Abstract
CsgD, the master regulator of biofilm formation, activates the synthesis of curli fimbriae and extracellular polysaccharides in Escherichia coli. To obtain insights into its regulatory role, we have identified a total of 20 novel regulation target genes on the E. coli genome by using chromatin immunoprecipitation (ChIP)-on-chip analysis with a high-density DNA microarray. By DNase I footprinting, the consensus CsgD-binding sequence predicted from a total of 18 target sites was found to include AAAAGNG(N2)AAAWW. After a promoter-lacZ fusion assay, the CsgD targets were classified into two groups: group I genes, such as fliE and yhbT, are repressed by CsgD, while group II genes, including yccT and adrA, are activated by CsgD. The fliE and fliEFGH operons for flagellum formation are directly repressed by CsgD, while CsgD activates the adrA gene, which encodes an enzyme for synthesis of cyclic di-GMP, a bacterial second messenger, which in turn inhibits flagellum production and rotation. Taking these findings together, we propose that the cell motility for planktonic growth is repressed by CsgD, thereby promoting the switch to biofilm formation.
INTRODUCTION
When Escherichia coli cells switch their growth mode from single planktonic cell growth to biofilm mode, flagellum formation is turned off, and in turn the production of curli fimbriae and extracellular polysaccharides for cell-cell adhesion is switched on (4, 13). In E. coli, motility for planktonic growth is under the positive control of a regulatory forward cascade, including the master regulator FlhDC and RpoF sigma factor (15, 48). In contrast, biofilm formation is under positive control of another regulatory cascade, including the master regulator CsgD and RpoS-RpoE sigma factors (5, 16, 19).
The FixJ/LuxR family protein CsgD is a key regulator for curli production, and it modulates the expression of not only the csg operon, encoding components and assembly of curli (22), but also a set of genes for adaptation of cell physiology to the biofilm lifestyle (9, 14), including adrA, which encodes the enzyme for synthesis of cyclic di-GMP, a bacterial second messenger (46) for enhancement of cellulose production (38, 57). c-di-GMP induces biofilm formation and inhibits flagellum production and rotation (1, 7, 52). The expression of curli is cryptic in laboratory E. coli strains due to the lack of CsgD expression, but an ompR mutation restores CsgD expression (41). By using this ompR234 mutant for expression of CsgD, Brombacher et al. (9) performed transcriptome analysis and predicted a set of regulation targets of CsgD, including csgB and adrA (yaiC). Recently, Zakikhany et al. (56) reported experimental evidence of CsgD binding to promoters of the hitherto-predicted CsgD targets csgBA and adrA in Salmonella enterica serovar Typhimurium, and they predicted another six possible target genes on the Salmonella genome based on knowledge of the CsgD recognition sequence on the two promoters. In good agreement with the master regulator function of CsgD, transcription of csgD is under the control of more than 10 transcription factors, each monitoring a specific and different factor or condition in the environment (24, 36, 37). After analysis of the regulatory modes of these transcription factors, we noticed a unique interplay, i.e., collaboration between positive factors and negative factors (37). As an extension of this line of research, we analyzed in this study the regulation network downstream of CsgD.
In order to obtain insights into the regulatory roles of CsgD as the master regulator of biofilm formation in E. coli, it is essential to identify the whole set of regulation target genes and operons on the E. coli genome under the direct control of CsgD. For the purpose of identification of regulation targets by a transcription factor, transcriptome analysis of genes affected after disruption of the gene coding for the test transcription factor is a widely used method, but the majority of genes thus detected represent those affected indirectly (24). A total of approximately 300 species of transcription factors in E. coli form complex hierarchic networks of regulation, and thus knockout of a gene for one specific transcription factor indirectly influences a large number of genes. To overcome this problem and identify the genes under the direct control of a test regulator, we have developed the genomic SELEX screening system for search of the recognition and binding sequences by use of a test transcription factor (43), and we have successfully employed this system for identification of whole sets of regulation targets by a number of transcription factors, such as PdhR (35), RutR (YcdC) (44), and Dan (YgiP) (50). However, this improved genomic SELEX system cannot be applied for a regulator such as CsgD, which may require an as-yet-unidentified effector for function. Thus, in this study, we performed a ChIP-chip (ChIP and microarray) analysis using a high-density microarray to identify the CsgD-associated sites on the E. coli genome (17, 18, 45), and we identified more than 20 novel targets of regulation. Based on detailed analysis of some of the novel targets, we propose dual roles for CsgD, i.e., promotion of biofilm formation and inhibition of flagellum production.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table 1 lists the strains used in this study. Escherichia coli BL21(DE3) [F− ompT hsd(rB− mB−) dcm gal λ(DE3)] (49) was used for expression and purification of all the transcription factors used in this study. E. coli K-12 BW25113 and its otherwise-isogenic mutant strain lacking CsgD were products of the Keio collection (3) and obtained from the National Bio-Resource Center (National Institute of Genetics, Japan). E. coli KP7600 (28), a derivative of wild-type W3110, was used for cloning the csgD gene. E. coli MC4100 (12) was used for construction of the promoter-lacZ reporter fusion vectors (see below). Cells were cultured in LB medium or YESCA (yeast extract-Cosamino Acids) medium (40) at 28°C. When necessary, 100 μg/ml ampicillin and 50 μg/ml kanamycin were added to the medium.
Table 1.
E. coli strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BL21(DE3) | F−ompT hsd(rB− mB−) dcm gal(DE3) | Studier and Moffatt (49) |
| MC4100 | F− Δ(argF-lac) U169 araD139rpsL150ptsF25fibB5301rbsRdeoC relA1 | Casadaban (12) |
| BW25113 | F−lacIqrrnB3lacZ4787hsdR514 Δ(araBAD)567 Δ(rhaBAD)568rph-1 | Baba et al. (3) |
| JW1023 | BW25113 ΔcsgD | Baba et al. (3) |
| BWcsgD | Km resistance marker of JW1023 eliminated with pCP20 | This study |
| BWWF1D | BW25113 λcsgD (F1)-lacZ | This study |
| BWcsgDF1D | BWcsgD λcsgD (F1)-lacZ | This study |
| BWWnlpA | BW25113 λnlpA-lacZ | This study |
| BWcsgDnlpA | BWcsgD λnlpA-lacZ | This study |
| BWWwrbA | BW25113 λwrbA-lacZ | This study |
| BWcsgDwrbA | BWcsgD λwrbA-lacZ | This study |
| BWWfliE | BW25113 λfliE-lacZ | This study |
| BWcsgDfliE | BWcsgD λfliE-lacZ | This study |
| BWWfliF | BW25113 λfliF-lacZ | This study |
| BWcsgDfliF | BWcsgD λfliF-lacZ | This study |
| BWWyccT | BW25113 λyccT-lacZ | This study |
| BWcsgDyccT | BWcsgD λyccT-lacZ | This study |
| BWWyhbT | BW25113 λyhbT-lacZ | This study |
| BWcsgDyhbT | BWcsgD λyhbT-lacZ | This study |
| BWWadrA | BW25113 λadrA-lacZ | This study |
| BWcsgDadrA | BWcsgD λasrA-lacZ | This study |
Plasmid constructions.
