Abstract
The genus Agrobacterium includes plant-associated bacteria and opportunistic human pathogens. Taxonomy and nomenclature within the genus remain controversial. In particular, isolates of human origin were all affiliated with the species Agrobacterium (Rhizobium) radiobacter, while phytopathogenic strains were designated under the synonym denomination Agrobacterium tumefaciens. In order to study the relative distribution of Agrobacterium strains according to their origins, we performed a multilocus sequence-based analysis (MLSA) on a large collection of 89 clinical and environmental strains from various origins. We proposed an MLSA scheme based on the partial sequence of 7 housekeeping genes (atpD, zwf, trpE, groEL, dnaK, glnA, and rpoB) present on the circular chromosome of A. tumefaciens C58. Multilocus phylogeny revealed that 88% of the clinical strains belong to genovar A7, which formed a homogeneous population with linkage disequilibrium, suggesting a low rate of recombination. Comparison of genomic fingerprints obtained by pulsed-field gel electrophoresis (PFGE) showed that the strains of genovar A7 were epidemiologically unrelated. We present genetic evidence that genovar A7 may constitute a human-associated population distinct from the environmental population. Also, phenotypic characteristics, such as culture at 42°C, agree with this statement. This human-associated population might represent a potential novel species in the genus Agrobacterium.
INTRODUCTION
Members of the genus Agrobacterium are well-known plant-associated bacteria. Agrobacterium tumefaciens, Agrobacterium rhizogenes, Agrobacterium rubi, Agrobacterium vitis, and Agrobacterium larrymoorei may be phytopathogens, causing diverse tumors in plants (4, 45). Since A. tumefaciens is able to transform vegetal cells, it has been used to obtain genetically engineered plants (32, 40). In the past 2 decades, Agrobacterium (Rhizobium) radiobacter has been recognized as an opportunistic human pathogen responsible for nosocomial infections, mainly bacteremia, peritonitis, and urinary tract infections (1, 3, 10, 28). Bacteremias are usually secondary to the use of intravenous devices (3, 11, 17). A. radiobacter strains have also been found in the respiratory tracts of cystic fibrosis patients (5).
The genus Agrobacterium belongs to the family Rhizobiaceae in subphylum alpha of the proteobacteria (16). 16S rRNA gene-based phylogeny grouped the genus Agrobacterium and some fast-growing nitrogen-fixing bacteria of the genus Rhizobium in the same clade (12). The paraphyly of the two genera has led to diverse revisions of the nomenclature (12, 42, 43, 44), but expert controversies have received little attention in practice. Currently, A. tumefaciens, A. radiobacter, and R. radiobacter have authority to denominate the same bacterium, even if two type strains named A. tumefaciens CFBP 2413T and A. radiobacter CFBP 2414T, are deposited and validated. As a rule, but without clear argumentation, clinical strains of the genus Agrobacterium are usually affiliated with the species A. radiobacter, whereas A. tumefaciens includes environmental phytopathogenic or nonphytopathogenic strains. In this study, Agrobacterium (Rhizobium) strains will be named only Agrobacterium.
The strains of the genus Agrobacterium can be grouped into three biovars on the basis of biochemical and physiological tests and into 11 genomospecies defined by DNA-DNA hybridization (30). Finally, relationships could not be established among biovars, genomospecies, and species. In particular, biovar 1 corresponds to 9 genomospecies and contains the type strains of A. tumefaciens and A. radiobacter.
Previously reported clinical strains were assigned to the genus Agrobacterium and to the species A. radiobacter (R. radiobacter) on the basis of conventional methods used for the routine identification of oxidase-positive, nonfastidious, nonfermenting Gram-negative bacilli or, though less frequently, on the 16S rRNA gene sequence. However, these methods are not reliable for identification to the species level due to the absence of species other than A. radiobacter in the commercial systems' phenotypic databases and to low interspecific polymorphism within the 16S rRNA gene. Considering the taxonomic problems associated with the absence of suitable tools for species identification, the distribution of the clinical strains among the general population of Agrobacterium has never been described.
The availability of the complete genomic sequences of A. tumefaciens strain C58 (18), Rhizobium rhizogenes strain K84 (35) (formerly A. radiobacter, formerly A. rhizogenes [41]), and A. vitis strain S4 (35) allowed the use of multilocus based-methods to study the genetic diversity in Agrobacterium species populations. The aim of the present study was to analyze a large collection of clinical and environmental Agrobacterium strains using multilocus genetics and phenotypic traits in order to determine whether clinical isolates displayed specific characteristics.
