Abstract
The ability of bacteria to utilize ethanolamine (EA) as a carbon and nitrogen source may confer an advantage for survival, colonization, and pathogenicity in the human intestinal tract. Enterococcus faecalis, a Gram-positive human commensal organism, depends on a two-component signaling system (TCS-17) for sensing EA and regulating the expression of the ethanolamine utilization genes. Multiple promoters participate in eut gene expression in the presence of EA as the sole carbon source and cobalamin (CoB12), an essential cofactor in the enzymatic degradation process. By means of in vivo and in vitro approaches, this study characterized the transcriptional activity identified in the eutT-eutG intergenic region of the E. faecalis eut cluster. Two novel promoters in this region were shown to be active in vivo. The distal P2-1 promoter was associated with a B12 riboswitch that terminated transcription in the presence of CoB12. Transcription elongation from the proximal P2-2 promoter was regulated by antitermination mediated by the phosphorylated form of the response regulator of TCS-17 (RR17). 3′-Rapid amplification of cDNA ends (RACE) analyses of the terminated RNA products allowed precise identification of the hairpin loop structures involved in termination/antitermination. The results uncovered the role of the B12 riboswitch and RR17 in eut gene expression, adding to the complexity of this regulatory pathway and extending the knowledge of possible means of transcription regulation in Gram-positive organisms.
INTRODUCTION
Bacteria of the genus Enterococcus are normal inhabitants of the human intestinal tract (12) but have demonstrated the ability to be significant pathogens by recently emerging as one of the top three causes of nosocomial infections in the United States (19, 27, 43). Infections caused by enterococci include urinary tract infections, intra-abdominal infections, bacteremia, and endocarditis (27). These infections are complicated by a high level of resistance to multiple antibiotics, including vancomycin, which seriously increases the incidence of deaths among individuals infected by enterococci (20). In the majority of Enterococcus infections, E. faecalis is the etiological agent (27).
The switch from commensal living to pathogenesis requires significant changes in gene expression to adapt to a variety of environmental conditions. Bacteria commonly utilize two-component signal transduction systems (TCS) to control gene transcription for physiological adaptation (17). TCS consist of a pair of proteins, a sensor histidine kinase (HK), involved in signal recognition, and a response regulator (RR) that mediates the output of signal sensing, usually by binding to DNA of the target gene and regulating transcription (14).
The E. faecalis genome contains 17 TCS and one orphan RR (17). Among these, TCS-17 was found to be flanked by genes encoding proteins homologous to those described for ethanolamine utilization (eut operon) in Salmonella enterica serovar Typhimurium (Fig. 1) (11). The eut operon is also found in the genomes of several other bacteria that are human commensal and/or associated with food poisoning, including Listeria monocytogenes and Clostridium perfringens (23, 35, 39). Ethanolamine (EA) is found in the human intestinal tract and in processed foods as a constituent of an abundant class of lipids (2, 9, 22, 25). The ability to use EA as a nutrient source may provide these organisms with a selective advantage over other bacteria in the intestinal tract and contribute to their pathogenic potential (6, 22, 39).
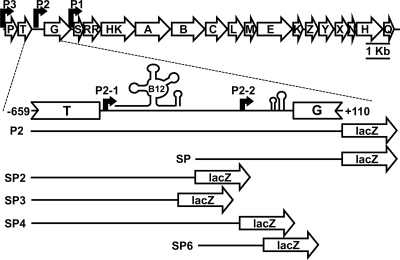
Fig. 1.
Schematic representation of the eut operon in E. faecalis with a magnified view of the eutT-eutG intergenic region. RR and HK indicate the rr17 and hk17 genes (aka eutV and eutW, respectively [15]) encoding the response regulator and histidine kinase, respectively. Shown is the approximate location of the B12 riboswitch, followed by the distal terminator loop, the promoters (bent arrows), and the antiterminator-terminator loops preceding the eutG gene. The contents of the fragments cloned in plasmid pTCVlacSpec and used as templates in the transcription analyses are also illustrated by the lines.
Comparative genomic analysis of genes for EA utilization revealed major differences in the regulation of eut operon expression (39). S. Typhimurium has a single transcription factor, EutR, that is the DNA-binding transcription activator of the eut operon (32). EutR requires both EA and cobalamin (CoB12), a cofactor for the enzyme ethanolamine-ammonia lyase in the EA degradation pathway, for activity (16, 32).
In contrast, in E. faecalis, a two-component signal transduction system, TCS-17, is responsible for activation of the eut operon (11). We previously established that EA acts as the signal ligand that induces autophosphorylation of the histidine kinase of TCS-17 (HK17, aka EF1632 and EutW) that in turn phosphorylates the response regulator/transcription factor, RR17 (aka EF1633 and EutV) (11, 15). The C-terminal domain of RR17 contains a putative ANTAR domain that generally regulates gene expression by an antitermination mechanism (8, 11, 34).
The ability of E. faecalis to grow in medium containing only EA as a carbon source depends on the presence of CoB12 (11). It was not surprising, then, that a B12 riboswitch was identified in the eutT-eutG intergenic region of E. faecalis (Fig. 1) and shown to bind CoB12 (5, 11, 15). Riboswitches are untranslated RNA elements that regulate gene expression in response to the concentration of their ligand in the cell (42). Riboswitches are composed of two functional domains, an aptamer, which binds a given metabolite, and the expression platform, whose structure changes in response to ligand binding, thus controlling transcription termination or translation initiation (42). CoB12 binding to the aptamer domain of B12 riboswitches in Gram-positive bacteria generally leads to premature termination of transcription of the downstream genes which have a physiological relationship to the binding metabolite (40).
Transcription in the eut operon differs substantially between Gram-negative and Gram-positive bacteria. In S. Typhimurium, EutR induces transcription from two promoters: one is located at the beginning of the eut operon and is activated by EutR, and the other is positioned immediately upstream from eutR and is constitutively expressed (32). While the eut operon of E. faecalis is similar to that of S. Typhimurium in that multiple promoters have already been identified, their positioning is different (11, 15). One promoter (P1) was found in the eutG-eutS intergenic region (Fig. 1) (11). Transcription induction from P1 only occurred in medium containing EA and CoB12 (11). The other promoter (P3) was found located upstream from eutP (Fig. 1) (15). Transcription from P3 was detected in serum, but whether its induction was dependent on EA and CoB12 was not investigated (15).
In the current study, we have identified and characterized a third transcriptionally active region (P2) in the eutT-eutG intergenic region (Fig. 1), comprised of two distinct promoters, the distal P2-1 and the proximal P2-2. By means of in vivo and in vitro approaches, we rigorously determined that the B12 riboswitch located immediately downstream from the distal P2-1 promoter caused transcription termination in the presence of CoB12. The phosphorylated form of the response regulator RR17 was required for transcription antitermination at a Rho-independent stem-loop structure located downstream from the P2-2 promoter. The mRNA transcript generated from P2-1 reduced transcription from P2-2 and, as a result, lowered the combined transcriptional activity from the eutT-eutG intergenic region. The results presented extend the results of previous studies (15), clarifying the role of the B12 riboswitch and the RR17 response regulator in eut gene expression and confirming the existence of a unique and complex regulatory pathway for EA utilization in E. faecalis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table 1 and 2, respectively. The initial manipulation of plasmids was performed in Escherichia coli strain DH5α or TG1. E. coli cells were cultured in Luria-Bertani medium (Miller formulation; Difco) at 37°C (DH5α) or 30°C (TG1). The correctness of each plasmid was confirmed by sequence analysis. Transformation of E. faecalis was carried out by electroporation using glycine-treated competent cells (10). E. faecalis strains were grown in Todd-Hewitt broth (THB; Difco) medium at 37°C or 30°C. Spectinomycin (Spec; 150 and 500 μg/ml), chloramphenicol (Cm; 7.5 and 15 μg/ml), and ampicillin (Amp; 100 μg/ml) were used for plasmid selection in E. coli and E. faecalis, respectively.
Table 1.
Enterococcus faecalis strains used in this study
| Strains | Relevant characteristicsa (reference) |
|---|---|
| V583 | Parental strain; clinical isolate (33) |
| rr17 mutant | V583 with nt +85 to +522 of rr17 deleted |
| hk17 mutant | V583 with nt +106 to +1377 of hk17 deleted |
| rho mutant | V583 with nt +46 to +1200 of rho deleted |
Positions are relative to the translational start site of each gene. nt, nucleotides.