For the construction of lacZ reporter vectors, DNA fragments, each containing the CsgD target promoter region, were prepared by PCR using E. coli KP7600 genome DNA as a template and a pair of gene-specific primers (see Table S1 in the supplemental material). After digestion with EcoRI and BamHI, the PCR-amplified fragments were inserted into pRS551 (47) at the corresponding sites to generate the promoter assay vectors.
For construction of an arabinose-inducible CsgD expression plasmid, a DNA fragment containing the csgD coding sequence was prepared by PCR using E. coli KP7600 genomic DNA as a template along with a pair of primers, csgD-BAD-EcoRI-F and csgD-BAD-XbaI-R (see Table S1 in the supplemental material). After digestion with EcoRI and XbaI, the PCR-amplified fragment was inserted at the corresponding site of pBAD18 (21) to generate the plasmid pBADcsgD.
For construction of the CsgD expression plasmid pCsgD for CsgD purification, a DNA fragment corresponding to the csgD coding region was amplified by PCR using E. coli KP7600 genome DNA as a template and a pair of primers, TF030S (5′-TAACGTTTCAGCGGCCGCTCGCCTGAGGTT-3′) and TF030T (5′-GCGGGGTTTCCATATGTTTAATGAAGTCCA-3′), and after digestion with NdeI and NotI, cloned into pET21a(+) (Novagen) at the corresponding sites.
Expression and purification of CsgD protein.
Construction of plasmid pCsgD for expression of His-tagged CsgD was carried out as described previously (34, 35, 54). His-tagged CsgD was expressed in pCsgD-transformed E. coli BL21(DE3) and affinity purified according to the standard purification procedure (34, 35, 54).
ChIP-chip analysis.
ChIP-chip analysis was employed to determine the chromosome-wide DNA-binding profile of CsgD, using the standard assay system developed by Grainger et al. (18). An arabinose-inducible CsgD expression plasmid pBADcsgD was constructed according to the methods described by Guzman et al. (21). E. coli BWcsgD transformed with either pBADcsgD or reference vector pBAD18 was grown at 28°C in YESCA medium up to the stationary phase (12 h). ChIP-chip analysis was performed as described previously (17, 18, 45). In brief, cells were treated with 1% formaldehyde for protein-DNA cross-linking and then disrupted by sonication for preparation of cleared lysates. CsgD cross-linked DNA fragments were immunoprecipitated from cleared lysates by using anti-CsgD rabbit polyclonal antiserum. After dissociation of protein-DNA cross-linking, DNA samples isolated from E. coli BWcsgD (pBADcsgD) and E. coli BWcsgD (pBAD18) were labeled with Cy5 and Cy3, respectively, and subjected to DNA microarray analysis. The array consisted of 43,450 species of 60-mer oligonucleotide probe, which are designed to cover the entire E. coli MG1655 genome at a 105-bp interval (Oxford Gene Technology, Oxford, United Kingdom). The Cy5/Cy3 ratio was measured for each peak and plotted against the corresponding position on the E. coli genome.
Polyclonal anti-CsgD antibody was produced in rabbits after injection of purified CsgD protein.
Measurement of promoter activity.
The test promoter-lacZ fusion plasmid was transformed into E. coli MC4100 (12), and the transformant was used as the host for preparation of λRS45. The recombinant phage-containing promoter-lacZ fusion was isolated from the resulting phage lysate and used to infect E. coli BW25113 and E. coli BWcsgD for screening of kanamycin-resistant and Lac+ colonies. Single-copy promoter-lacZ fusion strains were grown in YESCA medium and subjected to β-galactosidase activity measurements with o-nitrophenyl-β-d-galactopyranoside as described by Miller (30).
Gel shift assay of CsgD-DNA complexes.
Probes, each carrying the csgD, wrbA, nlpA, yccU-yccT, yhbT-yhbU, fliE-fliF, ydhO, or yaiC promoter region, were generated by PCR amplification by using a pair of primers (5′-fluorescein isothiocyanate [FITC]-labeled Lac30R-FITC and 551SQ-F), plasmids (pRScsgD, pRSwrbA, pRSnlpA, pRSyccT, pRSyhbU, pRSfliF, pRSydhO, and pRSyaiC; 100 ng) as the templates, and Ex Taq DNA polymerase. For generation of the csgB promoter probe, a pair of primers (5′-FITC-labeled csgB-FITC-R2A primer plus csgB-S1F1), E. coli KP7600 genome DNA template (100 ng), and Ex Taq DNA polymerase were used. Sequences of all the primers used are described in Table S1 of the supplemental material. All the PCR products with FITC at their 5′ termini were purified by polyacrylamide gel electrophoresis and then used for a gel shift assay performed under the standard conditions (34, 35).
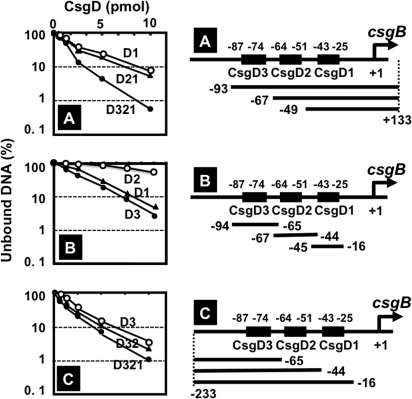
For construction of csgB promoter segment probes for determination of the CsgD-binding activity of each CsgD box (see Fig. 7), the following primer pairs and templates were used for PCR amplification: 5′ deletion segments (set A in Fig. 7), 551SQ-F plus lac30R-FITC primers and pRScsgBF1, -BF2, and -BF3 templates; 3′ deletion segments (set C in Fig. 7), csgB-FITC-R1, -R2, and -R3 plus csgB-S1F2A primers, and genome DNA template. Internal segment probes (set B in Fig. 7) were prepared by hybridization of the following pairs: csgB-FITC-R1 plus csgB-F1 Hybri; csgB-FITC-R2 plus csgB-F2 Hyrbi; and csgB-FITC-R3 plus csgB-S1F2A (see Table S1 in the supplemental material for sequences).
Fig. 7.
CsgD-binding activity of three CsgD boxes on the csgB promoter. Three sets of csgB promoter probes were constructed and examined for CsgD-binding activity by gel shift assay. (A) 5′ deletion set of the csgB promoter; (B) internal segments, each carrying one of three CsgD boxes; (C) 3′ deletion set of the csgB promoter. Construction of 5′ deletion (A) and 3′ deletion (C) probes was carried out by PCR using the primer pairs described in Materials and Methods, while the internal segment probes (B) were prepared by hybridization of complementary single-stranded DNA segments (see Table S1 for sequences).
