Abstract
Prokaryotes have developed multiple strategies to survive phage attack and invasive DNA. Recently, a novel genetic program denominated the CRISPR/Cas system was demonstrated to have a role in these biological processes providing genetic immunity. This defense mechanism is widespread in the Archaea and Bacteria, suggesting an ancient origin. In the last few years, progress has been made regarding the functionality of the CRISPR/Cas system; however, many basic aspects of the system remain unknown. For instance, there are few studies about the conditions and regulators involved in its transcriptional control. In this work, we analyzed the transcriptional organization of the CRISPR/Cas system as well as the positive and negative regulators involved in its genetic expression in Salmonella enterica serovar Typhi. The results obtained show that in S. Typhi the CRISPR/Cas system is a LeuO-dependent operon silenced by the global regulator LRP, in addition to the previously known nucleoid-associated protein H-NS; both LRP and H-NS bind upstream and downstream of the transcriptional start site of casA. In this study, relevant nucleotides of the casA regulatory region that mediate its LeuO transcriptional activation were identified. Interestingly, specific growth conditions (N-minimal medium) were found for the LeuO-independent expression of the CRISPR/Cas system in S. Typhi. Thus, our work provides evidence that there are multiple modulators involved in the genetic expression of this immune system in S. Typhi IMSS-1.
INTRODUCTION
In nature, bacteria and archaea are exposed to multiple viral infections; therefore, several strategies have evolved to inhibit phage predation. The mechanisms to avoid phage infection include the prevention of phage adsorption and DNA injection; the abortive infection and modification-restriction systems are also relevant (20, 37). Recently, a novel genetic program designated the CRISPR/Cas system was shown to provide immunity against virus and exogenous DNAs (7, 9, 42). In general, this system consists of a cluster of proteins (Cas) with functional domains of nucleases, helicases, polymerases, and polynucleotide binding proteins (24, 32, 41); a leader of A/T-rich noncoding sequences (31) is located immediately upstream of a singular cluster of regularly interspaced short palindromic repeats (CRISPR) that are composed of direct repeats separated by spacers that, in some cases, resemble viral or plasmid DNA (8, 49, 51). Accumulating evidence suggests that the CRISPR sequences are transcribed as a large mRNA which is cleaved in the repeat sequences by a Cas endonuclease protein (9, 11, 26), generating small transcripts (crRNAs) that are used by the Cas proteins to target exogenous genetic materials, leading to their degradation (9, 21, 25, 43). Thus, CRISPR/Cas provides immunity against foreign genetic material. The incorporation of new spacers into the CRISPR during a phage challenge has been shown to confer specific resistance against the invading virus in future events (7). The presence of a large assortment of short invader sequences in the CRISPR cluster shows the evolutionary history of multiple viral infections in the host, providing evidence of the rapid evolution of this dynamic program (6, 27).
Significant progress has been made regarding the functionality of the CRISPR/Cas system (30, 34, 44, 61). However, this genetic program is not fully characterized; for instance, there are few studies about its transcriptional organization and genetic control (1, 28, 38, 55, 58, 64). In this regard, we previously reported that in Salmonella enterica serovar Typhi IMSS-1, the casA gene (STY3070) is regulated positively by LeuO; footprinting experiments demonstrated that LeuO binds upstream of the casA transcriptional start site, and by use of transcriptional fusions it was shown that casA is induced in an H-NS-deficient strain (28). This finding was later observed in Escherichia coli K-12, as electrophoretic mobility shift assays (EMSAs) and Northern blot experiments showed that LeuO directly and positively regulates the casA (ygcL) gene, and in an H-NS-deficient strain overexpressing LeuO, the transcriptional casA levels were higher than in the wild type overexpressing this regulator (58). Furthermore, Pul et al. (55) determined the σ70-dependent promoters in the 5′ region of casA, including a Pcas promoter and a divergent anti-Pcas promoter as well as another one identified in the leader region upstream of the CRISPR I array (Pcrispr1). Interestingly, the three promoters are repressed by H-NS. Recently, Westra et al. (64) showed that H-NS-mediated casA repression can be relieved by the LeuO transcriptional regulator. Together, these data support the positive and negative roles of LeuO and H-NS in the genetic control of the CRISPR/Cas program in E. coli K-12. In the present work, we determined that in S. Typhi IMSS-1, the complete CRISPR/Cas system is driven by a single LeuO-dependent promoter, which is directly repressed by H-NS and LRP; both proteins bind upstream and downstream of the casA transcriptional start site. In addition, we have identified nucleotides of the regulatory region of casA that mediate its LeuO-dependent activation. Specific growth conditions that induce the CRISPR/Cas program in S. Typhi IMSS-1 in a LeuO-independent manner are also reported, thus demonstrating that multiple global regulators and specific conditions modulate the transcriptional expression of this genetic program in Salmonella enterica serovar Typhi.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used are listed in Table S1 in the supplemental material. S. Typhi IMSS-1 was grown in LB (10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter), liquid MA medium (7 g nutrient broth, 1 g yeast extract, 2 g glycerol, 3.75 g K2HPO4, and 1.3 g KH2PO4 per liter) (35), or N-minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 100 mM Tris-HCl (pH 7.5), 200 μM MgCl2, 0.5% glycerol, and 0.1% Casamino Acids] (15). When required, the following antibiotics were added: kanamycin (Km), 30 μg/ml; tetracycline (Tc), 12 μg/ml; and ampicillin (Ap), 100 μg/ml. S. Typhi and E. coli strains were grown aerobically at 37°C.
DNA manipulations.
Plasmid and genomic DNA isolation was performed according to published protocols (57). Primers for PCR amplifications were provided by the oligonucleotide synthesis facility at our institute (see Table S2 in the supplemental material). Restriction enzymes, ligase, nucleotides, and polymerases were obtained from New England BioLabs or Gibco BRL. For sequencing, double-stranded DNA was purified with a High Pure plasmid isolation kit (Boehringer Mannheim, Germany), and sequencing was performed with an automatic Perkin Elmer/Applied Biosystems 377-18 system.
Northern analysis.
Total RNA was collected from 10 ml of culture by acid-phenol extraction. RNAs were separated in 1% agarose gels, transferred to nylon membranes (GE Healthcare), and probed with PCR probes labeled with a Rediprime kit using [α-32P]dCTP. The membranes were prehybridized and hybridized at 65°C, using Rapid Hyb buffer (GE Healthcare), and were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% SDS for 15 min, 1× SSC plus 0.1% SDS for 15 min, 0.5× SSC plus 0.1% SDS for 15 min, and, finally, 0.1× SSC plus 0.1% SDS at 65°C for 30 min (57). To determine the molecular weight of the mRNA, a 0.24- to 9.5-kb RNA ladder (Invitrogen) was used.
Construction of transcriptional reporter fusions.
Oligonucleotides (see Table S2 in the supplemental material) were designed to PCR amplify the complete 5′ intergenic region as well as shorter fragments of different lengths of the casA regulatory region. PCR fragments were double digested with BamHI-XhoI or BamHI-KpnI and ligated into pKK232-8 or pKK232-9 (see Table S1 in the supplemental material), which contains the promoterless cat gene. Fusions were sequenced to verify the correct DNA sequences of the PCR fragments.
CAT assay.
