under the same conditions (Fig. 4B). This result might help explain the inability of Gp5 to support an efficient lysis of E. coli when it is coexpressed with different endolysins as the ability of holin molecules to oligomerize is essential for the lytic step in holin function (8, 9, 43).
Fig. 4.
Ms6 Gp4 (A) or Gp5 (B) oligomerization. Proteins from membranes of E. coli BL21(DE3) expressing Gp4 or Gp5 were extracted with Triton X-100 and treated with different BS3 concentrations as described in Materials and Methods. Proteins were detected by Western blotting with an anti-His6 antibody. Predicted molecular masses are indicated to the left of the panels. Oligomerization bands are indicated by arrows.
Evidence for an interaction between Gp4 and Gp5.
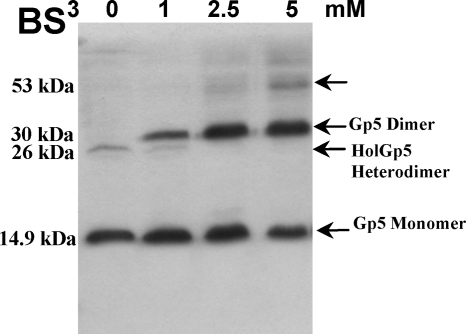
Despite the toxicity observed after expression induction of Gp4 or Gp5 membrane proteins in E. coli, simultaneous expression of these proteins attenuates the lethal effect (Fig. 1A). For bacteriophage lambda, it has been proposed that the lysis inhibitor S107 (a protein of 107 amino acids encoded by the S gene) inhibits lysis through intermolecular interaction with the lysis effector S105 (10). Accordingly, the ability of Gp5 to inhibit the Gp4 lethal effect suggests that it may interact with the holin. To demonstrate this, both proteins were expressed from the same vector in E. coli with Gp4 fused to an S tag at the N terminus and Gp5 fused to a His6 tag at the C terminus. At 60 min postinduction, cell membranes were collected by centrifugation, proteins were extracted with 1% Triton X-100, and this fraction was subjected to chemical cross-linking with BS3. In the absence of the cross-linker, we detected a band with a molecular mass of 14.9 kDa, corresponding to the size predicted for Gp5-His6 monomer, and a faint band of ∼26 kDa corresponding to an interaction between an S-tagged Gp4 monomer and a Gp5-His6 monomer (Fig. 5). Cross-linking using 1 mM BS3 revealed an additional band of 30 kDa corresponding to the Gp5 dimer. The increase in BS3 concentration to 5 mM led to the appearance of a band of ∼53 kDa as a result of oligomer formation between the Gp5 homodimer and Gp4 homodimer. This result suggests that Gp5 interacts with the Ms6 holin in some cooperative fashion to effect lysis and suggests that it may control Gp4 function during the lytic cycle.
Fig. 5.
Interaction between Gp4 and Gp5 of mycobacteriophage Ms6. Cross-linking and sample preparation for Western blot analysis was performed as described in the legend of Fig. 4. Proteins were detected with an anti-His6 antibody. In the absence of BS3, Gp5 monomer and Gp4-Gp5 heterodimer are detected. Predicted molecular masses are indicated to the left of the panels. Oligomerization bands are indicated by arrows.
Construction of Ms6 mutants defective for lysis.
