Abstract
Green fluorescent protein (GFP) fusions, immunofluorescence microscopy, and cryo-electron tomography revealed that the chemoreceptors of the Lyme disease spirochete Borrelia burgdorferi form long, thin arrays near both cell poles. These arrays are in close proximity to the flagellar motors. This information provides a basis for further understanding motility, chemotaxis, and protein localization in spirochetes.
TEXT
Bacterial chemotaxis is a complex sensory transduction pathway that enables cells to sense and respond to environmental stimuli. Although chemotaxis has been well studied in the paradigm models of Escherichia coli, Salmonella enterica serovar Typhimurium, and Rhodobacter sphaeroides (17, 33, 35), the understanding of this mechanism in spirochetes is at an early stage (4, 7, 10, 16, 24, 28, 30). Spirochetes have two bundles of periplasmic flagella (PF) that are subterminally attached at each cell pole (7, 8, 15, 18). Due to this unique geometry, the PF necessarily rotate asymmetrically during translational motility, with one bundle rotating counterclockwise (CCW) and the other rotating clockwise (CW) (7, 24, 30). One enigmatic question is how spirochetes coordinate the directional rotation (CCW or CW) of the two bundles of PF during chemotaxis (7, 24). Although several models have been proposed, the mechanism involved still remains unknown (7, 16, 24, 30).
Flagellar rotation is modulated by chemotaxis (3, 35). Methyl-accepting chemotaxis proteins (MCPs) form clusters that reside at or near the cell poles in most motile bacteria, and the spatial organization and polar positioning of chemotaxis arrays are extremely important for the process of tactic responses (2, 5, 6, 19, 32, 33). However, the location of chemotaxis arrays in spirochetes is still obscure (7, 24). Here we asked whether the MCPs are located at only one cell pole or both cell poles. Although Gestwicki et al. found using fluorescent antibodies that the MCPs in Spirochaeta aurantia were localized at either one or both cell poles, no quantitative data were presented with respect to the percentage of cells that have MCPs at one or both cell ends (14). We also asked whether the MCPs are in close proximity to the subterminally located flagellar motors, as Briegel et al., using cryo-electron tomography (cryo-ET), found that the MCPs were subpolarly localized in Borrelia burgdorferi (6). To address these two questions, we first used green fluorescent protein (GFP) and immunofluorescence assays (IFA) to determine the approximate cellular locations of two different MCPs and then applied cryo-ET to reveal the precise cellular position of the chemoreceptor arrays and their spatial orientation to the flagellar motors.
The strategy to localize the chemotaxis arrays of B. burgdorferi.
MCPs typically interact with each other and other chemotaxis proteins to form arrays at cell poles (5, 17, 29, 33, 34). There are five putative MCPs in B. burgdorferi, including MCP1 (BB0578), MCP2 (BB0596), MCP3 (BB0597), MCP4 (BB0680), and MCP5 (BB0681) (7, 12). Among these proteins, MCP3 and MCP5 are most similar to those of E. coli Trg and Tar (MCP3 shares 25% identity to Trg and Tar; MCP5 shares 33% identity to Trg), and both belong to the major MCP class referred to as the 34H class (1). In addition, our preliminary studies suggest that these two proteins are more abundant than the other MCPs (data not shown). Thus, these two MCPs were selected as markers to determine the cellular location of the chemotaxis arrays by using GFP and IFA.
Construction of GFP fusion proteins.
A strategy similar to one previously described was applied to construct MCP3-GFP and MCP5-GFP fusions (25, 34). Briefly, the flgB promoter (13), gfp (9), and the MCP3 gene were each amplified by PCR. For DNA cloning, BamHI, NdeI, NruΙ, and PstΙ cut sites were engineered in the respective primers (Table 1; see also Fig. S1a in the supplemental material). The obtained PCR products were further cloned into the pGEM-T Easy vector (Promega, Madison, WI). The flgB promoter and the MCP3 gene were first fused at an NdeI site, and then gfp was inserted in frame into the 3′ end of the MCP3 gene at NruΙ and PstΙ sites. The flgB-MCP3 gene-gfp fragment was then subcloned into the shuttle vector pKFSS1 (11) at BamHI and PstI sites, generating pKFSS1/flgBmcp3gfp (Fig. S1a). The same method was used to construct the pKFSS1/flgBmcp5gfp vector. The fusion genes in the final constructs were confirmed by DNA sequencing analysis. A control vector, in which gfp was fused to the flgB promoter, was also constructed.