Chloramphenicol acetyltransferase (CAT) assays were performed as follows (46). S. Typhi strains were grown in MA medium supplemented when required with Ap or Km, with or without IPTG (isopropyl-β-d-thiogalactopyranoside; 50 μM), to optical densities at 595 nm (OD595) of 0.4, 0.6, 0.8, 1, and 1.3; at these times, 1.5 ml of bacterial culture was collected by centrifugation and washed with 0.8 ml of TDTT buffer (50 mM Tris-HCl, pH 7.8, 30 μM dl-dithiothreitol). Bacterial cells were resuspended in 0.6 ml of TDTT and sonicated on ice for 10-s intervals with 10-s rest periods until the extract was clear. The homogenate was centrifuged, and the supernatant was used for activity measurement. For CAT assays, 5 μl of each extract was added in duplicate to a 96-well enzyme-linked immunosorbent assay (ELISA) plate, followed by the addition of 0.2 ml of a reaction mixture containing 1 mM DTNB [5,5′-dithiobis(2-nitrobenzoic acid)], 0.1 mM acetyl-coenzyme A (acetyl-CoA), and 0.1 mM chloramphenicol in 0.1 M Tris-HCl, pH 7.8. The absorbance at 412 nm was measured every 5 s for 5 min, using a Ceres 900 scanning autoreader and microplate workstation. The protein concentration of the cell extracts was determined using the bicinchoninic acid (BCA) protein assay reagent (Pierce). Protein values and the mean rate of product formation by CAT were used to determine CAT specific activity, in μmol/min/mg protein. To determine the transcriptional activity in N-minimal medium, the bacteria were grown for 16 h, 1 ml was collected, and the CAT protocol described above was used. The results presented in the figures are the means for three independent experiments performed in duplicate.
Purification of H-NS and LRP proteins.
H-NS and LRP proteins were purified as described previously (16). Briefly, the genes were cloned into the arabinose-inducible vectors pBAD/Myc-His and pMPM-T6, respectively (Invitrogen) (47). E. coli BL21(DE3) harboring the cloned gene was grown overnight in LB medium. One milliliter of this preculture was inoculated into 100 ml LB medium and allowed to grow to an OD600 of 0.4 before being induced for 4 h with l-arabinose (Sigma-Aldrich) to a final concentration of 1%. Samples were collected and centrifuged at 8,000 rpm (Thermo Scientific CL31R multispeed centrifuge) for 5 min at 4°C. The cell pellet was resuspended in binding buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl, pH 8.0), the cells were sonicated, and the suspension was centrifuged at 12,000 rpm for 5 min at 4°C and kept at 4°C.
A nickel affinity chromatography column (Ni-nitrilotriacetic acid [Ni-NTA] agarose; Qiagen) was equilibrated with binding buffer, the solution containing the protein was loaded onto the column and washed with 10 volumes of urea buffer at pH 8.0 and urea buffer at pH 6.0, and the bound protein was eluted with urea buffer, pH 4.5. Fractions containing H-NS-Myc-His6 or Lrp-His6 were resolved by SDS-PAGE. The selected fractions were loaded into a Slyde-A-Lyzer 10K cassette (Pierce) and gradually dialyzed at 4°C in a buffer containing 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 20% glycerol, 500 mM NaCl, 0.1% Triton X-100, and 4 M urea. The same buffer containing different amounts of urea (1 M and 0.2 M) was used to complete the dialysis. The proteins were stored in buffer containing 30 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 20% glycerol, 500 mM NaCl, 0.1% Triton X-100, and 3 mM EDTA. Protein concentrations were determined by the Bradford procedure.
Gel EMSA.
Nonradioactive EMSAs were performed according to a previously described protocol (16). The probes were obtained by PCR, using the primers described in Table S2 in the supplemental material. Each probe (100 ng) was mixed with increasing concentrations of purified protein in the presence of 10× H-NS binding buffer (400 mM HEPES, 80 mM MgCl2, 500 mM KCl, 10 mM DTT, 0.5% NP-40, and 1 mg/ml bovine serum albumin [BSA]) or 5× LRP buffer (100 mM Tris, pH 8.0, 2 mM EDTA, 250 mM NaCl, 5 mM MgCl2, 62.5% [vol/vol] glycerol, 500 mg/ml BSA, and 0.5 mM DTT). The mixture was incubated for 20 min at room temperature and then separated by electrophoresis in 6% or 5% native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer. The DNA bands were visualized by ethidium bromide staining.
Site-directed mutagenesis.
To analyze their role in the regulation of the CRISPR/Cas system in S. Typhi IMSS-1, LRP-, H-NS- (19), and double H-NS/LRP-deficient strains were obtained. The corresponding genes were deleted by a one-step nonpolar gene inactivation procedure and replaced with selectable antibiotic resistance gene markers (14). Different primer sets were used to verify the presence of the antibiotic resistance gene cassette. Each mutation was further characterized by sequencing to verify the authenticity of the deletion and the presence of the resistance marker.
Nucleotide sequence accession numbers.
The casA-cas2 and CRISPR sequences of S. Typhi IMSS-1 were deposited in the DDBJ database under accession number HQ655820.
RESULTS
A LeuO-dependent promoter is necessary to transcribe the complete CRISPR/Cas operon in Salmonella enterica serovar Typhi IMSS-1.
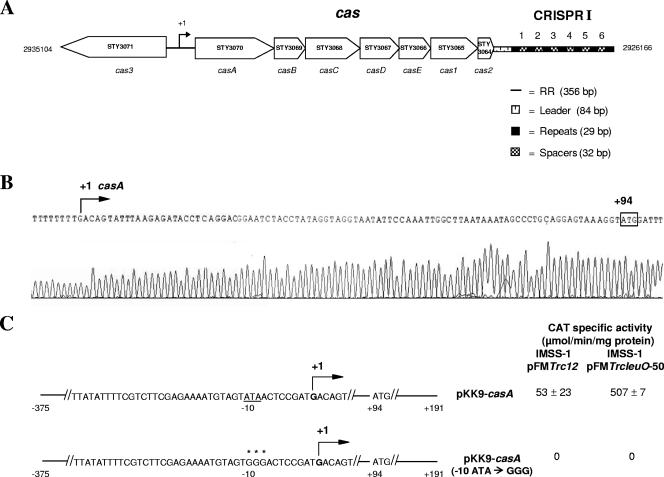
The CRISPR/Cas cluster is found in many prokaryotic genomes, including those of the Enterobacteriaceae. In Salmonella enterica serovar Typhi IMSS-1, a Mexican clinical strain (52), we detected the presence of a CRISPR/Cas system that resembles the type E (Cse) system found in S. Typhi CT18 and S. Typhi Ty2 (22, 24), and in this study we analyzed its transcriptional expression. The IMSS-1 cluster contains a 356-bp intergenic 5′-regulatory region and eight cas genes (STY3071 and STY3070-STY3069-STY3068-STY3067-STY3066-STY3065-STY3064) that are orthologous to cas3 and casA-casB-casC-casD-casE-cas1-cas2 in E. coli K-12 (9). It also has an 84-bp leader and the CRISPR I sequences, which contain seven 29-bp repeats and six 32-bp spacers (Fig. 1 A); the repeats are identical to those found in S. Typhi CT18 and E. coli K-12. Analysis of the spacers by use of the Blastn algorithm (3) against a database of phage and virus sequences downloaded from the DDBJ data bank (33) showed no significant similarity with any sequence in the database. In this work, the S. Typhi cas genes were named according to their orthologs in E. coli.
Fig. 1.