MATERIALS AND METHODS
Bacterial strains, culture, and phenotype.
A total of 89 strains of Agrobacterium spp., including 49 clinical and 40 environmental isolates, were analyzed (Tables 1 and 2). Clinical strains were sampled over a 37-year period in four countries in Europe, North America, and Oceania. However, most of them were recovered over a 14-year period from patients admitted to three French university hospitals. Nonclinical or nonhuman strains were collected in 14 countries and four continents over a 103-year period. They were representative of the 3 biovars of Agrobacterium and originated from soil, plants, or nematodes. A. tumefaciens C58, A. rhizogenes K84 (formerly A. radiobacter K84), A. radiobacter CFBP 2414T, and A. vitis LMG 8750T were included as the reference and type strains. All isolates were cultivated on nutrient glucose agar (NGA) at 28°C for 24 h. They were identified to the genus level by routine phenotypic tests, including Gram stain, oxidase production, and a API 20NE system (bioMérieux, Marcy l'Etoile, France). Genus identification was confirmed by 5′-end sequencing of the 16S rRNA gene as previously described (37). The ability to grow at 35°C and 42°C was tested on NGA incubated for 24 h. Two of the assays usually used for biovar identification (3-ketolactose production from lactose and differential acid production) were performed as described previously (2). The strains from the LMG collection identified at the biovar level in the LMG database (n = 16) were used for the validation of the assays based on the following criteria: 3-ketolactose production positive in biovar 1 strains and negative in biovar 2 strains and acid production from glucose positive only in biovar 2 strains (2).
Table 1.
| Strainb | Genovar within clade A | ST | Allelic profilei |
Growth (42°C)f | 3-Ketolactose productionf | PDA CaCO3f,h | Origin of strain | Place, yr of isolation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| atpD | dnaK | glnA | groEL | rpoB | trpE | zwf | ||||||||
| AGR01 | A7 | 3 | 3 | 5 | 7 | 3 | 31 | 2 | 5 | + | − | − | Scalp wound | Montpellier, Fr, 2003 |
| AGR35 | A7 | 3 | 3 | 5 | 7 | 3 | 31 | 2 | 5 | + | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR38 | A7 | 3 | 3 | 5 | 7 | 3 | 31 | 2 | 5 | + | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR40 | A7 | 3 | 3 | 5 | 7 | 3 | 31 | 2 | 5 | − | − | − | Blood | Montpellier, Fr, 2009 |
| AGR02 | A7 | 4 | 3 | 6 | 10 | 4 | 1 | 3 | 6 | + | − | − | Urine | Montpellier, Fr, 2004 |
| AGR03 | A7 | 5 | 18 | 7 | 10 | 5 | 2 | 5 | 7 | + | + | − | Urine | Montpellier, Fr, 2004 |
| AGR04 | A7 | 5 | 18 | 7 | 10 | 5 | 2 | 5 | 7 | + | + | − | Undocumentede | Nimes, Fr, 2004 |
| AGR17 | A7 | 5 | 18 | 7 | 10 | 5 | 2 | 5 | 7 | + | + | − | Sputumd | Montpellier, Fr, 2006 |
| AGR24 | A7 | 5 | 18 | 7 | 10 | 5 | 2 | 5 | 7 | + | + | − | Sputumd | Montpellier, Fr, 2008 |
| AGR33 | A7 | 5 | 18 | 7 | 10 | 5 | 2 | 5 | 7 | − | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR41 | A7 | 5 | 18 | 7 | 10 | 5 | 2 | 5 | 7 | + | + | − | Glans penis | Montpellier, Fr, 2009 |
| AGR05 | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | − | − | Fiberscope rinse fluide | Toulouse, Fr, 1997 |
| AGR07 | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | + | − | Undocumentede | Toulouse, Fr, 1998 |
| AGR09 | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | − | − | Collyree | Montpellier, Fr, 1995 |
