Abstract
We show that Escherichia coli DinB polymerase, which creates single-base deletions, prefers to extend slipped DNA substrates with the skipped base at the −4 position. A DinB(Y79L) variant, which extends these substrates less efficiently in vitro, allows the proofreading function of polymerase III to reverse their formation in vivo.
TEXT
The Y family of DNA polymerases catalyze replication on damaged DNA templates in a process called translesion DNA synthesis (TLS), which allows cells to tolerate DNA damage (7). Members of the Y family make relatively few contacts with the substrate DNA and incoming deoxynucleoside triphosphate (dNTP), which allows these enzymes to accommodate DNA lesions within their active sites (7). Several Y-family DNA polymerases have a marked preference for particular classes of DNA lesions (11, 14, 15), not only because of their open active sites but also due to their evolutionary divergent little-finger domains (for a review, see reference 17). In particular, DinB orthologs, which are the most ubiquitous of this polymerase class, are capable of bypassing certain N2-deoxyguanosine (N2-dG) adducts efficiently and accurately (9, 10). Escherichia coli DinB, the founding member of this subfamily, inserts dCTP across from N2-furfuryl-dG with 15-fold more catalytic efficiency than that found when dCTP is inserted across from an undamaged dG (10). DinB also extends from a dC:N2-furfuryl-dG base pair with a 25-fold increase in catalytic efficiency compared to a C·G base pair (9). However, the DinB variant DinB(Y79L) stalls 3 nucleotides downstream of the N2-furfuryl-dG lesion in vitro and, when expressed in vivo, results in death when cells are challenged with nitrofurazone, a compound which produces N2-dG adducts (9). Therefore, successful TLS requires not only insertion of the correct nucleotide opposite the lesion, but also, as suggested by our DinB(Y79L) results, efficient extension from it to avoid cell death (9).
Due to their accommodating active sites and fewer enzyme-DNA contacts, DinB, and all TLS polymerases, have lower replication fidelity on undamaged DNA than replicative polymerases. We have recently shown that Escherichia coli DinB (Pol IV) creates −1 frameshifts via a template-slippage mechanism on homopolymeric nucleotide runs (6). The template-slippage mechanism, originally proposed by Streisinger et al. (18), suggests that the polymerase can misalign the primer and template strand, thereby resulting in an unpaired “extrahelical” base within the template strand (Fig. 1 A). We proposed that DinB binds DNA templates containing homopolymeric nucleotide runs in a equilibrium between slipped and nonslipped DNA conformations, consistent with the template-slippage model (6). The incoming dNTP then displaces the equilibrium in favor of the slipped DNA conformation, thereby potentially resulting in a single-base deletion. However, it is still unclear which base within a homopolymeric run is most likely to become extrahelical during a DinB-mediated template-slippage reaction.
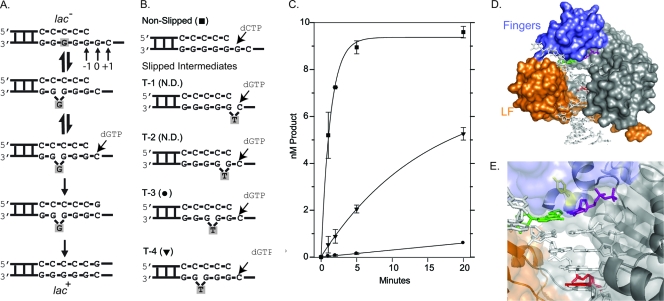
Fig. 1.
DinB prefers to extend DNA substrates where the “extrahelical” base is located at the −4 position. (A) Using the CC108 lac mutant (lac−) target as an example DNA substrate, the Streisinger template-slippage mechanism is shown. The DNA substrate is in equilibrium between nonslipped and slipped structures (top two structures). The equilibrium can be displaced in favor of the slipped substrate by a dNTP complementary to the +1 base. Further polymerization cycles by DinB will result in the CC108 strain reverting to lac+ as a result of this specific −1 frameshift. The skipped base is shaded throughout the mechanism. Our numbering system is shown. (B) The sequence around the primer terminus of the synthetic oligonucleotide substrates used in our reactions is shown. The primer (20 bp) and the template (33 bp) have the same sequence as the CC108 lac mutant target, except that a dT is positioned within the template strand (indicated) to mimic an extrahelical base. The dNTP used to initiate the reactions are shown. The symbols used to plot the results of the primer extension time course assay in panel C are shown. N.D. indicates that no dNTP incorporation was detected within 20 min. (C) A primer extension time course assay plotting product formation versus time suggests that DinB prefers to extend the T-4 substrate as opposed to the other slipped substrates. (D) A surface representation of our DinB homology model based on a crystal structure of Dpo4 (10). The fingers (blue), little-finger (LF; orange), palm and thumb (gray) domains are shown bound to a DNA substrate. Some solvent-exposed space between the fingers and little-finger domains where the extrahelical base is hypothesized to be located during a template-slippage reaction is visible in the surface representation. (E) A close-up of the active site showing the side chain of Y79 in a stick representation (yellow) as well as the templating base (green), incoming nucleotide (purple), and −4 base (red). The surface representation is semitransparent to reveal the secondary structure of the little-finger domain (orange) as well as that of the active site located in the palm domain. The secondary structure of the fingers domain is not shown for clarity, as it partially encloses the active site.