Table 2.
Plasmids used in this study
| Plasmids | Relevant characteristicsa | Primers usedb | Reference(s) or source |
|---|---|---|---|
| pLT06 | Integrational vector, contains P-pheS*; Cmr | 24, 37 | |
| pRR17Δ | pLT06 derivative containing flanking regions of RR17: upstream, 663-bp region including 84 bp of the 5′ end of RR17; downstream, 532-bp region including 52 bp of the 3′ end of RR17 | Upstream, RR17del5′BamHI, RR17del5′ overlap; downstream, RR17del 3′overlap, RR17del3′SalI | This study |
| pHK17Δ | pLT06 derivative containing flanking regions of HK17: upstream, 779-bp region including 105 bp of the 5′ end of HK17; downstream, 786-bp region including 57 bp of the 3′ end of HK17 | Upstream, RR17Bam25′, HK17del5′overlap; downstream, HK17del3′SalI, HK17del3′ overlap | This study |
| pRhoΔ | pLT06 derivative containing flanking regions of Rho: upstream, 689-bp region including 45 bp of the 5′ end of Rho; downstream, 788-bp region including 51 bp of the 3′ end of Rho | Upstream, Rhodel5′BamHI, Rhodel5′overlap; downstream, Rhodel 3′overlap, Rhodel3′PstI | This study |
| pML28 | pAT28 containing the aphA-3 promoter | 18 | |
| pML28-RR17 | pML28 derivative containing the RR17 sequence | 11 | |
| pUC19 | High-copy-number cloning vector; Ampr | 44 | |
| pTCVlacSpec | lacZ transcriptional fusion vector; Specr | 11 | |
| pTCV-RR17p | pTCVlacSpec derivative; contains nt −722 to +122 of RR17 | 11 | |
| pP2 | pTCVlacSpec derivative; contains nt −659 to +109 of eutG | EutTEco5′, EutGBam3′ | This study |
| pSP | pTCVlacSpec derivative; contains nt −248 to +109 of eutG | EutTGEco5′, EutGBam3′ | This study |
| pSP2 | pTCVlacSpec derivative; contains nt −659 to −254 of eutG | EutTEco5′, EutTGBam3′ | This study |
| pSP3 | pTCVlacSpec derivative; contains nt −659 to −296 of eutG | EutTEco5′, EutTG2Bam | This study |
| pSP4 | pTCVlacSpec derivative; contains nt −659 to −139 of eutG | EutTEco5′, EutTG3Bam | This study |
| pSP6 | pTCVlacSpec derivative; contains nt −248 to −84 of eutG | EutTGEco5′, EutTG4Bam | This study |
nt, nucleotides.
Sequences of primers are reported in Table 3.
The activity of the eut operon promoters was tested in M9HY medium plus essential vitamins and amino acids as previously described (11, 30). A carbon source (100 mM glucose, 100 mM ethanolamine, or 100 mM glycerol) and/or 40 nM adenosylcobalamin (CoB12; Sigma) was added to the M9HY medium as indicated in the text and figure legends. For transcription analysis, the method for anaerobic growth described by Del Papa and Perego (11) was followed except that the overnight cultures were grown aerobically in THB plus spectinomycin at 37°C without agitation in a flask with ample air space; cells were harvested by centrifugation, washed in M9HY medium, and then diluted 1:15 in the supplemented M9HY.
Construction of markerless rr17 (eutV), hk17 (eutW), and rho deletion strains.
The flanking regions of the rr17 (eutV), hk17 (eutW), and rho genes were amplified by PCR using primers listed in Tables 2 and 3. The upstream and downstream PCR products for each deletion were combined into one fragment by overlapping recombinant PCR. The final products were digested with BamHI and SalI (rr17 and hk17) or PstI (rho) and then cloned into pLT06 (Table 2) (37) to generate pRR17Δ, pHK17Δ, and pRhoΔ (Table 2). The clones were transformed into E. faecalis V583 and grown in THB plus chloramphenicol at the permissive temperature of 30°C, and then the protocol of Thurlow et al. (37) was followed to construct rr17, hk17, and rho markerless deletion strains (Table 1). Clones were screened for their ability to grow in EA plus CoB12 (for rr17 and hk17 strains only), and the presence of the deletion was verified by diagnostic PCR.
Table 3.
Oligonucleotide primers used in this study
| Primers | Sequencea |
|---|---|
| RR17del 5′BamHI | 5′-CCT GTGGAT CCA ACA GCA GCA GAA G |
| RR17del 5′overlap | 5′-ACT CAT ACG TGC GCG TTC GTA CCC TGC TTC |
| RR17del 3′overlap | 5′-GAA GCA GGG TAC GAA CGC GCA CGT ATG AGT |
| RR17del 3′SalI | 5′-ATG CGTCGA CTT GAC TAG TTG |
| RR17Bam25′ | 5′-GGG AGGATC CAA GCG GTG ATT GAA GGT TT |
| HK17del5′overlap | 5′-GGT TTT AGT TCC CTG ATA GAG TGT CGT CGA AG |
| HK17del 3′overlap | 5′-CTT CGA CGA CAC TCT ATC AGG GAA CTA AAA CC |
| HK17del 3′ SalI | 5′-GGC TTG TCG ACC AAG GTG CAA GGT TAG C |
| Rhodel5′BamHI | 5′-GGA TGG GAT CCG GAT TGA AGG AGT CGA AG |
| Rhodel5′overlap | 5′-CAA ACG AAA CAT CAT GGA ATA ACG TAC TGT TTT CCA AT |
| Rhodel3′overlap | 5′-ATT GGA AAA CAG TAC GTT ATT CCA TGA TGT TTC GTT TG |
| Rhodel3′PstI | 5′-TAC TGC AGT ATG CTT GTA CGG TCT CCA T |
| EutTEco5′ | 5′-CAT CAG AAT TCG TTT AAA TTG GAC AG |
| EutGBam3′ | 5′-CAG GGA TCC CCA TGA ATG GAT CTG TG |
| EutTG-Eco5′ | 5′-TTG TAG AAT TCC AAT TAA TGG CAC GAG GA |
| EutTG Bam3′ | 5′-TTG GGGATC CAC AAA TAT CTT GGG CTG GAT TTT CA |
| EutTG2Bam | 5′-CGT TGGATCCAA ACA TCC TGT TTT GCC T |
| EutTG3Bam | 5′-TTA AGGATCCGT CAA TAA AAA AAG GGA ATA TAG C |
| EutTG4Bam | 5′-CTT CGGATCCTA TAC GCC ATT GTA CAC G |
| dRR17p RNA adapter | 5′-TGC ACG GTT ACG ATG CGC CCA GGA TCC GAC-idTb |
| dRR17p RT 3′ | 5′-AGT CGG ATC CTG GGC GCA TCG TAA CCG TGC A |
| dRR17pA RT 5′ | 5′-CAT CAG AAT TCT GAA TGG AAG CCA GTG AGA A |
| dRR17pB RT 5′ | 5′-CAT CAG AAT TCT CGT GTA CAA TGG CGT ATA C |
Restriction sites are in boldface.
idT, inverted deoxythymidine.
Complementation study of the rr17 strain.
pML28-RR17 (11) and pML28 (18) (Table 2) were transformed into the V583 and rr17 strains (Table 1). The strains were grown as described above for transcription analysis in M9HY medium supplemented with EA and CoB12.
β-Galactosidase assays for transcription analysis.