Genetic organization of the CRISPR/Cas immune system in S. Typhi IMSS-1 and identification of the transcriptional start site of casA (STY3070). (A) The CRISPR/Cas system is composed of a 5′-regulatory region (RR) of 356 bp, eight genes (cas3 and casA-casB-casC-casD-casE-cas1-cas2), a leader of 84 bp, and the CRISPR I array, containing seven repeats and six spacers. The bent arrow indicates the STY3070 (casA) transcriptional start site. The divergent STY3071 (cas3) gene is also represented. (B) Mapping of the chromosomal transcriptional start site of the casA gene. A 5′-RACE experiment was performed using total RNA from S. Typhi harboring the pFMTrcleuO-50 plasmid and induced with 50 μM IPTG. The nucleotide immediately after the polynucleotide tail (T) corresponds to the transcriptional start site (bent arrow), located 94 bp upstream of the ATG translational start codon of casA. (C) Site-directed mutagenesis of the −10 box of casA was performed by use of a QuikChange Multi site-directed mutagenesis kit (Stratagene), and the wild-type ATA sequence (underlined) was replaced by a GGG sequence (asterisks); the mutation was verified by sequencing. The CAT activities of the wild-type plasmid pKK9-casA and the pKK9-casA (−10 ATA → GGG) plasmid were determined in the wild-type S. Typhi strain overexpressing the pFMTrcleuO-50 plasmid with 50 μM IPTG as well as in S. Typhi containing the pFMTrc12 plasmid.
Previously, we reported the results of primer extension experiments showing that the S. Typhi casA (STY3070) regulatory region contains two LeuO-dependent transcription initiation sites, located 94 and 84 bp upstream of the translational ATG start codon (28). To confirm the primer extension results, 5′ rapid amplification of cDNA ends (5′-RACE) was performed according to the method of Mendoza-Vargas et al. (48). Briefly, total RNA from the S. Typhi IMSS-1 wild-type strain harboring the pFMTrcleuO-50 plasmid (pFMTrc12 derivative plasmid carrying leuO behind an IPTG-inducible Ptrc promoter [see Table S1 in the supplemental material]) (18) and induced with 50 μM IPTG was randomly amplified, and in the resulting cDNA, an adapter (homopolynucleotide of A nucleotides) was incorporated by a terminal transferase. This cDNA was used with an oligonucleotide of T nucleotides and specific casA primers in a PCR, and the PCR products were purified and sequenced. The results showed that a single chromosomal transcriptional site located 94 bp upstream of the ATG was the only one detected by 5′-RACE (Fig. 1B). Site-directed mutagenesis was performed to modify the −10 sequence (ATA → GGG) of the casA initiation site. PCR fragments that contained the entire casA regulatory region as well as the −10 substitutions were fused to the cat reporter gene, producing plasmids pKK9-casA and pKK9-casA (−10 ATA → GGG), respectively, which were introduced into the S. Typhi IMSS-1 strain containing pFMTrc12 (a pTrc99A[5] derivative containing the p15A1 origin of replication) or pFMTrcleuO-50 (see Table S1). Experiments with these fusions showed that the overexpression of LeuO induced the transcriptional activity of casA, while no CAT activity in the presence of LeuO was observed with the −10 box substitutions, indicating that a unique LeuO-dependent promoter controls the genetic expression of casA in S. Typhi IMSS-1 (Fig. 1C).
These results prompted us to determine whether the complete CRISPR/Cas system is a LeuO-dependent operon. To resolve this issue, a large DNA fragment harboring the casA promoter region and the entire casA-to-cas2 gene cluster (6.15 kb) was first fused to the cat reporter gene, creating plasmid pKK9-RR-casA-cas2. This fusion was evaluated in an S. Typhi IMSS-1 wild-type strain harboring the control vector pFMTrc12 or the pFMTrcleuO-50 plasmid. The transcriptional results with pKK9-RR-casA-cas2 showed minimal activity in the absence of LeuO, while in the presence of LeuO the transcriptional activity increased notably (Table 1). The fusion plasmid pKK9-ΔRR-casA-cas2, which contains the same genetic elements but is devoid of the casA promoter region, showed null CAT activity in either the absence or presence of LeuO (Table 1). These data support the notion that casABCDE12 is a LeuO-dependent operon. To elucidate whether CRISPR is part of the casABCDE12 LeuO-dependent operon, the fusion plasmid pKK9-RR-casA-CRISPR (6.7 kb), containing the casA promoter region, the casA to cas2 genes, and the CRISPR sequences, was constructed and independently introduced into IMSS-1/pFMTrc12 and IMSS-1/pFMTrcleuO-50. In the absence of LeuO, only residual activity was observed, which notably contrasted with the transcriptional activity detected with LeuO overexpression (Table 1). A fusion of the complete 5′ intergenic region of the CRISPR was constructed, and no activity was observed in the presence or absence of LeuO (data not shown). These results indicate that the complete CRISPR/Cas system in S. Typhi comprises a LeuO-dependent operon. The pKK9-casA, pKK9-RR-casA-cas2, and pKK9-RR-casA-CRISPR reporter fusions showed 507 (Fig. 1), 272 (Table 1), and 136 (Table 1) activity units, respectively, with LeuO overexpression. Interestingly, a gradual decrease in activity from casA to cas2 was also found in the presence of LeuO in E. coli K-12 (64). The data presented above support the notion that in S. Typhi IMSS-1 a single LeuO-dependent promoter drives CRISPR/Cas expression.
Table 1.
The CRISPR/Cas cluster comprises a LeuO-dependent operon in S. Typhia
| Plasmid | CAT sp act (μmol/min/mg protein) |
|
|---|---|---|
| IMSS-1/pFMTrc12 | IMSS-1/pFMTrcleuO-50 | |
| pKK9-RR-casA-cas2 | 36 ± 3 | 273 ± 5 |
| pKK9-ΔRR-casA-cas2 | 0 | 0 |
| pKK9-RR-casA-CRISPR | 8 ± 11 | 137 ± 0.6 |
Transcriptional values for the LeuO-dependent casABCDE12-CRISPR operon were determined in MA medium. The plasmids pKK9-RR-casA-cas2, pKK9-ΔRR-casA-cas2, and pKK9-RR-casA-CRISPR were introduced into IMSS-1 harboring plasmid pFMTrc12 and IMSS-1 containing plasmid pFMTrcleuO-50. The CAT specific activity of each strain was determined using samples collected at OD595 values of 0.4, 0.6, 0.8, and 1 and at 12 h; the values presented are the results obtained at 12 h. IPTG (50 μM) was used to induce the pFMTrcleuO-50 plasmid. Data are means ± standard deviations for three independent experiments performed in duplicate.
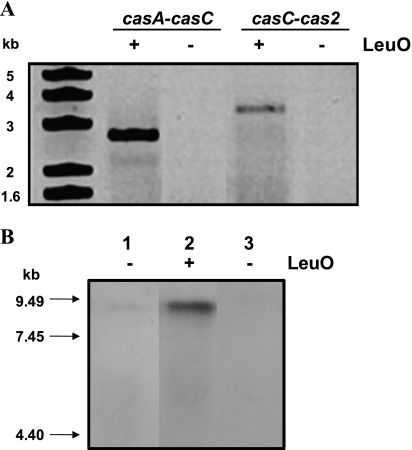
The expression of the LeuO-dependent CRISPR/Cas operon was also analyzed by chromosomal reverse transcription-PCR (RT-PCR). Total RNA isolated from the wild-type strain harboring pFMTrcleuO-50 and induced with IPTG was used with a specific CRISPR primer (CRISPR–R) located at the end of repeat 7 (see Table S2 in the supplemental material) to obtain a cDNA (6 kb) that included the complete casABCDE12-CRISPR operon. Using this template, casA-casC- and casC-cas2-specific primers were used to amplify the corresponding DNA fragments. The results showed that 2.9-kb and 3.4-kb fragments, containing the casA-casC and casC-cas2 regions, respectively, were amplified (Fig. 2 A), further supporting the LeuO-dependent operon structure. Northern experiments were also performed to support the CRISPR/Cas operon organization. Total RNAs from the bacterial strains IMSS-1/pFMTrc12, IMSS-1/pFMTrcleuO-50, and IMSS-1Δ(casA-CRISPR) were obtained. The RNAs were transferred, and independent membranes were hybridized with an internal fragment of the first gene of the operon, the casA locus, and with the last part of the operon, the CRISPR sequences. The results showed a null hybridization signal for the Salmonella Δ(casA-CRISPR) strain, which was devoid of the complete CRISPR/Cas operon; for the IMSS-1/pFMTrc12 strain, a weak signal was detected. In contrast, for the IMSS-1/pFMTrcleuO-50 strain induced with IPTG, a signal of 7 to 9 kb was detected with the casA or CRISPR probe. This result demonstrates that the complete CRISPR/Cas system comprises a LeuO-dependent operon. In Fig. 2B, we present only the results obtained with the CRISPR probe, since identical results were obtained with the casA probe.