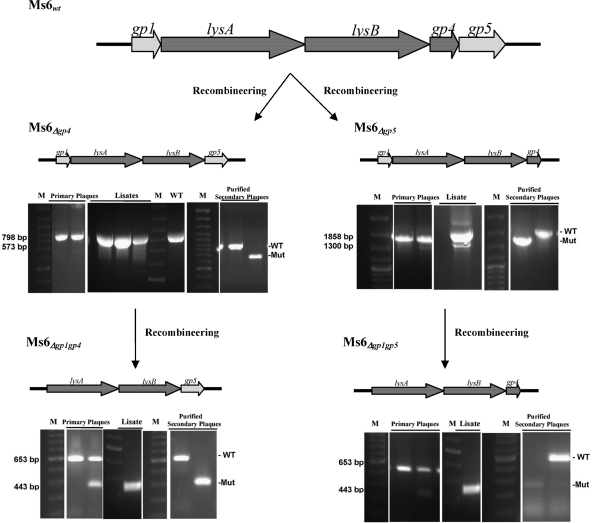
Concerning the possibility that Ms6 Gp4 and Gp5 may behave differently in the heterologous E. coli host and in the Ms6 natural host, M. smegmatis, we constructed Ms6 mutants defective for synthesis of Gp4, Gp5, or both proteins. Using the bacteriophage recombineering of electroporated DNA (BRED) system (19), we constructed internal in-frame deletions of Ms6 gp4, gp5, or gp4 and gp5 in the Ms6 wild-type phage and, in a second step, of the gp1 gene in the previously constructed mutants defective for Gp4 or Gp5 synthesis (Ms6Δgp4 or Ms6Δgp5) by allelic gene replacement. Even though we have not yet been able to recover a purified mutant derivative of phage Ms6Δgp4 gp5, probably reflecting poor viability of the mutant, pure mutants of Ms6Δgp4 and Ms6Δgp5 were readily identified in high frequencies and in the absence of any selection (Fig. 6). In view of the fact that the absence of Gp4 or Gp5 from the Ms6 virion has no apparent effect on phage viability and based on our recent observations that the accessory lysis protein Gp1 is required for a normal burst of infective phage particles (2), we further investigated its function during the Ms6 lytic cycle. We constructed an internal in-frame deletion of the gp1 gene in defective phage Ms6Δgp4 and Ms6Δgp5 using the same recombineering strategy described above, and pure mutants of Ms6Δgp1 gp4 and Ms6Δgp1 gp5 were isolated after PCR screening of secondary individual plaques (Fig. 6).
Fig. 6.
Strategy for construction of Ms6 lysis gene deletion mutants. A 200-bp dsDNA substrate that has 100-bp homology flanking the deletion was designed. Following coelectroporation of the 200-bp substrates and genomic DNA (of Ms6wt to obtain Ms6Δgp4 and Ms6Δgp5 mutants or of Ms6Δgp4 and Ms6Δgp5 to obtain Ms6Δgp1 gp4 and Ms6Δgp1 gp5 mutants), primary plaques were recovered to identify a mixed plaque containing wild-type and mutant phages. The mixed primary plaque was diluted and plated; the lysate was screened to check for phage viability, and purified secondary plaques were screened to identify homogenous deletion mutants.
These results demonstrate that Ms6Δgp4, Ms6Δgp5, Ms6Δgp1 gp4, and Ms6Δgp1 gp5 are viable, that neither gp4 nor gp5 is essential for plaque formation, and that an Ms6 mutant phage lacking the gp4 and gp5 lysis genes is able to infect and lyse mycobacteria even though it could not be isolated yet.
Ms6 holin-like proteins are not required for M. smegmatis lysis.