A series of overlapping fragments (P2, SP, SP2, SP3, SP4, and SP6) spanning the eutT-eutG intergenic region (Fig. 1) were amplified by PCR using primers listed in Tables 2 and 3. PCR products were digested with EcoRI and BamHI and then cloned into pTCVlacSpec (Table 2) (11). The plasmids were transformed into the V583, rr17, and hk17 strains (Table 1). Strains carrying the promoter fusion plasmids were cultured as described above in supplemented M9HY medium. Duplicate samples were withdrawn hourly for a period of 8 h, and the optical density at 525 nm (OD525) was measured. The collected samples were assayed for β-galactosidase activity using a protocol similar to that described by Ferrari et al. (13) with the following modification: the cells were resuspended in Z buffer, and 0.1-mm zirconia/silica beads (Biospec) were added in a 1:4 ratio of beads to buffer. Cells were lysed by shaking in 96-deep-well plates using a Mini-Beadbeater 96 (Biospec) for 9 min in 3-min intervals. The plates were placed on ice between intervals. After lysis was completed, the beads were allowed to settle for 10 min and the samples were transferred to a microtiter assay plate. Assay reagent volumes, as compared to those described by Ferrari et al. (13), were adjusted for use in microtiter plates. The OD525 and OD420 for each set of duplicate samples were averaged, and the mean values were used to calculate the Miller units (29). Each set of experiments was carried out at least twice to ensure reproducibility.
In vitro termination assays with CoB12.
Templates (P2, SP2, SP3, and SP4) (Fig. 1) for the in vitro termination assays were generated by PCR with primers listed in Tables 2 and 3. The PCR products were purified using a QIAquick PCR purification kit (Qiagen). Synchronized in vitro termination assays with CoB12 were performed using a method similar to that of Sudarsan et al. (36), with some modifications. In brief, two 50-μl reaction mixtures containing 1× transcription buffer (40 mM Tris-HCl, pH 8, 10 mM MgCl2, 0.1 mM EDTA, pH 8, 50 mM KCl), 70 nM PCR template, 1× initiation nucleoside triphosphates (NTPs) (1 μM ATP, 1 μM GTP, 0.4 μM UTP [lithium salt; Roche Applied Science]), 10 μg/ml bovine serum albumin, 1 mM 1-4-dithiothreitol, 30 U RNaseOut (Invitrogen), 66 nM [α-32P]UTP (3,000 Ci/mmol, 10 mCi/ml) (Perkin Elmer), and 68 nM E. coli RNA polymerase (Epicentre) were set up for each template and incubated for 10 min at 37°C. Elongation was initiated by immediately adding 6.25 μl of 50 mM CoB12 or diethylpyrocarbonate (DEPC)-treated water and 6.25 μl of elongation mix (60 μl of elongation mix contained 6 μl of 10× transcription buffer, 12 μl of 20 mg/ml heparin [grade 1-A; Sigma], 6 μl each of 1 mM ATP, CTP, GTP, and UTP, and 18 μl of DEPC-treated water). The reaction mixtures were incubated at 37°C in the dark, and 25-μl samples were withdrawn after 15 min and 2 h. To stop the reaction, 25 μl of DEPC-treated water and 5 μl of 0.5 M EDTA, pH 8, were added. Samples were extracted once with phenol and three times with chloroform and then ethanol precipitated using 10 μg of salmon sperm DNA as a carrier. The resulting pellets were resuspended in 10 μl of DEPC-treated water plus 10 μl of urea loading dye (10 M urea, 0.1% bromophenol blue, 1× Tris-acetate-EDTA [TAE] buffer). The samples (20 μl), along with 15 μl of the marker (described below), were resolved on a denaturing 7 M urea, 6% PAGE (29:1; Fisher) gel in TAE buffer. The gel was dried, exposed to a screen, and then scanned on a Storm 840 phosphorimager (Molecular Dynamics) for analysis with ImageQuant software.
Riboladder 100b RNA standard (Fisher), used as molecular weight marker, was dephosphorylated with rAPID Alkaline Phosphatase (Roche). T4 polynucleotide kinase (New England BioLabs) was used to label the ladder with [γ-32P]ATP (6,000 Ci/mmol, 150 mCi/ml) (Perkin Elmer). The radiolabeled RNA ladder was treated as described above for the in vitro transcription reactions and also resuspended in urea loading dye.
In vitro termination assays of SP with RR17 and HK17.
The SP template was generated by PCR with primers listed in Tables 2 and 3. The purification of the HK17 and RR17 proteins was described by Del Papa and Perego (11). Protein phosphorylation was carried out in a 50-μl reaction mixture containing 3 μM HK17 and 22.5 μM RR17 in 1× phosphorylation buffer A (11). The reaction was initiated by the addition of 20 μM EA and 2 mM ATP (Roche). Reaction mixtures were incubated for 1.5 h at room temperature. For the controls without HK17 and/or RR17, protein storage buffer (11) was added in place of the protein. For the unphosphorylated sample, EA and ATP were not added to the reaction mixture. In vitro termination assays were carried out as described above with CoB12, except that 10 μl of the phosphorylation reaction mixture was added along with the elongation mixture.
3′-RACE of SP4 and SP.
3′-Rapid amplification of cDNA ends (RACE) experiments were performed as described by Argaman et al. (3) with some changes. PCR-amplified SP4 and SP fragments were transcribed in vitro as described above for the termination assay, with 5 mM CoB12 or RR17/HK17, respectively. The RNA products were phenol extracted and separated on a 7 M urea, 6% PAGE (29:1) gel. The 3 main RNA products of the reaction on SP4 and the 2 RNA products for SP were excised from the gel and then passively eluted in 1× TE (10 mM Tris-HCl, pH 8, 1 mM EDTA) for 24 h with one change of buffer. The elutions were ethanol precipitated, and the concentrated RNAs were dephosphorylated with rAPid alkaline phosphatase (Roche Applied Science). The RNAs were ligated with T4 RNA ligase (NEB) to a primer, dRR17p RNA adapter (Table 3), that had been phosphorylated on the 5′ end with T4 polynucleotide kinase. The RNA/DNA hybrid was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen) using oligonucleotide primer dRR17p RT 3′ (Table 3). The cDNA was amplified by PCR using primers dRR17p RT 3′ and dRR17pA RT 5′ (for SP4) or dRR17pB RT 5′ (for SP) (Table 3) and cloned into pUC19 (Table 2), and then two clones were sequenced for each RNA product to determine the 3′ end of the transcripts.
RESULTS
EA- and CoB12-dependent transcription activity from two promoters in the eutT-eutG intergenic region.
Previous studies identified a promoter region (P1) upstream from the genes encoding the RR17 response regulator and HK17 histidine kinase in the eutG-eutS intergenic region (Fig. 1) (11). Because the eut gene cluster extends upstream from eutG, it was predictable that additional promoters existed (11). One potential promoter site was the eutT-eutG intergenic region, which is the largest intergenic region in the cluster (Fig. 1).
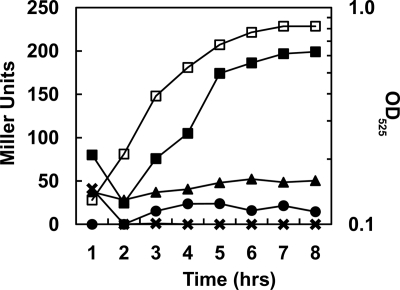
To determine whether the eutT-eutG intergenic region was transcriptionally active, a fragment spanning from within the eutT gene to within the eutG gene (Fig. 1) was fused to the promoterless lacZ reporter gene in vector pTCVlacSpec (Table 2) to produce plasmid pP2. The pP2 plasmid was transformed into E. faecalis V583, and the cells were analyzed for transcriptional activity by means of β-galactosidase assays. The results showed that transcription from the fragment in plasmid pP2 occurred but only in medium supplemented with EA and CoB12 (Fig. 2). When only one or none of the inducers was present, transcription was comparable to the background level (∼5 to 20 Miller units) obtained with vector pTCVlacSpec alone (11; data not shown). The presence of glucose in the medium repressed the transcription induced by EA and CoB12 (Fig. 2), indicating that P2 was carbon catabolite repressed, as was also shown for the P1 promoter (11) and for the main promoter of the eut operon in Salmonella (32).
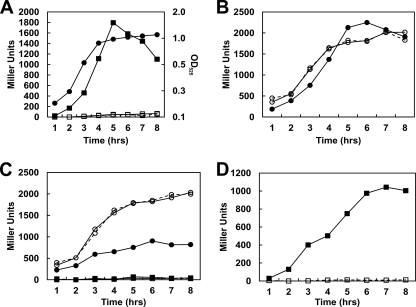
Fig. 2.
Transcription analysis of promoter P2. Time courses of β-galactosidase activity were carried out on E. faecalis V583 carrying the pP2 plasmid. M9HY medium was supplemented with 100 mM EA and 40 nM CoB12 (▪), 100 mM EA ( ), 40 nM CoB12 (•), or 100 mM glucose, 100 mM EA, and 40 nM CoB12 (▴). A representative growth curve in medium containing EA and CoB12 is shown (□), and the OD525 is indicated on the right axis.
