Abstract
The upstream intergenic regions for each of four genes encoding Ser/Thr kinases, all2334, pknE (alr3732), all4668, and all4838, were fused to a gfpmut2 reporter gene to determine their expression during heterocyst development in the cyanobacterium Anabaena (Nostoc) sp. strain PCC 7120. PpknE-gfp was upregulated after nitrogen step-down and showed strong expression in differentiating cells. Developmental regulation of pknE required a 118-bp upstream region and was abolished in a hetR mutant. A pknE mutant strain had shorter filaments with slightly higher heterocyst frequency than did the wild type. Overexpression of pknE from its native promoter inhibited heterocyst development in the wild type and in four mutant backgrounds that overproduce heterocysts. Overexpression of pknE from the copper-inducible petE promoter did not completely inhibit heterocyst development but caused a 24-h delay in heterocyst differentiation and cell bleaching 4 to 5 days after nitrogen step-down. Strains overexpressing pknE and containing PhetR-gfp or PpatS-gfp reporters failed to show developmental regulation of the reporters and had undetectable levels of HetR protein. Genetic epistasis experiments suggest that overexpression of pknE blocks HetR activity or downstream regulation.
INTRODUCTION
The filamentous cyanobacterium Anabaena (Nostoc) sp. strain PCC 7120 (here called Anabaena) is capable of fixing atmospheric nitrogen in specialized cells called heterocysts. During growth on nitrate- or ammonium-containing medium, Anabaena filaments possess only photosynthetic vegetative cells. When combined nitrogen becomes insufficient in the growth medium, approximately 10% of the vegetative cells differentiate into heterocysts semiregularly spaced along each filament. Genome-wide expression analysis revealed global changes in gene expression in Anabaena filaments upon nitrogen depletion, with upregulation of approximately 10% of chromosomal genes during heterocyst development (5, 6). Several genes related to the regulation of heterocyst development have been identified, including hetF, hetN, hetP, hetR, nrrA, ntcA, patA, and patS (8, 11, 17, 36).
Heterocyst development requires the key regulators NtcA and HetR. The ntcA gene encodes a transcriptional regulator of the cyclic AMP receptor protein (CRP) family that controls expression of genes involved in nitrogen metabolism in cyanobacteria and is involved in the early regulation of heterocyst development (8, 10, 17, 38). The hetR gene encodes a transcription regulator that is essential for heterocyst development (11, 12, 17, 25, 37). Deletion of hetR or mutations that replace amino acids at residues D17, G36, and S179 result in a Het− phenotype (26), and hetR overexpression or an R223W mutation results in a multiple-contiguous-heterocyst (Mch) phenotype (15). The hetR gene is regulated by a complex promoter, which requires HetR, NrrA, and, indirectly, NtcA (3, 4, 21, 22). NtcA upregulates the expression of nrrA during combined nitrogen limitation, and NrrA upregulates the expression of hetR (4). The regulation of ntcA and hetR shows a mutual dependence that requires the two protein phosphatases PrpJ1 and PrpJ2 (13, 22).
Protein phosphorylation, one of the most common posttranslational modifications, plays a pivotal role in the regulation of cellular functions from gene expression and signal transduction to metabolism and movement in both prokaryotes and eukaryotes. Phosphorylation is catalyzed by several classes of protein kinases differing in their modes of regulation and the target amino acid. Serine/threonine kinases are key components of signal transduction systems in eukaryotic organisms. Genome sequences show that many bacterial species, including cyanobacteria, possess eukaryotic-type Ser/Thr kinases (23, 28, 35). However, the possible function of these enzymes in signal transduction pathways during heterocyst development remains unclear. The Anabaena pknE gene encodes a Ser/Thr kinase, and a pknE inactivation mutant produces heterocysts with aberrant appearance and diminished nitrogenase activity, although the mutant has a normal pattern of heterocysts 24 h after nitrogen step-down (34). The pknE mutant initially shows diazotrophic growth, but growth slows after 5 or 6 days and filaments show defective cell morphology. In the wild type, the level of PknE protein drops at 3 h after nitrogen step-down and then slowly increases again back to the original levels (34). Microarray data showed upregulation of pknE by 8 h after nitrogen step-down (5, 6).
In this study, transcriptional gfp gene fusions were used to examine the expression of four genes encoding serine/threonine kinases during heterocyst development. The regulation and phenotypes of inactivation and overexpression strains of one of these genes, pknE, were further characterized.
MATERIALS AND METHODS
Strains and growth conditions.
The Anabaena (Nostoc) sp. strain PCC 7120 and Escherichia coli strains used in this study and their relevant genotypes are summarized in Table 1. Wild-type Anabaena and its derivatives were grown in BG-11 medium, which contains 17.5 mM NaNO3; BG-110, which lacks combined nitrogen; or BG-110 containing 2.5 mM NH4Cl and 5 mM morpholinepropanesulfonic acid (MOPS) (pH 8.0). Cultures were incubated at 30°C under continuous white-light illumination of approximately 75 μmol photons m−2 s−1 (9). Anabaena strains containing shuttle plasmids or integrated suicide plasmids were grown in liquid medium with appropriate antibiotics at the concentrations indicated: neomycin (Nm), 12.5 μg ml−1, or spectinomycin (Sp) and streptomycin (Sm), 1.25 μg ml−1 each. For agar-solidified medium in plates, antibiotic concentrations were doubled. Heterocyst development was induced by nitrogen step-down by transfer of filaments from combined-nitrogen-replete growth medium to BG-110 medium exposed to dinitrogen in air. Plates contained 40 ml of medium solidified with 1.5% agar; for BG-110 plates, the agar was washed to remove nitrogen compounds (29). The heterocyst frequency was scored per a previous description (33).
Table 1.