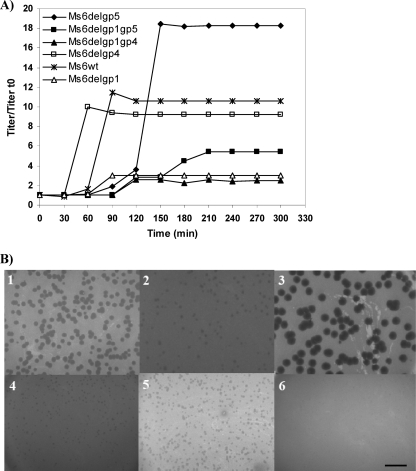
One-step growth curves and determination of phage growth parameters (latent period, rise period, and burst size) were carried out to compare the Ms6 mutant replication cycle. Results are summarized in Table 2. The one-step growth experiment (Fig. 7A) shows that Gp4 and Gp5, although nonessential for lysis, have an effect on the lysis timing since an Ms6 gp4 deletion mutant caused slightly accelerated lysis (80 min), whereas an Ms6 gp5 deletion mutant delayed lysis (170 min), which is consistent with holin function. These lysis times correspond to the latent time represented in Fig. 7A in addition to the initial 50 min of adsorption and were compared to the Ms6 wild-type phage (110 min) under the same experimental conditions. Thus, the absence of gp4 or gp5 in the infecting virion has an evident effect on the timing of lysis. Single-burst experiments were done to compare the viable progeny released from single cells infected with Ms6wt or the phage mutants. The number of infective particles released after infection with the Ms6Δgp4 phage is lower than in an Ms6wt infection, whereas after infection with the Ms6Δgp5 phage, an increase in the burst size was detected. Under our experimental conditions, when Ms6wt infects M. smegmatis mc2155, there is an average of 149 viable phage released from one bacterium while infection with Ms6Δgp4 or Ms6Δgp5 yielded an altered burst size of approximately 115 or 221 viable phage per infected cell, respectively. Deletion of the gp1 gene from Ms6Δgp4 or Ms6Δgp5 drastically reduced the burst size to ∼28 phage/infected cell or ∼77 phage/infected cell for Ms6Δgp1 gp4 or Ms6Δgp1 gp5 phage, respectively, which is in agreement with our previous results, which showed that Gp1 is essential to achieve the wild-type burst (2). When we analyzed the plating ability and the plaque size of the lysis-defective phage, we observed that all except Ms6Δgp5 produced smaller plaques, with no size variation, than those produced by Ms6wt phage (Fig. 7B). The size of the plaques produced by Ms6Δgp1 gp4 was <1 mm, whereas the plaques produced by the Ms6Δgp5 mutant were very large, with diameters of 4 to 5 mm, in agreement with a lower and a larger burst size, respectively, than Ms6wt phage. Taken together, these results suggest that in addition to gp4, gp5 encodes a holin-like protein, and they must act in concert to control the timing of lysis. Furthermore, as previously observed (2), the presence of Gp1 is crucial to obtain a normal burst of infective phage although it has no influence on duration of the latent time of the lytic cycle.
Table 2.
Viability and phage growth parameters of mycobacteriophage Ms6 and lysis gene deletion derivatives
| Phage | Plaque-forming ability | Burst size (no. of phage) | Latent time (min) | Plaque size (mm) |
|---|---|---|---|---|
| Ms6wt | Yes | 149 ± 32 | 110 | 2–3 |
| Ms6Δgp1 | Yes | 45 ± 13 | 110 | ∼1 |
| Ms6Δgp4 | Yes | 115 ± 39 | 80 | 1–2 |
| Ms6Δgp5 | Yes | 221 ± 56 | 170 | 4–5 |
| Ms6Δgp1 gp4 | Yes | 28 ± 6 | 140 | 1–2 |
| Ms6Δgp1 gp5 | Yes | 77 ± 27 | 200 | <1 |
| Ms6Δgp4 gp5 | Yes | |||
| Ms6Δgp1 gp4 gp5 | No |
Fig. 7.
(A) One-step growth curves of mycobacteriophage Ms6 and lysis gene deletion derivatives. For each curve the titers measured were divided by the titer at time zero (t0) for normalization (titer/titer t0). Results are averages of three independent experiments. (B) Plating ability of the different lysis gene mutant bacteriophage. Images show results of M. smegmatis infection with the following: frame 1, Ms6wt phage; frame 2, Ms6Δgp4; frame 3, Ms6Δgp5; frame 4, Ms6Δgp1; frame 5, Ms6Δgp1 gp5; frame 6, Ms6Δgp1 gp4. Scale bar, 1 cm.