Fig. 2.
The CRISPR/Cas cluster comprises a LeuO-dependent operon in S. Typhi. (A) The genetic organization of the LeuO-dependent casABCDE12-CRISPR operon was determined by chromosomal RT-PCR. S. Typhi IMSS-1 containing either the pFMTrcleuO-50 or pFMTrc12 plasmid was grown in MA medium with and without 50 μM IPTG, and total RNA was obtained. A 2.9-kb fragment was generated with casA- and casC-specific primers, and a 3.4-kb fragment was obtained with casC- and cas2-specific primers. PCRs using RNAs obtained from the strains mentioned above as templates and specific primers for the amplification of the cas2 and 16S rRNA genes were performed in order to verify the presence of DNA. As expected, no PCR amplification was observed, indicating the absence of DNA in the RNA purification. The left lane shows a DNA molecular size marker. (B) Detection of CRISPR/Cas mRNA by Northern blotting. Chromosomal expression patterns of CRISPR/Cas mRNAs from the bacterial strains IMSS-1/pFMTrc12 (lane 1), IMSS-1/pFMTrcleuO-50 (lane 2), and IMSS-1Δ(casA-CRISPR) (lane 3) are shown. Eight micrograms of total RNA from each sample was loaded and hybridized with an internal CRISPR probe. Sizes of the RNA molecular marker are shown on the left.
In E. coli, the CRISPR/Cas system contains two promoters: the first is a LeuO-dependent promoter (64) located upstream of the casA gene, and the second is located in the leader sequence of the CRISPR (55), suggesting that in E. coli this system is encoded by two transcriptional units. A comparison between the leader and CRISPR sequences of E. coli and S. Typhi shows that the leaders differ in length and share a low level of conservation. In fact, the −10 promoter sequence determined for E. coli (55) is missing in S. Typhi IMSS-1. The low level of conservation in the leader sequences could explain the lack of a CRISPR promoter in S. Typhi IMSS-1. It is worth mentioning that in S. Typhi and E. coli the transcriptional organization of the CRISPR/Cas systems is not conserved, suggesting that the signals or conditions that activate or repress this genetic program could be different even in these two closely related bacteria.
Determination of the nucleotides involved in LeuO activation of the CRISPR/Cas genetic system in S. Typhi IMSS-1.
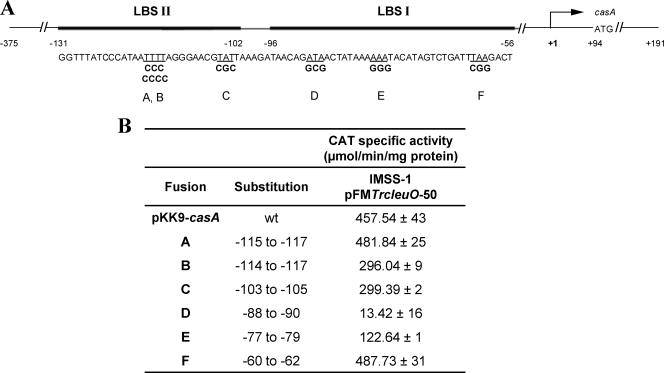
As mentioned above, LeuO is a positive activator of the CRISPR/Cas operon in S. Typhi; by footprinting experiments, we showed previously that LeuO binds to nucleotides −56 to −96 (LeuO binding site I [LBS I]) and −102 to −131 (LBS II) with respect to the transcriptional start site of casA (STY3070) (28). To determine which nucleotides mediate LeuO activation, a collection of substitutions (A, B, C, D, E, and F) were constructed in LBS I and LBS II (Fig. 3 A). Several regions in LBS I and LBS II with high A+T content were replaced by G+C nucleotides, considering that LeuO binds preferentially to sites with high A+T content (12). Reporter gene fusions containing three or four substitutions were introduced into the S. Typhi IMSS-1 wild-type strain harboring the pFMTrcleuO-50 plasmid. The transcriptional results showed that in the presence of LeuO, the A and F substitutions (nucleotides −115 to −117 and −62 to −60, respectively) gave similar expression levels to those with the wild-type fusion (Fig. 3B). However, a gradual decrease in activity was detected with the B and C substitutions (nucleotides −114 to −117 and −103 to −105, respectively) (Fig. 3B). Interestingly, a decrease of 76% was observed with the E substitution (nucleotides −77 to −79), and only 3% of wild-type activity remained with the D substitution (nucleotides −88 to −90) (Fig. 3B). The same constructs were introduced into the S. Typhi ΔleuO strain harboring pFMTrcleuO-50, and CAT activity was determined. The results of a single experiment (data not shown) gave the same level of expression for each mutation as that obtained with the wild-type S. Typhi strain, indicating that the chromosomal leuO gene did not affect the expression of the fusions evaluated. These data indicate that substitutions B, C, D, and E affected the transcriptional casA induction mediated by LeuO, suggesting that both LBS I and LBS II are relevant for casA LeuO-dependent induction. It will certainly be interesting to determine the impact of base changes on binding of the LeuO protein to elucidate whether the decrease in activity is because the protein is effectively unable to bind to the DNA or binds it in an ineffective manner.
Fig. 3.
Mapping of the LeuO recognition sequences in the regulatory region of casA. (A) Schematic representation of LBS I and LBS II in the promoter region of casA. The entire regulatory region of casA was used as a template to obtain mutants in LBS II (A, B, and C) and LBS I (D, E, and F). The substitutions (in bold) were sequenced to verify the correct mutations. The wild-type sequences are underlined. (B) Transcriptional analysis of casA promoter fusions with mutations in LBS I and LBS II. S. Typhi strain IMSS-1, containing each of the mutations individually as well as the pFMTrcleuO-50 plasmid and induced with 50 μM IPTG, was evaluated for CAT activity. The assays were performed in MA medium at different OD595 values, and the activities at 12 h are presented. As a positive control, we used the complete wild-type casA promoter region. The data are means ± standard deviations for three independent experiments performed in duplicate.
H-NS partially represses transcriptional expression of the CRISPR/Cas genetic program in S. Typhi IMSS-1.