), 40 nM CoB12 (•), or 100 mM glucose, 100 mM EA, and 40 nM CoB12 (▴). A representative growth curve in medium containing EA and CoB12 is shown (□), and the OD525 is indicated on the right axis.
A B12 riboswitch was predicted to be downstream from eutT (nucleotides −455 to −278 from the translational start of eutG) by the RibEx analysis program (P = 3.38 × 10−10) and by Barrick and Breaker (1, 5). The location of the riboswitch had subsequently been corroborated by Fox et al. with in-line probing experiments that proved the binding of CoB12 (15). In general, B12 riboswitches in Gram-positive organisms regulate the transcription of downstream genes through the stabilization of a terminator hairpin upon binding of CoB12 (40). An exception is a B12 riboswitch in Listeria monocytogenes that acts as a terminator for upstream genes (38). In order to determine whether the promoter activity in the eutT-eutG intergenic region was upstream or downstream from the riboswitch, the sequence of the eutT-eutG intergenic region was analyzed with the Softberry BPROM program. Analysis predicted two possible promoter sites, one located 470 bp upstream from the eutG translational start codon (TTGCTT-17 bp-TAAATT), hereinafter referred to as P2-1, and the other located 130 bp upstream from eutG (TTGACA-17 bp-TACTCT), hereinafter referred to as P2-2 (Fig. 1). Thus, P2-1 is upstream from the B12 riboswitch, while P2-2 is downstream from the riboswitch and immediately upstream from a potential weak antiterminator hairpin (nucleotides −83 to −53 upstream from eutG), as well as a strong, Rho-independent terminator (nucleotides −69 to −14 upstream from eutG) (Fig. 1; also see Fig. 8) predicted by RibEx and also proposed by Fox et al. (1, 15, 26).
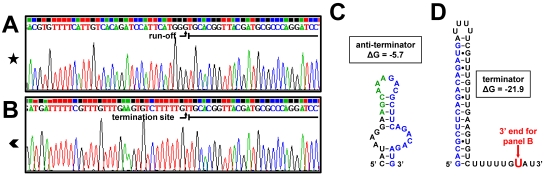
Fig. 8.
Identification of the 3′-end nucleotides of the in vitro RNA transcription products obtained from promoter P2-2. The RNA products of the in vitro reactions carried out on the SP template containing the P2-2 promoter (see Fig. 7A) were purified from the gel and used in 3′-RACE reactions as described in Materials and Methods. Underlined nucleotides in the sequencing graphs indicate the dRR17p RNA adapter sequence. Symbols refer to the RNA products in Fig. 7A. (A) Runoff RNA (★); shown are nucleotides +78 to +110 downstream from the translational start of eutG. (B) Terminated RNA ( ); shown are nucleotides −55 to −23 upstream from eutG. (C) Potential weak antiterminator hairpin. (D) Terminator hairpin. The structures in panels C and D are located upstream from eutG and were predicted by RibEx (1). The energy of the antiterminator and terminator hairpin is expressed in kcal/mol. Nucleotides in blue are shared by the antiterminator and terminator structures. Nucleotides in green are the proposed recognition sequence for RR17.
); shown are nucleotides −55 to −23 upstream from eutG. (C) Potential weak antiterminator hairpin. (D) Terminator hairpin. The structures in panels C and D are located upstream from eutG and were predicted by RibEx (1). The energy of the antiterminator and terminator hairpin is expressed in kcal/mol. Nucleotides in blue are shared by the antiterminator and terminator structures. Nucleotides in green are the proposed recognition sequence for RR17.
Transcription from P2-1 is repressed by CoB12.
To test whether the two potential promoters, P2-1 and P2-2, were active, the P2 fragment was subcloned into pTCVlacSpec to create pSP2 and pSP (Table 2 and Fig. 1). The activity from these constructs was also assayed by β-galactosidase activity. To focus on the effects of EA, CoB12, and TCS-17 on transcription, glycerol was added to the bacterial cultures as a carbon source to allow some growth even in the absence of EA. Glycerol was determined to have no effect on transcription and, as expected, did not cause carbon catabolite repression (data not shown).
The SP2 fragment in plasmid pSP2 includes the putative promoter P2-1 and the B12 riboswitch (Fig. 1). In the strain carrying this construct, a significant level of transcription was observed in glycerol-only medium, while the presence of EA plus CoB12 or CoB12 alone reduced transcription to a background level of ∼250 Miller units (Fig. 3A). This result suggested that the SP2 fragment contains an active promoter and the entire B12 riboswitch, including the expression platform, and this likely functions as a standard B12 riboswitch, causing termination of transcription when CoB12 is present (40). However, a background level of ∼250 Miller units, which is 10 times higher than the activity generated by plasmid pTCVlacSpec, suggested that considerable transcription read-through occurred even in the presence of CoB12.
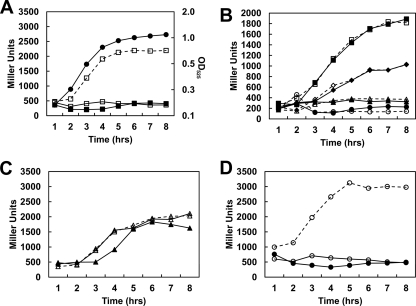
Fig. 3.
Transcription analysis of promoter P2-1. (A, C, and D) Time courses of β-galactosidase activity were carried out on E. faecalis V583 carrying plasmids pSP2 (A; squares), pSP3 (C; triangles), and pSP4 (D; circles); dashed lines with open symbols represent cultures grown in 100 mM glycerol; solid lines with closed symbols are results for cultures grown in 100 mM glycerol, 100 mM EA, and 40 nM CoB12; and solid lines with open symbols are results for cultures grown in 100 mM glycerol and 40 nM CoB12. Growth cultures and assays whose results are shown in panels A, C, and D were performed at the same time, but the graphs are separated for clarity. A representative growth curve in medium containing glycerol, EA, and CoB12 is shown in panel A with closed circles, and the OD525 is indicated on the right axis. (B) Transcription activities from pSP2 in strain V583 compared to those in the rr17 strain. Growth conditions were as follows: 100 mM glycerol, 100 mM EA, and 40 mM CoB12 (circles); 100 mM glycerol and 40 nM CoB12 (triangles); 100 mM glycerol and 100 mM EA (diamonds); 100 mM glycerol (squares); solid lines with closed symbols represent results for V583; dashed lines with open symbols represent results for the rr17 strain.
To determine whether the RR17 response regulator was controlling transcription from P2-1, pSP2 was assayed in the rr17 mutant strain (Table 1). Equivalent levels of transcription occurred under all growth conditions for both the parental V583 and the mutant strain, indicating that RR17 had no role in regulating P2-1 (Fig. 3B). Interestingly, EA caused a decrease in the level of transcription (2-fold) which was not dependent on the presence of CoB12 (Fig. 3B). The decrease was not due to EA's role as a signal for HK17 (11), since the absence of RR17 did not alter the level of transcription. Thus, EA seems to act by an unknown mechanism to regulate transcription from P2-1 independently of the TCS-17.
To determine the sequence boundaries of the riboswitch, two other clones, pSP3 and pSP4 (Table 2 and Fig. 1), were constructed and V583 strains carrying these constructs were assayed for β-galactosidase activity. Notably, the 3′ end of the SP3 fragment is the predicted last base-paired nucleotide of the B12 riboswitch (5, 15) (see Fig. S1 in the supplemental material). With the pSP3 construct, constitutive transcription from P2-1 was detected under all growth conditions (Fig. 3C). Therefore, the CoB12 repression was lost, indicating that an element in the 42 nucleotides that is missing in SP3 with respect to the sequence of SP2 is the basis for the CoB12 repression. Notably, a clone with a fragment that is only 12 nucleotides longer than SP3 showed repression by CoB12 equivalent to that seen with plasmid pSP2 (data not shown). There are no obvious Rho-independent terminators in this small region between the end of the B12 riboswitch and the end of the SP2 fragment. When the SP2 construct was tested in a rho deletion strain, there were no differences in the amount of transcription compared to the level of transcription in V583, indicating that termination was not occurring by a Rho-dependent mechanism (data not shown).
