Abstract
Bacterial strategies of innate immune evasion and essential metabolic functions are critical for commensal-host homeostasis. Previously, we showed that Sap translocator function is necessary for nontypeable Haemophilus influenzae (NTHI) behaviors that mediate diseases of the human airway. Antimicrobial peptide (AP) lethality is limited by binding mediated by the Sap complex. SapA shares homology with the dipeptide-binding protein (DppA) and the heme-binding lipoprotein (HbpA), both of which have previously been shown to bind the iron-containing compound heme, whose acquisition is essential for Haemophilus survival. Computational modeling revealed conserved SapA residues, similarly modeled to mediate heme binding in HbpA. Here, we directly demonstrate that SapA bound heme and was essential for heme utilization by iron-starved NTHI. Further, the Sap translocator permease mediated heme transport into the bacterial cytoplasm, thus defining a heretofore unknown mechanism of intracytoplasmic membrane heme transport in Haemophilus. Since we demonstrate multiple ligand specificity for the SapA-binding protein, we tested whether APs would compete with heme for SapA binding. We showed that human β-defensins 2 and 3, human cathelicidin LL-37, human neutrophil protein 1, and melittin displaced heme bound to SapA, thus supporting a hierarchy wherein immune evasion supercedes even the needed iron acquisition functions of the Sap system.
INTRODUCTION
The sap gene products share homology with a family of ATP-binding cassette (ABC) transporters that are diverse in substrate binding and uptake (1, 12). Based upon known ABC transporter structures, the Sap transporter is predicted to share a four-domain architecture comprised of two transmembrane proteins (SapB and SapC) and two membrane-associated nucleotide-binding proteins (SapD and SapF) that provide energy for ATP-dependent translocation of substrate across the bacterial inner membrane (33, 34). The SapA substrate-binding protein is predicted to localize to the bacterial periplasm due to the presence of a signal sequence and homology to other periplasmic solute-binding proteins involved in peptide transport (15).
SapA shares homology with a family of periplasmic binding proteins (DppA, OppA, NikA, HbpA) that mediate the uptake of dipeptides, oligopeptides, nickel, and the iron-containing compound heme (11, 36, 39, 41). These proteins primarily mediate the recognition of substrates that are then targeted for transport across the inner membrane into the bacterial cytoplasm.
Genes that encode the Sap transporter are conserved in the Gram-negative bacteria Escherichia coli, Salmonella, Vibrio, Pasteurella, Erwinia, Actinobacillus, and Haemophilus species, which suggests an important function for this transporter among bacterial species. In all studies, Sap transporter proteins provide a mechanism of resistance to antimicrobial peptides (APs), key components of host innate immunity, often with significant bactericidal activity (9). APs are typically small, cationic peptides that have affinity for the negatively charged bacterial membrane surface and mediate lethality via membrane insertion and disruption of membrane potential and electrochemical gradients (4, 31, 35). We recently demonstrated a novel AP resistance mechanism in nontypeable Haemophilus influenzae (NTHI) whereby SapA directly binds AP, signals increased sap gene expression, and subsequently enhances a bacterial AP resistance phenotype (22–24). Further, SapA was essential for survival of NTHI in vivo in an experimental mammalian model of human airway disease (22, 23). Importantly, this work identified the SapA substrate as a small peptide that was cationic in nature, similar in character to metabolic substrates bound by other members of this periplasmic binding protein family.
NTHI is a Gram-negative nasopharyngeal commensal microorganism, yet it can also mediate human airway disease. NTHI predominates in otitis media (OM) and other localized respiratory diseases such as acute sinusitis and community-acquired pneumonia and has important consequences for patients with chronic obstructive pulmonary disease or cystic fibrosis (17, 30, 38, 40, 42). The pathogenic potential of NTHI is dictated by the micronutrient environment of the host and the ability to resist innate immune clearance mechanisms. Microbes residing on the mucosal surface require iron for survival and key intracellular reactions (21). Thereby, microbes adapt to dynamic host environments by developing mechanisms to compete with their host for essential iron. Haemophilus, a strictly obligate human commensal, lacks the ability to synthesize the iron-containing moiety heme and therefore must possess multiple mechanisms to acquire heme for use in cytoplasmic enzymes (44). Proteins that function in heme acquisition include homologues of hemoglobin- and hemoglobin-haptoglobin-binding proteins (20, 25, 37), a heme-hemopexin utilization system (6), the heme utilization protein Hup (29), and the heme-binding lipoprotein HbpA (11, 26). The presence of numerous proteins involved in heme acquisition suggests that there is tight regulation of iron homeostasis in Haemophilus, yet the heme permease required for transport of heme across the cytoplasmic membrane into the cytoplasm remains unknown.
Since SapA shares homology with E. coli DppA and NikA, both of which have been shown to bind heme yet are absent from all sequenced Haemophilus strains, we investigated a potential role for the Sap transporter in the binding, utilization, and transport of heme in this prototype strain. Recognition of heme by SapA would equip this benign commensal with the ability to respond to iron limitation concomitant with a mechanism to resist AP lethality, as previously shown (22, 23). Our results indicated that recombinant SapA [(r)SapA] bound heme and a SapA-deficient strain was unable to utilize heme for growth following iron starvation. Importantly, we showed that the Sap translocator permease was required for heme transport across the Haemophilus inner membrane, thus providing a heretofore undescribed mechanism for heme acquisition and uptake for this important human pathogen. That APs compete for heme binding to SapA further suggests a shared substrate-binding site and a multifunctional role for the Sap transporter in both the metabolic needs of Haemophilus and its ability to resist effectors of innate immunity.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The parental NTHI strain 86-028NP is a minimally passaged clinical isolate obtained at Nationwide Children's Hospital. This strain has been extensively characterized in chinchilla models of OM and has been sequenced and annotated (13, 14). The sapA mutant was constructed by insertional mutagenesis as previously described (22). Complementation was performed with vector pSPEC1 as previously described (22). The sapBC deletion mutant was constructed as described below.
