Abstract
HflX GTPases are found in all three domains of life, the Bacteria, Archaea, and Eukarya. HflX from Escherichia coli has been shown to bind to the 50S ribosomal subunit in a nucleotide-dependent manner, and this interaction strongly stimulates its GTPase activity. We recently determined the structure of an HflX ortholog from the archaeon Sulfolobus solfataricus (SsoHflX). It revealed the presence of a novel HflX domain that might function in RNA binding and is linked to a canonical G domain. This domain arrangement is common to all archaeal, bacterial, and eukaryotic HflX GTPases. This paper shows that the archaeal SsoHflX, like its bacterial orthologs, binds to the 50S ribosomal subunit. This interaction does not depend on the presence of guanine nucleotides. The HflX domain is sufficient for ribosome interaction. Binding appears to be restricted to free 50S ribosomal subunits and does not occur with 70S ribosomes engaged in translation. The fingerprint 1H-15N heteronuclear correlation nuclear magnetic resonance (NMR) spectrum of SsoHflX reveals a large number of well-resolved resonances that are broadened upon binding to the 50S ribosomal subunit. The GTPase activity of SsoHflX is stimulated by crude fractions of 50S ribosomal subunits, but this effect is lost with further high-salt purification of the 50S ribosomal subunits, suggesting that the stimulation depends on an extrinsic factor bound to the 50S ribosomal subunit. Our results reveal common properties but also marked differences between archaeal and bacterial HflX proteins.
INTRODUCTION
GTPases of the translation factor-related (TRAFAC) class regulate a broad range of ribosome-associated processes, from the assembly of ribosomal subunits to the control of the mature ribosome during all phases of translation (5, 19, 23). The TRAFAC class is named after the family of classical translation factor GTPases, which includes the two elongation factors EF-Tu and EF-G. Homologs of these two GTPases are found in every organism from the three domains of life (Archaea, Bacteria, and Eukarya). Their molecular functions in translation are well known. Several other TRAFAC GTPase families are present in organisms from all three domains of life, although they are less ubiquitous than the classical translation factor GTPases (4, 6, 23). The biological function of these widespread GTPases is often not well understood. Among these is the family of HflX GTPases.
Bacterial and eukaryotic HflX GTPases are three-domain proteins. They are composed of a central G domain flanked by two putative ligand-binding domains. Escherichia coli HflX was originally presumed to be involved in phage lambda lysogeny, but recent experiments have dismissed this hypothesis (10). E. coli HflX was shown to interact with the 50S ribosomal subunit (18), suggesting that HflX might have a ribosome-associated function, similar to the observations made for members of the related family of Obg GTPases (14, 24, 39, 42). Both the N-terminal and the C-terminal domain are required for stable ribosome binding in vitro (18). The ribosome binding of E. coli HflX is nucleotide dependent (18). Surprisingly, E. coli HflX can bind and hydrolyze not only GTP but also ATP (10, 18, 34). This finding was unexpected because HflX GTPases encompass a sequence motif for specific recognition of the guanine base that is conserved in the vast majority of the various GTPase families. The GTPase and ATPase activities of Escherichia coli HflX are activated upon binding to the 50S ribosomal subunit. However, the precise biological function of HflX and the role of GTP hydrolysis therein remain unknown. In addition, hflX deletion strains of several bacterial species did not exhibit any phenotype, indicating that HflX is nonessential under the conditions tested (10, 11, 28).
Archaeal HflX GTPases are two-domain proteins; they lack the C-terminal domain that is present in bacterial and eukaryotic HflX GTPases. The recently published structure of the archaeal HflX ortholog from Sulfolobus solfataricus (SsoHflX) revealed that the N-terminal domain (hereinafter termed “HflX domain”) exposes a positively charged surface that might be involved in nucleic acid binding (40). This is analogous to the RNA binding domain present in other multidomain TRAFAC GTPases (22, 36). The two flexible regions of the G domain, termed switch 1 and switch 2, are positioned at the interface of the G domain and the HflX domain. Switches 1 and 2 usually undergo conformational changes in response to the nucleotide-bound state of the G domain. In SsoHflX, the switch 1 and switch 2 regions might therefore alter the position of the two domains relative to each other upon the exchange of nucleotides (17, 40).
To gain insight in the function of the archaeal HflX GTPases, we investigated whether the archaeal HflX homolog SsoHflX interacts with the large ribosomal subunit and how this interaction is influenced by the presence of guanine nucleotides. Our data suggest that interaction with the large ribosomal subunit is a conserved feature of HflX GTPases. However, considerable differences between archaeal and bacterial HflX GTPases were found concerning the ribosome dependence of the GTPase activity.
MATERIALS AND METHODS
Heterologous expression and purification of SsoHflX and SsoHflX-H.
Plasmids for the heterologous expression of SsoHflX and the truncated mutant protein encompassing the isolated HflX domain (SsoHflX-H) have been described previously (40). Heterologous expression was carried out in E. coli Rosetta (DE3) cells (Novagen) in LB medium according to standard methods, and cells were stored at −80°C until further use. For the production of 15N-labeled SsoHflX, LB medium was replaced by M9 medium containing 15NH4Cl (Cambridge Isotope Laboratories) as the sole nitrogen source and 0.4% (wt/vol) glucose as the carbon source. For the purification of SsoHflX, 1 g cell paste was resuspended in 4 ml buffer G (20 mM Tris-HCl, pH 7.4, 300 mM KCl, 10 mM imidazole, 1 mM dithiothreitol [DTT], 7.5% glycerol) and passed thrice through a French pressure cell at 16,000 lb/in2. Cell debris was removed by centrifugation (37,000 × g for 30 min at 4°C), and the supernatant was incubated at 70°C for 20 min, allowing the removal of the heat-unstable host proteins by centrifugation. The heat-stable cell extract was loaded on a Ni affinity chromatography column. After washing with at least 10 column volumes of buffer G, SsoHflX was eluted with buffer G containing 500 mM imidazole. The protein was further purified by gel filtration using a Superdex 75 HR 16/60 column (GE Healthcare) in buffer G containing no imidazole. SsoHflX-H was purified likewise, but the heat incubation was omitted and, after Ni chromatography, the protein was purified by heparin-agarose chromatography with a gradient from 0.2 to 2 M KCl. Protein concentrations were determined using a protein assay (Bio-Rad) based on the method by Bradford.