DISCUSSION
Even though the mechanisms underlying mycobacteriophage lysis of mycobacteria are poorly understood, recent work has contributed significantly to the progress in the field (2, 5, 6, 14, 25). Nonetheless, the exact mechanism by which the lysis effectors LysA and LysB are localized to their substrates remains elusive in the majority of the mycobacteriophages. Very recently, we have identified the product of Ms6 gp1 gene as a chaperone-like protein that specifically interacts with the endolysin and is involved in its translocation across the cytoplasmic membrane (2). Moreover, removal of Gp1 function in mycobacteriophage Ms6 showed that, although not essential for plaque formation, the protein is required for efficient phage release. Similarly to what has been reported for phages possessing endolysins endowed with signal sequences or SAR domains, Ms6 LysA translocation in E. coli also involves the host Sec system (2). These data, together with the previous reported absence of lysis when Ms6 LysA and Gp4 were coexpressed in E. coli (4), led us to investigate the function of the previously identified holin protein in the mycobacteriophage Ms6 infection context. The gp4 gene is localized downstream of lysB and encodes a protein with structural characteristics of class II holins with the ability to complement a λS defect. In phages like the lambdoid 21, where the endolysin possesses a SAR domain and translocation is holin independent, holins belong to a recently discovered class of proteins, the pinholins, that make small holes in the host membrane that are sufficient to depolarize it and allow membrane release of SAR endolysins even though they are not large enough to allow escape of canonical cytoplasmic endolysins. In these cases the holin function is restricted to regulation of the timing of lysis (21, 23).
Reexamination of the predicted amino acid sequence of Ms6 Gp4 showed that its TMD1 has characteristics of a SAR domain with a high percentage of hydrophobic residues, a characteristic described for the pinholin of the lambdoid phage 21 (22). Although Gp4 was unable to support the Ms6 LysA-mediated lysis of E. coli cells (4), unlike the pinholin of phage 21 S21 (23), it was able to promote the release of R, the cytosolic endolysin from phage λ (Fig. 2B), but not the release of Ms6 LysA or D29 LysA. Of note is the fact that the predicted molecular mass of the Ms6 endolysin is 43 kDa while λR is a protein of 17.8 kDa, which suggests that the passage of endolysins through holin holes is dependent on membrane pore size.
Gene organization in bacteriophage lysis cassettes may be extremely diverse: for the majority of phages, lysis genes are clustered and transcribed in the order of holin first and then endolysin, as exemplified by lambda phage (39). An inverted organization (lys upstream of hol) was reported for the Oenococcus oeni bacteriophage fOg44 (24), and in many cases the genes are not even linked (e.g., T4) (29). In many phages of Streptococcus thermophilus (30) and in phage Av-1, which infects the Gram-positive bacteria Actinomyces naeslundii (3), two putative holin genes precede the endolysin gene. In Bacillus subtilis prophage PBSX, it was proposed that two open reading frames preceding the endolysin xlyA, designated xhlA and xhlB, encode polypeptides that associate in the membrane to form a functional holin complex that allows XlyA access to the peptidoglycan (15, 18). In Staphylococcus aureus phage P68, a putative holin gene, hol15, was identified in the −1 reading frame at the 3′ and of the endolysin gene lys16. A second putative holin gene, hol12, was later identified at the end of the structural genes (33). In some phages, like λ and 21, the holin gene presents a dual-start motif producing two proteins by virtue of alternate translational starts, the holin and the antiholin, while in other phages these two proteins are encoded by separate genes (e.g., P1 and T4) (29, 42). Such diversity is also observed in mycobacteriophages: in addition to the endolysin LysA, the majority of mycobacteriophages sequenced so far encodes an additional enzyme with lipolytic activity, LysB, that targets the outer membrane of mycobacteria (5, 6). In phages belonging to cluster A2, like D29, the holin gene is positioned between lysA and lysB while in phages belonging to cluster F1, as in the Ms6 case, the holin gene is localized immediately downstream of lysB. For many mycobacteriophages a holin gene has not been identified yet, while in others (Ms6 and other members of subcluster F1) an additional lysis gene (gp5) encoding a predicted membrane protein is positioned immediately downstream of the gp4 gene. Gp5 encodes a 124-amino-acid protein possessing a single TMD and a very highly charged and hydrophilic C-terminal domain, and we hypothesized that it might function as a holin-like protein. Gp5 was found to be unable to support both LysA- or λR-mediated lysis in E. coli, and, in