DNase I footprinting assay.
A DNase I footprinting assay was carried out under the standard reaction conditions (34, 35). In brief, 1.0 pmol each of FITC-labeled probes was incubated at 37°C for 30 min with purified CsgD in 25 μl of 10 mM Tris-HCl (pH 7.8), 150 mM NaCl, 3 mM magnesium acetate, 5 mM CaCl2, and 25 mg/ml bovine serum albumin (BSA). After incubation for 30 min, DNA digestion was initiated by adding 5 ng DNase I (Takara). After digestion for 30 s at 25°C, the reaction was terminated by the addition of 25 μl phenol. DNA was precipitated from the aqueous layer by ethanol, dissolved in formamide dye solution, and analyzed by electrophoresis on a 6% polyacrylamide gel containing 8 M urea.
Primer extension analysis.
Overnight cultures of E. coli were diluted 100-fold in 30 ml of YESCA medium and incubated for 12 or 24 h at 28°C. RNA purification was carried out as described previously (34, 35). Primer extension analysis was performed using fluorescently labeled probes under the standard reaction conditions (34, 53). In brief, 20 μg of total RNA and 1 pmol of 5′-FITC-labeled primer (lac30R-FITC-R, 5′-AGGGTTTTCCCAGTCACGACGTTGTAAAAC-3′) were mixed in 20 μl of 10 mM Tris-HCl (pH 8.3 at 37°C), 50 mM KCl, 5 mM MgCl2, 1 mM (each) dATP, dTTP, dGTP, and dCTP, and 20 U of RNase inhibitor. The primer extension reaction was initiated by the addition of 5 U of avian myeloblastosis virus reverse transcriptase. After incubation for 1 h at 50°C, DNA was extracted with phenol, precipitated with ethanol, and subjected to electrophoresis on a 6% polyacrylamide sequencing gel containing 8 M urea, using a DSQ-500L apparatus (Shimadzu) for direct detection of fluorescence.
Western blotting assay.
The intracellular concentration of CsgD was determined by a Western blotting assay under the standard conditions (26). In brief, E. coli whole-cell lysates were prepared by sonication after lysozyme treatment and directly subjected to SDS-PAGE. After transfer of proteins onto filters, the protein-blotted filter was treated with anti-CsgD antibody, which was raised in rabbits against purified CsgD. Antibodies were detected with fluorescently labeled mouse anti-rabbit IgG antibody.
RESULTS
Genome-wide search for CsgD-associated sites on the E. coli genome.
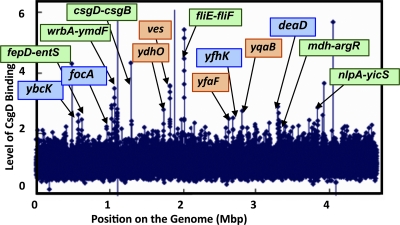
As an attempt to identify the regulation target genes or operons of CsgD on the entire genome of E. coli, we performed a ChIP-on-chip analysis (17, 18, 45) using the csgD deletion strain transformed with an arabinose-inducible csgD-expressing plasmid (pBADcsgD) or control plasmid (pBAD18). After induction of CsgD expression by adding arabinose, cells were treated with formaldehyde for protein-DNA cross-linking. CsgD-DNA complexes were recovered from sonicated cell lysates with anti-CsgD antibodies, and the precipitated DNA from CsgD-expressing cells was labeled with Cy5, while DNA fragments isolated from CsgD-lacking cell lysates were labeled with Cy3. Fluorescently labeled DNA mixtures were hybridized to a high-density microarray. Peaks showing high Cy5/Cy3 levels represented the locations where CsgD was associated under the culture conditions employed. For identification of specific positions of CsgD binding, we first screened peaks, which exhibited a high fluorescence level for two or more consecutive probes at the cutoff of 2. The center of each peak was defined as the center of the probe within the peak that had the highest Cy5/Cy3 signal. On the tiling array used, 60-nucleotide-long probes were aligned at 105-bp intervals on the E. coli MG1655 genome sequence, and thus approximately 500-bp-long ChIP fragments would bind to at least two or more consecutive probes, and so we employed this criterion for identification of positive peaks. However, we detected several one-point peaks, which may have mainly originated from a partial homology of ChIP fragments with the tiling array probes.
A total of more than 20 positive peaks were identified (Fig. 1 and Table 2), of which seven (wrbA-ymdF, yccT-yccU, csgD-csgB, ves, fliE-fliF, yhbT-yhbU, and nlpA-yicS) showed 2.8-fold higher-than-background level. The total of 20 CsgD-binding sites thus identified can be classified into two groups (Table 2; possible targets are shown in light green in Fig. 1). Thirteen group 1 CsgD-binding sites are located within spacer regions between two neighboring genes, while the remaining six sites, classified in group B, are located within open reading frames. Among the 14 group A peaks identified within the spacer regions, 8 were located between divergently transcribed operons, including the spacer between csgBAC and csgDEFG for curli formation. In prokaryotes, transcription factor-binding sites are generally located upstream or near promoters of regulation target genes or operons, and therefore when possible target genes would be located on both sides of the CsgD-binding site, we estimated that only one could be the target. In rare cases, however, the spacer-bound transcription factors regulate both of the divergent promoters on the E. coli genome (24; T. Shimada, K. Yamamoto, and A. Ishihama, submitted for publication). The remaining five CsgD-binding sites in group 1A are located upstream of one operon but downstream of another operon. In these cases, the gene or operon located downstream of the CsgD-binding site may be the regulation target with the highest possibility. The regulation targets for the CsgD-binding sites include genes for transporters (entS, focA, yojI, and fepD) and for lipid carrier and membrane lipoproteins (yhbT, nlpA, and ydhO). Transcriptome analysis in the presence of overexpressed CsgD indicated that the iron-sensing proteins FecR and FhuE are under the control of CsgD (10). Likewise, the fepDGC operon, encoding iron-enterobactin transporters, was predicted to be regulated by CsgD (Table 2).
Fig. 1.
Maps of CsgD-binding sites on the E. coli genome. CsgD-associated sites on the genome were identified by using the ChIP-chip system (17, 18, 45). Peaks that were more than 2-fold higher than the background are indicated by arrows. For group 1 targets CsgD-binding sites are located within spacer regions, while CsgD-binding sites are located on open reading frames for group 2 targets (for classifications, see Table 2). Targets shown with blue and green backgrounds indicate group 1A and group 1B, respectively. On the other hand, group 2 targets are shown with an orange background.
Table 2.