An additional construct, called SP4, consisted of a fragment longer than the SP2 fragment at the 3′ end which, nevertheless, did not include the −10 region of the P2-2 promoter (Fig. 1). With the pSP4 construct, transcription activity was similar to that of pSP2 (Fig. 3D). These results confirmed that the whole B12 riboswitch, aptamer domain and expression platform, was within the SP2 fragment.
Overall, these in vivo results indicated that the P2-1 region identified in silico contains a promoter that is active in vivo and that the riboswitch blocks downstream transcription in the presence of CoB12.
Promoter P2-2 is induced by EA plus CoB12 and requires TCS-17 for transcription.
The other subclone of the P2 fragment, pSP, included the putative promoter P2-2 and the Rho-independent terminator hairpin that is upstream from eutG (Fig. 1). β-Galactosidase assays carried out on the V583 strain harboring plasmid pSP showed that EA alone was not sufficient to induce transcription from the P2-2 promoter, as CoB12 was also required (Fig. 4A and data not shown). This was not due to the requirement for CoB12 for growth (11), because there was glycerol in the medium and the cells did not have to rely on EA as the only carbon source. A similar requirement was also observed for the P1 and P2 (the combined P2-1 and P2-2) promoters (data not shown). Comparison of the transcription from the P2-2 promoter on pSP and the combined activities of P2-1 and P2-2 on the pP2 construct (Fig. 4A and 2) showed a 5.5-fold reduction in the level of transcription, indicating the existence of a negative regulatory effect on the overall level of transcription occurring from the eutT-eutG region.
Fig. 4.
Transcription analysis of promoter P2-2. (A to C) Time courses of β-galactosidase activity were carried out for E. faecalis strains carrying plasmids pSP (A; squares) and pSP6 (B; circles). Panels A and B show the results obtained with the parental strain V583, and panel C shows the results obtained with the rr17 mutant strain. Dashed lines with open symbols are results for cultures grown in 100 mM glycerol; solid lines with closed symbols are results for cultures grown in 100 mM glycerol, 100 mM EA, and 40 nM CoB12; solid lines with open symbols are results for cultures grown in 100 mM glycerol and 40 nM CoB12. The assays for both strains were performed at the same time, but the graphs are separated for clarity. A representative growth curve in medium containing glycerol, EA, and CoB12 is shown in panel A with closed circles, and the OD525 is indicated on the right axis. (D) Results of transcription activity from plasmid pSP carried by the hk17 mutant strain; solid line with closed symbols shows the results for V583; dashed line with open symbols shows the results for the hk17 strain.
Since EA is the signal for autophosphorylation of HK17 and subsequent activation by phosphoryl transfer to RR17 (11), the pSP construct was tested for β-galactosidase activity in the rr17 and hk17 mutant strains. Little or no transcription occurred in the rr17 (Fig. 4C) or hk17 strains (Fig. 4D) even when both EA and CoB12 were present. This suggested that, because EA is required as an inducer in vivo and is the activating ligand of HK17 in vitro, phosphorylated RR17 was necessary for transcription elongation from P2-2. It should be noted that the growth defect caused by the deletion of rr17 when cells were grown in M9HY supplemented with EA and CoB12 was complemented by a replicative plasmid expressing RR17; therefore, there were no polar effects on the downstream genes of the eut cluster due to the markerless deletion (see Fig. S2 in the supplemental material; also data not shown).
The RR17 protein contains a C-terminal ANTAR domain (11). The AmiR protein from Pseudomonas aeruginosa, which has a similar ANTAR domain, binds to an RNA sequence upstream from a strong, Rho-independent terminator that results in antitermination of transcription (41). Bioinformatic analysis by Fox et al. of the sequence preceding the terminator hairpin in the amidase operon (regulated by AmiR) and the eut operon of Enterococcus, Listeria, and Clostridium revealed a consensus sequence that was predicted to be the binding site of the ANTAR domain-containing proteins (15).
To determine whether the terminator hairpin and its surrounding sequences were responsible for the regulation of P2-2 by EA and CoB12, plasmid pSP6 (Table 2 and Fig. 1), which contains a shortened SP fragment, was constructed and assayed in strains V583 and rr17. Deletion of the terminator hairpin, including the sequence that forms the putative antiterminator, from the SP fragment (pSP6) resulted in constitutive transcription under all growth conditions and eliminated the requirement for RR17 (Fig. 4B). However, in the rr17 mutant strain, the level of transcription from pSP6 in medium containing EA was reduced by 2-fold (Fig. 4C). As observed with P2-1, EA also regulated transcription from P2-2 by an unknown mechanism independent of RR17.
Overall, these in vivo results indicated that the P2-2 promoter identified in silico is also active and dependent on the TCS-17 for transcription elongation in the presence of EA and CoB12.
In vitro termination assays confirm that CoB12 causes transcription termination at the B12 riboswitch.
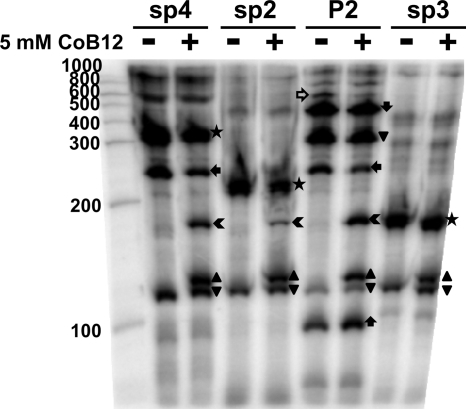
β-Galactosidase assays with the SP2- and SP4-carrying plasmids suggested that CoB12 repressed transcription elongation from the P2-1 promoter. To provide additional evidence that the B12 riboswitch in the eut operon terminates transcription in the presence of CoB12, in vitro transcription assays were performed using PCR-amplified fragments SP4, SP2, P2, and SP3 as templates. The reactions were carried out with E. coli RNA polymerase in the absence and in the presence of CoB12. The samples were run alongside a RNA marker to obtain the approximate size of each RNA product. The results are shown in Fig. 5. The expected full-length runoff product of the SP4 template (∼320 bases) was strongly present in the reaction mixtures without CoB12, together with a smaller RNA product (∼250 bases) that, based on its size, was likely due to pausing of the RNA polymerase at approximately 80 nucleotides downstream from the riboswitch. An additional RNA product was also estimated to be ∼120 bases long and, therefore, would represent RNA terminating within the B12 riboswitch. This small pausing product was also present in the mixtures for reactions carried out on the other templates (SP2, P2, and SP3), and its concentration was not affected by the presence of CoB12.
Fig. 5.
In vitro termination assay with CoB12. In vitro transcription assays were carried out on the SP4, SP2, P2, and SP3 templates. The results shown were obtained after incubation of the reaction mixtures for 15 min. RNA products: full-length runoff (★); terminated ( ); CoB12-induced pause (▴); pause product (▾); pause in SP4 and P2 reactions (
); CoB12-induced pause (▴); pause product (▾); pause in SP4 and P2 reactions ( ); RNA from P2-2 (
); RNA from P2-2 ( ); RNA from P2-1 to terminator in front of eutG (
); RNA from P2-1 to terminator in front of eutG ( ). The open horizontal arrow indicates the ∼500-base product in the P2 reaction mixture that is likely to be the full-length runoff product. The gel with the 120-min reaction products showed essentially identical results (data not shown).
). The open horizontal arrow indicates the ∼500-base product in the P2 reaction mixture that is likely to be the full-length runoff product. The gel with the 120-min reaction products showed essentially identical results (data not shown).
In the presence of CoB12, the reaction with the SP4 template generated two additional products, an ∼130-base product likely to terminate within the B12 riboswitch and an ∼160-base product which would correspond to an RNA terminated immediately after the riboswitch. Notably, in the presence of CoB12, there was a reproducible reduction in the amount of runoff RNA produced and of the longer paused-RNA product (the ∼320 bases and the ∼250 bases of RNA, respectively), as quantitated with ImageQuant software (data not shown). This correlated with the appearance of the CoB12-induced (∼130 bases) product and, more relevant, the putative terminated product of ∼160 bases.