contrast to Gp4, oligomerization appeared to be blocked at the trimer stage in detergent (as for Gp4, the final degree of oligomerization is not yet known). Despite the toxicity observed when the integral membrane proteins Gp4 and Gp5 are independently expressed in E. coli, simultaneous expression of these proteins attenuates the lethal effect, which suggested that they may interact. For bacteriophage lambda, it has been proposed that the lysis inhibitor S107 inhibits lysis through dimeric interactions with the lysis effector S105 (10). Indeed, using chemical cross-linking, we were able to obtain biochemical evidence for a direct interaction between Gp4 and Gp5; however, the exact mechanism by which Gp5 acts to control Gp4 function remains elusive. Owing to the concerns that exist when holin genes are expressed from strong inducible promoters, we constructed different Ms6 mutants with deletions of the holin-like genes. Indeed, it is well known that a membrane protein overproduced from a multicopy plasmid can insult the membrane sufficiently to cause release of cytoplasmic endolysins (42). Although more time-consuming, this strategy has two important advantages: (i) it allows the function of these proteins to be examined in their natural host, and (ii) each gene product in the cell, resulting from phage infection, is produced at physiological levels. When we analyzed the holin gene-deleted mutant infection cycle by one-step growth curves, we observed that the gene products of gp4 and gp5, although nonessential for phage viability, appear to play a role in controlling the timing of lysis. Ms6Δgp4 caused accelerated lysis, whereas Ms6Δgp5 delayed lysis, which is consistent with holin function. We also considered whether Gp5 could act as an antiholin. Antiholins generally delay phage lysis in order to optimize progeny phage production. This should have been observed if Gp5 acts as an antiholin, and its absence would result in earlier lysis and smaller plaques due to premature lysis. Unexpectedly, this phenotype was observed when Gp4 was deleted from the lytic cassette. In contrast, deletion of Gp5 delayed lysis and resulted in very large plaques due to an increase in the burst size. Not all dsDNA phages utilize an antiholin to regulate lysis timing since some apparently simply rely on delaying expression of their holin genes (16). These results suggest that mycobacteriophage Ms6 gp4 and gp5 encode holin proteins whose combined action could play the role of a holin and that expression of both proteins is necessary to effect host cell lysis at the correct and programmed timing, as described for other phages such as the A. naeslundii phage Av-1 (3) and the B. subtilis PBSX phage (15). Moreover, interaction of Gp5 with Gp4 may contribute to very precise adjustment of the timing of hole formation and to keeping the infected cell productive, allowing the assembly of more virions. The ubiquity of holin-mediated lysis systems results from the ability of phages to rapidly evolve to shorter or longer infection cycles to adjust to changes in host quality or density (37). Deletion of the chaperone-like protein gp1 in both deletion mutants Ms6Δgp4 and Ms6Δgp5 was catastrophic for lysis, with more than 3-fold reduction of the burst size, even though the mutant phage are viable and could be isolated. Remarkably, although Gp5 was unable to allow endolysin-mediated lysis in E. coli, a mutant phage lacking both gp1 and gp4 was able to infect M. smegmatis cells and undergo lysis, so it is expected that there will be alternative pathways to release phage progeny (13). We conclude that for mycobacteriophage Ms6 and related mycobacteriophages, the presence of the endolysin in addition to one of the lytic genes, gp1, gp4, or gp5, is sufficient for a lysis phenotype. However, this results in dramatic changes in the infective cycle and in lower viability of the mutant phage. The presence of the mycobacterium-specific lysis factors Gp1 and Gp5 that are restricted to mycobacteriophages (4, 12) may confer a selective advantage not only for fitness under different environmental conditions but also as an alternative to exclusively holin-dependent lysis; it has been shown that single missense changes within the holin proteins can have a profound effect on both the process of host lysis and its timing, unpredictably resulting in dramatic shortening or lengthening of the infection cycle (8, 42). In addition, holin membrane holes have different sizes, and for holes too small to allow the passage of endolysins, phages must evolve in order to survive. Some phages evolved by synthesizing secreted endolysins endowed with signal sequences, while mycobacteriophages throughout their evolution acquired additional lysis genes which may confer host lysis benefits and successful phage propagation and replication. With this study we hope to have contributed to a better understanding of lysis timing regulation by mycobacteriophages.