| AGR15 | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | − | − | Sputumd | Montpellier, Fr, 2005 |
| AGR20 | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | − | − | Sputumd | Montpellier, Fr, 2007 |
| AGR21 A | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | − | − | Sputumd | Montpellier, Fr, 2007 |
| AGR21 B | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | − | − | Sputumd | Montpellier, Fr, 2007 |
| CCUG 48648 | A7 | 6 | 3 | 6 | 12 | 4 | 1 | 3 | 6 | + | + | − | Blood | Stockholm, Sweden, 2004 |
| AGR06 | A7 | 7 | 4 | 8 | 5 | 6 | 3 | 22 | 8 | + | + | − | Culture mediume | Toulouse, Fr, 1997 |
| AGR10 | A7 | 8 | 5 | 9 | 9 | 7 | 4 | 9 | 9 | + | + | − | Sputumd | Montpellier, Fr, 2005 |
| AGR18 | A7 | 8 | 5 | 9 | 9 | 7 | 4 | 9 | 9 | + | − | − | Sputumd | Montpellier, Fr, 2006 |
| AGR11 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | − | − | Sputum | Nimes, Fr, 2005 |
| AGR16 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | − | − | Feces | Montpellier, Fr, 2006 |
| AGR26 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | − | − | Sputum | Montpellier, Fr, 2008 |
| AGR27 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | ND | ND | Sputum | Montpellier, Fr, 2008 |
| AGR37 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR39 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | − | − | Sputum | Montpellier, Fr, 2009 |
| CCUG 49619 | A7 | 9 | 10 | 10 | 9 | 8 | 5 | 6 | 6 | + | − | − | Bronchus brush rinse | Göteborg, Sweden, 2004 |
| AGR12 | A7 | 10 | 3 | 5 | 7 | 3 | 32 | 2 | 5 | + | − | − | Toe wound | Montpellier, Fr, 2005 |
| AGR14 | A7 | 11 | 6 | 11 | 6 | 8 | 7 | 8 | 11 | + | − | − | Sputumd | Montpellier, Fr, 2006 |
| AGR23 | A7 | 11 | 6 | 11 | 6 | 8 | 7 | 8 | 11 | + | − | − | Blood | Toulouse, Fr, 2006 |
| AGR43 | A7 | 11 | 6 | 11 | 6 | 8 | 7 | 8 | 11 | + | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR19 | A7 | 12 | 7 | 5 | 11 | 16 | 5 | 10 | 12 | + | + | − | Blood | Montpellier, Fr, 2007 |
| CCUG 12509 | A7 | 13 | 4 | 22 | 12 | 16 | 14 | 5 | 21 | + | + | − | Lesion | Texas, 1974 |
| AGR25 | A7 | 13 | 4 | 22 | 12 | 16 | 14 | 5 | 21 | + | − | − | Feces | Montpellier, Fr, 2008 |
| AGR32 | A7 | 15 | 8 | 25 | 8 | 19 | 3 | 9 | 23 | + | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR34 | A7 | 16 | 9 | 8 | 5 | 8 | 28 | 8 | 5 | + | − | − | Sputumd | Montpellier, Fr, 2009 |
| AGR08 | A7 | 18 | 3 | 3 | 7 | 3 | 33 | 1 | 5 | + | − | − | Sputumd | Montpellier, Fr, 2005 |
| LMG 361.1 | A7 | 37 | 30 | 32 | 24 | 25 | 27 | 29 | 11 | + | + | − | Sputum | Missouri, NA |
| LMG 378 | A7 | 38 | 7 | 5 | 9 | 16 | 5 | 10 | 12 | + | + | − | Blood | Georgia, NA |
| LMG 399 | A7 | 39 | 9 | 8 | 5 | 26 | 28 | 8 | 7 | + | + | − | Infected eye | Louisiana, NA |
| LMG 355 | A7 | 48 | 3 | 5 | 7 | 3 | 36 | 2 | 5 | − | + | − | Bronchial washings | Hawaii, NA |
| AGR28 | A2 | 14 | 11 | 23 | 13 | 17 | 15 | 11 | 22 | − | + | − | Blood | Montpellier, Fr, 2008 |
| AGR29 | A2 | 14 | 11 | 23 | 13 | 17 | 15 | 11 | 22 | − | + | − | Blood | Montpellier, Fr, 2008 |
| AGR30 | A2 | 14 | 11 | 23 | 13 | 17 | 15 | 11 | 22 | − | + | − | Blood | Montpellier, Fr, 2008 |
| AGR13 | A4 | 2 | 17 | 4 | 3 | 9 | 6 | 7 | 10 | − | + | − | Sputumd | Montpellier, Fr, 2005 |
| AGR36 | A4 | 17 | 17 | 4 | 3 | 9 | 23 | 7 | 10 | − | + | − | Sputumd | Montpellier, Fr, 2009 |
| LMG 227c | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | Blood | Denmark, before 1972 |
Results are presented by genovar within clade A and then by ST.