Anabaena sp. strain PCC 7120 and E. coli strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Anabaena PCC 7120 | ||
| AMC236 | ntcA::Ω Spr/Smr cassette | 32 |
| AMC451 | ΔpatS::Ω Spr/Smr cassette | 32 |
| AMC484 | WTa carrying shuttle plasmid pAM1951 (PpatS-gfp) | 32 |
| AMC485 | WT carrying shuttle plasmid pAM1956 (promoterless gfp) | 32 |
| AMC1256 | WT carrying shuttle plasmid pAM3264 (PhetR-gfp) | 30 |
| AMC1289 | WT hetR replaced by hetRR223W mutant allele | 15 |
| AMC1535 | WT Anabaena sp. strain PCC 7120 | R. Haselkorn |
| AMC1552 | WT carrying shuttle plasmid pAM3853 (PpknE-P-gfp) | This study |
| AMC1553 | WT carrying shuttle plasmid pAM3855 (PpknE-P1-gfp) | This study |
| AMC1554 | WT carrying shuttle plasmid pAM3857 (PpknE-P2-gfp) | This study |
| AMC1555 | WT carrying shuttle plasmid pAM3859 (PpknE-P3-gfp) | This study |
| AMC1556 | AMC236 carrying shuttle plasmid pAM3857 (PpknE-P2-gfp) | This study |
| AMC1557 | AMC236 carrying shuttle plasmid pAM3859 (PpknE-P3-gfp) | This study |
| AMC1558 | UHM103 carrying shuttle plasmid pAM3857 (PpknE-P2-gfp) | This study |
| AMC1559 | UHM103 carrying shuttle plasmid pAM3859 (PpknE-P3-gfp) | This study |
| AMC1560 | WT carrying shuttle plasmid pAM3826 (Pall2334-gfp) | This study |
| AMC1561 | WT carrying shuttle plasmid pAM3828 (PpknE-gfp) | This study |
| AMC1562 | WT carrying shuttle plasmid pAM3830 (Pall4668-gfp) | This study |
| AMC1563 | WT carrying shuttle plasmid pAM3832 (Pall4838-gfp) | This study |
| AMC1566 | WT carrying shuttle plasmid pAM4266 (rpoB-6His) | This study |
| AMC1702 | AMC236 carrying shuttle plasmid pAM1956 (promoterless gfp) | This study |
| AMC1704 | UHM103 carrying shuttle plasmid pAM1956 (promoterless gfp) | This study |
| AMC1759 | WT carrying shuttle vector pAM3919 (PpknE-PmutntcA-gfp) | This study |
| AMC1760 | WT carrying shuttle vector pAM3920 (PpknE-P2mutntcA-gfp) | This study |
| AMC1761 | UHM103 carrying shuttle vector pAM4375 (native-promoter-driven hetR with C-terminal 6His tag) | This study |
| AMC1762 | AMC1761 carrying pAM4012 (pAM1956-pknE-P with Ω Spr/Smr) | This study |
| AMC1763 | Knockout of pknE by insertion of pAM2178 through homologous single recombination in WT strain | This study |
| AMC1764 | WT carrying pAM4378 (PpetE-pknE) | This study |
| AMC1765 | WT carrying pAM4146 (native-promoter-driven pknE) | This study |
| AMC1766 | AMC1761 carrying pAM4171 (native-promoter-driven pknE) | This study |
| AMC1767 | AMC451 carrying pAM4146 (native-promoter-driven pknE) | This study |
| AMC1768 | AMC1289 carrying pAM4146 (native-promoter-driven pknE) | This study |
| AMC1769 | WT carrying shuttle plasmids pAM3264 (PhetR-gfp) and pAM4171 (native-promoter-driven pknE) | This study |
| AMC1770 | WT carrying shuttle plasmids pAM1951 (PpatS-gfp) and pAM4171 (native-promoter-driven pknE) | This study |
| AMC1771 | WT carrying pAM3318 (PpetE-hetRR223W) | This study |
| AMC1772 | AMC1771 carrying pAM4146 (native-promoter-driven pknE) | This study |
| AMC1780 | AMC1763 carrying shuttle plasmid pAM3264 (PhetR-gfp) | This study |
| UHM103 | ΔhetR; Het− (also AMC1537) | 1 |
| E. coli | ||
| AM1359 | DH10B carrying pRL623 and pRL443, for biparental conjugations with Anabaena | 32 |
| AM3626 | BL21(DE3) carrying pGEX-4T1-hetR, for IPTG-inducible expression of GST-HetR | 27 |
| AM3883 | BL21(DE3) carrying pQE30-2339, for IPTG-inducible expression of His-HetR | This study |
| AM3924 | AM4361 carrying pAM1951 (PpatS-gfp), for in vivo HetR activity assay, positive control | This study |
| AM3925 | AM3883 carrying pAM1951 (PpatS-gfp), for in vivo HetR activity assay, positive control | This study |
| AM3926 | AM4361 carrying pAM3857 (PpknE-P2-gfp), for in vivo assay | This study |
| AM3927 | AM3883 carrying pAM3857 (PpknE-P2-gfp), for in vivo assay | This study |
| AM4156 | AM4361 carrying pAM1954 (PrbcL-gfp), for in vivo assay, negative control | This study |
| AM4158 | AM3883 carrying pAM1954 (PrbcL-gfp), for in vivo assay, negative control | This study |
| AM4361 | BL21(DE3) carrying pQE30 vector alone, as negative-control strain for HetR activity | This study |
| AM4370 | BL21(DE3) carrying pGEX-4T1 vector alone, source of negative-control protein GST | This study |
| BL21(DE3) | Expression host and in vivo assay host | Invitrogen |
| DH10B | Cloning host | Invitrogen |
| GeneHogs | Cloning host | Invitrogen |
WT, wild type.
The copper-inducible expression of pknE from the PpetE promoter on shuttle plasmid pAM4378 was determined on filter-sterilized copper-free medium prepared using plasticware, on standard BG-11 medium, which contains 316 nM copper, or medium adjusted to 1 μM copper.
E. coli strains were maintained and grown in LB (Lennox L) liquid or agar-solidified medium at 37°C. Antibiotics and other additions were used, when required, at the concentrations indicated: ampicillin, 100 μg ml−1; chloramphenicol, 17 μg ml−1; kanamycin, 50 μg ml−1; spectinomycin and streptomycin, 50 μg ml−1 each; isopropyl-β-d-thiogalactoside (IPTG), 1 mM; and 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal), 40 μg ml−1. The strains DH10B and GeneHogs were used for DNA cloning and maintenance of plasmid constructs, strain AM1359 was used for biparental conjugations, and strain BL21(DE3) was used for IPTG-inducible protein expression and in vivo reporter assays.
Anabaena genetics.
Shuttle or suicide plasmids were transferred from E. coli to Anabaena by conjugation (7, 19). For biparental matings, the cargo plasmids were transferred by electroporation into the E. coli strain AM1359, which contains the conjugation plasmid pRL443, and the helper plasmid pRL623, which carries a mob (ColK) gene and three methylase genes to protect cargo plasmids from Anabaena restriction enzymes (7, 19).
Strains that contained two different plasmids were grown on standard concentrations of antibiotics to maintain selection for both plasmids. These strains showed normal levels of antibiotic resistance and growth characteristics, which indicate approximately normal copy numbers for each plasmid.
Segregation of pknE mutant chromosomes was confirmed by PCR amplification using primer pairs that flank the insertion site (AMO-1671 and AMO-1824) to detect wild-type chromosomes and using primer pairs that flank a chromosome-plasmid junction (AMO-1686 and AMO-1571) to detect mutant chromosomes (data not shown).
Plasmid constructions.
The plasmids and oligonucleotides used in this study are summarized in Table 2. Plasmid isolation, restriction endonuclease digestions, ligations, and transformation of E. coli were performed according to standard procedures. Total DNA from cyanobacterial strains was isolated as previously described (9). All plasmid constructs were confirmed by DNA sequencing.
Table 2.