CsgD target genes identified with the ChIP-chip systema
| Gene group and map coordinate | Gene function | Gene | Spacer or ORF | Gene | Gene function |
|---|---|---|---|---|---|
| Group 1A | |||||
| 12.24 | emrE> | >ybcK | DPL12 prophage recombinase | ||
| 20.54 | Formate transporter | focA< | <ycaO | ||
| 49.68 | ABC superfamily multidrug tranporter | yojI< | <alkB | ||
| 57.93 | TCS sensor kinase | yfhK< | <glmY | ||
| 71.21 | ATP-dependent RNA helicase | deaD< | <nlpI | ||
| Group 1B | |||||
| 13.39 | Iron-enterobactin transporter | fepD< | >entS | Predicted transporter | |
| 22.13 | Conserved protein | yccT< | >yccU | Predicted coenzyme A-binding protein | |
| 22.99 | NAD(P)H:quinone oxidoreductase | wrbA< | >ymdF | Conserved protein | |
| 23.76 | csg regulon regulator | csgD< | >csgB | Curlin nucleator protein | |
| 43.34 | Flagellum basal body component | fliE< | >fliF | Flagellum basal body MS ring protein | |
| 71.11 | Predicted lipid carrier protein | yhbT< | >yhbU | Predicted peptidase | |
| 72.89 | Malate dehydrogenase | mdh< | >argR | arg operon regulator | |
| 82.71 | Cytoplasmic membrane lipoprotein 28 | nlpA< | >yicS | Predicted protein | |
| Group 2 | |||||
| 37.34 | Conserved protein | grxD< | >ydhO | >sodB | Superoxide dismutase |
| 39.28 | cho> | <ves | <spy | ||
| 56.57 | ppx> | <yfgF | >yfgG | Predicted protein | |
| 81.13 | rhaA> | >yibA | >yibJ | Predicted Rhs family protein | |
| 60.67 | Conserved inner membrane protein | yqaA< | <yqaB | <raQ | |
| 62.03 | Predicted protein | ygcI< | <ygcJ | ygcK | |
| 71.51 | argG> | <yhbX | <leuU |
CsgD-binding sites identified by ChIP-chip analysis are located either within spacer regions (group 1) or on open reading frames of the genes shown (group 2). The genes with functions reported represent the regulation targets of CsgD that were predicted based on the criterion that the CsgD-binding sites are located upstream of the respective open reading frames. Regulation of some targets was confirmed in vivo in a csgD-lacZ reporter assay (for details, see the text).
At least seven CsgD-binding sites, classified in group 2, are located within open reading frames (Table 2). In E. coli, transcription factor-binding sites are sometimes located within open reading frames of upstream genes. Thus, the yfgG and ygcI operons could be regulation targets of CsgD. It is noteworthy that YgcI (CasD) is a component of the CRISPR (clusters of regularly interspersed short palindromic repeats) complex, which is involved in defensive digestion of harmful foreign DNA, including viral DNA (11).
Identification of CsgD-binding site on the predicted target promoters.
For detailed analysis of the regulatory roles of CsgD, we focused on six targets: two targets, fliE-fliF and csgD-csgB, related to two typical bacterial habitats, i.e., planktonic growth and biofilm formation; two targets, yccT-yccU and wrbA-ymdF, related to metabolism for energy production; and two targets, yhbT-yhbU and nlpA-yicS, related to membrane functions (Table 2). Except for the csgDEFG and csgBAC operons, the participation of CsgD in transcription regulation has not been identified for the other six operons, fliE (encoding the flagellum basal body protein), fliFGHIJK (encoding flagellum components and assembly proteins), yccT, yccU, wrbA-yccJ, and ymdF. In addition, we also analyzed cho-ves-spy, a representative target for CsgD binding on an open reading frame (Table 2). The sequences included in these segments are illustrated in Fig. S1 of the supplemental material.
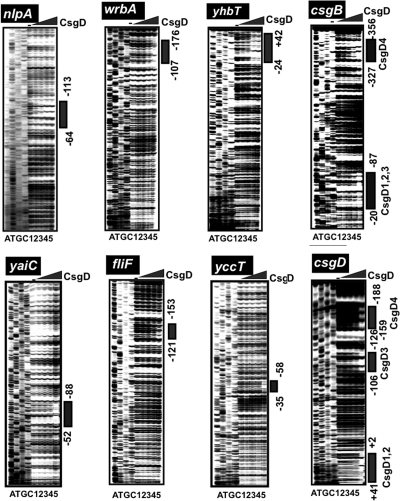
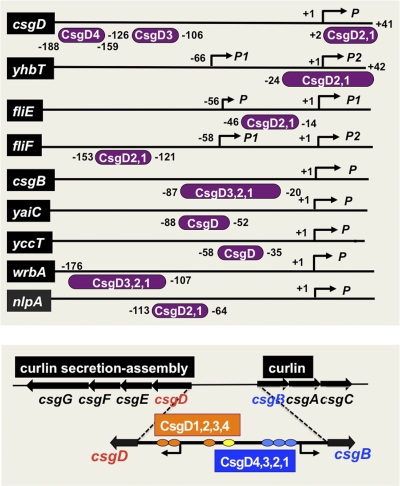
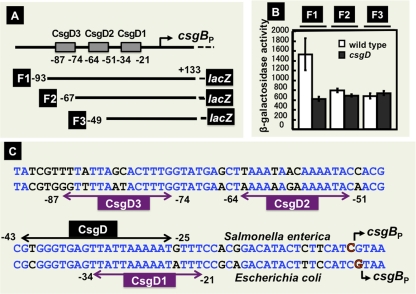
First we determined the CsgD-binding sequences within DNA fragments containing these promoters by DNase I footprinting. CsgD binding was detected for all six spacer regions, csgD-csgB, fliE-fliF, wrbA-ymdF, nlpA-yicS, yccU-yccT, and yhbT-yhbU (Fig. 2). Under the same reaction conditions, we also examined CsgD binding to the adrA (renamed from yaiC) promoter, the second known target besides csgBAC (Fig. 2). For two promoters, yaiC and yccT, a single CsgD-binding site was detected, while multiple CsgD-binding sites were identified for the other four promoters, nlpA, wrbA, yhbT, and fliF (Fig. 3). Within the spacer between the csgBAC and csgDEFG operons, we identified a total of seven sites of CsgD binding (Fig. 3). In the promoter-proximal region of the csgBAC operon, four CsgD-binding sites (CsgD1, CsgD2, and CsgD3) were identified between −21 and −87, and the CsgD4 site was mapped between −327 and −356 relative to the csgB transcription initiation site. On the promoter-proximal region of the csgDEFG operon, four CsgD-binding sites were identified: CsgD1-CsgD2 (+41 to +2), CsgD3 (−106 to −126), and CsgD4 (−159 to −188) (Fig. 2 and Fig. 3). Deletion analysis suggested that the three csgB promoter-proximal CsgD sites participate in csgB control while the three csgD promoter-proximal CsgD sites are involved in csgD control (see below). Among the seven CsgD-binding sites within the spacer between the csgDEFG and csgBAC operons, however, the role of the CsgD-binding site in the center is not yet clear, and thus it is tentatively referred to as CsgD4 on both the csgD and csgB promoters (Fig. 3, bottom). The promoter-proximal CsgD1 (−20 to −34) on the csgB promoter overlaps with the CsgD-binding site (−25 to −43) in Salmonella enterica (56).