All reference strains were purchased as Agrobacterium (Rhizobium) radiobacter (A. tumefaciens) strains from the corresponding collection.
Biovar 1.
From cystic fibrosis patients.
Probable human origin.
+, positive for the test; −, negative for the test; ND, not determined.
CCUG, Culture Collection University of Göteborg; LMG, Laboratorium voor Microbiologie Universiteit Gent; Fr, France; NA, not available.
PDA CaCo3, differential production of acid from a glucose-containing medium associated with differential dissolution of CaCo3.
For each locus, each different allele was assigned an arbitrary number.
Table 2.
| Strain/biovarb | Genovar within clade A | ST | Allelic profileg |
Growth (42°C)d | 3-Ketolactose productiond | PDA CaCO3d,e | Phytopathogenicityc,d | Origin of strain | Place, yr of isolationf | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| atpD | dnaK | glnA | groEL | rpoB | trpE | zwf | |||||||||
| 181 | A3 | 26 | 21 | 14 | 23 | 12 | 10 | 24 | 15 | − | − | − | + | Populus, tumor | New York, UD |
| RTG2 | A3 | 27 | 15 | 15 | 17 | 10 | 11 | 15 | 16 | − | + | − | − | Lycopersicon esculentum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTE4 | A3 | 27 | 15 | 15 | 17 | 10 | 11 | 15 | 16 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTB12 | A3 | 27 | 15 | 15 | 17 | 10 | 11 | 15 | 16 | − | + | − | − | Lycopersicon esculentum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| LMG 232/bv1 | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | − | Beta vulgaris, rhizosphere | United Kingdom, 1963 |
| RTB1 | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTB8 | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | − | Lycopersicon esculentum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTP7 | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTP1 | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTP2 | A3 | 28 | 16 | 16 | 18 | 10 | 12 | 16 | 4 | − | ND | ND | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTB7 | A3 | 30 | 24 | 18 | 16 | 10 | 12 | 18 | 18 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| RTA10 | A3 | 31 | 22 | 19 | 17 | 10 | 12 | 19 | 19 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| LMG 142/bv1 | A3 | 34 | 22 | 19 | 17 | 10 | 12 | 19 | 29 | − | + | − | − | soil | Netherlands or Germany, 1904 |
| LMG 197/bv1 | A3 | 35 | 28 | 12 | 19 | 10 | 8 | 13 | 13 | − | + | − | + | Malus sp., tumor | Wisconsin, 1935 |
| LMG 175/bv1 | A3 | 35 | 28 | 12 | 19 | 10 | 8 | 13 | 13 | − | ND | ND | + | Prunus cerasifera, tumor | North Bulgaria, 1959 |
| LMG 90/bv1 | A3 | 40 | 31 | 15 | 25 | 23 | 29 | 27 | 30 | + | + | − | + | Rosa sp. cv. Golden Rapture, tumor | New York, UD |
| A6 | A3 | 41 | 23 | 12 | 19 | 10 | 8 | 13 | 13 | − | + | − | + | apple tree, tumor | United States, UD |
| R10 | A3 | 41 | 23 | 12 | 19 | 10 | 8 | 13 | 13 | − | + | − | + | NA, tumor | United States, UD |
| LMG 167/bv1 | A3 | 45 | 35 | 16 | 18 | 10 | 12 | 16 | 4 | − | + | − | + | Prunus sp., tumor | Gembloux, Belgium, UD |
| LMG 303/bv1 | A3 | 45 | 35 | 16 | 18 | 10 | 12 | 16 | 4 | − | ND | ND | + | Chrysanthemum frutescens, tumor | Germany, 1927 |
| C58 | A5 | 20 | 1 | 1 | 20 | 1 | 20 | 1 | 1 | − | + | − | + | Prunus sp. cv. Montmorency tumor | New York, 1958 |