Plasmids and oligonucleotides used in this study
| Plasmid or primer name (description) | Primer used, 5′-3′ sequence; reference or source and/or other detailsa |
|---|---|
| pAM505 | Conjugal shuttle plasmid vector (32) |
| pAM1951 (PpatS-gfp) | 32 |
| pAM1956 (promoterless gfp) | 32 |
| pAM2178 | Conjugal suicide plasmid vector (16) |
| pAM3264 (PhetR-gfp) | 30 |
| pAM3826 (pAM1956-all2334) | AMO-1561, GTCGTCGACTCGTACATTCTAGAGCTTG |
| AMO-1562, GGTGGTACCTTGACGACACAGAGATTA | |
| pAM3828 (pAM1956-pknE) | AMO-1563, GTCGTCGACTATCCTCGCTACAGAGGAA |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3830 (pAM1956-all4668) | AMO-1565, GTCGTCGACTTGCCCACCCTAAAGATA |
| AMO-1566, GGTGGTACCGTCTTCCGCTAGATAGGT | |
| pAM3832 (pAM1956-all4838) | AMO-1567, GTCGTCGACGAAGATAGGAGATTGGGGAA |
| AMO-1568, TTCACGGTACCTATTGATGC | |
| pAM3851 (pK18-PpknE) | AMO-1563, GTCGTCGACTATCCTCGCTACAGAGGAA |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3853 (pAM1956-pknE-P) | AMO-1668, GTCGTCGACGCCTATGATAAAGACAGC |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3855 (pAM1956-pknE-P1) | AMO-1669, GTCGTCGACAATAGTCTTACTTTTTCA |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3857 (pAM1956-pknE-P2) | AMO-1670, GTCGTCGACAAGTTCTTAATTTTGGAT |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3859 (pAM1956-pknE-P3) | AMO-1671, GTCGTCGACAGCACGCAATCAAGAG |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3882 (pQE30-alr2339) | AMO-1675, GGAGGATCCAGTAACGACATCGATCTG |
| AMO-1490, GAGGAGCTCTTAATCTTCTTTTCTACCA | |
| pAM3918 (pK18-SDM-PpknE) | AMO-1684, TTAGCACGCAATCAAGAGAAT |
| AMO-1685, TTCCGGAAAGAGAAATTACTCTAAG | |
| pAM3919 (pAM1956-PpknE-PmutntcA) | AMO-1668, GTCGTCGACGCCTATGATAAAGACAGC |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM3920 (pAM1956-PpknE-P2mutntcA) | AMO-1670, GTCGTCGACAAGTTCTTAATTTTGGAT |
| AMO-1564, GGTGGTACCTTCAGTTTCTGTCAGAAAA | |
| pAM4004 (pAM2178-pknE) | AMO-1686, GGAGGATCCATCGGTAAAGTACTGCAA |
| AMO-1767, AAGAAGCTTACGTCTGATTAAATTTTCTGG | |
| pAM4012 (pAM1956-pknE-P with Ω Spr/Smr) | Ω Spr/Smr XbaI cassette from pDW9 cloned into pAM3853 |
| pAM4146 (pAM505-pknE-OE) | AMO-1768, GAGGAGCTCTAGAAAGTGAACCCG |
| AMO-1824, GTCGTCGACTACTAGCGAATATTTACTTCT | |
| pAM4171 (pAM505-pknE-OE with Ω Spr/Smr) | Ω Spr/Smr XbaI cassette from pDW9 cloned into pAM4146 |
| pAM4375 (pAM505-hetR-6His) | AMO-1763, CCCCCCGGGACTTATAATCAAAACAAATG |
| AMO-1764, AGGAGCTCTTAACGATGGTGATGGTGATGGTGATCTTCTTTTCTACCAAACACC | |
| pAM4378 (pAM2770-pknE-OE) | AMO-1762, CATCATATGATCGGTAAAGTACTGCAA |
| AMO-1672, GGTGGTACCTAGCGAATATTTACTTCTG | |
| pGEX-4T1 | IPTG-inducible GST protein overexpression; GE Healthcare |
| pGEX-4T1-hetR | IPTG-inducible GST-HetR protein overexpression (27) |
| Primers for PpknE | AMO-1926, TAGCGTGCTTGCAGTACTTT |
| AMO-1670, GTCGTCGACAAGTTCTTAATTTTGGAT | |
| Primers for PhetR | AMO-1135, TTCTCGAGCAGATAAGTTCCGGATAATAG |
| AMO-1136, TCAGAGCTCCGTAATTTATGGCATATAAC | |
| Primers for noncoding fragment from pAM505 for EMSAb | AMO-367, ACCCGTCGAACTGCGCGC |
| AMO-369, CGCTCTGCTGAAGCCAG | |
| pQE30 | IPTG-inducible overexpression of His-tagged proteins; Qiagen |
Nucleotides in italics indicate a restriction site, and underlined nucleotides indicate a site-directed mutation.
EMSA, electrophoretic mobility shift assay.
Transcriptional fusions to gfp (pAM3826, pAM3828, pAM3830, pAM3832, pAM3853, pAM3855, pAM3857, and pAM3859) were constructed with shuttle plasmid pAM1956. The upstream regions of the four kinase genes all2334, pknE, all4668, and all4838 and different lengths of the pknE upstream region were obtained by PCR. The resulting fragments contained engineered SalI and Acc65I sites and were inserted upstream of the promoterless gfpmut2 gene at the same sites in pAM1956. Reporter plasmid pAM4012 was obtained by inserting the Spr/Smr cassette as an XbaI fragment from pDW9 into the NheI site in pAM3853, which contains PpknE-P-gfp.
The reporter plasmids pAM3919 and pAM3920 with altered potential NtcA-binding sites were obtained by site-directed PCR mutagenesis using FideliTaq DNA polymerase (USB-GE Healthcare). The template for mutagenesis was pAM3851 (10 pg), which was obtained by cloning the PCR-amplified upstream fragment PpknE at Acc65I and SalI sites of pK18. Phosphorylated mutant primers (AMO-1684 and AMO-1685) were used to obtain pAM3918. PCR products were treated with DpnI prior to ligation and transformation into DH10B. The mutagenized upstream region of pknE was PCR amplified from pAM3918 to obtain fragments PpknE-PmutntcA and PpknE-P2mutntcA, which were inserted into pAM1956 at the Acc65I and SalI sites, resulting in gfp transcriptional fusion plasmids pAM3919 and pAM3920.
To construct pAM4004 for single homologous recombination knockout of pknE, a PCR-amplified internal fragment of pknE (453 bp) was cloned into suicide plasmid pAM2178 at the Acc65I and SalI sites.
For copper-regulated overexpression of pknE, the pknE open reading frame (ORF) was PCR amplified and cloned into the NdeI and Acc65I sites of shuttle vector pAM2770 to make pAM4378. To obtain overexpression of pknE from its native promoter, plasmid pAM4146 was constructed by cloning the PCR-amplified pknE gene with its upstream region at SacI and SalI sites of pAM505. pAM4146 was modified by inserting a Spr/Smr cassette from pDW9 at the NheI site to obtain pAM4171 for selection in certain genetic backgrounds.
The plasmid pAM3882 was constructed by inserting a PCR-amplified fragment containing the hetR ORF into the BamHI and SacI sites of pQE30 (Qiagen). The construct contains the hetR gene with an N-terminal 6His tag for overexpression of 6His-HetR in E. coli BL21(DE3).
Plasmid pAM4375 contains hetR with its native promoter fused with a C-terminal 6His tag and was obtained by cloning a PCR-amplified hetR gene and upstream region into SacI and XmaI sites of shuttle plasmid pAM505, with the C-terminal 6His tag added with the reverse primer. This construct complemented the phenotype of a ΔhetR mutant and produced a slightly increased frequency of heterocysts after nitrogen step-down.
Growth determination.
Triplicate flasks containing 100 ml of liquid BG-11 or BG-110 medium were inoculated with 1 ml of actively growing cultures (optical density at 750 nm [OD750], 0.3 to 0.4) for growth rate determination. Appropriate antibiotics were used at standard concentrations. Culture flasks were incubated at 30°C on a shaking platform at 150 rpm under continuous light at 75 μmol photons m−2 s−1. Two-milliliter culture samples were obtained on days 0, 1, 3, 5, and 7 to determine chlorophyll content (20).
E. coli in vivo assay of HetR activation of the pknE promoter.
E. coli strain BL21(DE3) or DH10B carrying one of three reporter plasmids and the hetR expression plasmid pAM3882 or the empty vector pQE30 was grown on LB medium containing ampicillin and kanamycin to maintain selection for both plasmids. The strains were incubated at 22°C, 30°C, and 37°C to an OD600 of 0.6 to 0.7, and then expression was induced with 0.5 mM IPTG for 4 h or longer before samples were analyzed by fluorescence microscopy for green fluorescent protein (GFP) reporter expression.
Microscopy.
Microscopy and photomicrography were carried out using a Zeiss Axioplan II microscope with a Hamamatsu camera (C5810) or a DeltaVision Core imaging system with an Olympus IX71 inverted microscope and Photometrics CoolSNAP HQ2 camera. Photomicrographs were obtained using differential interference contrast (DIC) optics or appropriate fluorescence filter sets. All fluorescence microscopy for an experiment was performed in a single sitting with identical microscope settings. To avoid bleaching, E. coli cells or Anabaena filaments were first framed and focused with DIC optics, and then fluorescence images were captured. The outer polysaccharide layer of developing heterocysts was stained with Alcian blue as described previously (19). Briefly, a solution of 0.5% Alcian blue (Sigma) in 50% ethanol-water was mixed with an equal volume of Anabaena cells and allowed to stain for 10 min before microscopic observations.
Partial purification of HetR-6His from Anabaena.