Table 1.
Oligonucleotide primers used in this study
| Primer | Description/functiona | Sequence (5′-3′)b |
|---|---|---|
| P1 | flgB promoter (F) | GGATCCTAATACCCGAGCTTCAAG |
| P2 | flgB promoter (R) | CATATGACCTCCCTCATTTAAAATTGC |
| P3 | MCP3 gene-gfp fusion (F) | CATATGACAGATGAGAATTTAATTGATG |
| P4 | MCP3 gene-gfp fusion (R) | TCGCGAAAGAATATCTTTAATCTCATC |
| P5 | gfp (F) plus 5× glycine | TCGCGAACCTCCACCTCCACCAGTAAAGGAGAAGAACTTTTCACT |
| P6 | gfp (R) | CTGCAGTTTGTATAGTTCATCCATGCCATGTG |
| P7 | MCP5 gene-gfp fusion (F) | CATATGGTTAGTATGAAGCTTAAAGC |
| P8 | MCP5 gene-gfp fusion (R) | TCGCGATTCGATCTTAAAATAATCAACAG |
| P9 | MCP3 overexpression (F) | AGATCTTTTTTAAACAGTGTGTCTGC |
| P10 | MCP3 overexpression (R) | CTGCAGAGAATCTCTAACTGGAAAAC |
| P11 | MCP5 overexpression (F) | AGATCTTCAATGGAAGAGAAAGTTAG |
| P12 | MCP5 overexpression (R) | CTGCAGATTAATCCCTCTAAAAGACC |
MCP5-GFP resides at the cell poles of B. burgdorferi.
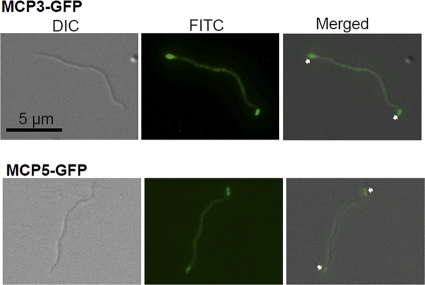
To determine the cellular locations of MCP3-GFP and MCP5-GFP, pKFSS1/flgBmcp3gfp, pKFSS1/flgBmcp5gfp, and a plasmid that expresses only gfp were transformed into the high-passage B. burgdorferi strain B31A (24, 25, 31). The transformants were confirmed by PCR. MCP3-GFP (112 kDa) and MCP5-GFP (99 kDa) were detected by Western blot analysis using a GFP-specific monoclonal antibody (Invitrogen, Carlsbad, CA), and the results showed that the full-length fusion proteins were expressed in the detected clones (see Fig. S1b in the supplemental material). The clones that express the fusion proteins were then examined by using a Zeiss Axiostar microscope at a wavelength of 480 nm, and images were captured and processed by using the Axiovision program (Zeiss, Germany). Intense polar clusters were observed at both cell poles in 94% of the cells (47 out of 50 cells) expressing MCP5-GFP (Fig. 1). These results suggest that MCP5 is located at both cell poles of B. burgdorferi. However, only 62% (31 out of 50 cells) of the cells expressing MCP3-GFP had fluorescence at both cell ends. In addition, because many of the cells containing MCP3-GFP fluoresced throughout the cells, there was evidently breakdown of the fusion protein, which is consistent with the Western blotting results (Fig. S1b). Thus, we could not definitively conclude the cellular location of MCP3 by using the GFP fusion.
Fig. 1.
Localizations of MCP3-GFP and MCP5-GFP. The two constructs, as described in Fig. S1 in the supplemental material, were independently transformed into B. burgdorferi. The micrographs were taken under differential interference contrast (DIC) light microcopy or fluorescence microscopy with a fluorescein isothiocyanate (FITC) emission filter (magnification, ×1,000), and the resulting images were then merged. Arrows point to the cellular locations of MCP3-GFP and MCP5-GFP.
The native MCP3 and MCP5 proteins reside at the cell poles of B. burgdorferi.