Fractionation of cell lysates on sucrose density gradients and in vitro translation.
S. solfataricus strain P2 cells were grown on modified Brock medium supplemented with 0.1% (wt/vol) tryptone and 0.4% (wt/vol) sucrose (41). Cell lysates were prepared as described previously (1). 70S ribosomes were obtained by chemical cross-linking of cell lysates programmed for translation as described previously (1), with the following modifications: 100 μl in vitro translation assays contained 480 μg cell lysate (referring to the protein concentration measured by the Bradford protein assay), 4 μg orf104 mRNA, 0.24 A260 units bulk Sulfolobus tRNA, 1.8 mM ATP, 0.9 mM GTP, 4 μl 1 mM amino acid mixture without methionine (Promega) in 20 mM triethanolamine (TEA)-KOH, pH 7.4, 20 mM magnesium acetate (MgOAc), 10 mM KCl. Samples were incubated at 73°C for 30 min and placed on ice. 70S ribosomes were stabilized by the addition of 1% (vol/vol) formaldehyde and further incubation for 30 min on ice. The samples were then loaded onto 10.5-ml linear 10%-to-30% sucrose gradients in 20 mM Tris-HCl, pH 7.4, 40 mM NH4Cl, 10 mM MgCl2, 1 mM DTT and centrifuged for 4 h at 36,000 rpm in a TST41.14 rotor (Kontron instruments). Gradients were fractionated, and proteins were concentrated by trichloroacetic acid-deoxycholate (TCA-DOC) precipitation. Pellets were dissolved in 25 μl 2× SDS-PAGE loading buffer.
Isolation of ribosomal subunits.
Ribosomal subunits from S. solfataricus were isolated as described previously (1). After separation on 10%-to-30% sucrose density gradients, the isolated subunits were concentrated and purified from sucrose by ultrafiltration (30,000 molecular-weight cutoff; Vivaspin). The concentration was determined based on the absorption at 260 nm using conversion factors of 60 pmol 50S ribosomal subunit per A260 unit.
Ribosome binding assays.
Amounts of 80 μl contained 80 pmol SsoHflX, 80 pmol of purified large ribosomal subunit, and 100 μM respective nucleotide in 20 mM Tris–HCl, pH 7.4, 40 mM NH4Cl, 10 mM MgOAc, 1 mM DTT, 5% glycerol. Samples were incubated at 50°C for 15 min and then loaded onto 10%-to-30% sucrose gradients. Further processing of the samples was as described above.
Generation of anti-SsoHflX antiserum and labeling of antibodies.
Rabbit antiserum against SsoHflX was produced at Eurogentec (Belgium). Protein A-agarose-purified antibodies were labeled with digoxigenin (DIG)-3-O-methylcarbonyl-ε-aminocaproic acid-N-hydroxysuccinimide ester (Roche) in a 1:25 molar ratio according to the manufacturer's protocol.
Immunodetection of SsoHflX.
After SDS-PAGE, proteins were blotted on 0.2-μm-pore-size nitrocellulose (Schleicher & Schuell) in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 11.0), 10% (vol/vol) methanol by wet transfer at 10 V overnight. Filters were blocked with 0.2% i-block (Applied Biosystems) in TBST buffer (20 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.1% [vol/vol] Tween 20). The DIG-labeled anti-SsoHflX antibodies were used in 500× dilution in combination with anti-DIG-alkaline phosphatase (AP) Fab fragments (Roche) as secondary antibody according to the manufacturer's protocol. Chemiluminescence was detected using CDP-Star reagent (New England BioLabs) and Biomax light films (Kodak).
NMR of SsoHflX.
Uniformly 15N isotopically labeled SsoHflX was used to assess binding to 50S ribosomal subunits using nuclear magnetic resonance (NMR) spectroscopy. Both the 15N-labeled SsoHflX and purified 50S ribosomal subunits were buffer exchanged to 10 mM HEPES-KOH, pH 7.5, 40 mM NH4Cl, 10 mM MgCl2, 1 mM DTT, with the addition of 10% D2O as an NMR lock solvent, and SsoHflX was concentrated to 26 μM. All NMR experiments were performed at 50°C. 15N-1H heteronuclear correlation spectra of SsoHflX were recorded using the SOFAST-heteronuclear multiple quantum coherence (HMQC) sequence (31), with 100 increments in the 15N dimension, a spectral width of 32 ppm, a recycling delay of 300 ms, and 256 scans. A 120° PC9 (2.454 ms at 700 MHz) shaped pulse was used for the excitation, and a REBURP shaped pulse (1.782 ms at 700 MHz) was used for the inversion. For heteronuclear stimulated echo (XSTE)–pulsed-field gradient (PFG) NMR (12), a 1-ms square gradient varying from 5% to 95% of the maximum gradient strength together with a 200-ms diffusion delay were employed. 1H spectra were recorded using an 18 μM sample of 50S ribosomal subunit with various concentrations of SsoHflX. Data processing was performed using Topspin 2.4 (Bruker) and NMRPipe (9). Proton transverse relaxation was measured using the spin echo T2 experiment (35).
Nucleotidase activity assays.