In vitro primer extension analysis on synthetic slipped DNA substrates, in which one base in the template strand lacked a base-pairing partner, demonstrated that Dbh, the archaeal DinB homolog from Sulfolobus acidocaldarius, preferred to extend from DNA substrates in which the unpaired base was located at the −3 position (20). Crystal structure analysis confirmed that Dbh stably binds a DNA substrate containing an unpaired base at the −3 position, with the base in an extrahelical conformation where it contacts residues on the surface of the little-finger domain (20). Similar analysis of Dpo4, the archaeal DinB homolog from Sulfolobus solfataricus, indicates that Dpo4 extends equally well from DNA containing an unpaired base at the −3 and −4 positions, but when given a choice during crystallization, Dpo4 prefers to bind when the base at the −4 position is in an extrahelical conformation, where it protrudes into the gap between the polymerase and little-finger domains (21). This difference in the positioning of the extrahelical base is potentially due to the amount of solvent-exposed area located between the finger and little-finger domains; the little-finger domain touches the finger domain in the Dpo4 structure (11, 21). To date, no full-length structural data for E. coli DinB exists to predict where the extrahelical base will be positioned. However, by using primer extension analysis that was conceptually similar to that used in the Dbh study (20), we sought to enhance our understanding of E. coli DinB-mediated single-base deletions by analyzing their preference for extending substrates, in which the extrahelical base is located at different positions relative to the primer terminus. We designed synthetic DNA substrates which mimic the mutated lacZ allele target used in the Cupples et al. CC108 reversion assay (4). This system has been used extensively to study DinB's propensity to create single-base deletions when overexpressed, and therefore, we were able to take advantage of both the in vivo and in vitro assay systems to further study DinB function (8, 12, 13). CC108 is a lac mutant that can convert to lac+ by a −1 frameshift mutation within a run of 6 dG nucleotides (Fig. 1A) (4). To mimic this system, our nonslipped control DNA substrate has the same sequence context as that found in CC108; however, the T-1, T-2, T-3, and T-4 substrates each have an unpaired deoxyribosylthymine (dT) within the template strand to mimic the extrahelical base at positions −1 to −4 (Fig. 1B). Similar to the Dbh and Dpo4 studies, we carried out a time course DNA primer extension analysis to determine which slipped DNA intermediate DinB extended the most efficiently. A solution of DinB (50 nM), purified by methods previously described (10), and the indicated DNA substrate (10 nM) in Tris reaction buffer (6) were mixed with an incoming dNTP (1 mM; dCTP or dGTP as indicated) to initiate the reaction. Reactions were then quenched at intervals during a 20-min period, resolved by electrophoresis, and quantitated as previously described (6). Consistent with the Dpo4 crystal structure, DinB displays the greatest preference for the T-4 substrate, extending ≈50% of the DNA primers within 20 min, which is at least 10-fold more than that of any of the other slipped intermediates (Fig. 1C). However, unlike Dpo4, DinB does not extend the T-3 substrate with the same efficiency as it does the T-4. At best, DinB extends the T-3 substrate modestly (≈10%), while no extension of the T-2 or T-1 substrate was observed under these conditions.
The preference for an unpaired base at the −4 position could suggest that the little-finger domain, which is attached to the remainder of the polymerase via a linker domain, is positioned in a manner that more closely resembles Dpo4 than Dbh. Our DinB homology model (10), which is based on the Dpo4 structure, positions the little-finger domain such that it touches the fingers domain (Fig. 1D). However, other architectural aspects of Dpo4 and DinB may result in a similar preference for the location of the extrahelical base. For example, residues that help stabilize the unpaired base may be more similarly positioned in Dpo4 and DinB. Preference for the position of the extrahelical base does not necessarily suggest that Dpo4 and DinB are more closely related. In fact, molecular modeling suggests that the active site of Dpo4 is a hybrid of the DinB and UmuD′2C (Pol V) active sites (3).