Cyanobacterial cultures (UHM103, AMC1761, and AMC1766) were harvested 12 h after nitrogen step-down and washed thoroughly with phosphate buffer (20 mM sodium phosphate, 10 mM NaCl, pH 7.6). For extraction of proteins, cells were resuspended in binding buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8) containing lysozyme (1 mg ml−1) and 1 mM phenylmethylsulfonyl fluoride (PMSF) and incubated at room temperature for 30 min. Cells were broken by vortexing (8 min; 1 min on, 1 min off at 4°C) with zirconia beads (Ambion) and glass beads (∼0.5-mm size; Sigma). The crude lysate was incubated on ice for 30 min with Benzonase nuclease (10 U ml−1; Novagen) and then centrifuged at 32,500 × g for 30 min at 4°C. The clear supernatant was bound to nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) in a spin column. After being washed with 20 column volumes of binding buffer containing 30 mM imidazole, the bound proteins were released with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 7.5). Total protein was determined by the Bradford assay (2) with Caliber protein determination reagent (Hoefer, Inc.) and bovine serum albumin as the standard.
SDS-PAGE and Western immunoassay.
Partially purified proteins were resolved by 10% SDS-PAGE, and duplicate samples were subjected to electrophoresis in parallel. One gel was stained with Coomassie brilliant blue G-250, and the other was transferred to a nitrocellulose membrane using a semidry blotting apparatus (Bio-Rad Trans-Blot SD) according to standard procedures. The membrane was blocked in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBS-T) and 5% nonfat dry milk for more than 1 h. The blocked membrane was incubated with penta-His antibody (Qiagen) (1:2,000 dilution in TBS-T with 5% nonfat dry milk) overnight at 4°C. The membrane was then washed twice with TBS-T for 8 min each and incubated for 2 h with anti-mouse IgG secondary antibody conjugated with horseradish peroxidase (1:10,000 dilution in TBS-T with 5% nonfat dry milk). The chemiluminescence signal was developed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). The gel images and chemiluminescent signals were captured with an Alpha Innotech FluorChem HD2 gel documentation system.
DNA gel mobility shift assay.
The upstream promoter region of pknE was PCR amplified from pAM3853 using primers AMO-1926 and AMO-1670 and end labeled with a biotin 3′-end DNA labeling kit according to the manufacturer's recommendations (Thermo Scientific). Similarly, a positive-control probe (PhetR; from pAM4375 using primers AMO-1135 and AMO-1136) and a negative-control probe (309-bp DNA fragment from the multiple cloning site region of pAM505 using primers AMO-367 and AMO-369) were PCR amplified and biotinylated. HetR-DNA complexes were formed at 30°C in 20-μl reaction mixtures containing 2 ng probe and increasing amounts of purified glutathione S-transferase (GST)-HetR (0 to 200 ng) in binding buffer (4 mM Tris-Cl [pH 7.5], 12 mM HEPES [pH 7.5], 50 mM NaCl, 10 mM MgCl2, 12% glycerol) for 30 min. Poly(dI-dC) was added to reaction mixtures at 5 to 25 times the amount of probe DNA; higher concentrations inhibited mobility shifts of positive controls. Purified GST protein (200 ng) was used as a negative control. Samples were separated on a 5% polyacrylamide native gel in 0.5× Tris-borate-EDTA (TBE) buffer at 10 mA of current in a Mini-Protean II (Bio-Rad) apparatus sitting on ice. For detection of biotinylated probes, gels were transferred to a nylon membrane using 0.5× TBE in a semidry blotting apparatus (Bio-Rad Trans-Blot SD) and developed using the Phototope-Star detection kit according to the manufacturer's instructions (New England BioLabs). Chemiluminescent images were collected for 10 to 15 min with an Alpha Innotech FluorChem HD2 gel documentation system.
RESULTS
Two serine/threonine kinase genes show developmental regulation.
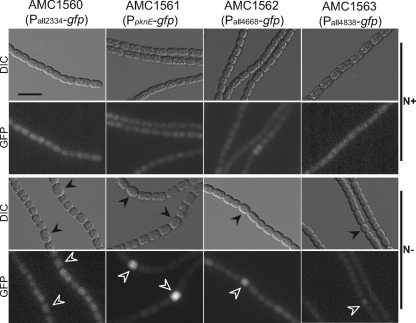
The reporter strains AMC1560, AMC1561, AMC1562, and AMC1563 were obtained by transferring transcriptional reporter plasmids, containing Pall2334-gfp, PpknE-gfp, Pall4668-gfp, and Pall4838-gfp, respectively, into wild-type Anabaena. Each reporter construct contained the upstream intergenic region of a kinase gene fused to a promoterless gfpmut2 reporter gene. All four reporter constructs produced low to moderate levels of GFP fluorescence in vegetative cells when the reporter strains were grown in BG-11 medium (Fig. 1). After nitrogen step-down, there was increased GFP fluorescence in differentiating cells from PpknE-gfp and Pall4668-gfp, but no change in fluorescence from Pall2334-gfp and very low fluorescence throughout the filaments from Pall4838-gfp. We chose to further study the regulation of pknE and its role in heterocyst development. A time course with strain AMC1561 (PpknE-gfp) showed an increase in GFP fluorescence in all cells 3 h after nitrogen step-down, and an emerging pattern of brighter cells was evident by 6 h (see Fig. S1 in the supplemental material). By 12 h, differentiating cells were bright while fluorescence from vegetative cells was reduced. The wild type and the control strain AMC485, which contains pAM1956 carrying promoterless gfp, showed no detectable fluorescence with our GFP filters and settings.
Fig. 1.
Analysis of developmental expression of four genes encoding Ser/Thr kinases. Each strain contains a plasmid carrying the upstream intergenic region of the indicated locus fused to a promoterless gfp reporter gene. Photomicrographs are of filaments of the indicated reporter strains grown on nitrate (N+; top panels) or induced for 24 h on combined-nitrogen free medium (N−; bottom panels). For each sample, the top panel shows a DIC image and the bottom panel shows the corresponding GFP fluorescence image. Photomicroscopy settings were the same for all strains, but increased brightness and contrast were applied to fluorescence images of strains AMC1560 and AMC1563 to more clearly show GFP reporter expression in different cells. Locus alr3732 is the pknE gene. Heterocysts are indicated by arrowheads. Bar, 10 μm.
RNA-seq data from total RNA samples obtained from filaments at 0, 6, 12, and 21 h after nitrogen step-down were consistent with the gfp reporter data, with RPKM (reads per kilobase of CDS [coding sequence] model per million mapped reads) values for pknE of 8.22, 4.70, 31.04, and 10.06, respectively (B. Flaherty, submitted for publication). The RNA-seq data at each time point showed a prominent stable 5′ end for pknE transcripts at −36 relative to the start codon, but significant numbers of staggered overlapping reads trailed upstream from this position, suggesting that this may be a processing site rather than a transcriptional start site. The low RPKM value for pknE at 6 h differs from the increased expression observed at that time point with a gfp reporter because of differences in growth conditions between these experiments. The nitrogen step-down protocol for the RNA-seq experiments resulted in an overall delay in heterocyst development of a few hours compared with the microscopy experiments.
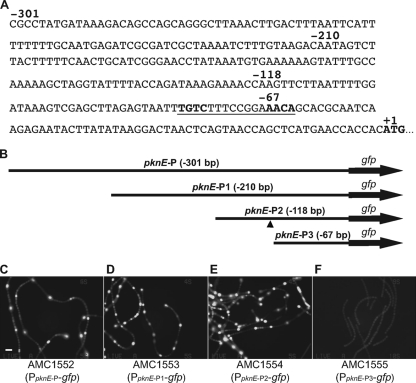
A 118-bp upstream region is required for pknE developmental regulation.
Four fragments containing different lengths of the pknE upstream region were used to drive expression of a gfp reporter gene (Fig. 2). Reporter plasmids containing PpknE-P-gfp (−301 bp), PpknE-P1-gfp (−210 bp), PpknE-P2-gfp (−118 bp), or PpknE-P3-gfp (−67 bp) were transferred into wild-type Anabaena to obtain the reporter strains AMC1552, AMC1553, AMC1554, and AMC1555, respectively. The reporter strain containing PpknE-P3-gfp (−67 bp) showed very low levels of GFP expression 12 h after nitrogen step-down, while the other three strains all showed developmental upregulation (Fig. 2). These data indicate that the cis-regulatory elements required for pknE upregulation are within a 118-bp upstream region.