Volumes of 20 μl contained the amounts of GTP or ATP (1.6 μM to 250 μM) indicated below, supplemented with a trace amount (8.25 nM) of [α-32P]GTP or [γ-32P]ATP in 20 mM Tris–HCl, pH 7.4, 50 mM NH4Cl, 10 mM MgOAc, 8% glycerol, 1 mM DTT. The amount of SsoHflX (0.24 μM to 7.52 μM) was adjusted in a manner such that approximately 10% of the nucleotides were hydrolyzed in the individual assays to ensure reliable quantification and minimize inhibition by GDP. Samples were incubated at 50°C for 20 min, after which 4 μl was withdrawn and mixed with 10 μl of ice-cold stop solution (20 mM EDTA). One microliter of this mixture was subsequently spotted onto polyethyleneimine (PEI)-cellulose thin-layer chromatography plates, resolved, and detected as described previously (40). Signals were detected on phosphor storage screens (Kodak), and the Quantity One software package (Bio-Rad) was used to quantify the spots. The amount of nucleotide hydrolysis was calculated for each individual lane, and average values were determined from at least two parallel experiments. All values were corrected for the amount of nucleotide hydrolyzed in samples of identical composition with buffer replacing SsoHflX.
RESULTS
Localization of endogenous SsoHflX.
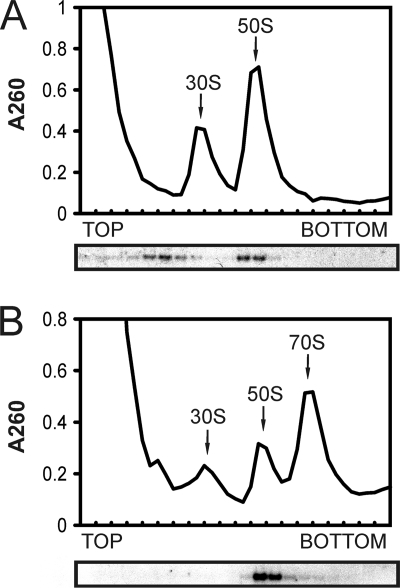
To establish initial insights into the physiological function of SsoHflX, we tested for comigration of endogenous SsoHflX with 50S ribosomal subunits during cell lysate fractionation on a sucrose density gradient because such an association had been observed previously for the E. coli HflX ortholog (18). Free 30S and 50S ribosomal subunits can readily be obtained from the S. solfataricus cell lysate using sucrose density gradient ultracentrifugation, whereas 70S ribosomes are not stable and dissociate into 30S and 50S ribosomal subunits during ultracentrifugation (1). S. solfataricus 70S ribosomes can be obtained by programming the cell lysate for translation using in vitro translation assays and subsequently cross-linking with formaldehyde. Without programming of the cell lysate, SsoHflX comigrated mainly with the free 50S ribosomal subunit. An additional peak of SsoHflX was observed further up in the gradient (Fig. 1A). This is approximately the position where the thermosome was detected on the Ponceau S-stained blot (not shown). The thermosome is the most abundant protein in S. solfataricus and sediments at 20S (30). This suggests that SsoHflX might be part of another high-molecular-weight complex besides the putative complex formed with the 50S ribosomal subunit.
Fig. 1.
Localization of endogenous SsoHflX in S. solfataricus cell lysates after sucrose density gradient centrifugation. The upper panels show the absorption profiles at 260 nm. The positions of ribosomal subunits are indicated. The lower panels show the immunodetection of SsoHflX. (A) Cell lysate after incubation at 70°C. (B) Cell lysate programmed for translation. 70S ribosomes were stabilized by formaldehyde cross-linking.
After the cell lysate was programmed for translation, most 30S and 50S ribosomal subunits were incorporated in 70S ribosomes (Fig. 1B). SsoHflX comigrated with the remaining free 50S ribosomal subunit but not with the 70S ribosomes. Furthermore, SsoHflX could not be detected any longer at the 20S position.
Reconstitution of SsoHflX-50S ribosomal subunit complexes.
The comigration of endogenous SsoHflX with the 50S ribosomal subunit in sucrose density gradients suggested that SsoHflX might bind directly to 50S ribosomal subunits. In order to test this, binding assays with recombinant SsoHflX and purified 50S ribosomal subunit were undertaken.
Complexes were reconstituted at 50°C, and free SsoHflX was separated from SsoHflX-50S ribosomal subunit complexes by sucrose density gradient centrifugation. SsoHflX was able to bind stably to the purified 50S ribosomal subunit in the presence of either GDP or the nonhydrolyzable GTP analog GppNHp (guanylylimidodiphosphate) (Fig. 2A and B). Apo-SsoHflX was able to bind to the 50S ribosomal subunit as well (Fig. 2C), but in a less stable manner than its binding in the presence of guanine nucleotides. Although it is possible that a fraction of the recombinant SsoHflX might still be in a nucleotide-bound state after purification, the fact that the apo-SsoHflX crystal structure was obtained from a protein preparation purified under comparable conditions indicates that such a fraction should be rather small. Similar results were obtained for endogenous SsoHflX when ribosomes were isolated from cell extracts incubated with the respective nucleotides (data not shown).
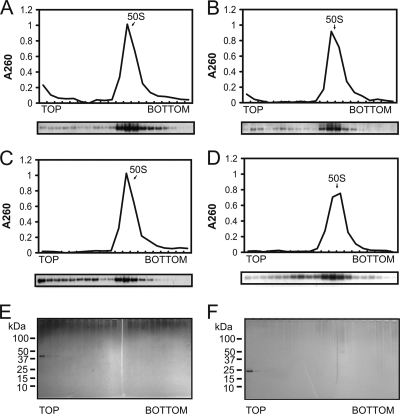
Fig. 2.