| CCUG 50385B | A5 | 22 | 27 | 30 | 20 | 22 | 25 | 26 | 28 | − | + | − | ND | Soil | Chile, 2004 |
| T37 | A5 | 25 | 12 | 13 | 20 | 11 | 9 | 14 | 14 | − | + | − | + | Juglans regia, tumor | California, 1926 |
| H100 | A5 | 25 | 12 | 13 | 20 | 11 | 9 | 14 | 14 | − | ND | ND | + | Humulus lupulus, tumor | United States, UD |
| AB35-9 | A4 | 2 | 17 | 4 | 3 | 9 | 6 | 7 | 10 | − | − | − | ND | Nematode | Avignon, Fr, 2007 |
| B10-2 | A4 | 2 | 17 | 4 | 3 | 9 | 6 | 7 | 10 | − | − | − | ND | Nematode | Avignon, Fr, 2007 |
| RTP8 | A4 | 32 | 13 | 20 | 4 | 14 | 6 | 20 | 20 | − | + | − | − | Nicotiana tabacum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| CFBP 2414T | A1 | 23 | 20 | 21 | 15 | 15 | 13 | 21 | 32 | − | + | − | − | Ditch water | Netherlands, before 1927 |
| LMG 306/bv1 | A1 | 36 | 29 | 31 | 22 | 24 | 26 | 28 | 31 | − | + | − | + | Prunus sp., tumor | Saitama, Japan, 1956 |
| RTH4 | A6 | 29 | 14 | 17 | 14 | 13 | 18 | 17 | 17 | − | + | − | − | Lycopersicon esculentum, rhizosphere | Gif-sur-Yvette, Fr, 2006 |
| A. vitis S4 | 1 | 25 | 26 | 2 | 20 | 22 | 25 | 24 | ND | ND | ND | + | Vitis vinifera, tumor | Hungary, UD | |
| LMG 109/bv2 | 19 | 34 | 35 | 28 | 30 | 21 | 32 | 2 | − | − | + | + | Rosa sp., tumor | Texas, UD | |
| LMG 29/bv2 | 21 | 33 | 38 | 27 | 28 | 35 | 31 | 33 | − | − | − | + | Rosa xanthina, tumor | United Kingdom, UD | |
| LMG 253/bv2 | 24 | 34 | 37 | 28 | 30 | 21 | 4 | 2 | − | − | + | + | Prunus persica, tumor | Greece, UD | |
| K84/bv2 | 33 | 2 | 2 | 1 | 2 | 21 | 4 | 2 | − | − | + | − | Prunus persic, soil around tumor | Australia, 1972 | |
| LMG 341/bv2 | 42 | 34 | 37 | 28 | 29 | 21 | 4 | 2 | − | − | + | + | Prunus dulcis, tumor | Israel, UD | |
| LMG 63/bv2 | 43 | 34 | 34 | 28 | 29 | 21 | 4 | 2 | − | − | + | − | Malus sylvestris, hairy root | United States, UD | |
| LMG 99/bv2 | 44 | 34 | 35 | 29 | 30 | 21 | 4 | 2 | − | − | + | + | Rosa sp., tumor | Pennsylvania, UD | |
| LMG 229/bv2 | 46 | 36 | 36 | 28 | 30 | 21 | 32 | 2 | − | − | + | + | Rosa sp., tumor | South Africa, UD | |
| A. vitis LMG 8750T | 47 | 37 | 27 | 30 | 31 | 34 | 33 | 34 | − | − | − | + | Vitis vinifera, tumor | Australia, 1977 | |
Results are presented by genovar within clade A and/or by ST.
All type and reference strains were purchased as Agrobacterium (Rhizobium) radiobacter (A. tumefaciens) strains from the corresponding collection, except for A. vitis S4, A. vitis LMG 8750T, and A. rhizogenes LMG 63.
Phytopathogenicity data for collection strains were obtained from the corresponding collection databases and from references 6, 22, 24, and 30.
+, positive for the test; −, negative for the test; ND, not determined.
PDA CaCo3, differential production of acid from a glucose-containing medium associated with differential dissolution of CaCo3.
LMG, Laboratorium voor Microbiologie Universiteit Gent; CCUG, Culture Collection University of Göteborg; CFBP, Collection Française des Bactéries Phytopathogènes; bv, biovar when available for Agrobacterium (Rhizobium) strains from LMG collection; Fr, France; UD, undocumented.
For each locus, each different allele was assigned an arbitrary number.
PFGE-restriction fragment length polymorphism (RFLP).