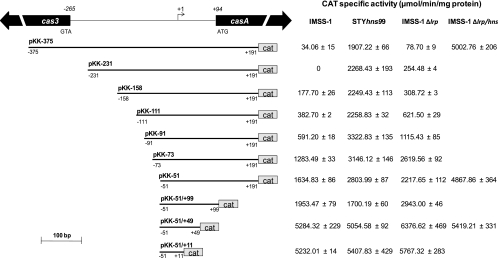
H-NS is a master regulator of the S. Typhi genome, and significant evidence has been compiled about the role of this protein in the expression of several genes belonging to the LeuO regulon (13, 16, 28, 59, 60). Previously, we used transcriptional fusions to show basal expression of the casA (STY3070) promoter region in the wild-type strain. However, transcriptional activity of this fusion was induced in the presence of LeuO as well as in an H-NS-deficient strain, supporting the notion that H-NS negatively regulates the expression of casA (STY3070) in S. Typhi IMSS-1 (28). To further characterize the casA regulatory region, a series of promoter fragments harboring progressively shorter upstream sequences of the entire promoter region were generated by PCR, and each of these DNA fragments was fused to the cat reporter gene in plasmid pKK232-8 (Fig. 4). The expression levels of these derivatives were examined by CAT activity assay of the wild-type S. Typhi IMSS-1 strain as well as the STYhns99 H-NS-deficient strain (19) (Fig. 4). The CAT activity levels of all of the single fusions analyzed (from pKK-375, which contains the entire casA 5′ intergenic regulatory region, to pKK-231, pKK-158, pKK-111, pKK-91, pKK-73, and pKK-51) were higher in the H-NS-deficient strain than in the wild-type strain. These results are consistent with the negative role of H-NS in the genetic expression of casA (Fig. 4).
Fig. 4.
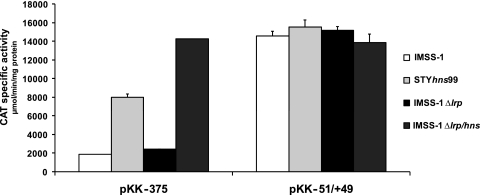
Promoter fusions harboring different lengths of the casA regulatory region. The bent arrow represents the transcriptional start site, located 94 bp upstream of the translational start codon of casA; the numbers in italics represent the localization of the casA and cas3 ATG initiation codons relative to the casA mRNA start site. Below the diagram of the regulatory region of casA, transcriptional fusions are shown. The fusions (in bold) were named according to the transcription initiation site; the numbers below each fusion represent the distances upstream and downstream of the mRNA start site. The right columns represent the expression of the casA fusions evaluated in the wild-type S. Typhi IMSS-1 strain, the single H-NS (STYhns99) and LRP (Δlrp) mutants, and the double Δlrp/hns mutant strain. The activities were determined at different ODs in rich medium (MA); the data presented correspond to the values obtained at an OD595 of 1.3. The values are the means ± standard deviations for at least three independent experiments performed in duplicate.
Interestingly, the CAT activity values for the H-NS-deficient strain with the pKK-375, pKK-231, pKK-158, and pKK-111 fusions were between 1,900 and 2,200 activity units, but the values with the pKK-91, pKK-73, and pKK-51 fusions were higher (2,800 to 3,300 activity units), indicating that in these three fusions regions of negative regulation were eliminated (Fig. 4). An increased activity in the wild-type strain was also observed with fusions pKK-91, pKK-73, and pKK-51 compared with the longer fusions pKK-375, pKK-231, pKK-158, and pKK-111, supporting the notion that nucleotides in the region from positions −111 to −51 are recognized by negative regulatory elements, in addition to H-NS, repressing casA expression (Fig. 4).
Additional fusions deleting downstream sequences relative to the transcriptional start site (pKK-51/+11 and pKK-51/+49) showed between 5,000 and 5,200 activity units for the wild type as well as the H-NS mutant strain. However, with the pKK-51/+99 fusion, 1,700 to 1,900 activity units were detected for the wild type and the H-NS mutant, indicating that between nucleotides +49 and +99 with respect to the transcriptional start site, negative regulatory elements repress casA expression (Fig. 4). In summary, systematic deletions upstream and downstream of the transcriptional start site of casA resulted in increased CAT activity when they were evaluated in an H-NS-deficient strain, indicating that this nucleoid-associated protein partially represses casA expression and that other genetic elements in addition to H-NS are involved in the silencing of casA in S. Typhi IMSS-1.
LRP is a negative regulator of the CRISPR/Cas operon in S. Typhi IMSS-1.
To further characterize the genetic expression of casA in S. Typhi IMSS-1 and to identify other putative genetic elements involved in the control of casA, a bioinformatic analysis was performed. A set of casA-orthologous proteins was constructed using the Blastp algorithm (3, 4); the highest E value and an upper threshold of 10−5 were considered to select the orthologous proteins. With this method, we detected a set of 21 casA-orthologous proteins, which are present in archaea and in alpha-, delta-, and gammaproteobacteria. The 400-bp sequences upstream of the putative translational start sites of the 21 casA-orthologous polypeptides were aligned to eliminate redundant sequences. The resulting 18 upstream sequences were analyzed using the Oligo-Analysis program (62). A search on both strands for motifs with a maximum length of 6 nucleotides was performed. After assembling the patterns discovered by the algorithm, we found diverse motifs. A conserved LRP motif (TAATAAA) was present in 10 of the sequences analyzed, and this motif was compared using the TOM TOM algorithm (23) with those reported in the DPINTERACT (56) and REGTRANSDB v4 (36) databases. The results showed that the TAATAAA pattern was preserved in close conservation with an LRP binding site of E. coli in both databases (P = 0.0080). Interestingly, LRP motifs were located 221 bp upstream and 66 nucleotides downstream of the casA transcriptional start site of S. Typhi IMSS-1. We also identified that the LRP pattern in S. Typhi Ty2 has the same distribution as that in S. Typhi IMSS-1. These findings suggest that the LRP protein is perhaps a casA negative regulator, especially considering the site found downstream of the transcriptional start site. In Methanospirillum hungatei, two LRP sites are present, 38 and 351 bp upstream of the casA ATG translational start codon. In the case of Granulibacter bethendens, an LRP motif was located 50 bp upstream of the casA translational start site. In E. coli K-12 and E. coli W3110, two LRP motifs were identified whose positions varied significantly, but all were located upstream of the transcriptional start site. In Desulfovibrio desulfuricans, Desulfococcus oleovorans, and Marinomonas MWYL1, an LRP motif was detected ∼400 bp upstream of the ATG translational start site. The presence of LRP motifs in the regulatory region of casA orthologs belonging to alpha-, delta-, and gammaproteobacteria and to archaea suggested that the LRP protein could be involved in the transcriptional regulation of the CRISPR/Cas system.
To evaluate the role of LRP in the genetic expression of the casA gene, we constructed an S. Typhi IMSS-1 strain deficient in lrp by using the method of Datsenko and Wanner (14). Transcriptional fusions of different lengths (Fig. 4) were introduced into the lrp-deficient strain, and CAT activity assays were performed. The results showed an increased CAT activity in the LRP-deficient strain relative to that in the wild-type strain for most of the fusions evaluated. The effect was most evident with the pKK-91, pKK-73, and pKK-51 fusions (Fig. 4), suggesting a negative role for LRP in the expression of casA.
The activities for the pKK-51/+11 and pKK-51/+49 fusions were 5,232 and 5,284 activity units, respectively, with the wild type, whereas with the LRP-deficient strain, the values obtained were 5,767 and 6,376 activity units, respectively. For the pKK-51/+99 fusion, around 1,900 and 2,900 activity units were observed with the wild type and the LRP mutant, respectively (see Fig. 4 for standard deviation values), again supporting the conclusion that negative regulatory elements between nucleotides +49 and +99 contribute to casA repression (Fig. 4). Thus, both LRP and H-NS repress the genetic expression of CRISPR/Cas. To evaluate the roles of both proteins, a double hns/lrp mutant was constructed, and transcriptional experiments were performed with the wild type, the single hns and lrp mutants, and the hns/lrp double mutant.