Binding assays for SsoHflX with purified 50S ribosomal subunits in the presence of different guanine nucleotides. (A to C) A 1 μM concentration of recombinant SsoHflX was mixed with 1 μM 50S ribosomal subunit in the presence of 100 μM GppNHp (A) or 100 μM GDP (B) and in the absence of nucleotides (C). (D) Binding assay carried out as described above with the C-terminal G domain deletion mutant protein SsoHflX-H in the absence of nucleotides. (E and F) Migration of SsoHflX (E) and SsoHflX-H (F) in the absence of 50S ribosomal subunits as detected by silver staining.
A truncated mutant protein encompassing only the isolated HflX domain (SsoHflX-H) was similarly able to form stable complexes with the 50S ribosomal subunit, indicating that the HflX domain contributes to the 50S ribosomal subunit binding interface of SsoHflX (Fig. 2D). The results for control samples lacking 50S ribosomal subunit particles demonstrated that the observed migration of SsoHflX and SsoHflX-H into the sucrose density gradient was entirely dependent on interaction with the 50S ribosomal subunit (Fig. 2E and F).
SsoHflX binds to the surface of the 50S ribosomal subunit.
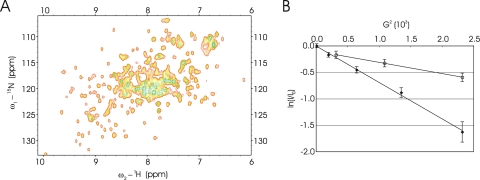
In order to investigate the dynamics of the 50S ribosomal subunit binding by SsoHflX in more detail, NMR spectroscopy experiments were carried out. Using pulsed-field gradient diffusion NMR experiments designed to determine the diffusion coefficient of the species that give rise to resonances, we found that the diffusion coefficient of 15N-labeled SsoHflX at 25°C was 1.14 × 10−10 ± 0.05 × 10−10 m2 s−1 (mean ± standard deviation) (Fig. 3B). Using the Stoke-Einstein equation (13), this corresponds to a particle with a hydrodynamic radius of 2 nm. The SsoHflX protomer has a molecular weight of 41.6 kDa, which if we assume a spherical particle would correspond to a radius of 2.3 nm. We therefore concluded that SsoHflX appears to be a monomer in solution, in line with previous results from native mass spectrometry experiments (40).
Fig. 3.
Examination of SsoHflX by NMR spectroscopy. (A) 2-D 1H-15N SOFAST-HMQC of SsoHflX at 50°C; (B) pulsed-field gradient diffusion NMR characterization of SsoHfIX (diamonds) compared to the 50S ribosomal subunit (open circles) at 25°C. The logarithm (y axis) of normalized resonance intensity (I/I0) is plotted against G2, the square of the gradient strength (x axis). I0 is the intensity at low gradient strength. The diffusion profile of SsoHflX is homogeneous, as indicated by the linear regression of ln(I/I0) as a function of G2. The diffusion coefficient extracted from the slope of ln(I/I0) as a function of G2 is 1.1 × 10−10 m2 s−1. The diffusion profile of the 50S resonances also appears to be homogeneous and corresponds to a diffusion coefficient of 4.2 × 10−11 m2 s−1. Error bars indicate the uncertainty of the signal intensity based on the spectral noise.
Two-dimensional (2-D) 1H-15N heteronuclear single quantum coherence (HSQC) spectra of SsoHflX were recorded at 25°C and 50°C. The spectrum at 25°C shows extensive broadening of resonances with an unresolved set of highly broadened peaks in the central region of the spectrum, but temperature elevation resulted in marked sharpening of the cross-peaks, and at 50°C, a large number (approximately 191 of 355) resolvable cross-peaks were observed (Fig. 3A), including a significant number in dispersed regions of the spectrum. Together with fast relaxation observed for the 1H signals (T2 ≈ 30 ms), the temperature effect and the heterogeneity in cross-peak linewidths are indicative of conformational exchange that the protein is undergoing on the ms-100 μs timescale. After the addition of 10 mM GTP, the GTP proton H8 was observed to undergo a progressive change over 12 h at 50°C, as expected from the slow GTP hydrolysis rate of SsoHflX (40). The 2-D 1H-15N HSQC spectra showed no changes in chemical shift or linewidth of the SsoHflX cross-peaks in the presence of GTP and GDP (data not shown).
The investigation of the possible interactions of SsoHflX with the 50S ribosomal subunits was performed at 50°C. The 1H spectrum of the S. solfataricus 50S ribosomal subunits is shown in Fig. 4A. Despite the high molecular weight of this particle, a significant number of resonances were observed, and by analogy with studies on the E. coli ribosome (8), these may arise from the L12 proteins in the GTPase-associated region of the 50S ribosomal subunit. In order to monitor the attachment of the observed protein to the 50S particle, a PFG-diffusion NMR experiment at 25°C was used to determine the diffusion coefficient of the observed signal, which was 4.3 ± 0.2 0.10−11 m2 s−1. This diffusion coefficient corresponds to a particle radius of 5.6 ± 0.3 nm, which corresponds well to a particle of the size of the 50S ribosomal subunit (radius of ca. 6 nm), indicating that the proteins that give rise to those resonances are attached to the 50S ribosomal subunits.