Genomic DNA was prepared in agarose plugs as previously described (39) and digested at 25°C with 40 U of SwaI (New England BioLabs, Hertfordshire, United Kingdom). SwaI fragments were separated by pulsed-field gel electrophoresis (PFGE) using a CHEF-DRII apparatus (Bio-Rad Laboratories, Hercules, CA) in a 1% agarose gel in 0.5× Tris-borate-EDTA (TBE) buffer at 150 V and at 10°C. Pulse ramps were 100 to 160 s for 35 h, followed by 40 to 80 s for 15 h. The gel was stained with ethidium bromide and photographed under UV light. PFGE profiles were interpreted visually.
Gene amplification and sequencing.
The complete genomic sequences of A. tumefaciens C58 (AE007869), R. rhizogenes K84 (formerly A. radiobacter, formerly A rhizogenes) (CP000628), and A. vitis S4 (CP000633) were used as references for gene selection and primer design. The primers are shown in Table 3. Genomic DNA was extracted using the MasterPure DNA Purification kit (Epicentre, Madison, WI). PCR was carried out in 50 μl of reaction mixture containing 200 nM (each) primer (Sigma Genosys), 200 μM (each) desoxynucleoside triphosphate (dNTP) (Euromedex, Mundolsheim, France), 2.5 U of Taq DNA polymerase (Promega, Madison, WI) in the appropriate reaction buffer, and 50 ng of genomic DNA as the template. The amplification conditions were as follows: initial denaturation for 3 min at 95°C, followed by 35 cycles with 1 min at 94°C; 1 min at 60°C (trpE and atpD), 65°C (dnaK, glnA, groEL, and zwf), or 68°C (rpoB); and 2 min 30 s (dnaK, glnA, groEL, trpE, and rpoB) or 1 min 30 s (zwf and atpD) at 72°C. The final extension was carried out at 72°C for 10 min. PCR products and a molecular weight marker (phage phiX DNA digested with HaeIII; New England BioLabs) were separated in a 1.5% (wt/vol) agarose gel in 0.5× TBE buffer. The amplification products were sequenced in both directions using forward and reverse amplification primers (Table 3) on an Applied Biosystems automatic sequencer (Beckman Coulter Genomics).
Table 3.
Primers used for gene amplification and sequencing
| Locus | Function | Putative gene product | Locus positiona | Gene size (bp) | Primersb | Primer sequence 5′-3′ | Sequence length (bp) |
|---|---|---|---|---|---|---|---|
| atpD | Energy metabolism | F0-F1 ATP synthase subunit beta | 2604717 | 1,454 | 800F | GGCCAGGACGTTCTGTTCTT | 465 |
| 1350R | CTTGAAGCCCTTGATCGTGT | ||||||
| dnaK | Stress response | Heat shock protein, 70 kDa | 126205 | 1,901 | 720F | GAAGACTTCGACATGCGTCT | 480 |
| 1400R | GCCGAGCAGCTTGTTGTC | ||||||
| glnA | Amino acid biosynthesis | Glutamine synthetase | 196923 | 1,358 | 144F | GTCATGTTCGACGGCTCCT | 474 |
| 1340R | CGCATGACTTCCTGCATCT | ||||||
| 900R | CCTTGGCATGCTTGATGAT | ||||||
| groEL | Stress response | Heat shock protein, 60 kDa | 676328 | 1,634 | 100F | GTGGTGATCAGCAGCGAAG | 504 |
| 1240R | AGGCCAAGGCCAAGAAGAT | ||||||
| 760R | CTGGAAGACATCGCCATCCT | ||||||
| rpoB polymerase | Transcription | Beta subunit RNA | 1927198 | 4,136 | 2040F | GAAAACGACGACGCCAAC | 534 |
| 3150R | TGGACCTTTTCGACCTTGTC | ||||||
| 2718R | GCGCAGAAGCTTTTCTTCC | ||||||
| trpE | Amino acid biosynthesis | Anthranilate synthase | 2262145 | 2,189 | 890F | CGCCCTATTCCTTCTTCATC | 510 |
| 2090R | ATCGATTCCGGGTGGAACT | ||||||
| 1630R | GAAATAATTCGCCAGCGTGT | ||||||
| zwf | Pentose phosphate pathway | Glucose-6-phosphate 1-dehydrogenase | 585849 | 1,475 | 530F | AGATCTTCCGCATCGACCA | 384c |
| 950R | CTTGATGGCGACGAAGGTT |
Gene start codon position on the circular chromosome sequence of A. tumefaciens C58 (accession number AE007869).