The ∼160-base RNA product, as well as the CoB12-induced ∼130-base product, were also obtained in the reactions carried out on the SP2 and P2 templates, and their presence also correlated with decreases in the concentrations of the respective highest-molecular-weight products (∼200 bases and ∼430 bases, respectively). The bands that corresponded to the terminated RNA of fragments SP4, SP2, and P2 in the in vitro termination assays were approximately the same size as the full-length (runoff) RNA obtained with fragment SP3 as the template, whose 3′ end is at the 3′ end of the riboswitch.
The ∼430-base product from the P2 template corresponded in size to a product terminated at the hairpin identified immediately upstream from eutG, while the ∼300-base product probably resulted from a pause of the E. coli RNA polymerase at a polyuridine tract that precedes the P2-2 −35 sequence (see Fig. S1 in the supplemental material). Notably, a fragment of >500 bases was present in the reaction mixture with the P2 template in the absence of CoB12, and its concentration decreased in the reaction mixture in the experiment carried out with CoB12. This could be the full-length RNA runoff product that originated at P2-1 and read through the P2-2 promoter and its terminator, despite the absence of RR17 in the reaction mixture (the expected size of such a product was 570 bases).
In the case of the P2 template, in addition to the seven RNA bands described above, a small RNA (∼90 bases) was observed, likely resulting from transcription from the P2-2 promoter present on the fragment. CoB12 had a small stimulatory effect on the 90-base RNA from P2-2, suggesting that the long RNA originating at P2-1 interfered with transcription initiation at P2-2.
These in vitro results are consistent with CoB12 inducing transcription termination from P2-1 at the B12 riboswitch.
Identification of the 3′-end nucleotides of the in vitro RNA products originating at the P2-1 promoter.
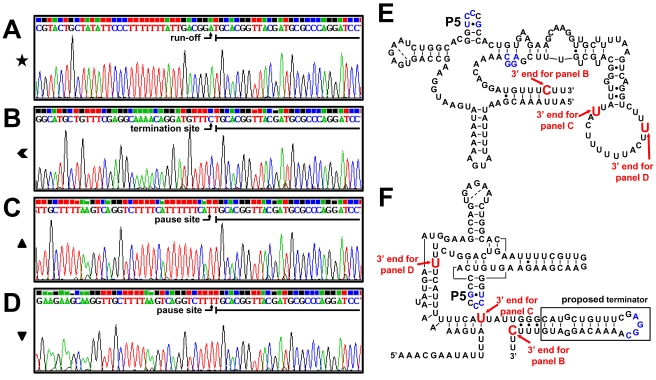
In order to unambiguously identify the products of the in vitro transcription assays, a 3′-RACE experiment was performed to ascertain the exact 3′-terminal nucleotides of the paused, the terminated, and the runoff in vitro-transcribed RNA products obtained from the P2-1 promoter on the SP4 fragment (see above) (as the paused and terminated RNA products were expected to be similar for all four templates analyzed in the assay whose results are shown in Fig. 5). The nucleotide at the 3′ end of the runoff product from fragment SP4 was identified as the last nucleotide of the template, as expected (Fig. 6A). The 3′ end of the terminated RNA product was identified at nucleotide −295 upstream from eutG (Fig. 6B), i.e., the C in the UUUC sequence that immediately follows a sequence with the potential of forming a terminator hairpin (Fig. 6F). Thus, the terminator hairpin of the B12 riboswitch is near the end of the riboswitch rather than further downstream as proposed by Fox et al. (15). The 3′ end of the CoB12-induced paused RNA product (Fig. 6C) and those of the paused RNA found in every sample (Fig. 6D) were all located within the riboswitch sequence (Fig. 6E and F). Both pause sites were near or within a polyuridine tract that may have formed an unstable DNA-RNA hybrid that caused the RNA polymerase to stall, resulting in prematurely terminated RNA.
Fig. 6.
Identification of the 3′-end nucleotides of in vitro transcription products from P2-1. The RNA products of the in vitro transcription reactions carried out on the SP4 fragment containing the P2-1 promoter (see Fig. 5) were purified from the gel and used in 3′-RACE reactions as described in Materials and Methods. Underlined nucleotides in the sequencing graphs indicate the dRR17p RNA adapter sequence. Symbols refer to the symbols used in Fig. 5. (A) Runoff RNA (★); shown are nucleotides −168 to −139 upstream from eutG. (B) Terminated RNA ( ); shown are nucleotides −327 to −295 upstream from eutG. (C) CoB12-induced paused RNA (▴); shown are nucleotides −365 to −333 upstream from eutG. (D) Paused RNA (▾); shown are nucleotides −377 to −345 upstream from eutG. (E) Representation of the B12 riboswitch secondary structure based on base pair predictions by Barrick and Breaker (5). (F) Representation of the B12 riboswitch secondary structure model developed by Fox et al. (15); the boxed hairpin loop is the B12 riboswitch terminator proposed in this study. In panels E and F, red nucleotides indicate the location of the 3′-end nucleotides from panels B to D, and blue nucleotides indicate potential base-paired sequences during termination.
); shown are nucleotides −327 to −295 upstream from eutG. (C) CoB12-induced paused RNA (▴); shown are nucleotides −365 to −333 upstream from eutG. (D) Paused RNA (▾); shown are nucleotides −377 to −345 upstream from eutG. (E) Representation of the B12 riboswitch secondary structure based on base pair predictions by Barrick and Breaker (5). (F) Representation of the B12 riboswitch secondary structure model developed by Fox et al. (15); the boxed hairpin loop is the B12 riboswitch terminator proposed in this study. In panels E and F, red nucleotides indicate the location of the 3′-end nucleotides from panels B to D, and blue nucleotides indicate potential base-paired sequences during termination.
Overall, these data show that the B12 riboswitch downstream from the P2-1 promoter terminates transcription in the presence of CoB12 in vivo, as well as in vitro.
Phosphorylated RR17 induced transcription antitermination at the Rho-independent terminator upstream from eutG.
A markerless deletion of the hk17 gene abolished EA- and CoB12-dependent expression from the P2-2 promoter on fragment SP in the in vivo assays (Fig. 4C). This implied that phosphorylation of RR17 was required for the induction of transcription. However, it was reported by Fox et al. that unphosphorylated RR17 bound to the promoter region of eutP (15), although transcription studies and binding studies with other promoters in the eut operon were not reported.
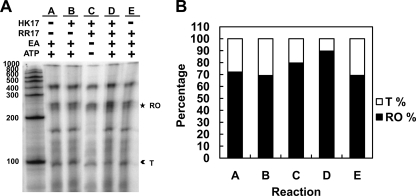
To investigate the possible discrepancy between the results of our in vivo assays and the results of the binding studies by Fox et al. (15), in vitro termination assays were carried out with the P2-2 promoter on the SP fragment using purified RNA polymerase from E. coli (Fig. 7A). Under all assay conditions tested, two main RNA products were observed, one of approximately 250 bases that corresponded to the expected size of the full-length runoff product and a 90-base product consistent in size with being the product terminated at the Rho-independent terminator (Fig. 7A). Quantification of the intensity of the RNA products indicated that the reaction mixture with phosphorylated RR17 (Fig. 7A and B, lane D) showed a higher percentage of antiterminated and 3-fold less terminated RNA than the other reaction mixtures. It should be noted that there were at least 3 bands in the runoff product of these reactions, probably due to strand separation of the ends of the PCR template that caused the RNA polymerase to stop prematurely. These results indicated that phosphorylated RR17 was required for the antitermination activity of this response regulator at the P2-2 promoter.
Fig. 7.
In vitro transcription assay of promoter P2-2 in the presence of RR17 and HK17. (A) Reactions were carried out on the SP template containing the P2-2 promoter. Shown are the products of the reaction mixtures incubated for 15 min. RNA products: runoff (RO; ★); terminated (T;  ). The gel with the products of the 120-min reactions showed essentially identical results (data not shown). (B) Quantification of RNA products from the in vitro termination assay shown in panel A. The percentages of RNA present in the RO and T bands were calculated as the fraction of the sum of the pixels of the RO and T bands from each reaction (4). Because the products are essentially end labeled (see Materials and Methods), the length of the transcripts is uninfluential in this calculation (4).