Fig. 2.
DNase I footprinting of CsgD-binding sites. Fluorescently labeled DNA fragments of the indicated CsgD target promoters (1.0 pmol each) were incubated in the absence (lane 1) or presence of increasing concentrations of purified CsgD (lanes 2 to 5 contain 2.5, 5, 10, and 20 pmol) and then subjected to DNase I footprinting assays. Lanes A, T, G, and C represent the sequence ladders. Bold lines on the right indicate the CsgD-binding sequences as detected by their protection pattern after DNase I treatment. The nucleotide numbers indicate the distances from the respective transcription start sites. The locations of CsgD-binding sites are illustrated in Fig. 3, below.
Fig. 3.
Location of the CsgD-binding site(s) on the regulation target promoters. The locations of CsgD-binding sites on the promoters examined in the footprinting assay (Fig. 2) are shown along the respective promoter sequences. Transcription initiation sites have been determined for nlpA (6), csgD (22, 35), yaiC (10), fliE (33, 48), fliF (33, 48), csgB (2), yccT (data not shown), yhbT (29), and wrbA (29). Numbers on each line represent the distance (in bp) from the respective transcription start site. Note that the CsgD-binding site of fliE (−46 to −14) is the same as that of fliF (−121 to −153).
Identification of the CsgD recognition sequence.
After sequence logo analysis of the CsgD-binding sequences among a total of 18 known and newly identified sites on eight target genes, a consensus 14-bp-long sequence AAAAGNGNNAAAWW (with W representing A/T) was identified (Fig. 4). (The CsgD box sequence in each segment is shown in Fig. S1 of the supplemental material.) This E. coli CsgD box sequence is slightly different from the Salmonella sequence, CGGGKGAGNKA, that was recently proposed, based on the experimentally determined CsgD-binding sequences of two targets, csgB and adrA (56). The CsgD-binding sites on both the E. coli csgB (−43 to −25) and adrA (−90 to −28) promoters overlap in part with the corresponding CsgD-binding sites in Salmonella enterica (Fig. 5C). The three CsgD box sequences on the E. coli csgB promoter differ at one to four positions from the corresponding sequences of the Salmonella csgB promoter (see Fig. 5D below). It is also noteworthy that the structure of the E. coli CsgD protein of 216 amino acids differs from that of the Salmonella CsgD protein: it is 8 residues longer and contains a 17-amino-acid substitution. We cannot exclude the possibility that this structural difference leads to the different recognition properties of the CsgD box sequence.
Fig. 4.
Consensus sequence of the CsgD box. CsgD-binding sites were identified in the wrbA, yhbT, yccT, csgB, fliF, yaiC, csgD and nlpA promoters, as shown in Fig. 2. The sequences of these promoter segments were analyzed to identify the consensus sequence of the CsgD box.
Fig. 5.
Analysis of functional roles of CsgD sites on the csgB promoter. (A) Three different segments (F1, F2, and F3) of the csgB promoter were inserted into pRS551 (47), and the resulting lacZ fusion plasmids were transformed into wild-type E. coli and csgD-defective mutant strains. (B) β-Galactosidase activities of the transformants. (C) Sequence of the E. coli csgB promoter and the locations of CsgD-binding sites are shown on the lower sequence, while the sequence for the Salmonella csgB promoter and the location of the CsgD-binding site (56) are shown in the upper sequence.
Influence of csgD deletion on expression of target genes.
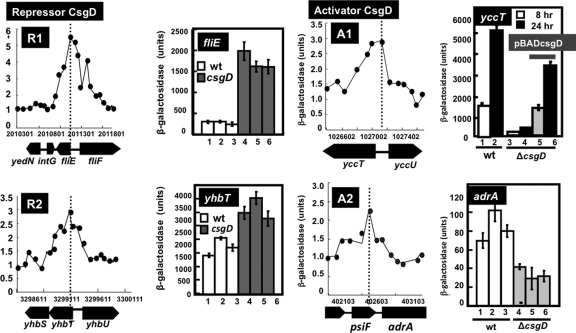
To examine the possible influence of CsgD on the activity of promoters with CsgD-binding sites, we constructed a set of E. coli strains carrying a single copy of target promoter-lacZ transcriptional fusions on the genome and performed LacZ reporter assays with both wild-type and csgD mutant strains. A group of promoters were activated in the csgD mutant, indicating that CsgD is involved as a repressor for this group of promoters. For instance, the promoter activity of fliE (encoding the flagellum basal body) and yhbT (encoding a putative lipid carrier protein) increased in the csgD mutant (Fig. 6, R1 and R2). For both promoters, the CsgD-binding site is located downstream of the respective transcription initiation sites (Fig. 3). This finding agrees well with the general rule regarding the transcription factor-binding site relative to its promoter and the regulatory mode of its action (24).
Fig. 6.
Classification of the regulation mode for CsgD. The promoter activity was measured in both the wild type and a csgD-defective mutant by using lacZ as a reporter. Based on the activity ratio between the wild type and csgD mutant, the test promoters were classified into repression mode (R1, fliE promoter; R2, yhbT promoter) and activation mode (A1, yccT promoter; A2, adrA promoter). For each promoter, both the ChIP-chip profile (left) and lacZ reporter activity (right) are shown. The sequences of peak regions are described in Fig. S1 of the supplemental material.
The activity of promoters carrying the CsgD-binding site upstream of the respective promoters decreased in the mutant lacking CsgD. In these cases, CsgD might be involved in activation of the target genes, including yccT, encoding a putative periplasmic protein, and adrA, encoding a putative diguanylate cyclase (GGDEF) with a regulatory role in curli formation (Fig. 6, panels A1 and A2). The activity of both yccT and adrA promoters increased in the stationary phase, suggesting the involvement of YccT and AdrA in stationary-phase adaptation. The transcription initiation site of yccT, shown in Fig. 3, was determined in a primer extension assay (data not shown). The CsgD box was thus found to be located upstream of the promoter at −35, indicating that CsgD is a class I activator (25).
Role of multiple CsgD sites on the csgB promoter.
The genes for all the essential components for the formation of curli fimbriae, including curlin subunits (CsgBAC), and components for curlin secretion and assembly (CsgEFG) are organized in the divergent operons csgDEFG and csgBAC. As noted above, we identified seven CsgD-binding sites within a 754-bp-long spacer between two csg operons (Fig. 3). The presence of seven CsgD-binding sites itself implies complex regulation for these operons. We then performed a more detailed analysis on these divergently transcribed csg operons. To obtain insights into the regulation of csg promoters, we constructed csgB-lacZ and csgD-lacZ fusions for monitoring the promoter activities.