Fig. 2.
The upstream promoter region of pknE (alr3732), construction of gfp reporter fusions, and GFP reporter expression. (A and B) DNA endpoints indicated in the sequence (A) correspond to the four reporter plasmids that contain different lengths of the pknE upstream region (B) driving expression of a gfp reporter gene. A potential NtcA-binding site is underlined in panel A and marked (▴) in panel B. (C to F) GFP fluorescence micrographs of filaments of the four reporter strains grown without combined nitrogen for 12 h. Bar, 10 μm.
pknE is not upregulated in ntcA or hetR mutant backgrounds.
To determine if either the ntcA or the hetR gene is required for the heterocyst-specific expression of pknE, reporter plasmids containing PpknE-P2-gfp or PpknE-P3-gfp (as an unregulated control) were transferred into ntcA (AMC236) or hetR (UHM103) mutant backgrounds to produce strains AMC1556, AMC1557, AMC1558, and AMC1559. No pattern or increase in GFP fluorescence from PpknE-P2 or PpknE-P3 was observed in either mutant background (not shown). Control strains AMC1702 and AMC1704, carrying a promoterless gfp reporter on plasmid pAM1956, also did not show any detectable GFP fluorescence, as expected. Because NtcA and HetR exhibit interdependent regulation and because the mutants are blocked at early stages of differentiation, these genetic data cannot show direct regulation and indicate only that pknE upregulation requires activation of the heterocyst differentiation pathway.
A potential NtcA-binding site is not required for upregulation of pknE.
The −118 upstream region that provides developmental regulation contains a potential NtcA-binding site, TGT-N10-ACA (14, 17, 24) (Fig. 2). Site-directed mutagenesis was used to modify this site from TGTCTTTCCGGAAACA to TCTCTTTCCGGAATTA in reporter plasmids containing the entire upstream intergenic region (PpknE-PmutntcA) and the −118 upstream region (PpknE-P2mutntcA). These plasmids were transferred into wild-type Anabaena to obtain strains AMC1759 and AMC1760. After nitrogen step-down, both strains showed increased GFP fluorescence in differentiating cells, similar to reporter strains containing the wild-type upstream regions (see Fig. S2 in the supplemental material). These results show that this potential NtcA-binding site is not required for the heterocyst-specific upregulation of pknE.
HetR may directly upregulate pknE.
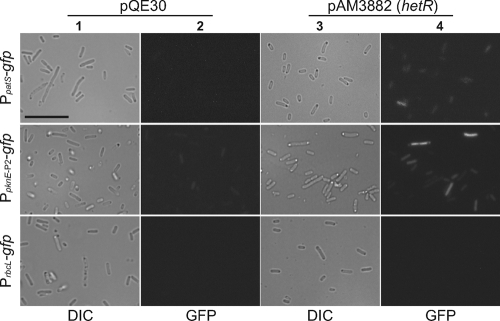
A heterologous transcription assay based in E. coli was used to investigate if HetR can directly upregulate pknE expression. We examined the effect of HetR on GFP fluorescence produced by E. coli BL21(DE3) carrying the reporter plasmid pAM3857, which contains the PpknE-P2 promoter driving gfp expression (Fig. 3). HetR was produced from pAM3882, which is based on the expression vector pQE30. As a positive control, we showed that expression of HetR from pAM3882 in E. coli cells also carrying a PpatS-gfp reporter on pAM1951 produced increased GFP fluorescence, while no fluorescence was detected in a strain carrying pAM1951 and the pQE30 vector (Fig. 3). E. coli cells that carried the reporter plasmid pAM3857 containing PpknE-P2-gfp and the HetR expression plasmid pAM3882 showed greater GFP fluorescence than did cells carrying pAM3857 and the control pQE30 vector. E. coli cells carrying a PrbcL-gfp reporter on pAM1954 and either pAM3882 or pQE30 showed no GFP fluorescence. Experiments performed at 30°C and those performed at 37°C produced similar results, while experiments performed at room temperature (22°C) produced very low GFP fluorescence, and experiments performed in E. coli strain DH10B yielded results identical to those obtained with strain BL21(DE3). These results suggest that HetR can directly activate the expression of the patS and pknE promoters in a heterologous E. coli host.
Fig. 3.
Upregulation of the pknE promoter by HetR in E. coli. Plasmids containing a gfpmut2 reporter gene driven by the upstream promoter regions of patS (pAM1951), pknE-P2 (pAM3857), or rbcL (pAM1954) were introduced into E. coli BL21(DE3) along with either pAM3882, which carries an IPTG-inducible hetR gene, or the empty expression vector pQE30 as a control. The GFP fluorescence images in columns 2 and 4 correspond to DIC images in columns 1 and 3. Photomicroscopy settings were the same for all strains, except that increased brightness and contrast were applied equally to fluorescence images of the strain carrying PpatS-gfp to more clearly show GFP expression in column 4. Bar, 10 μm.
Because HetR can increase expression of the pknE promoter in E. coli, we asked if pknE might show an altered pattern of expression in the presence of extracopy hetR in Anabaena. Anabaena strain AMC1761 carries plasmid pAM4375 in ΔhetR strain UHM103 and produced a mild Mch phenotype; pAM4375 contains hetR-6His driven by its native promoter on a multicopy shuttle plasmid. The PpknE-P-gfp reporter plasmid pAM4012 was conjugated into strain AMC1761 to produce strain AMC1762, which showed patterned GFP reporter expression in differentiating cells and heterocysts similar to that in a wild-type background and did not cause expression in vegetative cells (see Fig. S3 in the supplemental material). As a control, pAM4012 was conjugated into UHM103, and the resulting strain did not show any GFP fluorescence after nitrogen step-down (Fig. S3). These results show that the loss of PpknE-P-gfp reporter expression in the ΔhetR background was complemented by plasmid pAM4375 and that pknE promoter activity was restricted to differentiating cells, which should contain active HetR.
We used DNA gel mobility shift assays to determine if HetR protein interacts in vitro with DNA fragments upstream of the pknE gene. GST-HetR and GST (control) proteins were overexpressed in E. coli and purified (see Fig. S4 in the supplemental material). A positive-control fragment from the hetR upstream region (−136 to −321 bp) formed a complex with GST-HetR (Fig. 4). The pknE upstream fragment PpknE-P2, which produced developmentally regulated expression, also formed a complex with GST-HetR (Fig. 4). A single shifted band suggests the presence of a single HetR binding site in this fragment. In other experiments, a complex with the PpknE-P2 fragment could be detected with 50 and 100 ng of GST-HetR. A nonspecific DNA fragment incubated with 200 ng of GST-HetR did not produce a band shift (data not shown). These data indicate that HetR may directly activate expression of pknE in Anabaena.
Fig. 4.
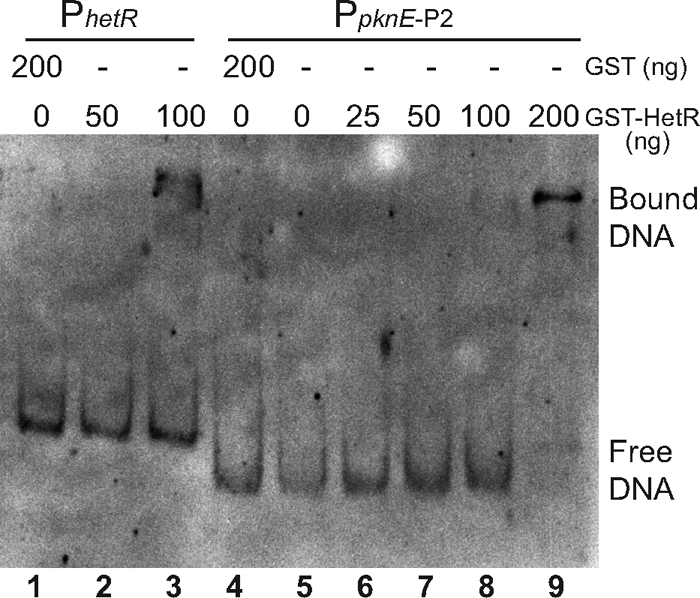
Binding of purified HetR to a pknE promoter fragment in a DNA gel mobility shift assay. The amounts of GST control and GST-HetR protein used in each reaction mixture are shown above the lanes. The biotin-labeled DNA probes were a 154-bp fragment containing the pknE upstream promoter region from PpknE-P2 (Fig. 2) and a 186-bp hetR promoter fragment as a positive control.