NTHI was grown on chocolate II agar (Becton Dickinson, Sparks, MD) or a chemically defined iron source medium (DIS) prepared as previously described (22). To monitor heme utilization, strain 86-028NP was grown overnight on chocolate agar at 37°C and 5% CO2, subcultured into nitric acid-washed 15-ml round-bottom glass tubes containing prewarmed iron-free DIS medium and 0 or 2 μg heme/ml, and then equilibrated to an optical density at 490 nm (OD490) of 0.65. The cultures were then diluted 10-fold and incubated statically for 24 h at 37°C and 5% CO2. This incubation in DIS (without heme) is sufficient to deplete internal stores of heme (iron starved), in contrast to growth in the presence of 2 μg heme/ml. Following incubation, the cultures were equilibrated to an OD490 of 0.37 and diluted appropriately to match cultures to 1 × 107 CFU/ml for use in each subsequent experiment.
Iron-starved or nonstarved bacteria prepared as described above (4 μl) were added to the wells of a clear, flat-bottom, 96-well MICROTEST 96 polystyrene plate (Falcon; Becton Dickinson) with a low-evaporation lid containing 100 μl of DIS supplemented with 0, 2, or 20 μg heme/ml, and the experiments were performed in triplicate. The plate was incubated at 37°C, and absorbance (optical density at 490 nm [OD490]) was monitored every hour for 16 h using the Synergy HT kinetic plate reader (BioTek, Bath, United Kingdom). The sapBC mutant strain was not iron starved but subcultured directly from chocolate agar into DIS medium in the absence or presence of heme and monitored for iron utilization by measuring bacterial growth for 10 h.
Nonpolar deletion mutation.
Construction of an unmarked, nonpolar deletion mutation of the sapB and sapC genes was performed by the recombineering strategy as described previously (46). Briefly, primers 5′-GCGCGGTGCCTTCTAAC-3′ and 5′-TCCTCCATTTTGTCGCATCT-3′ were used to amplify sapBC and 1 kb of surrounding DNA. The amplicon was then ligated into the pGEM-T-Easy (Promega, Madison, WI) vector, and the resulting plasmid was saved as pBSZ007. This plasmid was used to transform E. coli strain DY380, which expresses lambda recombination proteins. In parallel, primers 5′-GGTAGCTTAGATTTTTCCACCTTATATTTTCAGGAGAAACACTAATGATTCCGGGGATCCGTCGACC-3′ and 5′-AGGTTACAAATGTCTAAAAGTGCCATAGGCTATTCTTGATGTTGATTGATTGTAGGCTGGAGCTGCTTCG-3′, each containing 50-bp regions of homology to each end of sapBC, were used to amplify the spec-rpsl cassette from pRSM2832. This amplicon was electroporated into strain DY380/pBSZ007. A plasmid with the spec-rpsl cassette insertion in sapBC was saved as pBSZ008. This plasmid was then used to transform the streptomycin-resistant NTHI strain 86-028NPΔrpsl, and a sapBC mutant was selected on chocolate agar containing spectinomycin. To generate the sapBC unmarked deletion mutant, the temperature-sensitive plasmid pRSM2947 was used to transform the sapBC mutant strain, which was then grown at 32°C and FLP recombinase expression was induced using anhydrotetracycline. The cells were cured of the plasmid by growth at 37°C.
Computer modeling.
A theoretical model of SapA was constructed based on homology to the E. coli dipeptide-binding protein DppA by using SwissModel's default alignment against chain A of Protein Data Bank structure 1DPP (DppA with bound dipeptide ligand). To validate the predicted model, we performed a molecular dynamics (MD) simulation with heme inserted into the binding pocket of the structure modeled upon. To verify that this observation was not simply an expected property of the general DppA structure on which our SapA model is based, we also performed an MD analysis (200,000 steps, 400ps) of the 1DPP structure with the ligand removed from the simulation.
Nondenaturing polyacrylamide gel electrophoresis (PAGE) and heme detection by chemiluminescence.
Purified SapA and TrxA (thioredoxin) were prepared as previously described (22). Heme detection was performed as previously described (19). Briefly, 20 μl of purified SapA or TrxA protein (11.3 μM) was incubated at room temperature for 30 min with a 5-fold excess of heme (56.5 μM), with a mixture of heme and dipeptide AA (Leu-Gly, 11.3 μM; Sigma, St. Louis, MO) or heme and bovine serum albumin (BSA; 11.3 μM; Sigma), or with only buffer. Mixtures were separated by PAGE at 4°C in the absence of sodium dodecyl sulfate (SDS), and the proteins were transferred to nitrocellulose filters. Heme complexed with protein bands on the gel retains intrinsic peroxidase activity, which was detected by chemiluminescence (ECL; Amersham Pharmacia, Pittsburgh, PA).