). The gel with the products of the 120-min reactions showed essentially identical results (data not shown). (B) Quantification of RNA products from the in vitro termination assay shown in panel A. The percentages of RNA present in the RO and T bands were calculated as the fraction of the sum of the pixels of the RO and T bands from each reaction (4). Because the products are essentially end labeled (see Materials and Methods), the length of the transcripts is uninfluential in this calculation (4).
Identification of the 3′-end nucleotides of the in vitro RNA products that originated at the P2-2 promoter.
To precisely identity the 3′ ends of the terminated and runoff RNA products originating at the P2-2 promoter on fragment SP, a 3′-RACE experiment was performed with the RNA products obtained in the in vitro termination assay described above. Two clones obtained from the runoff RNA were sequenced. The 3′ end of the runoff product of one clone corresponded to the last nucleotide of the SP template (Fig. 8A), while the second clone was about 10 nucleotides shorter (data not shown), which is consistent with the multiple bands observed on the gel (Fig. 7A). The 3′ end of the terminated RNA (Fig. 8B) was identified within the polyuridine tract that is immediately downstream from the strong Rho-independent terminator (Fig. 8D). This is probably the structure that phosphorylated RR17 destabilizes in order to cause antitermination.
Thus, the results of these in vitro experiments, together with the results of the in vivo approaches, confirmed that the Rho-independent terminator upstream from eutG is the target for the antitermination activity of the phosphorylated form of the ANTAR-containing response regulator RR17 at the P2-2 promoter.
DISCUSSION
In E. faecalis, a unique and complex transcriptional regulatory pathway exists for the expression of the genes required for ethanolamine utilization. Previous studies predicted the existence of multiple regulatory mechanisms of gene expression and identified two promoters: P1, in the eutG-eutS intergenic region, and P3, upstream from eutP, activated when cells were grown in serum (11, 15). In this study, we demonstrated that there are two distinct promoters in the eutT-eutG intergenic region (P2-1 and P2-2) and that they are subject to posttranscriptional regulatory mechanisms. This extends previous findings by Fox et al. (15), whose in vivo transcription analysis in serum only detected activity from the P3 promoter and not from the eutT-eutG intergenic region.
The P2-1 promoter is immediately followed by a B12 riboswitch (1, 5, 15) that, in this study, was demonstrated in vivo, as well as in vitro, to regulate transcription termination based on the presence of CoB12. With more extensive approaches and combined experiments, our results prove that the B12 riboswitch in the eut operon terminates transcription from the upstream promoter upon binding CoB12, thus clarifying previous work by Fox et al. (15).
Termination of transcription at the B12 riboswitch was determined to occur at a Rho-independent terminator structure (distal terminator hairpin) (nucleotides −325 to −299 upstream from the translational start of eutG) that is within the structure of the B12 riboswitch as identified by Barrick and Breaker (5). Our in vitro analysis and sequence determination confidently identified the termination site of the B12 riboswitch in the C residue within a polyuridine tract that follows the distal terminator hairpin. Furthermore, the in vivo analysis showed that a truncation of this polyuridine tract, as in the pSP3 construct (Fig. 3C; also see Fig. S1 in the supplemental material), results in constitutive transcription from the P2-1 promoter, suggesting the possibility that this sequence may be required for CoB12-induced termination. The structure of the riboswitch and subsequent binding of CoB12 should not be affected by the truncation in the SP3 fragment because its 3′ end is the last nucleotide in the riboswitch that is base paired.
The location of this terminator hairpin, or expression platform, is unusual because the terminator is generally a few bases downstream from the riboswitch aptamer domain. Also unusual is the lack of a recognizable antiterminator structure. Possibly for these reasons, the RibEx server identified, and Fox et al. proposed (15), another Rho-independent terminator as the expression platform for the eut riboswitch. This other terminator (proximal terminator hairpin) is located immediately upstream from the eutG gene and, as discussed below, is involved in termination of transcription from the P2-2 promoter but has no role in regulating transcription from the P2-1 promoter.
Two potential secondary structures of the B12 riboswitch in the E. faecalis eut region have been proposed. The Barrick and Breaker structure is based on the classic secondary structure of the B12 riboswitch (5), which has an unpaired region upstream from the 3′-end nucleotide of the CoB12-terminated RNA (Fig. 6E). The structure proposed by Fox et al. was based on their in-line probing results and presents a hairpin upstream from the 3′-end nucleotide instead of an unpaired sequence (Fig. 6F) (15). Our results suggest that the hairpin at bases 343 to 369, shown in Fig. S1 in the supplemental material, is the terminator of the B12 riboswitch in the eut operon.
Notably, this hairpin has a low-stability structure (ΔG = −5.7 kcal/mol as determined by M-fold analysis [28, 45]), and the nucleotides of the loop were reported by Fox et al. to become more constrained in the presence of CoB12, indicating that CoB12 may cause the loop nucleotides to form base pairs with another part of the riboswitch sequence (15). A potential binding partner is a complementary sequence in the highly conserved hairpin located in the B12 aptamer core (labeled P5 in Fig. 6E and F) that also becomes constrained in the presence of CoB12 (5, 15).
The eutG-proximal part of the eutT-eutG intergenic region (SP fragment) contains the P2-2 promoter, which depends on the RR17 and HK17 proteins for activity in vivo and phosphorylated RR17 for antitermination in vitro. A strong, Rho-independent terminator (the proximal terminator hairpin) is present downstream from P2-2, and its deletion relieved transcription of the requirement for TCS-17. Termination of transcription in vitro was shown to occur at a U residue of the polyuridine tract that immediately follows this terminator (Fig. 8D). Also within the SP fragment, a sequence element potentially responsible for antitermination by phosphorylated RR17 was identified by RibEx (Fig. 8C). This proposed antiterminator loop is a low-stability structure (ΔG = −5.7 Kcal/mol). The nucleotides in the right arm of the antiterminator contribute to the formation of the left arm of the terminator loop (residues in blue in Fig. 8C and D). Immediately preceding are the residues belonging to the element found by Fox et al. to be conserved between the binding site of the AmiR protein and the eutG 5′ untranslated region (AGCAAA) and proposed to be the recognition sequence for the ANTAR regulatory protein RR17 (residues in green in Fig. 8C) (15). Notably, this consensus sequence does not seem to be shared by the NasR protein of Klebsiella oxytoca (8). Our results support the hypothesis that binding of RR17 to this sequence must prevent the formation of the proximal terminator hairpin, thereby allowing transcription to continue further downstream.
Our in vivo data using the hk17 mutant strain and the in vitro transcription data show that antitermination at the P2-2 promoter requires both RR17 and HK17. Thus, phosphorylation of RR17 can be inferred to be required for antitermination of transcripts initiating at P2-2. Phosphorylated RR17 is also inferred to be required for transcription antitermination at the P1 promoter located in the eutG-eutS intergenic region that also features a terminator structure with the consensus ANTAR recognition motif, like the one present in front of eutG (11, 15). This conclusion is potentially in contrast with the observation by Fox et al. that unphosphorylated RR17 bound the P3 promoter region in vitro, although support for this result by in vivo or in vitro transcription approaches was not presented (15).
The observation that the eutT-eutG region contains two promoters, both active and subject to distinct regulatory mechanisms, raises the question of whether there is any influence of one promoter on the activity of the other. The in vivo transcription assays carried out on the lacZ gene fused to the P2 fragment (P2-1 and P2-2 promoters) or the SP2 (P2-1 promoter) and SP (P2-2 promoter) individual fragments showed that the combined activity of P2-1 and P2-2 on the P2 fragment was at the same level as that of the P2-1 promoter alone in EA-plus-CoB12 growth conditions (Fig. 2 and 3), as if the P2-2 promoter in the P2 fragment were not active or strongly inhibited. It is unlikely that another protein factor is contributing to the repression of P2-2, since it is transcriptionally active when assayed alone under the same growth conditions. In vivo transcription analysis with an additional construct (pSP5) (see Fig. S1 in the supplemental material), similar to the pP2 construct but missing the antiterminator-terminator sequence at the 3′ end, showed the P2-1-plus-P2-2 combined level of transcription being one order of magnitude higher than that of the P2 construct when cells were grown in the presence of EA and CoB12 (see Fig. S3 in the supplemental material). This suggested that some read-through of the distal terminator at the B12 riboswitch from P2-1 must occur even in the presence of CoB12 and that this RNA interfered with antitermination by RR17 at the proximal terminator hairpin. Notably, the in vitro transcription assay carried out with the P2 template reproducibly generated a more intense band of ∼90 bases in the presence than in the absence of CoB12. Thus, the long RNA product resulting from P2-1 and read-through of the B12 riboswitch terminator also interfered with transcription initiation at the P2-2 promoter. It is possible that secondary structures formed by the read-through RNA from P2-1 sequester the P2-2 promoter and antitermination-termination region. This means that termination at the B12 riboswitch in the presence of CoB12 results in increased efficiency of both transcription initiation and antitermination by RR17 at the P2-2 promoter. This partially resolves the paradoxical situation of termination occurring at the B12 riboswitch in the eut operon when the required CoB12 cofactor for EA utilization is present. Nevertheless, the rationale for the presence of the P2-1 promoter and the B12 riboswitch remains unclear when an antitermination-termination mechanism dependent on EA and CoB12 is in place at the P2-2 promoter and could be serving the purpose of regulating eut gene expression.