The longer transcriptional fusion pKK-375, which contains the complete 5′ intergenic regulatory region of casA, and the pKK-51 and pKK-51/+49 fusions were introduced into the strains described above. The pKK-375 fusion in the wild-type strain showed a value of 34 activity units; in the single H-NS mutant, an activity of 1,907 activity units was observed; in the lrp single mutant, a value of 78 activity units was detected; and the double hns/lrp mutant showed 5,002 activity units, hence demonstrating the role of both LRP and H-NS in the repression of casA (see Fig. 4 for standard deviation values). Interestingly, the pKK-51 fusion had values of around 1,600, 2,800, 2,200, and 4,800 activity units in the wild type, the hns and lrp single mutants, and the double hns/lrp mutant, respectively. The activity values obtained with pKK-51/+49 were 5,200 for the wild type, 5,000 for the hns mutant, 6,300 for the lrp-deficient strain, and 5,400 for the double hns/lrp mutant strain (Fig. 4). This indicates that the sequence from nucleotides −51 to +49 contains the complete promoter region, without any negative regulatory elements, since with the wild-type strain we obtained the same activity as that observed with the double H-NS/LRP mutant strain. These data also confirm that the casA regulatory region contains two main zones of negative regulation, from nucleotides −51 to −375 and from nucleotides +49 to +99, and that H-NS and LRP are the genetic elements that repress casA expression. With the data mentioned above, we also showed that in the presence of H-NS, LRP has a minor effect; nevertheless, in the absence of H-NS, LRP mainly represses casA expression (Fig. 4), suggesting that LRP could compensate for the absence of H-NS.
It is interesting that previous studies by Pul et al. (55) indicated that in E. coli K-12 the LRP protein did not have a significant effect on the expression of the casA gene. Thus, in E. coli and S. Typhi, the negative control of casA expression is exerted differently, indicating that the environmental conditions or proteins that control CRISPR/Cas genetic expression in these two related bacteria could be different.
H-NS and LRP directly repress the expression of the casA promoter.
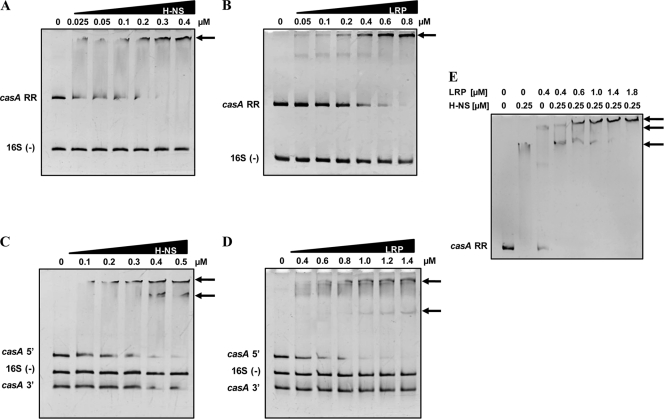
The data presented above showed that sequences upstream and downstream of the transcription initiation site of casA are involved in its silencing and that the genetic regulators involved in this repression are LRP and H-NS. To determine whether these proteins bind upstream and downstream of the initiation site, EMSAs were performed with the corresponding purified proteins and with various fragments of the casA promoter region. EMSA using the complete regulatory region of casA and the H-NS protein showed that H-NS interacted directly with this fragment, since a single complex was observed (Fig. 5 A). In the case of EMSA with LRP and the entire casA regulatory region, a predominant complex was observed, supporting the direct interaction of LRP with the casA promoter region. A second transient band was also visible; however, it disappeared with increased LRP amounts, and only the predominant complex remained in the retardation gel (Fig. 5B). Further EMSAs showed that H-NS and LRP bound to the 5′ casA region (nucleotides −51 to −375); interestingly, these proteins also interacted with the 3′ casA region (nucleotides +49 to +191), since we detected two complexes, the highest corresponding to the 5′ region and the lowest to the 3′ fragment's interactions with the respective proteins (Fig. 5C and D). That is, EMSAs with H-NS or LRP in the presence of only the 5′ casA fragment or the 3′ casA fragment rendered the complexes mentioned above (not shown). It is noteworthy that no strong binding was observed with LRP and the 3′ casA fragment in the presence of the 5′ casA fragment (Fig. 5D), suggesting that the latter has a higher affinity for LRP. The 3′ casA fragment did bind to LRP at 0.6 μM in the absence of the 5′ casA fragment (data not shown). As a negative control, in these assays we used a 242-bp coding region of the 16S rRNA gene of S. Typhi IMSS-1. These results support the hypothesis that H-NS and LRP bind independently to the 5′ and 3′ casA regulatory regions. However, in order to elucidate if the two regulators are able to interact simultaneously with the casA regulatory sequence, EMSAs with both proteins were performed, showing that the H-NS–casA complex in the presence of increasing amounts of LRP was modified, with a new band visualized. This novel complex was of a higher mass than those obtained with the H-NS and LRP proteins independently, indicating that both regulators bound simultaneously to the same fragment of DNA (Fig. 5E). It is relevant to mention that although the two proteins interacted with the same fragment, they did not appear to displace each other, thus demonstrating the simultaneous involvement of H-NS and LRP in the repression of the CRISPRS/Cas system in S. Typhi.
Fig. 5.
H-NS and LRP interact with the casA regulatory region. EMSAs were performed with the LRP and H-NS purified proteins and with various DNA fragments of the casA regulatory region to determine the H-NS and LRP recognition sites. The complete 5′ intergenic regulatory region of casA (RR) was incubated independently with H-NS (A) or with LRP (B). The casA 5′ (nucleotides −375 to −52) and 3′ (nucleotides +50 to +191) regions were incubated with H-NS (C) or LRP (D), and the casA RR was incubated with both proteins (E). Increasing concentrations of purified H-NS-Myc-His6 or LRP-His6 protein were incubated with the PCR-generated DNA fragments. The EMSA experiments were performed in 5% or 6% polyacrylamide gels, and gels were stained with ethidium bromide. 16S rRNA gene structural fragments of 242 bp were used as a negative control for the EMSAs. Arrows indicate DNA-protein complexes.
The CRISPR/Cas genetic system is induced in S. Typhi IMSS-1 grown in N-minimal medium.
The CRISPR/Cas immune system is widely distributed in prokaryotes; however, few studies point out the natural conditions that induce its expression. Therefore, it is of interest to investigate the signals or conditions that activate this genetic system. For S. Typhi, the selective capture of genes expressed in macrophages showed that the third component of the CRISPR/Cas system, STY3068 (casC ortholog), is induced under these conditions (17), and according to Deiwick et al. (15), a minimal medium deficient in magnesium and phosphate simulates the environmental niche encountered by Salmonella in the eukaryotic cell, since several genes of Salmonella pathogenicity island 2 that are relevant to the survival of this pathogen inside macrophages are induced in this medium (10).
To elucidate whether the S. Typhi CRISPR/Cas program is induced under conditions that simulate the environment inside the macrophage, the expression of the casA regulatory region was evaluated in N-minimal medium. The results showed that in N-minimal medium, the pKK-375 fusion gave 1,800 activity units in the wild-type strain, while its activity was only 34 activity units in rich MA medium (Fig. 4 and 6). Interestingly, in N-minimal medium, we observed that the expression values for the pKK-375 fusion in the hns and lrp single null and double hns/lrp null strains were 8,000, 2,400, and 14,000 activity units, respectively, supporting the negative role of H-NS and LRP in the transcriptional expression of casA in N-minimal medium (Fig. 6). In the wild type and the hns, lrp, and double hns/lrp mutants, the activity values for the pKK-51/+49 fusion (which contains the promoter region without cis-acting negative elements) were around 14,000 to 15,000 (Fig. 6), supporting the conclusion that H-NS and LRP are the main repressors of casA expression.
Fig. 6.
Transcriptional profile of the casA promoter region in N-minimal medium. The expression profiles of the pKK-375 and pKK-51/+49 fusions were evaluated in the wild-type S. Typhi IMSS-1, STYhns99, IMSS-1 Δlrp, and IMSS-1 Δlrp/hns strains. The activities were determined at 16 h in N-minimal medium. The values are the means and standard deviations for at least three independent experiments performed in duplicate.