To determine the specificity of heme binding, we added equimolar (11.3 μM) amounts of either TrxA or the periplasmic sialic acid-binding protein SiaBP (a kind gift from Robert S. Munson) to binding reaction mixtures that contained purified SapA and heme as described above. Since we have previously shown that numerous APs are ligands for SapA, we monitored the ability of APs to displace heme bound to SapA. SapA protein (11.3 μM) was incubated with heme (56.5 μM) in the presence of increasing concentrations (1, 10, and 100 μM) of human β-defensin 1 (hBD-1), hBD-2, hBD-3, cathelicidin LL-37, human neutrophil protein 1 (hNP-1), or melittin. Heme bound to SapA was detected as described above.
Coprecipitation of SapA with heme-conjugated agarose beads.
Purified, His-tagged SapA or TrxA (6.0 μM) was incubated with hemin-agarose (6.0 μM; Sigma) for 30 min at room temperature. Following incubation, beads were washed four times with phosphate-buffered saline (to remove unbound protein) and boiled in 5× SDS loading dye and captured protein was resolved by SDS-PAGE. Proteins were transferred to nitrocellulose filters and probed for the presence of His tagged protein by Western blot analysis.
Heme transport.
To eliminate environmental iron, glassware, perfluoroalkoxy (PFA) tubes (Savillex Corp.), and polypropylene tubes (Becton Dickinson) were prepared by overnight soaking in nitric acid, 4 washes with double-distilled H2O, air drying in a laminar-flow hood, and autoclaving. Iron-free DIS medium was prepared as previously described (22). Strain 86-028NP and the sapBC deletion mutant were grown overnight on chocolate agar, used to dilute 30 ml (iron-free) DIS medium to an OD490 of 0.65, added (1:10 dilution) to 300 ml DIS medium, and then statically incubated at 37°C in 5% CO2 for 3 h. Following incubation, the cultures were split into two halves to which either no heme or 2 μg/ml stable heme-iron (57Fe) isotope (Frontier Scientific, Logan, UT) was added and then incubated statically at 37°C in 5% CO2, for 1 h. Following incubation, each culture was sampled for OD490 and plated for determination of the number of CFU/ml. Cultures were then centrifuged at room temperature at 3,700 × g for 10 min. Pellets were washed twice with 25 ml (iron-free) DIS medium and then once with 10 ml Chelex-treated distilled H2O. Pellets were inverted to drain excess water and resuspended in 1 ml water. The washed cells were placed into iron-free glass vials (Pyrex, 20 by 125 mm; cleaned using nitric acid and perchloric acid and oven dried) and frozen at −80°C overnight.
Inductively coupled plasma mass spectroscopy (ICP-MS) analysis for absolute iron and iron isotope ratios.
Iron isotope measurements were performed by Huffman Laboratories (Golden, CO). Heme transport samples were thawed and digested with a mixture of nitric and perchloric acids, i.e., 20.0 ml concentrated reagent perchloric acid and 50.0 ml concentrated reagent nitric acid, to a final volume of 100.0 ml with type I reagent grade deionized water (Ultrapure Milli-Q; Millipore, Billerica, MA), ending in reflux with concentrated perchloric acid to ensure the total oxidation of all organic material. A volume of 1 ml of the acid mixture was added to each of the sample tubes, which were slowly heated in an aluminum block to a final temperature of 180°C, with perchloric acid vapors retained by condensation from air cooling of the upper portions of the tubes. Samples were diluted to a total volume of 20.0 ml with Milli-Q water in a final matrix of nominal 1% (vol/vol) perchloric acid. Measurement for concentrations and ratios of iron isotopes was performed using an Agilent 7500cx quadrupole inductively coupled plasma mass spectrometer with multigas collision cell interference abatement. In parallel, fortified samples using the normal (Sigma) and labeled (Frontier Scientific) heme solutions were prepared and digested at spiking levels corresponding to 0, 0.5, 1.0, 2.5, and 5.0 ng Fe/ml after dilution to 20.0 ml. The instrument was calibrated in hydrogen gas mode using dilutions of the National Institute of Standards and Technology traceable standard reference solution (high-purity standards, 1,000 ± 3 μg/liter Fe, 2% HNO3) with calibration points at concentrations of 0, 0.5, 2, 10, and 20 ng/ml total Fe. As the measured concentrations in some of the digested and diluted cell culture medium samples were in the range of 30 ng/ml for 56Fe and approaching 100 ng/ml for 57Fe, some samples exceeded instrument calibration and were diluted an additional 1:10 in 1% (vol/vol) HNO3 and then reanalyzed. Specific isotope concentrations were calculated using natural isotopic abundances in atom percent as follows: 54Fe, 5.845; 56Fe, 91.754; 57Fe, 2.119; 58Fe, 0.282. Best and analytical values were selected based on relative concentrations compared to calibration, and concentrations and isotope ratios are reported on the cell culture medium samples only for the56Fe and 57Fe isotopes.
RESULTS
SapA shares conserved secondary structure with known heme-binding proteins.