Supplementary Material
ACKNOWLEDGMENTS
We thank Graham Hatfull, Julia van Kessel, and Laura Marinelli (University of Pittsburgh, Pittsburgh, PA) for supplying plasmid pJV53 and for technical assistance with the recombineering experiments.
This work was supported by grant PTDC/SAU-FCF/73017/2006 from the Fundação para a Ciência e Tecnologia (FCT). M. J. Catalão and F. Gil are the recipients of FCT Ph.D. fellowships (SFRH/BD/24452/2005 and SFRH/BD/29167/2006).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Bläsi U., Young R. 1996. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol. Microbiol. 21:675–682 [DOI] [PubMed] [Google Scholar]
- 2. Catalão M. J., Gil F., Moniz-Pereira J., Pimentel M. 2010. A chaperone-like protein is involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 77:672–686 [DOI] [PubMed] [Google Scholar]
- 3. Delisle A. L., Barcak G. J., Guo M. 2006. Isolation and expression of the lysis genes of Actinomyces naeslundii phage Av-1. Appl. Environ. Microbiol. 72:1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcia M., Pimentel M., Moniz-Pereira J. 2002. Expression of mycobacteriophage Ms6 lysis genes is driven by two σ70-like promoters and is dependent on a transcription termination signal present in the leader RNA. J. Bacteriol. 184:3034–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gil F., et al. 2008. The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology 154:1364–1371 [DOI] [PubMed] [Google Scholar]
- 6. Gil F., et al. 2010. The mycobacteriophage Ms6 LysB specifically targets the outer membrane of Mycobacterium smegmatis. Microbiology 156:1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graschopf A., Bläsi U. 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33:569–582 [DOI] [PubMed] [Google Scholar]
- 8. Gründling A., Bläsi U., Young R. 2000. Biochemical and genetic evidence for three transmembrane domains in the class I holin, lambda S. J. Biol. Chem. 275:769–776 [DOI] [PubMed] [Google Scholar]
- 9. Gründling A., Bläsi U., Young R. 2000. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J. Bacteriol. 182:6082–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gründling A., Smith D. L., Bläsi U., Young R. 2000. Dimerization between the holin and holin inhibitor of phage lambda. J. Bacteriol. 182:6075–6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gründling A., Manson M. D., Young R. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. U. S. A. 98:9348–9352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hatfull G. F., et al. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heineman R. H., Bull J. J., Molineux I. J. 2009. Layers of evolvability in a bacteriophage life history trait. Mol. Biol. Evol. 26:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henry M., et al. 2010. In silico analysis of Ardmore, a novel mycobacteriophage isolated from soil. Gene 453:9–23 [DOI] [PubMed] [Google Scholar]
- 15. Krogh S., Jørgensen S. T., Devine K. M. 1998. Lysis genes of the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 180:2110–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loessner M. J., Gaeng S., Wendlinger G., Maier S. K., Scherer S. S. 1998. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 162:265–274 [DOI] [PubMed] [Google Scholar]
- 17. Loessner M. J. 2005. Bacteriophage endolysins: current state of research and applications. Curr. Opin. Microbiol. 8:480–487 [DOI] [PubMed] [Google Scholar]
- 18. Longchamp P. F., Mauël C., Karamata D. 1994. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology 140:1855–1867 [DOI] [PubMed] [Google Scholar]
- 19. Marinelli L. J., et al. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor NY [Google Scholar]