To determine the promoter that activates the expression of casA in N-minimal medium, 5′-RACE experiments were performed. RNA from the wild-type strain grown in N-minimal medium was obtained, and a specific primer (casA 5′-RACE) located 83 bp downstream of the ATG initiation codon of casA was used for the determination of the transcription initiation site (see Table S2 in the supplemental material). The experiment showed that for cells grown in N-minimal medium, a transcriptional start site was located 94 bp upstream of the ATG translational codon. Interestingly, this initiation site coincides with the LeuO-dependent transcriptional site observed in rich medium (MA medium). These data indicate that transcriptional expression in MA medium in the presence of overexpressed LeuO, as well as that in N-minimal medium in the absence of overexpressed LeuO, is driven by the same promoter. Additional experiments validated this assertion, since with a fusion that harbors a mutation in the Pribnow box [pKK9-casA (−10 ATA → GGG)] of the transcriptional start site of casA, null CAT activity was detected in the presence of LeuO in either MA medium or N-minimal medium (data not shown). Furthermore, a fusion of 62 bp (nucleotides −51 to +11), which contained 51 and 11 bp upstream and downstream, respectively, of the transcription initiation site of casA, showed activity values of 5,232 (Fig. 4) and 16,470 (data not shown) in rich and minimal medium, respectively, supporting the idea that a common promoter drives casA expression in both media. To determine whether LeuO is the casA activator in N-minimal medium, the pKK-375 fusion was introduced into the IMSS-1 ΔleuO strain: we found that LeuO does not induce casA expression in N-minimal medium, since the activity values in the leuO-deficient strain were the same as those in the wild-type strain (data not shown). However, LeuO could increase transcriptional activity in N-minimal medium, as overexpression of the leuO gene resulted in a 2-fold induction of casA transcription compared to an absence of overexpression in the wild-type Salmonella strain (data not shown). These data support the role of LeuO in N-minimal medium and MA rich medium as an inducer of casA expression. However, casA can also be expressed in a LeuO-independent mode in N-minimal medium, which reveals that additional regulators are needed to fully activate casA under these conditions. Future research is needed to identify the full repertoire of genetic elements that control casA activation in both N-minimal medium and MA medium.
DISCUSSION
Three distinct stages are involved in CRISPR/Cas functionality: integration of the spacers, expression of CRISPR/Cas, and interference with the foreign DNA. In this study, we analyzed the transcriptional expression of the CRISPR/Cas system in S. Typhi IMSS-1. Previously, we reported that two putative promoters control casA expression in S. Typhi IMSS-1 and that the initiation sites are located 94 and 84 bp upstream of the ATG translational start codon (28). In this work, we performed 5′-RACE experiments that showed a single chromosomal transcription initiation site (Fig. 1B). By site-directed mutagenesis of the −10 box, we demonstrated that a unique +1 mRNA is located 94 bp upstream of the ATG translational start codon of casA (Fig. 1C). The promoter region reported in this work is induced in the presence of the global regulator LeuO. Site-directed mutagenesis of the LeuO binding sites supports the positive role of this protein in the expression of casA, and we showed that both LBS I and LBS II are determinants of LeuO induction. Interestingly, the ATA bases located at positions −88 to −90 (LBS I) are key for LeuO activation (Fig. 3B). Previously, we demonstrated that a TATgTcATAT region located at positions −125 to −134 with respect to the transcriptional start site was necessary for the LeuO induction of the ompS1 gene (16). These data provide evidence that this regulator does not necessarily recognize a consensus sequence (28). Additional efforts are needed to elucidate the rules that LeuO follows for DNA recognition and transcriptional activation.
Using transcriptional fusions, RT-PCR, and Northern experiments, we demonstrated that the entire CRISPR/Cas system is induced as a large polycistronic mRNA in rich medium in the presence of LeuO, supporting an operon organization (Table 1; Fig. 2) that forms part of the LeuO regulon in S. Typhi. Interestingly, Northern blot experiments with E. coli showed that the casA-casB-casC-casD-casE-cas1-cas2 genes are part of one transcriptional unit (58). It has also been reported by Pul et al. (55) that the CRISPR sequences contain their own promoter in E. coli. Thus, in contrast to the CRISPR/Cas operon organization in S. Typhi, E. coli has two transcriptional units, one driving the genetic expression of casABCDE12 and the other inducing CRISPR transcription. This result provides evidence that the genetic control of the CRISPR/Cas program is highly regulated in the Enterobacteriaceae and that even in two closely related bacteria, such as S. Typhi and E. coli, the functional transcriptional organization is different, raising the possibility that the CRISPR/Cas system can be fine-tuned in response to multiple environmental inputs.
Various studies have shown that genes upregulated by LeuO are downregulated by H-NS (28, 29, 39, 40, 58, 64). A recent report showed that casA and CRISPR expression is repressed independently by H-NS in E. coli (55). In contrast, in the case of S. Typhi IMSS-1, a fusion containing the leader and the CRISPR locus showed no effect in an H-NS-deficient strain (data not shown), while H-NS repression was observed in the transcriptional expression of casA. These data again point out the differences in CRISPR/Cas transcriptional control in E. coli versus S. Typhi. Another notable difference is the finding that the global regulatory protein LRP is a negative repressor of casA in S. Typhi (Fig. 4), while Pul et al. (55) reported that LRP did not have a significant effect on the expression of the cas genes in E. coli.
The involvement of H-NS, LRP, and a LysR-type regulator in the control of other Salmonella genes has been documented. For instance, the spv virulence genes are positively regulated by the LysR-type regulator SpvR and repressed by H-NS and LRP (45, 50). A negative role of H-NS and LRP is also reported for the 16S rRNA genes, where it is postulated that these proteins bind upstream and downstream of the transcription initiation site and work together in a synergic fashion to repress the expression of the 16S rRNA genes (53, 54). Similar to these studies, we found that H-NS and LRP bind upstream and downstream of the transcription initiation site to repress casA (Fig. 5C and D). Interestingly, both proteins appear to simultaneously bind the casA regulatory region (Fig. 5E), and it is possible that a nucleosome structure formed between H-NS, LRP, and the target promoter is needed to exert repression of CRISPR/Cas, as suggested for the 16S rRNA repression mediated by H-NS and LRP (53, 54).
Future research is needed to elucidate the fine-tuning mechanism of casA repression mediated by H-NS and LRP, and it would also be interesting to determine how LeuO in the presence of these proteins induces casA activity. The transcriptional results in the absence of H-NS and LRP showed casA activity values of around 5,000 in MA medium (Fig. 4), while the expression value obtained in the presence of LeuO was only 500 (Fig. 1C), indicating that LeuO moderately induces casA transcriptional expression and that some additional positive regulatory elements or signals are needed to obtain its complete induction. We also found that the casA promoter region is highly induced in N-minimal medium, in a LeuO-independent manner (Fig. 6). Nevertheless, LeuO can also enhance casA activity in N-minimal medium if it is overexpressed from a synthetic promoter.
The participation of global regulatory proteins such as LeuO, H-NS, and LRP, which have been implicated in virulence metabolism and other biological processes, suggests that the CRISPR/Cas system responds to a wide variety of environmental cues. It was reported that the CRISPR/Cas program is involved in sporulation in Myxococcus xanthus (63) and in biofilm formation in Pseudomonas aeruginosa (65) and also that housekeeping genes can be targets of CRISPR interference (2). The fact that the CRISPR/Cas system is induced under stress conditions such as growth in N-minimal medium suggests that it could have a role in pathogenesis. Further studies of mutants inactivated in the cas genes are needed to evaluate their role in infection and virulence in Salmonella.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants to I.H.-L. from DGAPA/UNAM (IN214808) and CONACYT (89337 and 127298) and by grants from CONACYT, México (82383), and DGAPA-UNAM (216310) to E.C.