The regulatory roles of three promoter-proximal CsgD sites on the csgB promoter were analyzed using a set of lacZ fusions, each carrying different segments of the csgB promoter (Fig. 5A). After deletion of the upstream CsgD3 site, the promoter activity was essentially the same between the wild type and the csgD mutant (Fig. 5B), indicating that CsgD bound at the CsgD3 site plays a major regulatory role for control of the csgBC operon. In order to identify the role of downstream CsgD sites on the csgB promoter, we constructed three sets of csgB-lacZ fusions, each carrying a different segment of the csgB promoter, and we analyzed the binding activity in vitro of the CsgD protein (Fig. 7). Among the set of upstream deletion derivatives, the CsgD-binding activity was maximum for the longest segment containing all three CsgD boxes (the D321 segment), but after deletion of CsgD3, the activity of CsgD binding remained for the D21 and D1 segments, albeit at decreased levels of about 50% (Fig. 7A). We also constructed a set of downstream deletion derivatives and measured their CsgD-binding activities. Even though the activity was the highest for the D321 segment, the CsgD-binding activity was not so different between the three segments, supporting the prediction that CsgD3 plays a major role for CsgD binding to the csgB promoter (Fig. 7C). Finally, we examined the CsgD-binding activities of three segments, each carrying only one of the three CsgD boxes (Fig. 7B). Both CsgD3 and CsgD1 were equally active in CsgD binding, but the CsgD2 segment showed weak but significant CsgD binding activity. Based on these results taken together, we conclude that all three CsgD boxes on the csgB promoter carry CsgD-binding activity but at different levels, with CsgD3 being the strongest and CsgD2 being the weakest.
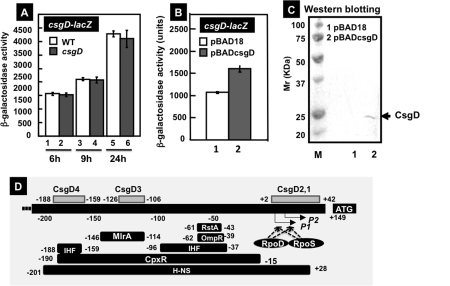
Regulatory role of CsgD on the csgD promoter.
To obtain insights into the regulation of the csgD promoter, we analyzed promoter activities by using a csgD-lacZ fusion. The csgD promoter activity in wild-type E. coli increased in the stationary phase (Fig. 8A), in agreement with the physiological role of CsgD as a master regulator of biofilm formation under stress conditions. However, the csgD promoter activity was essentially the same between the wild type and csgD mutant under the culture conditions employed (Fig. 8A). The failure to detect the influence of csgD knockout under steady-state growth in exponential phase may be due to the low-level expression of CsgD, as detected by Western blotting (37). In addition, a number of transcription factors are involved in regulation of the csgD promoter (36, 37), leading to masking of the influence of a single transcription factor. To overcome the masking effect by other transcription factors, CsgD was overexpressed by using an arabinose-inducible expression vector (Fig. 8C). We found significant enhancement of the csgD promoter (Fig. 8B), indicating that CsgD plays a role as an activator under high-level expression conditions. It should also be noted that the enhancement of csgD promoter activity after overproduction of CsgD is not simply due to the promoter activation but also to an indirect influence through enhanced synthesis of RpoS sigma factor by an increase in CsgD-dependent synthesis of YaiB, an inhibitor of RssB phosphorylation (19).
Fig. 8.
Effects of CsgD on the csgD promoter. (A) E. coli BWWF1D (white bars) and BWcsgDF1D (gray bars) were grown in YESCA medium for 6 (lanes 1 and 2), 9 (lanes 3 and 4), or 24 h (lanes 7 and 8) for measurement of β-galactosidase activities. (B) E. coli BWcsgDF1D was transformed with either pBAD18 (lane 1) or pBADcsgD (lane 2) and grown in YESCA medium at 28°C for 12 h, and then β-galactosidase activities were measured. (C) Confirmation of CsgD expression in pBADcsgD-transformed E. coli BWcsgDF1D by Western blot analysis. (D) Locations of CsgD-binding sites along the csgD promoter. The binding sites of other transcription factors on the cagD promoter have been reported previously (36, 37).
Taking these findings together, we conclude that different CsgD molecules when bound within the spacer region regulate separately the divergently transcribed csgDEFG and csgBC operons. The presence of seven CsgD-binding sites within this spacer region itself suggests the involvement of complex regulation systems of two transcription units in opposite directions, depending on the number and order of CsgD binding between these seven sites. Since a number of different transcription factors also bind to the same regions, the molecular interplay between transcription factors should be more complex than we proposed in earlier studies (36, 37).
Role of CsgD in flagellum synthesis and motility.
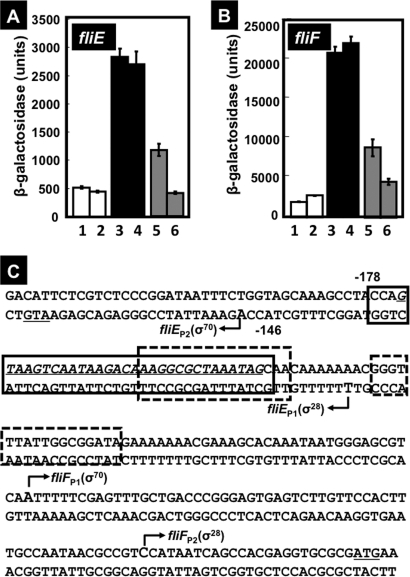
Among the list of regulation target candidates (Fig. 1 and Table 2), we determined that the spacer between the fliE and fliFGHIJK operons has a high affinity for CsgD. The fliE gene encodes a component of the flagellum basal body assembly, connecting the MS ring and rod, while the fliFGHIJK operon encodes essential components and functional modulators of the flagellaum, including components of the flagellum export apparatus (31, 33). Based on a DNase footprinting assay, multiple CsgD-binding sites were identified within this 215-bp-long spacer, which is located downstream (+10 to +42) of fliE promoter P1 (Fig. 2) or −95 to −63 upstream of fliF promoter P1 (Fig. 2). In good agreement with the location of the CsgD-binding site, the fliE promoter activity markedly increased in the csgD mutant (Fig. 9A), indicating repression of fliE by CsgD. In parallel, we also examined the possible influence of CsgD on the promoter of the fliF operon, which is transcribed in a different direction from the fliE gene.
Fig. 9.