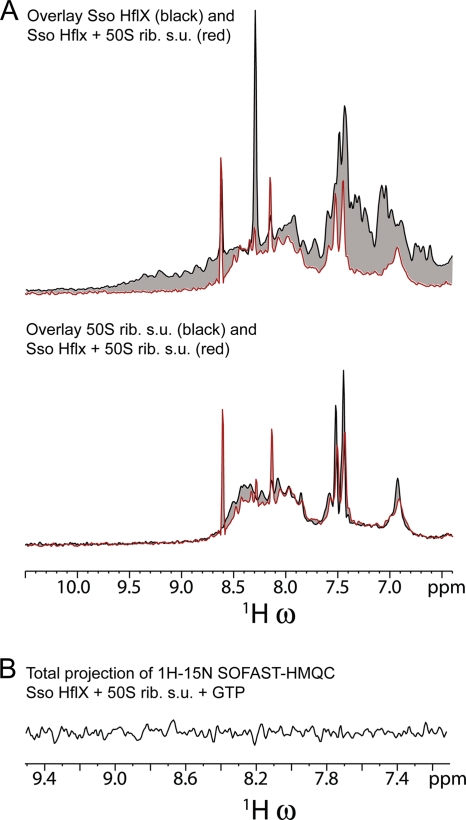
Fig. 4.
The interaction of SsoHflX with the 50S ribosomal subunit (rib. s.u.) results in the broadening of the SsoHflX resonances. (A) Overlays of the 1H one-dimensional spectra of SsoHflX in the presence of GTP (upper panel, black line), the S. solfataricus 50S ribosomal subunit (lower panel, black line), and SsoHflX plus the 50S ribosomal subunit in the presence of GTP (both panels, red line). (B) 1H projection of the 1H-15N SOFAST-HMQC of SsoHflX plus 50S ribosomal subunit in the presence of GTP at 50°C. The entire spectrum of SsoHflX is broadened beyond detection.
Upon the addition of SsoHflX to the 50S ribosomal subunit, the resonances in the 2-D spectrum of SsoHflX became broadened completely beyond detection, indicating an interaction of the protein with the 50S ribosomal subunit (Fig. 4A). The proton signals from flexible parts of the 50S ribosomal subunit were not affected by the binding of SsoHflX (Fig. 4A).
The addition of a 10-fold molar excess of SsoHflX over the 50S ribosomal subunit (in the absence of GTP or nonhydrolyzable GTP analogs) did not result in the detection of signal from SsoHflX (Fig. 4B and data not shown). The complete broadening indicates that all SsoHflX molecules in solution appeared to be interacting with the ribosome during the 50-ms acquisition time, i.e., there are no SsoHflX molecules that remain free in solution for the entire acquisition time. Therefore, the complete broadening of the 10-fold molar excess indicates a dissociation rate (koff) larger than 10 times 1/50 ms: 200 s−1. Cross-peaks from the GTPase were also broadened beyond detection in the presence of 10 mM nonhydrolyzable GTP analog GppNHp under conditions of a 2:1 molar ratio of SsoHflX over the 50S ribosomal subunit.
Stimulation of GTPase activity by the large ribosomal subunit.
Previously, we assessed the GTPase activity of SsoHflX by measuring Pi release by a malachite green assay (40). Here, we measured the GTPase activity of SsoHflX in a ribosome-compatible buffer system and detected GTP hydrolysis at 50°C by thin-layer chromatography. This method appeared to be more robust concerning elevated levels of background GTP hydrolysis occurring in the presence of ribosomal subunits of different purity.
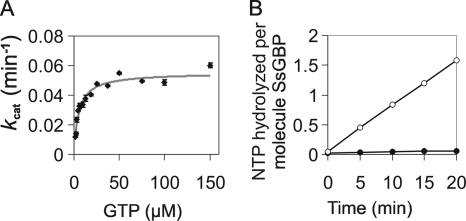
The kcat value for SsoHflX in the absence of ribosomes was 9.2 × 10−4 ± 0.03 × 10−4 s−1, and the Km value was determined to be 5.3 ± 0.6 μM (Fig. 5A). These values were similar to those measured previously using the malachite green assay (40), with a slightly increased substrate affinity that might be explained by the greater accuracy of the thin-layer chromatography system at low GTP concentrations. GTP hydrolysis by SsoHflX was linear during the 20-min incubation, and no sign of inhibition by the accumulating GDP was observed (Fig. 5B). E. coli HflX binds and hydrolyzes GTP, as well as ATP (10, 18, 34). In order to clarify whether SsoHflX also possesses ATPase activity, we tested SsoHflX for such an activity in a time course experiment. However, ATP hydrolysis was insignificant compared to the levels of GTP hydrolysis (Fig. 5B).
Fig. 5.
Basal nucleotidase activity of SsoHflX. Samples were incubated at 50°C for 20 min with the indicated concentrations of GTP. For all samples, GTP hydrolysis was corrected for background occurring in the absence of SsoHflX under otherwise identical conditions. (A) Substrate concentration-dependent GTPase activity of SsoHflX. (B) Time course experiment to test for ATPase activity of SsoHflX. Samples contained 7.5 μM SsoHflX and 100 μM ATP (filled circles) or GTP (open circles). NTP, nucleoside triphosphate. Error bars show 1 standard deviation.
The GTPase activity of E. coli HflX increases strongly in the presence of 50S ribosomal subunits, as well as empty and poly(U)-programmed 70S ribosomes (18, 34). In order to further investigate the functional conservation between bacterial and archaeal HflX orthologs, we studied the effects of different preparations of S. solfataricus ribosomes on the GTPase activity of SsoHflX. Sucrose cushion centrifugation at 500 mM NH4Cl is not sufficient to remove all extrinsic factors from the ribosomal subunits, but higher concentrations of NH4Cl lead to partial disintegration of the subunits and loss of poly(U)-directed translation activity (1, 25).