We then wondered if the preference of DinB to extend from slipped substrates with the unpaired base at the −4 position may be related to the propensity of the DinB(Y79L) variant to arrest with N2-furfuryl-dG at the −4 position (9). This hypothesis would further support our previous suggestion that some feature of DinB orthologs that enables them to bypass N2-dG adducts would also result in single-base deletion formation by a common mechanism (6). One potential architectural feature of DinB orthologs that could lead to a common mechanism might indeed be the pocket between the finger and little-finger domains, which is on the minor groove side of the active site (Fig. 1D). N2-furfuryl-dG is a minor groove adduct, and thus, the pocket may have evolved to accommodate N2-dG adducts for the final extension steps during TLS. Residue Y79 is located within the active site (Fig. 1E), and the DinB(Y79L) variant is ≈25-fold less efficient at extending DNA substrates when the N2-furfuryl-dG lesion is located at the −4 position compared to that of wild-type DinB (9). Therefore, we wondered if DinB(Y79L) would be similarly inhibited in extension from slipped intermediates using the same primer extension assay conditions described above. This variant is only modestly less efficient than the wild type at extending from the T-4 substrate (Fig. 2 A). There are ≈3-fold-fewer primers extended by DinB(Y79L) in 5 min than wild-type DinB, while the nonslipped control reactions are indistinguishable. Moreover, we did not observe any primer extension from the T-1, T-2, and T-3 substrates using DinB(Y79L). The absence of detectable dNTP incorporation on the T-3 substrate by DinB(Y79L) (none detected) is consistent with our conclusion that this variant is less efficient (≥3-fold) at extending slipped substrates than the wild type.
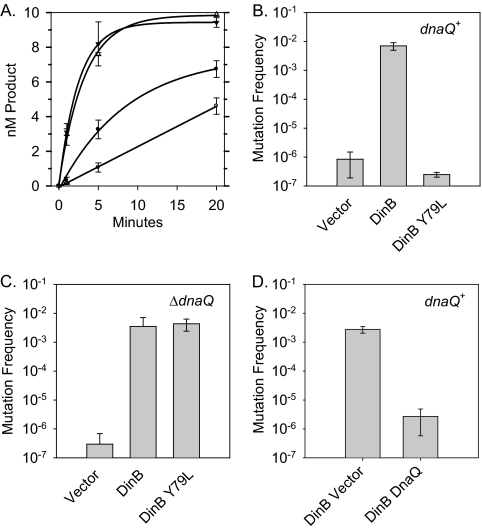
Fig. 2.
Inefficient extension of slipped DNA substrates leads to an opportunity for DnaQ to realign the primer-template junction. (A) In vitro primer extension analysis indicates that DinB(Y79L) (open circles) is modestly less efficient at extending T-4 substrates than DinB (closed circles), but the two polymerases are indistinguishable at extending a nonslipped control [DinB, closed triangles; DinB(Y79L), open triangles]. (B) Plot of the mutation frequency measured by the CC108 lac mutant reversion assay for strains overexpressing DinB (pGY782) or DinB(Y79L) (pGY782 Y79L) compared to that for a vector control (pWSK30). (C) Overexpression of DinB(Y79L) in CC108 ΔdnaQ::kan results in an increase in mutation frequency comparable to that of DinB overexpression. The mutation frequency and standard deviation reported is derived from 3 independent CC108 ΔdnaQ::kan isolates, which were freshly transformed with DinB-overexpressing plasmids. (D) Expression of DnaQ from a multicopy plasmid (pPF2) decreases the mutation frequency of a strain overexpressing DinB (pGY782).
In striking contrast, we saw a major difference between the abilities of DinB and DinB(Y79L) to cause −1 frameshifts when overexpressed in vivo. We measured the DinB- and DinB(Y79L)-mediated single-base deletion frequencies using the CC108 assay system with the method previously described (13) but with a slight modification. Overnight cultures were inoculated (5 to 10 independent cultures for each strain) into LB-ampicillin from fresh transformants before overnight incubation at 37°C. Strikingly, we saw a ≈104-fold reduction in the mutagenesis frequency when DinB(Y79L) was overexpressed (2.5 × 10−7) compared to when wild-type DinB was overexpressed (7.0 × 10−3). Moreover, the mutation frequency of the strain overexpressing DinB(Y79L) is similar to that of the empty vector control (8.4 × 10−7).