Inactivation of pknE caused short filaments with higher heterocyst frequency.
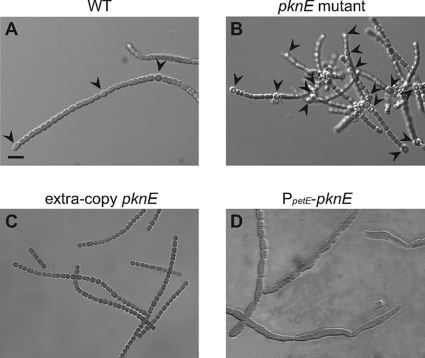
We inactivated the pknE gene by single homologous recombination with suicide plasmid pAM4004, producing strain AMC1763. This pknE mutant strain will produce a truncated form of PknE with only 151 amino acids from its N-terminal region, which lacks PknE's activation loop, ATP-binding pocket, and substrate-binding pocket. PCR analysis of several exconjugants showed the absence of the wild-type gene and the presence of the integrated suicide plasmid, indicating complete segregation.
The pknE mutant strain was similar to the wild type on nitrogen-replete medium, but on nitrogen-depleted medium, it had shorter filaments with an increased heterocyst frequency (14%) compared to the wild type (10%) (Fig. 5). Very short filaments with terminal heterocysts were often observed. Heterocysts formed 20 h after nitrogen step-down and had normal morphology and Alcian blue staining. The mutant strain grew similarly to the wild type on BG-11 medium, but on BG-110 the mutant grew slightly slower and cell pellets appeared lighter green (Fig. 6).
Fig. 5.
pknE knockout mutant and overexpression strains have altered heterocyst development. Photomicrographs of filaments showing heterocyst formation in the wild type (WT) (A) and pknE knockout mutant strain AMC1763 (B) 24 h after nitrogen step-down. Strain AMC1765 contains extra copies of pknE expressed from its native promoter on plasmid pAM4146 and showed no heterocyst formation 24 h after nitrogen step-down (C). Strain AMC1764 contains plasmid pAM4378, which carries pknE expressed from the copper-inducible petE promoter, and showed elongated misshapen cells after 4 to 5 days of growth on nitrate-containing medium (D). All panels are the same magnification. Arrowheads indicate heterocysts. Bar, 10 μm.
Fig. 6.
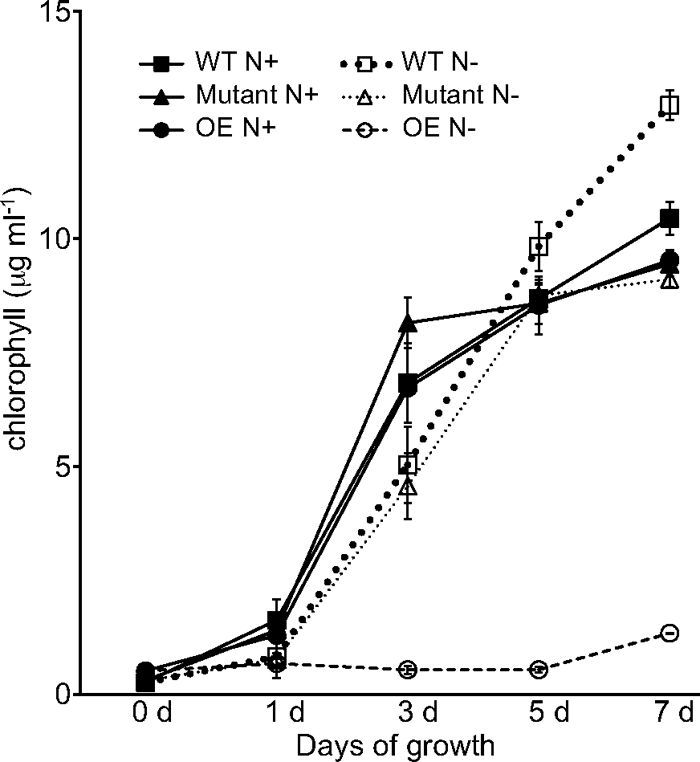
pknE overexpression strain AMC1765 is unable to grow diazotrophically. The growth of Anabaena wild-type (WT), pknE-knockout mutant AMC1763 (Mutant), and pknE overexpression AMC1765 (OE) strains on BG-11 (N+) and BG-110 (N−) is shown. Growth was followed for 7 days by determining chlorophyll (μg ml−1) content. Results are averages of three biological replicates (± standard deviations, n = 3).
Overexpression of pknE inhibited heterocyst development.
The pknE overexpression strain AMC1765 contains pAM4146, which carries extra copies of pknE driven by its native promoter. Shuttle plasmid pAM4146 contains a pDU1 replication origin, and a similar plasmid has been shown to be present in Anabaena at 17 copies per chromosome (18), which would be expected to result in moderate overexpression of PknE. AMC1765 grew similarly to the wild type on BG-11 medium but showed complete inhibition of heterocyst development (Fig. 5) and no growth on BG-110 medium (Fig. 6). AMC1765 filaments were shorter than those of the wild type and cells appeared more granular than the wild type on both BG-11 and BG-110 media. In contrast, strain AMC1764, which contains pAM4378 carrying pknE expressed from the copper-inducible petE promoter, did not completely inhibit heterocyst development but showed a 1-day delay in heterocyst differentiation. The copper-inducible petE promoter is expressed mainly in vegetative cells but may have some activity in differentiating cells (31). On BG-11 medium, AMC1764 formed pleomorphic vegetative cells and exhibited defective cell division that produced abnormally long cells in some filaments (Fig. 5). The vegetative cells were highly granular on BG-11 and BG-110 medium, and BG-110 cultures yellowed and stopped growing 4 to 5 days after nitrogen step-down.
Overexpression of pknE blocked hetR and patS expression.
Because the pknE-overexpressing strain was defective for heterocyst development, we determined the spatiotemporal expression of the hetR and patS genes in this background. Strain AMC1769 carries both PhetR-gfp and pknE on shuttle vectors maintained by antibiotic selection. AMC1769 showed a low level of GFP fluorescence in vegetative cells but failed to show a patterned increase in GFP reporter expression when induced by nitrogen step-down, indicating a block in developmental hetR expression. As a positive control, strain AMC1780, which contains PhetR-gfp in a pknE mutant background, showed increased GFP fluorescence in differentiating cells 6 h after nitrogen step-down, similar to that of a wild-type background (AMC1256).
The same result was obtained with a patS reporter. Strain AMC1770 carries both PpatS-gfp and extracopy pknE. Similar to AMC1769, AMC1770 failed to show a patterned increase in GFP reporter expression in differentiating cells upon nitrogen step-down. A positive-control strain for PpatS-gfp, AMC484, showed the expected developmentally regulated GFP expression.
These data indicated that overexpression of pknE blocked HetR activity; therefore, we measured HetR protein levels. Western immunoblotting experiments showed that HetR-6His protein levels in the pknE-overexpressing strain AMC1766 were undetectable, the same as in the ΔhetR strain UHM103 (Fig. 7). A positive-control strain, AMC1761 (ΔhetR strain containing hetR-6His on pAM4375), showed the expected band of approximately 35 kDa, corresponding to HetR-6His. A parallel gel stained with Coomassie blue showed the absence of the 35-kDa band in strains UHM103 and AMC1766; the band was excised from the AMC1761 lane and further confirmed as HetR by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis.
Fig. 7.