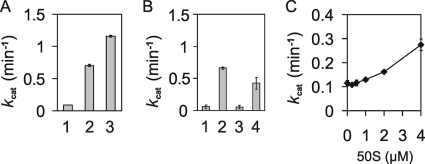
Unfractionated ribosomal subunits purified by sucrose cushion centrifugation at 500 mM NH4Cl did stimulate the GTPase activity of SsoHflX more than 10-fold at 1 μM concentration (Fig. 6A). Notably, the effect was less pronounced when the concentration of NH4Cl during ribosome purification was reduced to 100 mM. To determine whether the effect is caused by specific ribosomal subunits only, the ribosomal subunits were separated on sucrose density gradients. Although these ribosomal subunits were purified by sucrose cushion centrifugation under high-salt conditions for 6 h, some residual translation factors still seemed to be present in the preparations, as a relatively high background GTPase activity was observed (data not shown). In agreement with the interaction of SsoHflX with the 50S ribosomal subunit, stimulation of the GTPase activity of SsoHflX was observed for the 50S ribosomal subunit but not for the 30S ribosomal subunit (Fig. 6B). In the presence of both subunits, the stimulation of GTPase activity of SsoHflX was somewhat reduced compared to the activation by the 50S ribosomal subunit alone, suggesting that the 30S ribosomal subunit competes with the SsoHflX-50S ribosomal subunit complex formation. When the sucrose cushion purification of 50S ribosomal subunits was prolonged to 13 h, the background GTPase activity of the ribosome preparations was reduced significantly. Surprisingly, this coincided with a significant decrease of the stimulatory effect on the GTPase activity of SsoHflX (Fig. 6C). This result suggests that the stimulation of the GTPase activity of SsoHflX might depend on an extrinsic factor that binds tightly to the 50S ribosomal subunit and is only partially removed during the high-salt washing steps. Although rather unlikely, an alternative explanation would be that SsoHflX itself stimulated the activity of an extrinsic GTPase copurified with the 50S ribosomal subunit.
Fig. 6.
Stimulation of the GTPase activity of SsoHflX by ribosomal subunits in the presence of 250 μM GTP. (A) Effect of partially purified ribosomes on SsoHflX GTPase activity. GTPase activity of SsoHflX was measured in the absence of ribosomes (1) or with 1 μM ribosomes purified by 6-h sucrose cushion centrifugation at 100 mM NH4Cl (2) or 500 mM NH4Cl (3). (B) Stimulation of GTPase activity of SsoHflX by isolated ribosomal subunit GTPase activity was measured in the absence of ribosomal subunits (1) or the presence of 1 μM 50S (2), 0.6 μM 30S (3), or a mixture of 50S and 30S (4) ribosomal subunits. (C) Stimulation of GTPase activity of SsoHflX by increasing amounts of highly purified 50S ribosomal subunits after 13 h of sucrose cushion centrifugation at 500 mM NH4Cl. Error bars show 1 standard deviation.
DISCUSSION
GTPases of the TRAFAC class fulfill multiple functions associated with the process of translation. Recent studies of several bacterial TRAFAC GTPases revealed their function in ribosome biogenesis (3, 5, 16, 20, 21, 26–28, 33, 37, 38). Archaea possess several TRAFAC GTPases, but apart from the classical translation factors, SsoHflX from the HflX family is the first archaeal representative for which evidence for a translation-associated function has been provided. Our results reveal not only similarities but also marked differences between archaeal and bacterial HflX GTPases concerning their molecular function.
The binding of E. coli HflX to the 50S ribosomal subunit requires the presence of a nucleotide, but it occurs both in the “active” (GTP-bound) and “inactive” (GDP-bound) state of the GTPase (18). Here, we show that for both SsoHflX and E. coli HflX, the binding of guanine nucleotides strengthens the interaction with the 50S ribosomal subunit but that the regulation of binding is less strict in the case of SsoHflX. No conformational changes were observed between the structures of nucleotide-free and GDP-bound SsoHflX (17, 40). A possible explanation for the stabilizing effect of guanine nucleotides on the ribosome binding by SsoHflX would be that the guanine nucleotide is part of the interface of the 50S ribosomal subunit and SsoHflX.
We could not detect binding of endogenous SsoHflX to 70S ribosomes during in vitro translation. Similarly, no binding to 70S ribosomes or polysomes has been found for E. coli HflX (18). Therefore, the molecular role of HflX GTPases might be limited to free 50S ribosomal subunits, but it cannot be ruled out that the observed association of archaeal and bacterial HflX orthologs with free 50S ribosomal subunits only, as observed in cell lysates, does not reflect the in vivo situation. Shields et al. (34) recently reported stimulation of E. coli HflX nucleotidase activity by highly purified 70S ribosomes, as well as by poly(U)-programmed ribosomes, suggesting that E. coli HflX might also interact with 70S ribosomes during translation (34).
E. coli HflX requires both the N-terminal HflX domain and the relatively poorly conserved C-terminal domain for stable binding to the 50S ribosomal subunit (18). Similarly, the HflX domain of bacterial Chlamydophila pneumoniae HflX is required for stable 50S ribosomal subunit binding but not sufficient on its own (29). Because archaeal HflX GTPases only contain the HflX domain as a single putative ribosome-binding domain, it most likely provides the major surface for interaction with the archaeal 50S ribosomal subunit. The acquisition of additional domains was certainly a major driving force during evolution toward new biological functions of TRAFAC GTPases, as is obvious from the great variety of domain architectures occurring in this class of proteins (23). In the case of bacterial (and eukaryotic) HflX, the additional C-terminal domain might have taken over part of the ribosome binding and, hence, may also have contributed to its tighter regulation as observed for E. coli HflX. An alternative explanation would be that this effect is due to differences in the binding site between bacterial and archaeal 50S ribosomal subunits.
The binding site of SsoHflX on the 50S ribosomal subunit does not affect the resonances from the ribosome, which possibly arise from the L12 stalk region by analogy with the E. coli 50S (8). This is in contrast to the ribosome binding by the bacterial GTPase elongation factor EF-G. Upon the addition of EF-G to ribosomes, the L12 stalk resonances are broadened beyond detection, which is in line with the function of this flexible complex as a landing platform for translation elongation factors (8).