The results described above clearly indicate that DinB(Y79L) is not a mutator when overexpressed; however, it seemed unlikely that the very modest reduction in the ability of DinB(Y79L) to extend from slipped intermediates could account for a ≈104-fold reduction in the spontaneous mutation frequency. Therefore, we wondered if the decreased ability of the DinB(Y79L) derivative to extend would result in a greater opportunity for the proofreading function of the replicative polymerase, polymerase III (Pol III), to realign the primer and template strand, thus removing the extrahelical base and preventing the formation of a lac+ colony. The 3′-to-5′ exonucleolytic proofreading function of Pol III is provided by the ε subunit, encoded by dnaQ, which removes incorrectly incorporated nucleotides by melting the primer-template junction to expose the primer end to its active site (2, 16). Even without cleavage, the melting of the primer-template junction could remove the extrahelical base if the primer was to subsequently realign correctly with the template in a nonslipped conformation.
To test this hypothesis, we constructed a CC108 ΔdnaQ::kan strain by transducing the ΔdnaQ::kan allele from the Keio knockout collection by P1vir transduction into CC108 (1). Strikingly, when we overexpressed DinB(Y79L) in three independent CC108 ΔdnaQ::kan isolates, we observed a complete restoration of DinB(Y79L)'s mutator capability (Fig. 2). The mutation frequencies of DinB (3.5 × 10−3) and DinB(Y79L) (4.4 × 10−3) are virtually the same in a ΔdnaQ::kan strain. Moreover, the mutation frequencies caused by overexpression of DinB and DinB(Y79L) in a ΔdnaQ::kan strain are similar to that of DinB overexpressed in a dnaQ+ strain. Taken together, these results support the conclusion that efficient extension of slipped intermediates by DinB does not give DnaQ an opportunity to act and thus remove extrahelical bases via proper alignment of the primer terminus.
In principle, DnaQ could gain access to the primer terminus after DinB stalls by virtue of a polymerase switchback to Pol III, or it could gain access to the primer terminus without an interaction with the Pol III catalytic subunit DnaE. To begin to distinguish between these possibilities, we introduced the multicopy plasmid pPF2 (DnaQ+) (5) into a CC108 strain to see if it could suppress DinB-mediated mutagenesis when DinB was overexpressed. We observed a ≈1,000-fold reduction of DinB-mediated mutagenesis (2.7 × 10−6) when the multicopy plasmid carrying DnaQ was present rather than the empty vector control (pGD104; 2.7 × 10−3). Thus, it seems plausible that the effect of DnaQ on removing extrahelical bases is independent of its interaction with DnaE. However, since this phenomenon was observed with strains expressing higher-than-normal levels of DinB and DnaQ, it is difficult to absolutely eliminate a role for the Pol III catalytic subunit. Moreover, physical interactions with other proteins, such as the gene products of umuD (UmuD2 and UmuD′2), may potentially add to the complexity of the system (8, 19). DnaQ can suppress the umuDC-mediated cold sensitivity phenotype via a direct interaction with the umuD gene products, while UmuD can modulate the mutagenic capability of DinB. These results suggest that more careful studies are warranted before physiological roles can be fully defined.
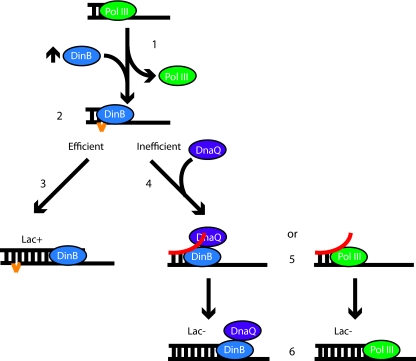
The data presented here further highlights the complex regulation present at the replication fork to ensure that the correct enzymes have access to primer terminus and is consistent with the following working model (Fig. 3). DinB overexpression results in a polymerase switch from Pol III to DinB (Fig. 3, step 1), which results in a greater opportunity for the creation of slipped substrates on homopolymeric nucleotide runs (step 2). If DinB efficiently extends these substrates, then a single-nucleotide deletion is generated, as measured by Lac+ colonies in CC108 (Fig. 3, step 3). If DinB inefficiently extends, then DnaQ, either as an independent subunit or as part of the Pol III holoenzyme, has a greater opportunity to regain access to the primer terminus (Fig. 3, step 4). Once there, DnaQ can melt the primer template terminus during its proofreading cycle, thereby removing the extrahelical base (Fig. 3, step 5). The primer then reanneals in a nonslipped conformation, thus resulting in a Lac− colony (step 6).