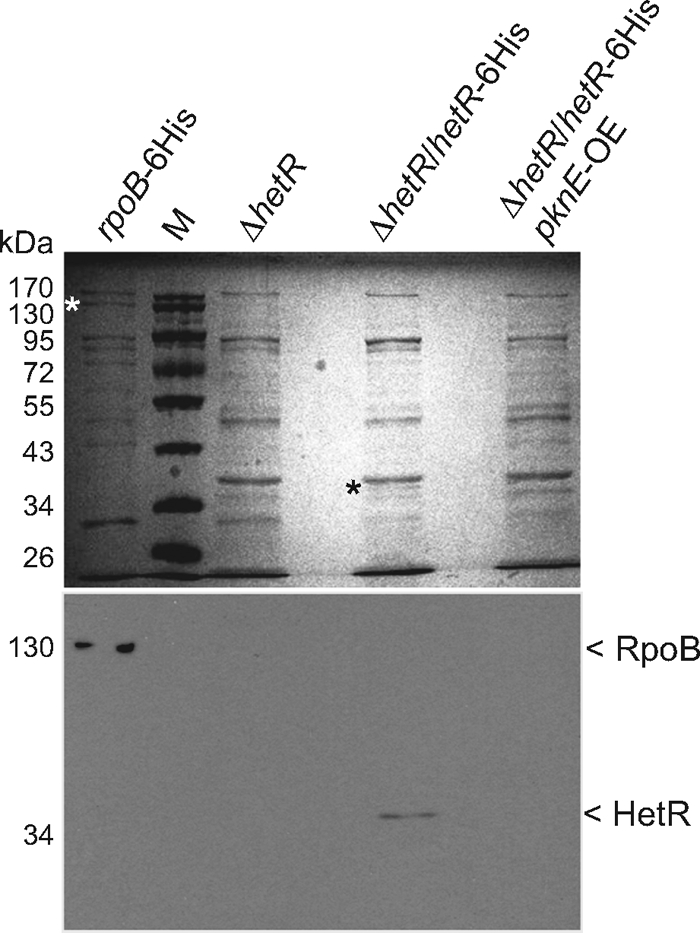
Immunoblot analysis of HetR. Partially purified proteins were probed with penta-His antibody (Qiagen) (lower panel) to detect HetR-6His in AMC1761 (ΔhetR/hetR-6His) and AMC1766 (ΔhetR/hetR-6His/pknE-OE), which overexpresses pknE. Partially purified proteins from UHM103 (ΔhetR) and AMC1566 (rpoB-6His) were used as controls. The samples were resolved on 12% SDS-PAGE gels, and the proteins were stained with Coomassie brilliant blue G-250 (upper panel). Lane M contains the PageRuler prestained protein ladder size marker (Fermentas). Asterisks in the Coomassie blue-stained gel mark the sizes of the bands detected in the immunoblot.
Overexpression of pknE blocks heterocyst differentiation downstream of HetR.
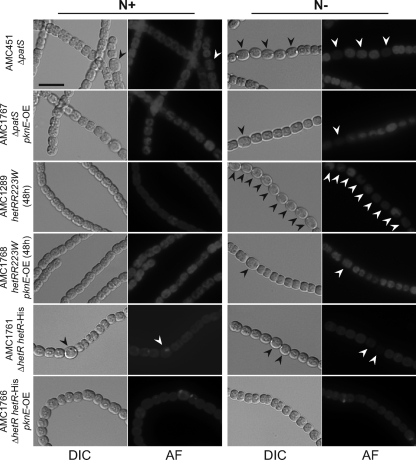
Because hetR is positively autoregulated, the pknE overexpression phenotype could be caused by a block in the production of HetR protein or blocked HetR activity. We used genetic epistasis experiments to help determine where PknE acts in the regulatory pathway (Fig. 8). A pknE-overexpressing plasmid was put into four genetic backgrounds that overproduce heterocysts: AMC451 (ΔpatS), AMC1289 (hetRR223W), AMC1761 (ΔhetR with PhetR-hetR-6His), and AMC1771 (PpetE-hetRR223W) to obtain AMC1767, AMC1768, AMC1766, and AMC1772, respectively. Overexpression of pknE caused inhibition of heterocyst formation on BG-110 medium in all of these strain backgrounds (Fig. 8). Strain AMC1772, which carries hetR driven by a heterologous promoter, produced no heterocysts in 80% of filaments upon nitrogen depletion, indicating that pknE overexpression blocks heterocyst differentiation downstream of hetR transcription.
Fig. 8.
Overexpression of pknE inhibits new heterocyst formation. Upon nitrogen step-down (N−), Anabaena strains AMC451 (ΔpatS), AMC1289 (hetRR223W), and AMC1761 (ΔhetR/hetR-6His) produce increased frequency of heterocysts. Overexpression of pknE (pknE-OE) in these genetic backgrounds inhibited the differentiation of new heterocysts. Note that these genetic backgrounds often produce some heterocysts in the presence of nitrate (N+). DIC images were obtained with 0.006-s exposures and 32% output light intensity. Autofluorescence (AF) images were obtained with 0.02-s exposures and 100% output excitation light intensity. Arrowheads indicate heterocysts. Bar, 10 μm.
The patS mutant strain AMC451 produces 5 to 6% heterocysts in BG-11 medium, and AMC1767 showed the same heterocyst frequency as did AMC451 in BG-11, indicating that PknE's strong inhibition of heterocysts in BG-110 is somehow dependent on the dynamics of regulation associated with nitrogen step-down.
DISCUSSION
Serine/threonine kinases have diverse functions in prokaryotic physiology, including the regulation of stress responses, pathogenicity, development, and cellular differentiation. The genome of Anabaena PCC 7120 possesses 52 genes annotated as encoding serine/threonine protein kinases (23). We selected four of these genes, all2334, pknE, all4668, and all4838, all of which have homologs in the sequenced genomes of other heterocystous cyanobacteria, for further analysis. Microarray data obtained by Ehira and Ohmori (5), as well as our unpublished microarray data, show differential expression of these genes after nitrogen step-down. In the present study, our analysis of spatiotemporal expression of the four kinase genes focused our attention on further studies of pknE because of its strong upregulation in differentiating cells.
We found that a 118-bp DNA fragment upstream of the pknE start codon provided developmentally regulated expression. This upstream region contains a possible NtcA-binding site; however, site-directed mutagenesis of this site indicated that it is not required for upregulation of pknE in differentiating cells (see Fig. S2 in the supplemental material).
In ΔhetR (UHM103) and ntcA::Ω Spr/Smr (AMC236) mutant backgrounds, a PpknE-P2-gfp reporter failed to show a normal patterned increase in GFP fluorescence after nitrogen step-down. Evidence that HetR might be directly involved in pknE regulation was obtained by gel mobility shift assays with purified HetR protein, demonstrating an interaction between HetR and the pknE promoter (Fig. 4). We also used a heterologous in vivo transcription assay in E. coli to support a direct role of HetR in the activation of a PpknE-gfp reporter.
To understand the role of PknE in heterocyst development, we created both knockout mutant and overexpression strains of pknE. The pknE mutant strain formed apparently normal heterocysts at 24 h after nitrogen step-down but showed some filament fragmentation and a slightly increased frequency of heterocysts. Its growth on nitrogen-replete or nitrogen-depleted medium was comparable to that of the wild-type strain over several days (Fig. 6). In contrast, overexpression of pknE from its native promoter completely inhibited heterocyst development and resulted in no growth on nitrogen-depleted medium (Fig. 5 and 6). Overexpression of pknE from the petE promoter produced a somewhat different result, producing delayed heterocyst formation and abnormal cell division and morphology in vegetative cells (Fig. 5).