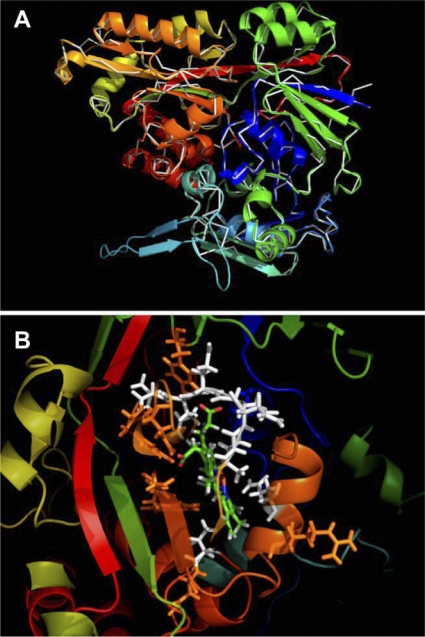
Comparison of the deduced SapA amino acid sequence with those of other periplasmic binding proteins revealed a cluster of proteins that were 100 residues larger than other binding proteins of this classification and predicted to contain three structural domains (7, 8, 19). SapA is predicted to be structurally similar to the heme-binding lipoprotein (HbpA) from Haemophilus and the E. coli dipeptide-binding protein (DppA) in both domain organization and size. DppA shares 55% identity with HbpA, which suggests a conserved functional relationship. The known crystal structure of DppA reveals a binding site for dipeptide ligands that also shares amino acid residues predicted to mediate binding to heme (7, 8, 19). An HbpA model built upon the DppA structure suggests a mode of heme binding by the formation of a heme-binding pocket (7, 8). Since SapA shares 31% identity (50% similarity) with HbpA and 29% identity (50% similarity) with DppA, we examined whether amino acid residues predicted to mediate the formation of a heme-binding pocket in both DppA and HbpA were also conserved in SapA. Toward this end, we threaded the SapA sequence on the crystal structure of the E. coli DppA protein when ligand bound (Fig. 1A). Despite the relatively low sequence conservation (29% identity), four SapA cysteine residues were aligned for disulfide bond formation and a secondary structure similar to that of DppA. Thus, SapA contained amino acid residues that were likewise modeled to bind heme in the periplasmic protein HbpA (Fig. 1B) and that appeared to line a structurally conserved pocket in SapA.
Fig. 1.
Comparative modeling of SapA sequence on the crystal structure of DppA. (A) SapA sequence was modeled on the crystal structure of DppA (white backbone) in the bound conformation. (B) Conservation of identical (orange residues) and similar (white residues) amino acids in the predicted substrate-binding site of SapA. The heme moiety is shown in green as positioned in the putative heme-binding pocket.
(r)SapA binds the iron-containing compound heme.
Analysis of three-dimensional models of SapA in silico indicates that the structurally conserved pocket will accommodate a single heme molecule. To directly demonstrate this capability, (r)SapA was purified as previously described (22) and heme binding was determined using a chemiluminescence detection method (19). We observed (r)SapA-heme complexes but did not detect heme complexed with the control proteins (r)TrxA (thioredoxin A) (Fig. 2A), BSA, and recombinant His-tagged sialic acid-binding protein [(r)SiaBP] (data not shown). Equimolar concentrations of recombinant proteins and relative migration of the native proteins were confirmed by staining an identically loaded gel that was run in parallel (Fig. 2A, right panel). The addition of these proteins likewise failed to dissociate SapA-heme complexes. We further confirmed (r)SapA-heme complexes by coprecipitation of (r)SapA with heme conjugated to agarose beads. The SapA-heme-agarose interactions were specific, as no complexes with any of the control proteins were observed (Fig. 2B and data not shown). Moreover, the His tag moiety did not mediate heme-agarose associations, as His-tagged (r)TrxA and (r)SiaBP did not coprecipitate with the agarose beads. We again confirmed that equimolar concentrations of recombinant protein were used in our experiments (Fig. 2B, right panel).
Fig. 2.
Heme binding by SapA protein. (A) SapA binds heme as determined by nondenaturing gel electrophoresis and transfer to nitrocellulose with detection of heme peroxidase activity by chemiluminescence. SapA or the control TrxA protein was incubated in the absence (−) or presence (+) of heme (Hm). The arrow indicates binding of heme to SapA. The presence of protein was confirmed by Coomassie staining of a gel run in parallel (right panel). (B) Heme complexed to agarose beads was able to immunoprecipitate His-tagged SapA but not the control protein His-tagged TrxA, as determined by an anti-His Western blot assay. Equimolar concentrations of recombinant protein were confirmed by His tag detection via Western blotting (right panel).
The biochemical properties of heme and the low affinity for complex formation make the use of standard methodologies ineffective in the elucidation of precise dissociation constants (47). Despite these limitations, we attempted to estimate the relative affinity of heme for SapA by various methods previously used to determine the heme-binding affinities of the dipeptide and heme-binding proteins DppA and HbpA (19, 47). These periplasmic binding proteins share homology and structural characteristics with SapA. Preliminary titration experiments of SapA-heme complexes resolved by native PAGE revealed that the KD of heme association with SapA was greater than 50 μM, similar to that described for the DppA-heme and HbpA-heme complexes (19, 47). Further, SapA-heme complexes were detected using isothermal titration calorimetry. However, as observed with other low-affinity complexes, we were unable to determine a relative KD by this method due to increased signal-to-noise ratio. Recently, Vergauwen et al. determined that the relative KD of the well-described heme-binding lipoprotein (HbpA)-heme complex was weak (655 μM) and that HbpA preferentially bound the glutathione substrate (47). Importantly, the authors determined the affinity of heme association by densitometric quantitation of apo-HbpA (unbound HbpA), which was observed to decrease with heme titration coincidently with the increase in the heme-bound HbpA complex. Thus, we utilized this method and determined the relative KD of the SapA-heme complex to be 56 μM, approximately 10-fold higher than that described for HbpA-heme complex formation (see Fig. S1 in the supplemental material). The increased affinity of heme for SapA suggests an important role for SapA in periplasmic heme trafficking.
SapA is required for heme utilization in NTHI.