- 21. Pang T., Savva C. G., Fleming K. G., Struck D. K., Young R. 2009. Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. U. S. A. 106:18966–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park T., Struck D. K., Deaton J. F., Young R. 2006. Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. U. S. A. 103:19713–19718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park T., Struck D. K., Dankenbring C. A., Young R. 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J. Bacteriol. 189:9135–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parreira R., et al. 1999. Gene organization in a central DNA fragment of Oenococcus oeni bacteriophage fOg44 encoding lytic, integrative and non-essential functions. Gene 226:83–93 [DOI] [PubMed] [Google Scholar]
- 25. Payne K., Sun Q., Sacchettini J., Hatfull G. F. 2009. Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 73:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Portugal I., Anes E., Moniz-Pereira J. 1989. Temperate mycobacteriophage from M. smegmatis. Acta Leprol. 7:243–244 [PubMed] [Google Scholar]
- 27. Ramanculov E., Young R. 2001. A ancient player unmasked: T4 rI encodes a t-specific antiholin. Mol. Microbiol. 41:575–583 [DOI] [PubMed] [Google Scholar]
- 28. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. São-José C., Parreira R., Santos M. A. 2003. Triggering of host-cell lysis by double-stranded DNA bacteriophages: fundamental concepts, recent developments and emerging applications. Recent Res. Dev. Bacteriol. 1:103–130 [Google Scholar]
- 30. Sheehan M. M., Stanley E., Fitzgerald G. F., van Sinderen D. 1999. Identification and characterization of a lysis module present in a large proportion of bacteriophages infecting Streptococcus thermophilus. Appl. Environ. Microbiol. 65:569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911–1919 [DOI] [PubMed] [Google Scholar]
- 32. Summer E. J., et al. 2010. Genomic and biological analysis of phage Xfas53 and related prophages of Xylella fastidiosa. J. Bacteriol. 192:179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takáč M., Witte A., Bläsi U. 2005. Functional analysis of the lysis genes of Staphylococcus aureus phage P68 in Escherichia coli. Microbiology 151:2331–2342 [DOI] [PubMed] [Google Scholar]
- 34. van Kessel J. C., Hatfull G. F. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147–152 [DOI] [PubMed] [Google Scholar]
- 35. Vukov N., Moll I., Bläsi U., Scherer S., Loessner M. J. 2003. Functional regulation of the Listeria monocytogenes bacteriophage A118 holin by an intragenic inhibitor lacking the first transmembrane domain. Mol. Microbiol. 48:173–186 [DOI] [PubMed] [Google Scholar]
- 36. Walker J. T., Walker D. H., Jr 1980. Mutations in coliphage P1 affecting host cell lysis. J. Virol. 35:519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang I.-N., Smith D. L., Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 38. Wang I.-N., Deaton J., Young R. 2003. Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J. Bacteriol. 185:779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Young R., Wang I.-N., Roof W. D. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120–127 [DOI] [PubMed] [Google Scholar]
- 41. Young R. 2002. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4:21–36 [PubMed] [Google Scholar]
- 42. Young R., Wang I.-N. 2006. Phage lysis, p. 104–125 In Calendar R. (ed.), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 43. Zagotta M. T., Wilson D. B. 1990. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J. Bacteriol. 172:912–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ziermann R., Bartlett B., Calendar R., Christie G. E. 1994. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J. Bacteriol. 176:4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.