Overexpression of pknE blocked expression of hetR and patS reporter constructs, which indicates inhibition of an essential early gene product such as HetR. Because the hetR gene is positively autoregulated, the pknE overexpression phenotype could be due to a block of either hetR gene expression or of HetR protein activity. Genetic epistasis experiments indicated that pknE overexpression blocks heterocyst differentiation downstream of HetR because the phenotype was observed even if the hetR gene was expressed from a heterologous promoter. Because pknE is upregulated in heterocysts and its overexpression appears to downregulate HetR activity, PknE may function to regulate the timing of HetR activity and act as a HetR shutoff switch during the later stages of differentiation. PknE could function to downregulate the HetR regulon after differentiation has been completed. The reported autokinase activity of PknE (35) and the timing of its expression during the middle phase of differentiation are consistent with the hypothesis that PknE could act as a switch to shut down genes after their expression is no longer needed. It seems likely that turning off the HetR regulon at the end of the differentiation process would be required for normal heterocyst formation and function.
A model of interactions, either direct or indirect, between HetR and PknE is shown in Fig. 9. Briefly, during the early phase of heterocyst differentiation, HetR induces the expression of pknE. Subsequently, increased PknE levels and activity lead to inhibition of HetR at the later stages of differentiation. In conclusion, the pknE gene is developmentally regulated in Anabaena PCC 7120 and its proper expression is required for normal vegetative cell morphology, control of cell division, and normal heterocyst development.
Fig. 9.
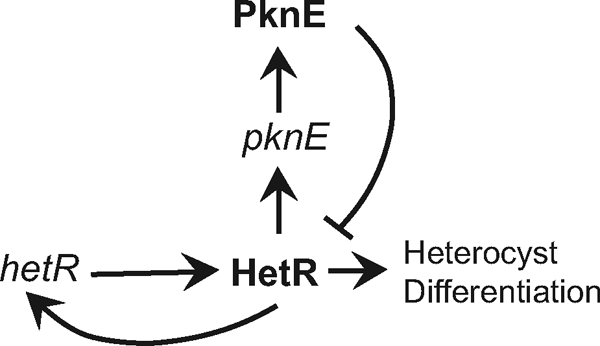
A model showing the proposed regulation of pknE and HetR in differentiating cells. The hetR gene is initially upregulated in differentiating cells and shows positive autoregulation. During the early phase of differentiation, HetR upregulates the expression of pknE. We propose that increased PknE activity during middle or late stages of heterocyst differentiation negatively regulates HetR abundance or activity, which may be required for normal heterocyst differentiation. Arrows indicate positive regulation, and the bar indicates negative regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sean Callahan for providing the ΔhetR strain UHM103 and Britt Flaherty for her unpublished RNA-seq data.
This work was supported by Department of Energy grant DE-FG03-ER020309 and National Science Foundation grant 0925126.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Borthakur P. B., Orozco C. C., Young-Robbins S. S., Haselkorn R., Callahan S. M. 2005. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 57:111–123 [DOI] [PubMed] [Google Scholar]
- 2. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 3. Buikema W. J., Haselkorn R. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U. S. A. 98:2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ehira S., Ohmori M. 2006. NrrA directly regulates the expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 188:8520–8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ehira S., Ohmori M. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692–1703 [DOI] [PubMed] [Google Scholar]
- 6. Ehira S., Ohmori M., Sato N. 2003. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97–113 [DOI] [PubMed] [Google Scholar]
- 7. Elhai J., Vepritskiy A., Muro-Pastor A. M., Flores E., Wolk C. P. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flores E., Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39–50 [DOI] [PubMed] [Google Scholar]
- 9. Golden J. W., Whorff L. L., Wiest D. R. 1991. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:7098–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herrero A., Muro-Pastor A. M., Valladares A., Flores E. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469–487 [DOI] [PubMed] [Google Scholar]
- 11. Higa K. C., Callahan S. M. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol. Microbiol. 77:562–574 [DOI] [PubMed] [Google Scholar]
- 12. Huang X., Dong Y., Zhao J. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101:4848–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jang J., Shi L., Tan H., Janicki A., Zhang C. C. 2009. Mutual regulation of ntcA and hetR during heterocyst differentiation requires two similar PP2C-type protein phosphatases, PrpJ1 and PrpJ2, in Anabaena sp. strain PCC 7120. J. Bacteriol. 191:6059–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang F., Wisen S., Widersten M., Bergman B., Mannervik B. 2000. Examination of the transcription factor NtcA-binding motif by in vitro selection of DNA sequences from a random library. J. Mol. Biol. 301:783–793 [DOI] [PubMed] [Google Scholar]
- 15. Khudyakov I. Y., Golden J. W. 2004. Different functions of HetR, a master regulator of heterocyst differentiation in Anabaena sp. PCC 7120, can be separated by mutation. Proc. Natl. Acad. Sci. U. S. A. 101:16040–16045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khudyakov I. Y., Golden J. W. 2001. Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6667–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar K., Mella-Herrera R. A., Golden J. W. 2010. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2:a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee M. H., Scherer M., Rigali S., Golden J. W. 2003. PlmA, a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J. Bacteriol. 185:4315–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu D., Golden J. W. 2002. hetL overexpression stimulates heterocyst formation in Anabaena sp. strain PCC 7120. J. Bacteriol. 184:6873–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacKinney G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140:315–322 [Google Scholar]
- 21. Muro-Pastor A. M., Olmedo-Verd E., Flores E. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 256:171–177 [DOI] [PubMed] [Google Scholar]
- 22. Muro-Pastor A. M., Valladares A., Flores E., Herrero A. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377–1385 [DOI] [PubMed] [Google Scholar]
- 23. Ohmori M., et al. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271–284 [DOI] [PubMed] [Google Scholar]
- 24. Ramasubramanian T. S., Wei T. F., Golden J. W. 1994. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J. Bacteriol. 176:1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Risser D. D., Callahan S. M. 2009. Genetic and cytological evidence that heterocyst patterning is regulated by inhibitor gradients that promote activator decay. Proc. Natl. Acad. Sci. U. S. A. 106:19884–19888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Risser D. D., Callahan S. M. 2007. Mutagenesis of hetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:2460–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi Y., Zhao W., Zhang W., Ye Z., Zhao J. 2006. Regulation of intracellular free calcium concentration during heterocyst differentiation by HetR and NtcA in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. U. S. A. 103:11334–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang L., Sun Y. P., Chen W. L., Li J. H., Zhang C. C. 2002. Genomic analysis of protein kinases, protein phosphatases and two-component regulatory systems of the cyanobacterium Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 217:155–165 [DOI] [PubMed] [Google Scholar]
- 29. Waterbury J. B., Willey J. M. 1988. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 167:100–105 [Google Scholar]
- 30. Wu X., Lee D. W., Mella R. A., Golden J. W. 2007. The Anabaena sp. strain PCC 7120 asr1734 gene encodes a negative regulator of heterocyst development. Mol. Microbiol. 64:782–794 [DOI] [PubMed] [Google Scholar]
- 31. Wu X., Liu D., Lee M. H., Golden J. W. 2004. patS minigenes inhibit heterocyst development of Anabaena sp. strain PCC 7120. J. Bacteriol. 186:6422–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoon H. S., Golden J. W. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935–938 [DOI] [PubMed] [Google Scholar]
- 33. Yoon H. S., Golden J. W. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 183:2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang C. C., Friry A., Peng L. 1998. Molecular and genetic analysis of two closely linked genes that encode, respectively, a protein phosphatase 1/2A/2B homolog and a protein kinase homolog in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 180:2616–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang C. C., Jang J., Sakr S., Wang L. 2005. Protein phosphorylation on Ser, Thr and Tyr residues in cyanobacteria. J. Mol. Microbiol. Biotechnol. 9:154–166 [DOI] [PubMed] [Google Scholar]
- 36. Zhang C. C., Laurent S., Sakr S., Peng L., Bedu S. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59:367–375 [DOI] [PubMed] [Google Scholar]
- 37. Zhang J. Y., Chen W. L., Zhang C. C. 2009. hetR and patS, two genes necessary for heterocyst pattern formation, are widespread in filamentous nonheterocyst-forming cyanobacteria. Microbiology 155:1418–1426 [DOI] [PubMed] [Google Scholar]
- 38. Zhao M. X., et al. 2010. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. U. S. A. 107:12487–12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.