SapA deficiency resulted in an iron-starved phenotype characterized by the upregulation of genes involved in multiple iron acquisition systems as determined by reverse transcription (RT)-PCR analysis (see Table S1 in the supplemental material). The Munson lab has constructed and characterized a mutant deficient in the expression of the ferric uptake regulator (fur) in the same NTHI background. Although the majority of the same iron acquisition systems were upregulated in both the sapA and fur mutants, sap gene mRNA levels were the same in the fur mutant and the parent grown under iron-replete conditions (A. Harrison and R. S. Munson, unpublished data). There is no identifiable Fur box 5′ of the sapA gene.
Together with the data demonstrating that heme is a substrate for SapA, we hypothesized that loss of SapA function would impair heme utilization in NTHI, resulting in the iron (heme)-starved phenotype. To test this, the parent strain, a SapA-deficient strain (23), or a SapA-complemented mutant strain was iron starved for 24 h in a chemically defined medium (DIS) and then subcultured in the presence of increasing concentrations of heme. The iron-starved parent strain was unable to grow when subcultured in heme-depleted medium, which indicated that the microorganism was sufficiently starved of all internal iron stores. Parent strain growth was restored, however, when the strain was subcultured in the presence of heme (Fig. 3A). Although the SapA-deficient strain demonstrated growth comparable to that of the parent in a heme-replete environment, following iron starvation, the cells were unable to utilize heme for growth (Fig. 3B). Complementation of the sapA mutation fully restored the growth of the iron-starved microorganism compared to that of the parent strain (Fig. 3C). These data indicated that SapA was critical for heme utilization in NTHI.
Fig. 3.
Role of SapA in heme utilization. The parent strain (A), the SapA-deficient strain (B), and the complemented SapA-deficient strain (C) were starved for heme (Hm)-iron for 24 h and then shifted to DIS cultures supplemented with or without increasing amounts of heme. Growth was assessed by monitoring the increase in culture OD490 for 16 h. Error bars represent the standard deviation of the mean of cultures assayed in triplicate from three independent assays (n = 3).
The Sap translocator is an inner membrane heme transporter.
Since we showed that heme utilization was dependent upon SapA interaction with heme, we investigated whether the Sap translocator permease is required for heme utilization by NTHI. We first showed that a functional Sap permease was required for normal growth of NTHI (Fig. 4A). Unlike the SapA-deficient strain that required rigorous iron starvation to reveal a deficiency in heme utilization, the SapBC permease-deficient strain was unable to utilize heme as an iron source even in the absence of previous iron starvation, suggesting that Haemophilus iron homeostasis is dependent upon Sap permease and its likely function to transport substrate across the cytoplasmic membrane. To determine whether this inability to utilize heme was due to a lack of transport across the cytoplasmic membrane via the SapBC permease, we passaged bacteria in iron-free DIS medium and then subcultured them in DIS medium supplemented with [57Fe]heme, a stable heme iron isotope that is not present in the environment. The ability to transport heme was dependent upon the Sap permease complex, suggesting that a failure to utilize heme was due to decreased Sap translocator function (Fig. 4B). Therefore, we demonstrated here, for the first time, that heme transport across the cytoplasmic membrane and into the bacterial cytoplasm requires the Sap translocator permease.
Fig. 4.
Role of SapBC membrane permease in heme utilization and intracytoplasmic membrane transport. (A) The parent strain or the SapBC permease-deficient strain was grown overnight on chocolate agar and placed into DIS cultures supplemented with heme or not supplemented with heme but was not previously iron starved. Growth was assessed over a 10-h period by monitoring the increase in culture OD490. Error bars represent the standard deviation of the mean of cultures assayed in triplicate from three independent assays (n = 3). There is a statistically significant (Student's t test) difference in growth between the parent and the sapBC mutant at each point from 4 to 10 h (P < 0.02). (B) The SapBC permease is required for intracytoplasmic membrane heme transport. To determine efficiency of heme transport, the parent and the sapBC mutant were placed in iron-free DIS medium containing the stable heme isotope (57Fe) and cultured for 1 h. Whole bacteria were subjected to IC-MS analysis for determination of iron isotope ratios of endogenous environmental iron to heme isotope iron, and the ratio per viable cell (56Fe/57Fe) was calculated. An increase in the ratio indicates a deficiency in the ability to transport heme. Error bars represent standard deviations of duplicate samples from three independent experiments (n = 3).
Heme and APs compete for substrate binding to SapA.
We have shown that heme is a substrate for SapA. Previously, we demonstrated that SapA binds APs and is required for resistance to killing by these effectors of innate immunity (22), suggesting a promiscuity in the substrate recognition and biological function of SapA. To determine if APs might also compete with heme for substrate binding, thereby limiting the ability of SapA to bind heme, we incubated SapA with heme in the presence of increasing concentrations of LL-37, hBD-3, or control proteins. We showed that both hBD-3 and LL37 competed for binding to SapA, whereas matched concentrations of dipeptide, soluble thioredoxin (Trx) protein, or a soluble periplasmic binding protein (SiaBP) similar in molecular weight to SapA did not compete with heme binding by SapA (Fig. 5A and data not shown). hBD-2, hNP-1, and melittin could also displace heme (Fig. 5C), which indicated that substrate binding was not restricted to a particular peptide structure but was likely dependent upon the net charge of the substrate bound. In fact, we observed a hierarchy of heme displacement that was proportional to the relative charge of the AP used for competition; the most efficient being hBD-3, a highly cationic AP (+11). We observed an increase in the apparent molecular mass of the SapA-AP complex. This increase in apparent molecular mass corresponded to a loss of heme peroxidase activity due to displacement of heme binding (Fig. 5A). Further, by Western blot analysis, hBD-3 was detected in this ligand-bound complex, which confirmed that SapA bound this AP (Fig. 5B). Similar results were observed with LL37, which also bound and displaced heme (Fig. 5B). Collectively, our data indicate that both APs and heme are substrates for SapA binding.