Effects of CsgD on the fliE and fliF promoters. (A) E. coli BWWfliE (lanes 1 and 2) and BWcsgDfliE (lanes 3 to 6) strains were transformed with either pBAD18 (lanes 1 to 4) or pBADcsgD (lanes 5 and 6), grown in YESCA medium at 28°C for 8 h (lanes 1, 3, and 5) or 24 h (lanes 2, 4, and 6), and β-galactosidase activities were determined. (B) E. coli BWWfliF (lanes 1 and 2) and BWcsgDfliF (lanes 3 to 6) strains were transformed with either pBAD18 (lanes 1 to 4) or pBADcsgD (lanes 5 and 6), grown in YESCA medium at 28°C for 8 h (lanes 1, 3, and 5) or 24 h (lanes 2, 4, and 6), and β-galactosidase activity was assayed. (C) Locations of CsgD-binding sites within the spacer region between fliE and fliF. The CsgD-binding site of CsgD is indicated by the solid-lined box, while two binding sites of FlhDC, the activator for the fliE and fliF operons, are indicated by the boxes with dotted lines. The fliE and fliF operons are both transcribed by both σ70 and σ28 RNA polymerases.
The activity of the fliF promoter was also enhanced in the csgD mutant (Fig. 9B), even though the CsgD-binding site is located upstream from both the P1 and P2 promoters (Fig. 3, fliF lane). In limited cases, upstream bound transcription factors repress transcription initiation due to interference with promoter escape by a high-affinity protein-protein interaction between transcription factors and the RNA polymerase alpha subunit (55). The genes for flagellum formation are considered to be under a complex regulation system, but at the present time, little is known regarding transcription factors involved in regulation of these flagellum operons, except that the FlhDC complex is the master regulator of the hierarchy of the regulation network (15, 48). RpoF, one of the seven E. coli sigma factors, is synthesized under the control of FlhDC. The CsgD-binding site between the fliE and fliF operons overlaps with the binding site of FlhDC, the master regulator of genes for flagellum formation (Fig. 9C). Thus, the apparent repression of the fliF promoter by CsgD may also be attributable to interference of FlhDC in binding to its target site.
DISCUSSION
Formation of a biofilm begins with the attachment of free-floating microorganisms to a solid surface and cell-cell contact using adhesion structures such as curli fimbriae. CsgD (curlin subunit gene D) is a FixJ/LuxR/UhpA family transcription factor that regulates the csgBAC and csgDEFG operons for the synthesis, secretion, and assembly of curli components (10, 20, 22, 27, 41, 42). Besides the csgBAC and csgDEFG operons, CsgD is known to regulate yaiC (renamed adrA), which encodes the diguanylate cyclase for the synthesis of c-di-GMP (14, 38) and plays dual roles in the enhancement of biofilm formation and inhibition of cell motility through repression of flagellum production and rotation (52). The C-terminal domain of CsgD contains a potential helix-turn-helix DNA-binding motif, while its N-terminal domain contains the receiver domain (51). Changes in various environmental conditions, such as low osmolarity, low temperature, starvation, and high cell density, influence, directly or indirectly, the expression of CsgD (8, 10, 20). Based on the responsibility of CsgD expression in responding to various external stresses, a number of transcription factors, each monitoring a different factor or condition, are involved in the regulation of csgD promoter (24, 36, 37). At present, however, the specific effector(s) or condition(s) that affects CsgD function has not yet been identified.
To obtain insights regarding the whole network of gene regulation involving CsgD, the identification of a whole set of regulation targets by CsgD is an initial and essential step. In the absence of knowledge of effectors affecting CsgD activity, we searched for the regulation targets by analysis of CsgD-associated genes in vivo using the ChIP-chip system (17, 18, 45), and we identified more than 20 CsgD-binding sites along the E. coli genome (Fig. 1). Noteworthy is that the eight members of the CsgD regulon so far identified in this study carry one to four CsgD-binding sequences that are located at various positions from the respective transcription start position (Fig. 3). Such a unique organization of CsgD-binding sites has been reported in several other cases, such as for the E. coli TyrR regulon (39) and the PhhR regulon of Pseudomonas putida (23). All these transcription factors are dual regulators, acting as an activator or a repressor, and the mode of transcription regulation correlates with the location of regulator binding relative to the promoter. It should be noted, however, that the functional form of CsgD remains unidentified under the culture conditions employed. For instance, CsgD is supposed to be phosphorylated through an as-yet-unidentified mechanism (56).
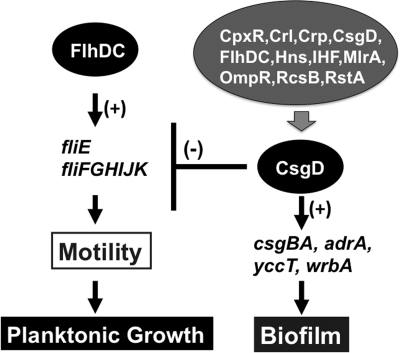
In the promoter assay utilizing lacZ fusions, CsgD exhibited activation of a set of genes and repression of another set of genes, suggesting the unique nature of CsgD as a transcription factor with a dual role, similar to IclR family transcription factors (32). However, it cannot be ruled out that there were two forms of CsgD, i.e., phosphorylated and unphosphorylated, under the experimental conditions employed. The genes related to biofilm formation are activated by CsgD, but here we found that CsgD repressed the genes related to flagellum formation and rotation. CsgD directly represses transcription of both the fliE and fliFGHIJK operons by binding to the spacer region. The binding site of the CsgD repressor in this region overlaps with that of the FlhDC complex, the master regulator of flagellum formation (15), implying competition in DNA binding between these two master regulators, CsgD for biofilm formation and FlhDC for flagellum formation. Interestingly, the binding consensus sequences are also similar for these two regulators. In addition to the fliE-fliFGHIJK spacer region, CsgD associates at the spacer regions between flgB and flgA within the flgBAMN operon, between fliC and fliD within the fliCD operon, and on the fliL gene within the fliFGHIJK operon, altogether leading to inhibition of flagellum formation and assembly; these findings are in concert with the proposal that CsgD is involved in regulation of flagellum-related operons (56). Along this line, it is noteworthy that c-di-GMP, the metabolic product of CsgD-activated AdrA diguanylate cyclase, is an inhibitor of cell motility (38), acting by interfering with flagellar motor speed through the c-di-GMP-binding protein YcgR (1, 7, 52). Accordingly, CsgD interferes with the formation and function of flagella, leading to inhibition of planktonic growth for the switch to biofilm formation (Fig. 10).
Fig. 10.
The regulatory roles of CsgD in biofilm formation and cell motility. The master regulator CsgD for biofilm formation represses the genes for flagellum formation and motility, thereby switching the bacterium's lifestyle to the biofilm mode.
Supplementary Material
ACKNOWLEDGMENTS
We thank David C. Grainger, Tomohiro Shimada, and Stephen J. W. Busby for technical advice on the ChIP-chip assay and Ayako Kori and Kayoko Yamada for CsgD protein purification.