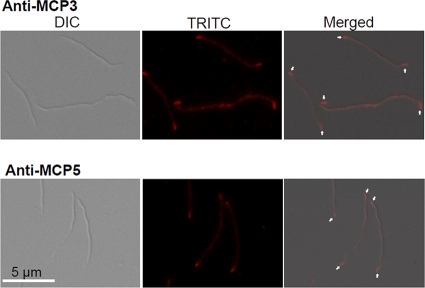
To further confirm the observations with MCP5 described above and to determine the location of MCP3, the cells were analyzed by using IFA. To generate a specific antibody against MCP3 or MCP5, the regions that most likely encode the periplasmic domains of these two proteins (MCP3, 55 to 210 aa; MCP5, 62 to 218 aa) were cloned and overexpressed in pQE30 (Qiagen, Valencia, CA). The recombinant proteins were purified and injected into rats for immunization (24, 25). Western blotting revealed that the obtained antibodies specifically reacted to the target proteins (see Fig. S2 in the supplemental material). IFA was carried out as previously described (34). Briefly, the cells were fixed with methanol, treated with lysozyme, and then probed with antibodies against MCP3 and MCP5. The resulting samples were then incubated with the secondary goat anti-rat Texas Red antibody (Invitrogen). Texas Red images were taken using a Zeiss Axioimager Z1 Axiophot microscope with an excitation filter (541 to 569 nm) and an emission filter (581 to 654 nm). As shown in Fig. 2, brighter loci were observed at both cell poles, and such a pattern was observed in >94% of the cells (47 out of 50) probed with anti-MCP3 antibody and >97% of the cells (39 out of 40) probed with anti-MCP5 antiserum. The results shown here, along with the GFP fusion assay, demonstrate that MCP3 and MCP5 reside at both cell poles.
Fig. 2.
Localizations of MCP3 and MCP5 by using IFA. Wild-type cells were fixed with methanol, stained with either anti-MCP3 or anti-MCP5 antibody, and counterstained with anti-rat Texas Red antibody. The micrographs were taken under DIC light microcopy or fluorescence microscopy with a tetramethylrhodamine isothiocyanate (TRITC) emission filter (magnification, ×1,000), and the resulting images were then merged. Arrows point to the cellular locations of MCP3 and MCP5.
Position of the chemoreceptors relative to the flagellar motors.
To determine the ultrastructure of the chemoreceptors and their spatial relationship to the flagellar motors, cryo-ET analysis was performed as previously described (6, 27). Briefly, the spirochete cells were first deposited onto holey carbon grids and rapidly frozen in liquid ethane. The frozen-hydrated specimens were then imaged by using a Polara electron microscope (FEI Company) equipped with a 4,096- by 4,096-pixel charge-coupled-device (CCD) camera (GMBH, Gauting, Germany). The microscope was operated at 300 kV and ×31,000 magnification, resulting in an effective pixel size of 5.6 Å after 2-by-2 binning. A total of 30 high-quality tomograms were reconstructed through fiducial alignment with the IMOD package (21). A typical reconstruction of a cell end was visualized by using the three-dimensional (3-D) modeling software Amira (Visage Imaging). 3-D segmentation of the PF, outer and inner cell membranes, outer surface proteins, and chemoreceptor arrays was manually constructed. The surface model from B. burgdorferi flagellar motors (27) was computationally mapped back into the original cellular context as previously described (26).
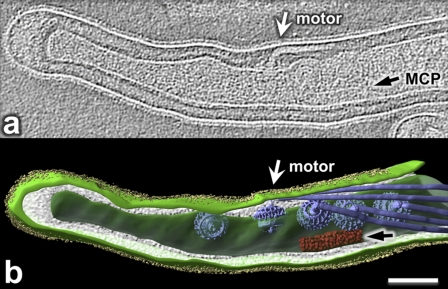
As illustrated in Fig. 3 a and Fig. S3 in the supplemental material, long arrays of chemoreceptors were found in the subpolar regions of approximately 80% of the B. burgdorferi cells (15 out of 19 cells), and the mean distance between the center of the observed arrays and the cell tips was 406 ± 175 nm (range, 172 to 817 nm). Thus, although IFA and GFP analysis indicated that the MCPs were polarly located, the cryo-ET analysis revealed that they were actually subpolarly localized, in agreement with the results of Briegel et al. (6) and those recently described for Treponema pallidum (26). The average size of these arrays was 29.2 ± 1.1 nm in width and 283 ± 91 nm in length (Table 2). The observed arrays appear in clusters of pillar-like densities that extended from the inner membrane into the cell (Fig. 3b and c), which is consistent with what is found in other bacteria, in which these proteins complex with CheA and CheW and form a concave “basal plate” (6, 20, 23, 36, 37).
Fig. 3.