Fig. 5.
Monitoring of the ability of APs to displace heme bound to SapA. (A) SapA (11.5 μM) was incubated with heme (Hm; 50 μM) in the presence of decreasing concentrations (100, 10, and 1 μM) of LL-37, hBD-3, or BSA, and complexes were resolved by nondenaturing gel electrophoresis and transferred to nitrocellulose. Heme was detected by peroxidase activity as measured using chemiluminescence. Those fractions that demonstrated loss of heme binding were then probed for the presence of AP (B). Matched concentrations of BSA and dipeptide did not block or displace bound heme, yet different classes of AP compounds could displace heme from low (+)- to high (+++)-affinity interactions (C). N/A, not applicable.
DISCUSSION
In this work, we showed that the Haemophilus SapA periplasmic binding protein binds heme and requires the SapBC transporter permease for transport of heme into the bacterial cytoplasm, thus describing, for the first time, a mechanism of heme acquisition and transport across the bacterial cytoplasmic membrane. Previously, we demonstrated that the Sap translocator inner membrane components and the SapA-binding protein are essential for bacterial resistance to APs, potassium homeostasis, and survival in vivo (22, 23). Together, these data support a critical multifunctional role for this transporter in Haemophilus survival strategies.
Recently, it was shown that the Dpp permease of E. coli functions along with two optional peptide-binding proteins for heme utilization (19), in addition to its role in the transport of small peptides. Similarly, the E. coli nickel-binding protein NikA binds heme at a location independent of its nickel-binding site (41). Collectively, these data suggest that these periplasmic transport proteins are not restricted in substrate specificity. It is clear that the SapA-binding protein binds multiple ligands for transport and metabolic purposes. Comparative modeling of SapA with the known crystal structure of the SapA homologue DppA revealed that amino acid residues and side chains were universally conserved and consistent with their predicted location in the heme-binding pocket. The shape and volume of the ligand-binding sites would allow the binding of heme, and modeling further fostered the identification of amino acid residues of SapA that likely mediate ligand interaction and support heme orientation in the binding pocket. This mode of heme binding is distinct from that described for proteins of the human host or for other bacterial heme and hemoglobin receptors on the cell surface (3). Thus, we show here that SapA belongs to a class of heme-binding proteins also shown to bind additional ligands which, in association with specific ABC peptide permease components in the inner membrane, appear to be multifunctional.
Due to the strong peroxidase activity of heme, heme interference with Coomassie staining, and the low affinity of heme binding by this family of substrate-binding proteins, better methods to accurately determine the affinity of heme binding are needed. However, it is clear that although its affinity is weak, the role of HbpA in heme acquisition has been well described and shown to be biologically significant (10, 11, 26, 27). In fact, DppA, HbpA, and the oligopeptide-binding protein OppA have all been shown to be promiscuous in substrate binding (2, 19, 47), consistent with our observations for SapA. This promiscuity appears to be important for the related biological function of these proteins in bacterial metabolic activities, all of which are essential for bacterial growth and virulence.
Periplasmic heme-binding protein constituents of ABC permeases belong to two classes (19). The large multifunctional binding proteins (DppA, MppA, NikA, and HbpA) recognize and transport substrates other than heme. The second class of smaller proteins, such as HemTUV from Serratia marcescens, ShuTUV from Shigella dysenteriae, and PhuTUV from Pseudomonas aeruginosa, are involved in high-affinity heme transport (5, 16, 18). No HemTUV orthologs exist in Neisseria or Haemophilus species, but these bacteria have functional outer membrane heme receptors. As these species require the use of heme as an iron source and for enzyme function, transport of heme must occur via multifunctional inner membrane peptide permeases. In fact, hemTUV gene inactivation in Yersinia and Vibrio species does not abolish heme uptake, which suggests the use of additional mechanisms for heme uptake by these strains (32, 43, 45). Our studies showed that the relative affinity of heme binding to SapA is comparable to that described for heme binding to DppA, the peptide/heme-binding protein in E. coli (19).
The DppA periplasmic heme-binding protein functions along with the Dpp permease to facilitate heme transport in E. coli (19). Since E. coli K-12 lacks a heme-binding outer membrane receptor, it lacks the ability to utilize exogenous heme as an iron source. The role of Dpp peptide/heme permease may thus involve endogenous heme recycling in E. coli. Further, MppA and NikA, a nickel-binding protein, have also been shown to bind heme in E. coli (19, 41). The assembly of anaerobic hydrogenase isoenzymes may explain why NikA has evolved to bind both heme and nickel. NikA may function to deliver heme for the assembly of cytochromes associated with the hydrogenase isozymes of E. coli. Interestingly, single mutations in genes that encode the DppA periplasmic binding protein homologues SapA, NikA, OppA, and MppA did not affect heme utilization by E. coli. These data demonstrate that the function of SapA in E. coli is not restricted to heme binding and transport via the Sap permease but that it may function primarily as an innate immune defense mechanism to mediate AP resistance, particularly since E. coli does not have an absolute requirement for heme. Thus, SapA ligand binding and subsequent transport via the Sap permease appear to be multifaceted.