HflX GTPases possess a canonical G4 motif in the amino acid sequence of the G domain. The G4 motif normally provides binding specificity for guanine nucleotides. C. pneumoniae HflX possesses GTPase activity that is not inhibited by ATP, indicating specificity for GTP (29). In contrast, E. coli HflX has been shown to hydrolyze both GTP and ATP (10, 18, 34). SsoHflX possesses specificity for GTP, indicating that the ATPase activity of E. coli HflX is not a general feature of HflX GTPases. The rate of GTP hydrolysis by SsoHflX in the absence of ribosomes is similar to that reported for E. coli HflX (kcat = 8.4 × 10−4 s−1), but the extent of the stimulation of GTPase activity by ribosomes may differ, as the stimulation observed for E. coli HflX was about 1,000-fold (34). Although the data presented here might underestimate the extent of GTPase activity stimulation for SsoHflX, they strongly suggest that the GTPase activity of SsoHflX can be stimulated only to a much lesser extent.
Two alternative mechanisms might explain the observed stimulation of GTP hydrolysis by SsoHflX after binding of the 50S ribosomal subunit. The crystal structures of SsoHflX revealed extensive interactions between the HflX and the G domain, including several key elements of the G domain involved in guanine nucleotide binding and hydrolysis. Deletion of the HflX domain led to a significantly increased rate of GTP hydrolysis (24-fold) (40). It therefore seems possible that the binding of SsoHflX to the 50S ribosomal subunit loaded with an unidentified extrinsic factor causes alterations in the interdomain interactions, providing the G domain with greater structural flexibility required for more efficient GTP hydrolysis. Unfortunately, the expression of the isolated G domain and its purification using the buffer conditions described here led to a very unstable protein that could not be detected after sucrose density gradient centrifugation and appeared to have only a 3-fold-increased GTPase activity in comparison to that of SsoHflX (data not shown).
Alternatively to an interference with the interdomain interactions of SsoHflX, the 50S ribosomal subunit loaded with some unknown extrinsic factor might play a direct role in GTP hydrolysis by SsoHflX, analogous to the role GTP-activating proteins play in GTP hydrolysis by p21 Ras-related GTPases. The fact that this unidentified extrinsic factor appears to bind strongly to the 50S ribosomal subunit and can be removed completely only by several consecutive high-salt washes suggests that SsoHflX might be required for the release of this factor. Such a function has been described, for example, for the GTPase Lsg1 from yeast, which is needed for the release of the ribosome export factor Nmd3 (15). In archaea, a possible candidate is the translation factor aIF6, absent in bacteria but present in eukaryotes. This protein is a ribosome anti-association factor that binds strongly to the 50S subunits and may control their entry into the translation cycle or help in ribosome recycling after termination (2). In eukaryotes, IF6 release from the 60S subunits requires either protein modification (in mammals) or the intervention of a GTPase (in yeast). It is therefore conceivable that archaeal HflX may control the dissociation of aIF6 from the 50S subunits.
Since the binding of SsoHflX to the 50S ribosomal subunit appears to be only weakly regulated by guanine nucleotides, the highly transient binding that we observed at 50°C would still allow for efficient scanning of the 50S ribosomal subunit and factor release.
In bacteria, TRAFAC GTPases are thought to play an essential role in coordinating ribosome assembly with the energy state of the cell by sensing the concentrations of guanine nucleotides, including the “alarmone” ppGpp (guanosine 3′,5′-bispyrophosphate) (5). This coupling allows bacterial cells to quickly redirect the cellular metabolism during the stringent response. The stringent response in archaea apparently differs from that in bacteria, as in Sulfolobus it is not linked to decreasing GTP concentrations (7), and the bacterial alarmone ppGpp is probably generally absent in archaea (7, 32). An important question is how archaea link ribosome biogenesis with the energy state of the cell. It is likely that TRAFAC GTPases play a key role in the regulation of ribosome assembly in archaea as well (4). Here, we provide the first experimental evidence for this in the case of the archaeal HflX homolog, one of the few TRAFAC GTPases that archaea share with bacteria. Besides conventional sucrose density gradient centrifugation, we employed NMR to test for ribosome binding by SsoHflX, thereby providing insight into the fast dynamics of the SsoHflX-50S ribosomal subunit interaction at close-to-physiological temperatures. We believe that this approach is highly valuable for studies on the interaction of proteins with archaeal ribosomes, as many archaeal species are thermophiles or hyperthermophiles and their ribosomes exhibit considerable rigidity at lower temperatures.
ACKNOWLEDGMENTS
We thank Jasper Akerboom (Howard Hughes Medical Institute Janelia Farm Research Campus) for critical reading of the manuscript. We thank John Kirkpatrick (UCL) for assistance with NMR experiments.
This work was supported by NWO (ALW-Vici project 865.05.001 to J.V.D.O.) J.C., L.D.C., and H.L. acknowledge support from the BBSRC (9015651/JC), and J.C. acknowledges an HFSP Young Investigator Award (RGY67/2007).