This work was supported by a grant-in-aid for Scientific Research on Priority Area System Cell Engineering by Multi-Scale Manipulation (17076016) to A.I., grants-in-aid for Scientific Research A (21241047) and B (18310133) to A.I., and a Grant-in-Aid for JSPS Fellows (218850) to H.O. from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We also acknowledge the support from the Micro-Nano Technology Research Center of Hosei University. H.O. is a recipient of a JSPS postdoctoral fellowship.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Armitage J. P., Berry R. M. 2010. Time for bacteria to slow down. Cell 141:24–26 [DOI] [PubMed] [Google Scholar]
- 2. Arnqvist A., Olsen A., Normark S. 1994. Sigma S-dependent growth-phase induction of the csgBA promoer in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021–1032 [DOI] [PubMed] [Google Scholar]
- 3. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnhart M. M., Chapman M. R. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beloin C., et al. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 6. Bodero M. D., Pilonieta M. C., Munson G. P. 2007. Repression of the inner membrane lipoprotein NlpA by Rns in enterotoxigenic Escherichia coli. J. Bacteriol. 189:1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boehm A., et al. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 8. Bougdour A., Lelong C., Geiselmann J. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279:19540–19550 [DOI] [PubMed] [Google Scholar]
- 9. Brombacher E., Baratto A., Dorel C., Landini P. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 188:2027–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brombacher E., Dorel C., Zehnder A. J., Landini P. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847–2857 [DOI] [PubMed] [Google Scholar]
- 11. Brouns S. J., et al. 2008. Small CRISP RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Casadaban M. J. 1976. Transcription and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 13. Chapman M. R., et al. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chirwa N., Herrington M. B. 2003. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology 149:525–535 [DOI] [PubMed] [Google Scholar]
- 15. Claret L., Hughes C. 2002. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J. Mol. Biol. 321:185–199 [DOI] [PubMed] [Google Scholar]
- 16. Gerstel U., Park C., Romling U. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49:639–654 [DOI] [PubMed] [Google Scholar]
- 17. Grainger D. C., Busby S. J. 2008. Methods for studying global patterns of DNA binding by bacterial transcription factors and RNA polymerase. Biochem. Soc. Trans. 36:754–757 [DOI] [PubMed] [Google Scholar]
- 18. Grainger D. C., Hurd D., Harrison M., Holdstock J., Busby S. J. 2005. Study of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 102:17693–17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gualdi L., Tagliabue L., Landini P. 2007. Biofilm formation-gene expression relay system in Escherichia coli: modulation of sigma S-dependent gene expression by the CsgD regulatory protein via sigma S protein stabilization. J. Bacteriol. 189:8034–8043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gualdi L., et al. 2008. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 154:2017–2024 [DOI] [PubMed] [Google Scholar]
- 21. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hammar M., Arnqvist A., Bian Z., Olsen A., Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661–670 [DOI] [PubMed] [Google Scholar]
- 23. Herrera M. C., Krell T., Zhang X., Ramos J.-L. 2009. PhhR binds to target sequences at different distances with respect to RNA polymerase in order to activate transcription. J. Mol. Biol. 394:576–586 [DOI] [PubMed] [Google Scholar]
- 24. Ishihama A. 2010. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 34:628–645 [DOI] [PubMed] [Google Scholar]
- 25. Ishihama A. 1993. Protein-protein communication within the transcription apparatus. J. Bacteriol. 175:2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jishage M., Ishihama A. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 177:6832–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loferer H., Hammar M., Normark S. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 16:11–23 [DOI] [PubMed] [Google Scholar]
- 28. Makinoshima H., et al. 2003. Growth phase-coupled alterations in cell structure and function of Escherichia coli. J. Bacteriol. 185:1338–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendoza-Vargas A., et al. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 31. Minamino T., Moriya N., Hirano T., Hughes K. T., Namba K. 2009. Interaction of fliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol. Microbiol. 74:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molina-Henares A. J., Krell T., Guazzaroni M. E., Segura A., Ramos J. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30:157–186 [DOI] [PubMed] [Google Scholar]
- 33. Muller V., Jones C. J., Kawagishi I., Aizawa S., Macnab R. M. 1992. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J. Bacteriol. 174:2298–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ogasawara H., et al. 2007. Genomic SELEX search for target genes under the control of PhoQP-RstBA signal relay cascade. J. Bacteriol. 189:4791–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogasawara H., Ishida Y., Yamada K., Yamamoto K., Ishihama A. 2007. PdhR (pyruvte dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 189:5534–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogasawara H., Yamamoto K., Ishihama A. 2010. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 312:160–168 [DOI] [PubMed] [Google Scholar]
- 37. Ogasawara H., Yamada K., Kori A., Yamamoto K., Ishihama A. 2010. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483 [DOI] [PubMed] [Google Scholar]
- 38. Pesavento C., et al. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pittard J., Camakaris H., Yang J. 2005. The TyrR regulon. Mol. Microbiol. 55:16–26 [DOI] [PubMed] [Google Scholar]
- 40. Pratt L. A., Silhavy T. J. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225–1236 [DOI] [PubMed] [Google Scholar]
- 41. Prigent-Combaret C., et al. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213–7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Romling U., Sierralta W. D., Eriksson K., Normark S. 1998. Multicellular and aggregative behavior of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 43. Shimada T., Fujita N., Maeda M., Ishihama A. 2005. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells 10:907–918 [DOI] [PubMed] [Google Scholar]
- 44. Shimada T., Hirao K., Kori A., Yamamoto K., Ishihama A. 2007. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimdines. Mol. Microbiol. 66:744–757 [DOI] [PubMed] [Google Scholar]
- 45. Shimada T., Ishihama A., Busby S. J. W., Grainger D. C. 2008. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 36:3950–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simm R., Morr M., Kader A., Nimtz M., Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic d-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 47. Simons R. W., Houman F., Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 48. Stafford G. P., Ogi T., Hughes C. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 151:1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Studier F. W., Mofatt B. A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 50. Teramoto J., Yoshimura S., Takeyasu K., Ishihama A. 2010. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res. 38:3605–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Volz K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741–11753 [DOI] [PubMed] [Google Scholar]
- 52. Wolfe A. J., Visick K. L. 2008. Get the message out: cyclic-Di-GMP regulates multiple level of flagellum-based motility. J. Bacteriol. 190:463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamada M., Izu H., Nitta T., Kurihara K., Sakurai T. 1998. High-temperature, nonradioactive primer extension assay for determination of a transcription initiation site. Biotechniques 25:72–75 [DOI] [PubMed] [Google Scholar]
- 54. Yamamoto K., et al. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448–1456 [DOI] [PubMed] [Google Scholar]
- 55. Yamamoto K., Ishihama A. 2003. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol. Microbiol. 47:183–194 [DOI] [PubMed] [Google Scholar]
- 56. Zakikhany K., Harrington C. R., Nimtz M., Hinton N., Romling U. 2010. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 77:771–786 [DOI] [PubMed] [Google Scholar]
- 57. Zogaj X., Nimtz N., Rohde M., Bokranz W., Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.