NTHI strain 86-028NP lacks DppA, MppA, and NikA yet encodes the HbpA and SapA homologous heme-binding proteins (10, 11). It has recently been demonstrated that the dppBCDF gene cluster constitutes part of the periplasmic heme acquisition system of H. influenzae, likely dependent upon the delivery of heme to the DppBC inner membrane permease via the independently expressed heme-bound periplasmic protein HbpA (28). However, since heme utilization was only partially impaired, the authors indicated that additional periplasmic systems must be available to transport heme. Similar to these studies, we demonstrated that heme utilization by the DppC permease mutant strain was only partially impaired when iron-starved cultures were subsequently monitored for growth when supplied with increasing amounts of heme (see Fig. S2 in the supplemental material). In fact, recent work indicates that HbpA, which has been shown to bind glutathione with a higher affinity than that with which it binds its heme substrate, may function to deliver glutathione for transport via the Haemophilus DppBCDF inner membrane transporter (47). It is unknown whether this function is required primarily to counteract oxidation stress in Haemophilus in vivo, but our findings place the roles of HbpA and the Dpp transporter in heme utilization in question. In fact, our data demonstrate that the SapA periplasmic protein is a heme-binding protein and further that delivery of heme to the SapBC inner membrane permease is both necessary and sufficient for heme transport in H. influenzae, since loss of SapA completely abrogates heme utilization. In our studies, functional HbpA and Dpp transporter expression was not sufficient to rescue the growth of the Sap-deficient strains when the microorganisms were iron starved. The presence of multiple heme acquisition and cytoplasmic membrane transport systems does not appear to be functionally redundant, but rather they constitute metabolic systems that are regulated environmentally and are multifunctional yet essential for Haemophilus survival.
Commensal microorganisms must sense and adapt to various microenvironmental cues in their mammalian host. Bacterial strategies to resist mechanisms of host defense and the meeting of metabolic needs are required for sustained commensal-host homeostasis and colonization. Here, we showed that the Sap transporter permease is both necessary and sufficient for intracytoplasmic membrane transport of heme. Intriguingly, we observed that many APs displaced heme from SapA. Taken together, these data support multifunctional roles for the Sap transporter, i.e., in AP resistance and heme transport, yet the physiological relevance of this multifunctional nature remains unknown. It is clear, however, that perturbations of Sap function significantly decrease the survival of NTHI in the host (22, 23). We hypothesize that Sap-dependent recognition and subsequent transport of APs for targeted degradation are initially important for Haemophilus to establish itself as a commensal or its transition to a pathogen in a disease state. In fact, recent evidence from our lab supports active intracytoplasmic membrane transport of APs and susceptibility to cytoplasm-localized peptidase activity (unpublished data). Collectively, these data suggest a clear hierarchy wherein immune evasion trumps even the needed iron acquisition functions of the Sap system (Fig. 6). To compensate for this trumping of iron uptake functionality, NTHI has multiple means to acquire iron. These redundant iron uptake and storage mechanisms would not only benefit early-stage colonization in the iron-restricted host environment but also direct bacterial functions to meet the immediate survival needs of the microbe, particularly during disease development. This concept of multifunctional system balance, as shown here, is applicable to the study of many human pathogens and the diseases they cause and will likely inform us as to improved ways to treat or prevent these diseases. A better understanding of factors, both host and microbe expressed, that support commensal behavior will help define the bacterial factors utilized during opportunistic disease so as to exploit these as targets for the design of better methods to treat or prevent disease.
Fig. 6.
Model depicting the multifunctional role of the Sap transporter and its substrate-binding protein (SapA). The periplasmic SapA protein binds heme for SapBC-dependent intracytoplasmic membrane transport. Following AP interaction with and disruption of the bacterial outer membrane, SapA also mediates innate immune evasion by binding APs, which have been shown to displace heme and transport these cationic peptides for intracytoplasmic membrane degradation. Sap-dependent immune evasion alters the role of Sap in iron homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by R21 A1070825 from NIAID/NIH to K.M.M.