Ultrastructure of chemoreceptor arrays in B. burgdorferi. (a) Presence of chemotaxis receptor (CR) arrays near the tip of the wild-type cell. (b) Zoomed-in view of region b (inset) in panel a reveals the outer membrane (OM), cytoplasmic membrane (CM), peptidoglycan layer (PG), basal layer (CheA and CheW), and cytoplasmic domain of MCPs forming an area of relatively homogeneous density (array). Bar, 100 nm. (c) A cross-section view of the chemotaxis receptors in panel b.
Table 2.
Physical dimensions of the chemoreceptor arrays and the flagellar motors in B. burgdorferi
| Constructs | Cell no.b | Size (nm)e | Distance (nm)d,e |
|---|---|---|---|
| Chemoreceptors | 19 (15) | 29.2 ± 1.1 (width) | 406 ± 175 |
| 283 ± 91 (length) | |||
| Flagellar motorsa | 30 | 13.9 ± 1.8c | |
| 1 | 153 ± 78 | ||
| 3 | 369 ± 106 | ||
| 5 | 607 ± 138 | ||
| 7 | 872 ± 176 |
Motor numbers represent the sequential positions of PF, e.g., 1 is the PF closest to the cell end and 7 is the farthest.
Total number of cells examined (number with chemoreceptor arrays in parentheses).
Diameter of the PF.
The distance from the chemoreceptors or flagellar motors to the cell ends.
Values are means ± standard deviations.
Previous studies showed that the flagellar motors also reside at the subpolar regions of spirochete cells (6, 22). In B. burgdorferi, between 7 and 11 of these structures form an approximate linear arrangement on one side of the cell that is parallel to the axis of the cell (8). We found that the mean spacing between individual motors is approximately 118 nm (range, 102 to 132 nm), which is slightly larger than the previous mean measurement of 90.8 nm (8, 22). In the cells examined, we could detect only 7 motors at a given cell end, because of a limited field of view. To further determine the spatial relationship between the observed arrays and the flagellar motors, the distances from the cell tips to the flagellar motors were measured. Although the distances were quite variable between cells, the flagellar motors were adjacent to the MCPs but not within the line of the MCPs themselves (Fig. 4). The mean distances from the cell tips to the most proximal and the most distal flagellar motors were 153 nm (range, 50 to 326 nm) and 872 nm (583 to 1,276 nm), respectively. As noted above, the mean distance from the MCPs to the cell poles was 406 nm. Thus, the chemotaxis arrays are positioned adjacent to the central region of the flagellar motors. A 3-D model of one cell end illustrates the close proximity of the MCPs to the flagellar motors (Fig. 4; see also the movie in the supplemental material).
Fig. 4.
Cellular architecture of intact B. burgdorferi revealed by cryo-ET. (a) One central slice of a tomographic reconstruction from one organism displays the PF (white arrow) and MCP array (black arrow). (b) A 3-D model was generated by manually segmenting the outer membrane (light green), cytoplasmic membrane (green), flagellar filaments (blue), MCP array (red), and outer surface proteins (Osps) (yellow). 3-D maps of flagellar motors were computationally mapped back into the cytoplasmic membrane (26). Bar, 100 nm.
Conclusion.
Many spirochete species are much slimmer and longer (approximately 0.1 to 0.3 μm in diameter and approximately 10 to 20 μm in length) than other bacteria, and they have two bundles of PF attached near each end of the cells (7, 15). Consequently, these bacteria have swimming modalities that are unique (7, 24, 30). It still remains unknown how spirochetes respond to environmental stimuli and coordinate the rotation of the two bundles of PF and whether or not both ends of a spirochetal cell can be a leading end in response to chemotactic stimuli. In this report, the GFP fusion and IFA analyses showed that MCP3 and MCP5 chemoreceptors form patches that reside at both cell poles of spirochetal cells, suggesting that both ends can sense environmental stimuli during chemotactic responses. The cryo-ET analysis further revealed that the chemoreceptors form arrays in the subpolar regions, where they are parallel to and adjacent to the center of the line of motors (Fig. 4). This geometry enhances rapid transmission, via CheY, of environmental stimuli from the chemoreceptors to the flagellar motors that reside at a given cell end, which is consistent with what is found in other bacteria (33). It remains to be seen how the signal is presumably transmitted to the motors at the other end of the cell. Finally, one intriguing question remains as to what determines the subpolar localization of both the MCPs and the flagellar motors in spirochetes.