Fig. 3.
Model for DnaQ-mediated removal of slipped DNA substrates. (Step 1) DinB (blue) overexpression, as indicated by the up arrow, results in a polymerase switch that presumably removes the Pol III holoenzyme (green). (Steps 2 and 3) On homopolymeric nucleotide runs, DinB can generate slipped DNA substrates (step 2; the extrahelical base is indicated in orange) that, if extended efficiently, can result in a Lac+ colony in CC108 (step 3). (Step 4) If the slipped substrate is inefficiently extended, DnaQ (purple) can gain access to the primer terminus independently of DnaE or as part of the Pol III holoenzyme (green). (Step 5) Through the action of DnaQ, the DNA primer terminus is melted. (Step 6) If the primer is annealed in a nonslipped conformation, then extension will result in a Lac− colony.
Acknowledgments
We thank Janice Pata for critical reading of the manuscript and Steve Bell and Uttam Rajbhandary for the use of instruments. The pPF2 plasmid was a kind gift from Patricia Foster.
This work is supported by National Institutes of Health grants RO1 CA021615 (to G.C.W.), F32 GM079885 (to J.J.F.), and P30 ES002019 (to the MIT Center for Environmental Sciences). G.C.W. is an American Cancer Society Professor.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brutlag D., Kornberg A. 1972. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 247:241–248 [PubMed] [Google Scholar]
- 3. Chandani S., Loechler E. L. 2009. Y-family DNA polymerases may use two different dNTP shapes for insertion: a hypothesis and its implications. J. Mol. Graph. Model. 27:759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cupples C. G., Cabrera M., Cruz C., Miller J. H. 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foster P. L., Sullivan A. D., Franklin S. B. 1989. Presence of the dnaQ-rnh divergent transcriptional unit on a multicopy plasmid inhibits induced mutagenesis in Escherichia coli. J. Bacteriol. 171:3144–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Foti J. J., Delucia A. M., Joyce C. M., Walker G. C. 2010. UmuD(2) inhibits a non-covalent step during DinB-mediated template slippage on homopolymeric nucleotide runs. J. Biol. Chem. 285:23086–23095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedberg E. C., et al. 2005. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 8. Godoy V. G., et al. 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28:1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jarosz D. F., Cohen S. E., Delaney J. C., Essigmann J. M., Walker G. C. 2009. A DinB variant reveals diverse physiological consequences of incomplete TLS extension by a Y-family DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 106:21137–21142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jarosz D. F., Godoy V. G., Delaney J. C., Essigmann J. M., Walker G. C. 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225–228 [DOI] [PubMed] [Google Scholar]
- 11. Johnson R. E., Prakash S., Prakash L. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 283:1001–1004 [DOI] [PubMed] [Google Scholar]
- 12. Kim S. R., et al. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. U. S. A. 94:13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim S. R., Matsui K., Yamada M., Gruz P., Nohmi T. 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207–215 [DOI] [PubMed] [Google Scholar]
- 14. Masutani C., et al. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399:700–704 [DOI] [PubMed] [Google Scholar]
- 15. McCulloch S. D., et al. 2004. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature 428:97–100 [DOI] [PubMed] [Google Scholar]
- 16. Miller H., Perrino F. W. 1996. Kinetic mechanism of the 3′→5′ proofreading exonuclease of DNA polymerase III. Analysis by steady state and pre-steady state methods. Biochemistry 35:12919–12925 [DOI] [PubMed] [Google Scholar]
- 17. Pata J. D. 2010. Structural diversity of the Y-family DNA polymerases. Biochim. Biophys. Acta 1804:1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Streisinger G., et al. 1966. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb. Symp. Quant. Biol. 31:77–84 [DOI] [PubMed] [Google Scholar]
- 19. Sutton M. D., Murli S., Opperman T., Klein C., Walker G. C. 2001. umuDC-dnaQ interaction and its implications for cell cycle regulation and SOS mutagenesis in Escherichia coli. J. Bacteriol. 183:1085–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson R. C., Pata J. D. 2008. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol. Cell 29:767–779 [DOI] [PubMed] [Google Scholar]
- 21. Wu Y., Wilson R. C., Pata J. D. 2011. The Y-family DNA polymerase Dpo4 uses a template slippage mechanism to create single-base deletions. 193:2630–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]