We thank S. J. Jaime-Rodríguez, M. L. Zavala-García, N. Becerra-Lobato, F. J. Santana, M. Fernández-Mora, P. Gaytan, E. Bustos, S. Becerra, J. Yañez, and J. A. Ramirez-Trujillo for technical help and A. Gutierrez-Preciado, A. Medrano-López, J. M. Villarreal, M. A. De la Cruz, R. Oropeza, J. Caballero, M. Dunn, and J. Miranda for critical readings of the manuscript and stimulating discussions.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Agari Y., et al. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 395:270–281 [DOI] [PubMed] [Google Scholar]
- 2. Aklujkar M., Lovley D. R. 2010. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol. Biol. 10:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amann E., Birgit O., Karl-Josef A. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315 [DOI] [PubMed] [Google Scholar]
- 6. Andersson A. F., Banfield J. F. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050 [DOI] [PubMed] [Google Scholar]
- 7. Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 8. Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561 [DOI] [PubMed] [Google Scholar]
- 9. Brouns S. J., et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bustamante V. H., et al. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105:14591–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carte J., Wang R., Li H., Terns R. M., Terns M. P. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22:3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C. C., et al. 2003. LeuO-mediated transcriptional derepression. J. Biol. Chem. 278:38094–38103 [DOI] [PubMed] [Google Scholar]
- 13. Chen C. C., Chou M. Y., Huang C. H., Majumder A., Wu H. Y. 2005. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 280:5101–5112 [DOI] [PubMed] [Google Scholar]
- 14. Datsenko K., Wanner B. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deiwick J., Nikolaus T., Erdogan S., Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759–1773 [DOI] [PubMed] [Google Scholar]
- 16. De la Cruz M. A., et al. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 66:727–743 [DOI] [PubMed] [Google Scholar]
- 17. Faucher S. P., Curtiss III R., Daigle F. 2005. Selective capture of Salmonella enterica serovar Typhi genes expressed in macrophages that are absent from the Salmonella enterica serovar Typhimurium genome. Infect. Immun. 73:5217–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernández-Mora M., Puente J. L., Calva E. 2004. OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene. J. Bacteriol. 186:2909–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flores-Valdez M. A., Puente J. L., Calva E. 2003. Negative osmoregulation of the Salmonella ompS1 porin gene independently of OmpR in an hns background. J. Bacteriol. 185:6497–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forde A., Fitzgerald G. F. 1999. Bacteriophage defence systems in lactic acid bacteria. Antonie Van Leeuwenhoek 76:89–113 [PubMed] [Google Scholar]
- 21. Garneau J. E., et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71 [DOI] [PubMed] [Google Scholar]
- 22. Grissa I., Vergnaud G., Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta S., Stamatoyannopolous J. A., Bailey T. L., Noble W. S. 2007. Quantifying similarity between motifs. Genome Biol. 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hale C. R., et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haurwitz R. E., Jinek M., Wiedenheft B., Zhou K., Doudna J. A. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heidelberg J. F., Nelson W. C., Schoenfeld T., Bhaya D. 2009. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One 4:e4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hernández-Lucas I., et al. 2008. The LysR-type transcriptional regulator LeuO controls the expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 190:1658–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hommais F., et al. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20–36 [DOI] [PubMed] [Google Scholar]
- 30. Horvath P., Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 31. Jansen R., van Embden J. D., Gaastra W., Schouls L. M. 2002. Identification of a novel family of sequence repeats among prokaryotes. OMICS 6:23–33 [DOI] [PubMed] [Google Scholar]
- 32. Jansen R., van Embden J. D., Gaastra W., Schouls L. M. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575 [DOI] [PubMed] [Google Scholar]
- 33. Kaminuma E., et al. 9 November 2010. DDBJ progress report. Nucleic Acids Res. doi:10.1093/nar/gkq1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karginov F. V., Hannon G. J. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawaji H., Mizuno T., Mizushima S. 1979. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J. Bacteriol. 140:843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazakov A. E., et al. 2007. RegTransBase—a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35:D407–D412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Labrie S. J., Samson J. E., Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 38. Lillestøl R. K., et al. 2009. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72:259–272 [DOI] [PubMed] [Google Scholar]
- 39. Lucchini S., et al. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Madhusudan S., Paukner A., Klingen Y., Schnetz K. 2005. Independent regulation of H-NS-mediated silencing of the bgl operon at two levels: upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology 151:3349–3359 [DOI] [PubMed] [Google Scholar]
- 41. Makarova K. S., Grishin N. V., Shabalina S. A., Wolf Y. I., Koonin E. V. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marraffini L. A., Sontheimer E. J. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marraffini L. A., Sontheimer E. J. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marraffini L. A., Sontheimer E. J. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marshall D. G., Sheehan B. J., Dorman C. J. 1999. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol. Microbiol. 34:134–145 [DOI] [PubMed] [Google Scholar]
- 46. Martinez-Laguna Y., Calva E., Puente J. L. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153–166 [DOI] [PubMed] [Google Scholar]
- 47. Mayer M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41–46 [DOI] [PubMed] [Google Scholar]
- 48. Mendoza-Vargas A., et al. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mojica F. J., Díez-Villaseñor C., García-Martínez J., Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174–182 [DOI] [PubMed] [Google Scholar]
- 50. O'Byrne C. P., Dorman C. D. 1994. Transcription of the Salmonella typhimurium spv virulence locus is regulated negatively by the nucleoid-associated protein H-NS. FEMS Microbiol. Lett. 12:99–106 [DOI] [PubMed] [Google Scholar]
- 51. Pourcel C., Salvignol G., Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663 [DOI] [PubMed] [Google Scholar]
- 52. Puente J. L., Flores V., Fernández M., Fuchs Y., Calva E. 1987. Isolation of an ompC-like outer membrane protein gene from Salmonella typhi. Gene 61:75–83 [DOI] [PubMed] [Google Scholar]
- 53. Pul U., et al. 2005. LRP and H-NS cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol. Microbiol. 258:864–876 [DOI] [PubMed] [Google Scholar]
- 54. Pul U., Wurm R., Wagner R. 2007. The role of LRP and H-NS in transcription regulation: involvement of synergism, allostery and macromolecular crowding. J. Mol. Biol. 366:900–915 [DOI] [PubMed] [Google Scholar]
- 55. Pul U., et al. 2010. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 7:1495–1512 [DOI] [PubMed] [Google Scholar]
- 56. Robison K., McGuire A. M., Church G. M. 1998. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 284:241–254 [DOI] [PubMed] [Google Scholar]
- 57. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 58. Shimada T., Yamomoto K., Ishihama A. 2009. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J. Bacteriol. 191:4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stratmann T., Madhusudan S., Schnetz K. 2008. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 190:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ueguchi C., Ohta T., Seto C., Suzuki T., Mizuno T. 1998. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J. Bacteriol. 180:190–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van der Oost J., Jore M. M., Westra E. R., Lundgren M., Brouns S. J. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407 [DOI] [PubMed] [Google Scholar]
- 62. van Helden J., André B., Collado-Vides J. 1998. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281:827–842 [DOI] [PubMed] [Google Scholar]
- 63. Viswanathan P., Murphy K., Julien B., Garza A. G., Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 189:3738–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Westra E. R., et al. 2010. H-NS mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77:1380–1393 [DOI] [PubMed] [Google Scholar]
- 65. Zegans M. E., et al. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.