We thank B. Szelestey for growth curve analysis and S. Wallace Sharpe for assistance with figure preparation. We thank R. S. Munson for helpful discussions and A. Harrison and S. S. Justice for review of the manuscript. We thank R. S. Munson for (r)SiaBP and D. Morton for NTHI strain 86-028NP/dppC:spec.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Abouhamad W. N., Manson M., Gibson M. M., Higgins C. F. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035–1047 [DOI] [PubMed] [Google Scholar]
- 2. Berntsson R. P., et al. 2009. The structural basis for peptide selection by the transport receptor OppA. EMBO J. 28:1332–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bracken C. S., Baer M. T., Abdur-Rashid A., Helms W., Stojiljkovic I. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 5. Burkhard K. A., Wilks A. 2008. Functional characterization of the Shigella dysenteriae heme ABC transporter. Biochemistry 47:7977–7979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cope L. D., et al. 1994. The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13:863–873 [DOI] [PubMed] [Google Scholar]
- 7. Dunten P., Mowbray S. L. 1995. Crystal structure of the dipeptide binding protein from Escherichia coli involved in active transport and chemotaxis. Protein Sci. 4:2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunten P., Mowbray S. L. 1995. Modeling of the structure of the Haemophilus influenzae heme-binding protein suggests a mode of heme interaction. Protein Sci. 4:2335–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ganz T. 2004. Antimicrobial polypeptides. J. Leukoc. Biol. 75:34–38 [DOI] [PubMed] [Google Scholar]
- 10. Hanson M. S., Hansen E. J. 1991. Molecular cloning, partial purification, and characterization of a haemin-binding lipoprotein from Haemophilus influenzae type b. Mol. Microbiol. 5:267–278 [DOI] [PubMed] [Google Scholar]
- 11. Hanson M. S., Slaughter C., Hansen E. J. 1992. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect. Immun. 60:2257–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harms C., et al. 2001. Identification of the ABC protein SapD as the subunit that confers ATP dependence to the K+-uptake systems TrkH and TrkG from Escherichia coli K-12. Microbiology 147:2991–3003 [DOI] [PubMed] [Google Scholar]
- 13. Harrison A., et al. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 187:4627–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison A., et al. 2007. The OxyR regulon in nontypeable Haemophilus influenzae. J. Bacteriol. 189:1004–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152:205–210 [DOI] [PubMed] [Google Scholar]
- 16. Kaur A. P., Lansky I. B., Wilks A. 2009. The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J. Biol. Chem. 284:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klein J. O. 1997. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16:S5–S8 [DOI] [PubMed] [Google Scholar]
- 18. Létoffé S., Delepelaire P., Wandersman C. 2008. Functional differences between heme permeases: Serrati a marcescens HemTUV permease exhibits a narrower substrate specificity (restricted to heme) than the Escherichia coli DppABCDF peptide-heme permease. J. Bacteriol. 190:1866–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Létoffé S., Delepelaire P., Wandersman C. 2006. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U. S. A. 103:12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maciver I., et al. 1996. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect. Immun. 64:3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Markel T. A., et al. 2007. The struggle for iron: gastrointestinal microbes modulate the host immune response during infection. J. Leukoc. Biol. 81:393–400 [DOI] [PubMed] [Google Scholar]
- 22. Mason K. M., Bruggeman M. E., Munson R. S., Bakaletz L. O. 2006. The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and SapD-dependent potassium acquisition. Mol. Microbiol. 62:1357–1372 [DOI] [PubMed] [Google Scholar]
- 23. Mason K. M., Munson R. S., Jr., Bakaletz L. O. 2005. A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect. Immun. 73:599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mason K. M., Munson R. S., Jr., Bakaletz L. O. 2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71:3454–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morton D. J., et al. 2004. Reduced severity of middle ear infection caused by nontypeable Haemophilus influenzae lacking the hemoglobin/hemoglobin-haptoglobin binding proteins (Hgp) in a chinchilla model of otitis media. Microb. Pathog. 36:25–33 [DOI] [PubMed] [Google Scholar]
- 26. Morton D. J., et al. 2005. The heme-binding lipoprotein (HbpA) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol. Lett. 253:193–199 [DOI] [PubMed] [Google Scholar]
- 27. Morton D. J., et al. 2009. The heme-binding protein (HbpA) of Haemophilus influenzae as a virulence determinant. Int. J. Med. Microbiol. 299:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morton D. J., Seale T. W., Vanwagoner T. M., Whitby P. W., Stull T. L. 2009. The dppBCDF gene cluster of Haemophilus influenzae: role in heme utilization. BMC Res. Notes 2:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morton D. J., et al. 2004. Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology 150:3923–3933 [DOI] [PubMed] [Google Scholar]
- 30. Murphy T. F. 2003. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16:129–134 [DOI] [PubMed] [Google Scholar]
- 31. Nizet V. 2006. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 8:11–26 [PubMed] [Google Scholar]
- 32. Occhino D. A., Wyckoff E. E., Henderson D. P., Wrona T. J., Payne S. M. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493–1507 [DOI] [PubMed] [Google Scholar]
- 33. Parra-Lopez C., Baer M. T., Groisman E. A. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12:4053–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parra-Lopez C., Lin R., Aspedon A., Groisman E. A. 1994. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 13:3964–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peschel A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179–186 [DOI] [PubMed] [Google Scholar]
- 36. Poolman B., et al. 2005. Functional analysis of detergent-solubilized and membrane-reconstituted ATP-binding cassette transporters. Methods Enzymol. 400:429–459 [DOI] [PubMed] [Google Scholar]
- 37. Ren Z., Jin H., Morton D. J., Stull T. L. 1998. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect. Immun. 66:4733–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Román F., Canton R., Perez-Vazquez M., Baquero F., Campos J. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42:1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saier M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sethi S., Murphy T. F. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shepherd M., Heath M. D., Poole R. K. 2007. NikA binds heme: a new role for an Escherichia coli periplasmic nickel-binding protein. Biochemistry 46:5030–5037 [DOI] [PubMed] [Google Scholar]
- 42. St. Geme J. W., III 2000. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19(Suppl. 1):S41–S50 [DOI] [PubMed] [Google Scholar]
- 43. Stojiljkovic I., Hantke K. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719–732 [DOI] [PubMed] [Google Scholar]
- 44. Stojiljkovic I., Perkins-Balding D. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281–295 [DOI] [PubMed] [Google Scholar]
- 45. Thompson J. M., Jones H. A., Perry R. D. 1999. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67:3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tracy E., Ye F., Baker B. D., Munson R. S., Jr 2008. Construction of non-polar mutants in Haemophilus influenzae using FLP recombinase technology. BMC Mol. Biol. 9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vergauwen B., Elegheert J., Dansercoer A., Devreese B., Savvides S. N. 2010. Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA. Proc. Natl. Acad. Sci. U. S. A. 107:13270–13275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.