Supplementary Material
Acknowledgments
We thank J. Radolf for providing gfp cassettes, S. Samuels for the shuttle vectors, and the Confocal Microscope Facility in the School of Medicine and Biomedical Sciences, the State University of New York at Buffalo, for the assistance in fluorescence microscopy.
This research was supported by Public Health Service AI073354 and AI078958 and American Heart Association grants to C.L., grant AI29743 to N.W.C., grant AI087946 from the National Institute of Allergy and Infectious Diseases (NIAID), and grant AU-1714 from the Welch Foundation to J.L.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Alexander R. P., Zhulin I. B. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ames P., Studdert C. A., Reiser R. H., Parkinson J. S. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armitage J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229–289 [DOI] [PubMed] [Google Scholar]
- 4. Bakker R. G., Li C., Miller M. R., Cunningham C., Charon N. W. 2007. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: flow cytometry-based capillary tube chemotaxis assay. Appl. Environ. Microbiol. 73:1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bardy S. L., Maddock J. R. 2007. Polar explorations recent insights into the polarity of bacterial proteins. Curr. Opin. Microbiol. 10:617–623 [DOI] [PubMed] [Google Scholar]
- 6. Briegel A., et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charon N. W., Goldstein S. F. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spriochetes. Annu. Rev. Genet. 36:47–73 [DOI] [PubMed] [Google Scholar]
- 8. Charon N. W., et al. 2009. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J. Bacteriol. 191:600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eggers C. H., et al. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281–295 [DOI] [PubMed] [Google Scholar]
- 10. Fosnaugh K., Greenberg E. P. 1989. Chemotaxis mutants of Spirochaeta aurantia. J. Bacteriol. 171:606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frank K. L., Bundle S. F., Kresge M. E., Eggers C. H., Samuels D. S. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 185:6723–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fraser C. M., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 13. Ge Y., Old I. G., Saint Girons I., Charon N. W. 1997. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus σ70 promoter. J. Bacteriol. 179:2289–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gestwicki J. E., et al. 2000. Evolutionary conservation of methyl-accepting chemotaxis protein location in bacteria and archaea. J. Bacteriol. 182:6499–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldstein S. F., Buttle K. F., Charon N. W. 1996. Structural analysis of the Leptospiraceae and Borrelia burgdorferi using high-voltage electron microscopy. J. Bacteriol. 178:6539–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goulbourne E. A., Jr., Greenberg E. P. 1981. Chemotaxis of Spirochaeta aurantia: involvement of membrane potential in chemosensory signal transduction. J. Bacteriol. 148:837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hazelbauer G. L., Falke J. J., Parkinson J. S. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hovind-Hougen K. 1984. Ultrastructure of spirochetes isolated from Ixodes ricinus and Ixodes dammini. Yale J. Biol. Med. 57:543–548 [PMC free article] [PubMed] [Google Scholar]
- 19. Kentner D., Sourjik V. 2006. Spatial organization of the bacterial chemotaxis system. Curr. Opin. Microbiol. 9:619–624 [DOI] [PubMed] [Google Scholar]
- 20. Khursigara C. M., Wu X., Zhang P., Lefman J., Subramaniam S. 2008. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 105:16555–16560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kremer J. R., Mastronarde D. N., McIntosh J. R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 22. Kudryashev M., et al. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol. Microbiol. 71:1415–1434 [DOI] [PubMed] [Google Scholar]
- 23. Lefman J., et al. 2004. Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 186:5052–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li C., et al. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc. Natl. Acad. Sci. U. S. A. 99:6169–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li C., Xu H., Zhang K., Liang F. T. 2010. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol. Microbiol. 75:1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J., et al. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 403:546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J., et al. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J. Bacteriol. 191:5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lux R., Sim J. H., Tsai J. P., Shi W. 2002. Construction and characterization of a cheA mutant of Treponema denticola. J. Bacteriol. 184:3130–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maddock J. R., Shapiro L. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723 [DOI] [PubMed] [Google Scholar]
- 30. Motaleb M. A., et al. 2005. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J. Bacteriol. 187:7963–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samuels D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shapiro L., McAdams H. H., Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sourjik V., Armitage J. P. 2010. Spatial organization in bacterial chemotaxis. EMBO J. 29:2724–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sourjik V., Berg H. C. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740–751 [DOI] [PubMed] [Google Scholar]
- 35. Wadhams G. H., Armitage J. P. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 36. Weis R. M., et al. 2003. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 185:3636–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang P., et al. 2004. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J. Microsc. 216:76–83 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.