F, forward primer; R, reverse primer. Primers in boldface were used for gene sequencing in both directions. The primer denominations correspond to their hybridization regions in the gene according to the complete genome sequence of A. tumefaciens C58.
The size of the zwf sequences of the two strains of A. vitis was 381 bp.
Multilocus sequence-based phylogeny.
Gene sequences were codon aligned using CLUSTALW after translation with TRANSLATE (http://www.expasy.org). The sizes of the codon-aligned sequences used for further analyses are indicated in Table 3. Phylogenetic analyses were performed for each of the seven gene sequences and for the manually concatenated sequence. Evolutionary distance was analyzed using the Phylip package v3.66 (13) by neighbor joining (NJ) after distance matrix construction using DNADIST (with F84 as a substitution model). Bootstrap values were calculated using SEQBOOT and CONSENSE after 1,000 reiterations. Maximum-likelihood (ML) analysis was performed using phylogenetic analysis at http://www.phylogeny.fr (9). The general time-reversible (GTR) model plus gamma distribution and invariant sites was used as a substitution model. ML bootstrap support was computed after 100 reiterations. The sequences of Ochrobactrum anthropi ATCC 49188T (CP000758) were used as outgroup sequences in order to place an artificial tree root. The seven gene sequences from complete genomic sequences of A. vitis S4 (CP000633), Sinorhizobium fredii NGR234 (CP001389), Rhizobium etli CFN 42T (CP000133), and Rhizobium leguminosarum bv. trifolii WSM2304 (CP001191) were also used for phylogenetic analysis.
Multilocus sequence analysis (MLSA).
For each locus, each different allele was assigned to a different arbitrary number using a nonredundant database program available at http://linux.mlst.net/nrdb/nrdb.htm. The combination of allele numbers for each isolate defined the sequence type (ST). A distance matrix in nexus format was generated from the set of allelic profiles using a Web version of the program SplitsTree (http://www.pubmlst.org) and then used for decomposition analyses with SplitsTree 4.0 software (21). The program LIAN 3.1 (19) was used to calculate the standardized index of association (sIA) and to test the null hypothesis of linkage disequilibrium, as well as to determine the mean genetic diversity (H) and genetic diversity at each locus (h). The numbers of synonymous (dS) and nonsynonymous (dN) substitutions per site were determined on codon-aligned sequences using SNAP online software (http://www.hiv.lanl.gov).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank data library under accession numbers HM238283 to HM238884.
RESULTS
MLSA scheme design.
Seven genes encoding housekeeping proteins involved in transcription (rpoB), stress response (dnaK and groEL), amino acid biosynthesis (glnA and trpE), and energy metabolism (atpD and zwf) were selected from the complete genome sequence of A. tumefaciens C58 (Table 3). They were dispersed on the large circular chromosome of the bipartite genome of A. tumefaciens strain C58 (minimal distance between loci, 90 kb) in order to minimize the influence of localized linkage disequilibrium. Partial sequences of the seven loci were obtained for all 89 strains studied, showing that the MLSA scheme was suitable for analysis of strains belonging to the 3 biovars in the genus Agrobacterium, biovar 1, biovar 2, and A. vitis (biovar 3). The sequence size at each locus is given in Table 3.
Multilocus sequence-based phylogeny.