EA is the signal that triggers HK17 autophosphorylation and then phosphoryl transfer to RR17 (11, 15). However, the in vivo assays with strains carrying the pSP6 (Fig. 1 and 4B) and pSP2 (Fig. 1 and 3A) plasmids demonstrated that EA is also capable of inhibiting transcription independently of TCS-17. This raised the possibility that, in addition to being subject to carbon catabolite repression by glucose, the promoters in the P2 region are also carbon catabolite repressed by EA. Carbon catabolite repression generally refers to the mechanism used by bacteria to ensure that the most preferred carbon source, in most cases glucose, is catabolized first (7). In the presence of glucose, transcription of the utilization genes for alternative carbon sources in Gram-positive organisms is repressed by the catabolite control protein CcpA, which binds to the catabolite-responsive element (cre) in the promoter region of the regulated operon (7). However, transcription can be autoregulated by a nonpreferred carbon source when glucose is not present through a mechanism that utilizes CcpA, as shown for the sucrose utilization genes in Staphylococcus xylosus (7, 21). Autoregulation prevents transcription from being fully induced, so the bacteria do not exceed their catabolic capabilities (7, 21). Although examples of autoregulation involve alternative sugar or sugar-derivative sources, the possibility that a nonsugar carbon source, such as EA, could autoregulate its own transcription through CcpA or another regulator cannot be excluded.
Although the riboswitch is not present on the SP fragment (Fig. 4A), CoB12 was found to be required in vivo for induction of the transcription of P2-2. The requirement for CoB12 was also observed for the P1 promoter in the eutG-eutS intergenic region (11). It is known that CoB12 by itself is unable to induce phosphorylation of HK17 (11) and that CoB12 does not stimulate the EA-induced autophosphorylation of HK17 or phosphoryl transfer to RR17 (data not shown). We cannot exclude the possibility that CoB12 aids in the binding of RR17 to the RNA, but the mechanism remains to be determined.
With at least four promoters, a two-component transcription activator that requires EA and CoB12 for induction, and a standard B12 riboswitch, the system for transcription regulation of the eut operon in E. faecalis seems much more complex than that in S. Typhimurium. However, the physiological rational for such a complex regulatory pathway remains unclear: it may relate to the physiology of the organism in the mammalian host or/and it may be a consequence of the lack in E. faecalis of the TCA cycle (31). In the E. faecalis system, CoB12 has two antagonistic roles, since it is essential for inducing transcription through an unknown mechanism but also terminates transcription at a riboswitch affecting the transcription of at least one promoter downstream. We cannot exclude that an effect is also exerted on promoters downstream from eutG, such as P1, by CoB12 binding to the B12 riboswitch in the eutT-eutG intergenic region.
Additional rigorous studies are needed to fully unravel the complex interplay of transcriptional and posttranscriptional regulatory mechanisms controlling ethanolamine utilization in E. faecalis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grant AI52289 from NIAID and grant GM19416 from NIGMS, National Institutes of Health, USPHS.
We thank George B. Spiegelman (UBC-Vancouver, Canada) for helpful suggestions. We thank Lynn Hancock for the gift of plasmid pLT06.
This is Manuscript No. 21010 from the Department of Molecular and Experimental Medicine, The Scripps Research Institute.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Abreu-Goodger C., Merino E. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33:W690–W692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson R. 1988. Biogenic amines in lactic acid-fermented vegetables. Lebenson. Wiss. Technol. 21:68–69 [Google Scholar]
- 3. Argaman L., et al. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941–950 [DOI] [PubMed] [Google Scholar]
- 4. Artsimovitch I., Henkin T. M. 2009. In vitro approaches to analysis of transcription termination. Methods 47:37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrick J. E., Breaker R. R. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bertin Y., et al. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13:365–377 [DOI] [PubMed] [Google Scholar]
- 7. Bruckner R., Titgemeyer F. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141–148 [DOI] [PubMed] [Google Scholar]
- 8. Chai W., Stewart V. 1998. NasR, a novel RNA-binding protein, mediates nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader in vitro. J. Mol. Biol. 283:339–351 [DOI] [PubMed] [Google Scholar]
- 9. Collier P. D., Cromiue D. D. O., Davies A. P. 1991. Mechanism of formation of chloropropanols present in protein hydrolysates. J. Am. Oil Chem. Soc. 68:785–790 [Google Scholar]
- 10. Cruz-Rodz A. L., Gilmore M. S. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152–154 [DOI] [PubMed] [Google Scholar]
- 11. Del Papa M. F., Perego M. 2008. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 190:7147–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donskey C. J. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39:219–226 [DOI] [PubMed] [Google Scholar]
- 13. Ferrari F. A., Trach K. A., Hoch J. A. 1985. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J. Bacteriol. 161:556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foussard M., et al. 2001. The molecular puzzle of two-component signaling cascades. Microbes Infect. 3:417–424 [DOI] [PubMed] [Google Scholar]
- 15. Fox K. A., et al. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 106:4435–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garsin D. A. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hancock L. E., Perego M. 2002. Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184:5819–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hancock L. E., Perego M. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hidron A. I., et al. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 20. Huycke M. M., Sahm D. F., Gilmore M. S. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jankovic I., Bruckner R. 2002. Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus. J. Mol. Microbiol. Biotechnol. 4:309–314 [PubMed] [Google Scholar]
- 22. Kofoid E., Rappleye C., Stojiljkovic I., Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korbel J. O., et al. 2005. Systematic association of genes to phenotypes by genome and literature mining. PLoS. Biol. 3:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kristich C. J., Chandler J. R., Dunny G. M. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawhon S. D., et al. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 26. Lesnik E. A., et al. 2001. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 29:3583–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Low D. E., Keller N., Barth A., Jones R. N. 2001. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S133–S145 [DOI] [PubMed] [Google Scholar]
- 28. Mathews D. H., Sabina J., Zuker M., Turner D. H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911–940 [DOI] [PubMed] [Google Scholar]
- 29. Miller J. H. 1972. Experiments in Molecular Genetics, p. 352– 355 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30. Murray B. E., et al. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paulsen I. T., et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 32. Roof D. M., Roth J. R. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sahm D. F., et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shu C. J., Zhulin I. B. 2002. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27:3–5 [DOI] [PubMed] [Google Scholar]
- 35. Stojiljkovic I., Baumler A. J., Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sudarsan N., et al. 2006. Tandem riboswitch architectures exhibit complex gene control functions. Science 314:300–304 [DOI] [PubMed] [Google Scholar]
- 37. Thurlow L. R., Thomas V. C., Hancock L. E. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191:6203–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toledo-Arana A., et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 39. Tsoy O., Ravcheev D., Mushegian A. 2009. Comparative genomics of ethanolamine utilization. J. Bacteriol. 191:7157–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9:1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson S. A., Wachira S. J., Norman R. A., Pearl L. H., Drew R. E. 1996. Transcription antitermination regulation of the Pseudomonas aeruginosa amidase operon. EMBO J. 15:5907–5916 [PMC free article] [PubMed] [Google Scholar]
- 42. Winkler W. C., Breaker R. R. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59:487–517 [DOI] [PubMed] [Google Scholar]
- 43. Wisplinghoff H., et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 44. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the m13mp18 and pUC10 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 45. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.