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Benelli D., Londei P. 2007. In vitro studies of archaeal translational initiation. Methods Enzymol. 430:79–109 [DOI] [PubMed] [Google Scholar]
- 2. Benelli D., et al. 2009. Function and ribosomal localization of aIF6, a translational regulator shared by archaea and eukarya. Nucleic Acids Res. 37:256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bharat A., Jiang M., Sullivan S. M., Maddock J. R., Brown E. D. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 188:7992–7996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blombach F., Brouns S. J., van der Oost J. 2011. Assembling the archaeal ribosome: roles for translation factor-related GTPases. Biochem. Soc. Trans. 39:45–50 [DOI] [PubMed] [Google Scholar]
- 5. Britton R. A. 2009. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63:155–176 [DOI] [PubMed] [Google Scholar]
- 6. Caldon C. E., March P. E. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6:135–139 [DOI] [PubMed] [Google Scholar]
- 7. Cellini A., et al. 2004. Stringent control in the archaeal genus Sulfolobus. Res. Microbiol. 155:98–104 [DOI] [PubMed] [Google Scholar]
- 8. Christodoulou J., et al. 2004. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U. S. A. 101:10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delaglio F., et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 10. Dutta D., Bandyopadhyay K., Datta A. B., Sardesai A. A., Parrack P. 2009. Properties of HflX, an enigmatic protein from Escherichia coli. J. Bacteriol. 191:2307–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engels S., et al. 2005. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol. Microbiol. 57:576–591 [DOI] [PubMed] [Google Scholar]
- 12. Ferrage F., Zoonens M., Warschawski D. E., Popot J. L., Bodenhausen G. 2003. Slow diffusion of macromolecular assemblies by a new pulsed field gradient NMR method. J. Am. Chem. Soc. 125:2541–2545 [DOI] [PubMed] [Google Scholar]
- 13. Garcia de la Torre J., Huertas M. L., Carrasco B. 2000. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 147:138–146 [DOI] [PubMed] [Google Scholar]
- 14. Gradia D. F., et al. 2009. Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int. J. Parasitol. 39:49–58 [DOI] [PubMed] [Google Scholar]
- 15. Hedges J., West M., Johnson A. W. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Himeno H., et al. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 32:5303–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang B., et al. 2010. Functional study on GTP hydrolysis by the GTP-binding protein from Sulfolobus solfataricus, a member of the HflX family. J. Biochem. 148:103–113 [DOI] [PubMed] [Google Scholar]
- 18. Jain N., et al. 2009. E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem. Biophys. Res. Commun. 379:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karbstein K. 2007. Role of GTPases in ribosome assembly. Biopolymers 87:1–11 [DOI] [PubMed] [Google Scholar]
- 20. Kimura T., et al. 2008. Ribosome-small-subunit-dependent GTPase interacts with tRNA-binding sites on the ribosome. J. Mol. Biol. 381:467–477 [DOI] [PubMed] [Google Scholar]
- 21. Kimura T., et al. 2007. Interaction between RsgA and the ribosome. Nucleic Acids Symp. Ser. (Oxf.) 2007(51):375–376 [DOI] [PubMed] [Google Scholar]
- 22. Kukimoto-Niino M., et al. 2004. Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8. J. Mol. Biol. 337:761–770 [DOI] [PubMed] [Google Scholar]
- 23. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41–72 [DOI] [PubMed] [Google Scholar]
- 24. Lin B., Thayer D. A., Maddock J. R. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186:481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Londei P., Teichner A., Cammarano P., De Rosa M., Gambacorta A. 1983. Particle weights and protein composition of the ribosomal subunits of the extremely thermoacidophilic archaebacterium Caldariella acidophila. Biochem. J. 209:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuo Y., et al. 2006. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 281:8110–8117 [DOI] [PubMed] [Google Scholar]
- 27. Matsuo Y., Oshima T., Loh P. C., Morimoto T., Ogasawara N. 2007. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J. Biol. Chem. 282:25270–25277 [DOI] [PubMed] [Google Scholar]
- 28. Morimoto T., et al. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539–3552 [DOI] [PubMed] [Google Scholar]
- 29. Polkinghorne A., et al. 2008. Chlamydophila pneumoniae HflX belongs to an uncharacterized family of conserved GTPases and associates with the Escherichia coli 50S large ribosomal subunit. Microbiology 154:3537–3546 [DOI] [PubMed] [Google Scholar]
- 30. Ruggero D., Ciammaruconi A., Londei P. 1998. The chaperonin of the archaeon Sulfolobus solfataricus is an RNA-binding protein that participates in ribosomal RNA processing. EMBO J. 17:3471–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schanda P., Kupce E., Brutscher B. 2005. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 33:199–211 [DOI] [PubMed] [Google Scholar]
- 32. Scoarughi G. L., Cimmino C., Donini P. 1995. Lack of production of (p)ppGpp in Halobacterium volcanii under conditions that are effective in the eubacteria. J. Bacteriol. 177:82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma M. R., et al. 2005. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol. Cell 18:319–329 [DOI] [PubMed] [Google Scholar]
- 34. Shields M. J., Fischer J. J., Wieden H. J. 2009. Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli: a kinetic approach. Biochemistry 48:10793–10802 [DOI] [PubMed] [Google Scholar]
- 35. Sklenar V., Bax A. 1987. Spin-echo water suppression for the generation of pure-phase two-dimensional NMR-spectra. J. Magn. Reson. 74:469–479 [Google Scholar]
- 36. Teplyakov A., et al. 2003. Crystal structure of the YchF protein reveals binding sites for GTP and nucleic acid. J. Bacteriol. 185:4031–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tu C., et al. 2009. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uicker W. C., Schaefer L., Britton R. A. 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59:528–540 [DOI] [PubMed] [Google Scholar]
- 39. Wout P., et al. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 186:5249–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu H., et al. 2010. Structure of the ribosome associating GTPase HflX. Proteins 78:705–713 [DOI] [PubMed] [Google Scholar]
- 41. Zaparty M., et al. 2010. “Hot standards” for the thermoacidophilic archaeon Sulfolobus solfataricus. Extremophiles 14:119–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S., Haldenwang W. G. 2004. Guanine nucleotides stabilize the binding of Bacillus subtilis Obg to ribosomes. Biochem. Biophys. Res. Commun. 322:565–569 [DOI] [PubMed] [Google Scholar]