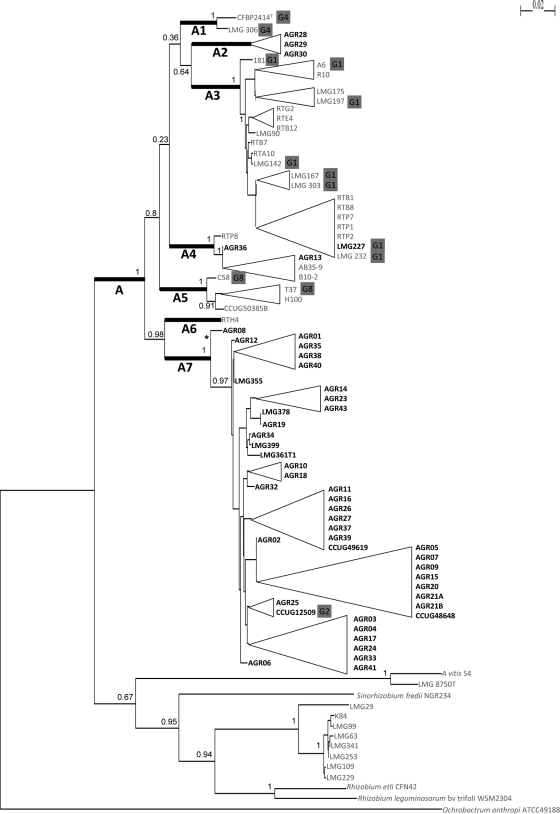
We applied distance and ML phylogenetic approaches to the concatenated sequences (3,351 nucleotides) of the seven loci. The two methods gave congruent trees, and the ML tree is presented in Fig. 1. Phylogenetic analysis clustered Agrobacterium strains in a major clade, clade A. Clade A included 79 of the 89 strains distributed into seven subclades, named genovars A1 to A7, and all the isolates of human origin (Fig. 1). The main genovars, A7 and A3, grouped 54% (n = 43) and 27% (n = 21) of the isolates of clade A, respectively. The 43 strains found in genovar A7 were clinical isolates, representing 88% (43 out of 49 strains) of the strains of human origin in this study. Agrobacterium sp. strains AGR28, AGR29, and AGR30, isolated from the same patient, presented the same sequence for the seven loci and formed, by themselves, genovar A2. Strains AGR13 and AGR36 belonged to genovar A4, together with three environmental strains, and A. radiobacter strain LMG 227 was the only clinical isolate among the 21 strains of genovar A3. Other minor genovars, A1, A5, and A6, did not include clinical strains. Outside of clade A, we observed Agrobacterium strains grouping together with Rhizobium sp. strains. This group, including only environmental Agrobacterium strains, was not well enough represented (n = 10), and therefore, we focused on clade A for further analyses.
Fig. 1.
Maximum-likelihood tree based on concatenated sequences of the seven housekeeping gene fragments indicating the relative placement of clinical (boldface) and environmental (lightface) strains in the genus Agrobacterium. The horizontal lines show genetic distance. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values of >0.5 are indicated. The scale bar indicates the number of substitutions per nucleotide position. O. anthropi ATCC 49188T was used as the outgroup organism. The sequences of S. fredii NGR234 (CP001389), R. etli CFN 42T (CP000133), and R. leguminosarum bv. trifolii WSM2304 (CP001191) were also included. Agrobacterium strains with identical concatenated sequences are presented on collapsed branches and are listed on the right of the triangular representations. The names of the clades and genovars discussed in the text are shown at the corresponding roots, indicated by bold lines. The node with an asterisk varied depending on the ML trees reconstructed with each of the seven loci. Genomic species G1 to G9 are indicated in gray boxes after the names of the corresponding strains. The data are from references 7, 8, and 30.
For each locus, phylogenetic trees reconstructed using NJ or ML methods were compared to the NJ or ML trees reconstructed from the seven concatenated sequences (data not shown). The genovars determined in the concatenated tree were also found in individual trees. Despite some conflicting topologies in terms of branching order observed among the trees, the distribution of strains in each genovar was not modified according to the gene used or the phylogenetic method, except for strain AGR08, which belonged to genovar A5 after trpE locus analysis and was placed in an intermediate position between genovars A5 and A7 by rpoB locus analysis.
Genomic species have been previously described in the genus Agrobacterium by determination of DNA-DNA hybridization (7, 8, 30). These data are available for 13 strains included in this study and are shown in Fig. 1. The population studied here contained strains representative of the genomic species G1, G2, G4, and G8 according to the nomenclature of Mougel et al. (26). We observed concordance between MLSA clusters and genomic groups for members of clade A: the eight strains of group G1 belonged to genovar A3, while strains of groups G4 and G8 were found in genovars A1 and A5, respectively. The only strain representative of group G2, A. radiobacter CCUG 12509, was located in the “human” genovar, A7. It was noteworthy that this clinical strain, isolated in the United States in 1974, had the same sequence for the seven loci as the clinical strain AGR25, isolated in France in 2008. Genovars A2, A4, and A6 in clade A did not contain strains previously affiliated with a genomic group, and therefore, the relationship between these genovars and genomic species could not be established.
Multilocus and genomic typing.
Forty-eight unique sequence types (STs) were observed; 33 of them (69%) were identified only once, suggesting an overall high level of genetic diversity among the studied population (Tables 1 and 2). In clade A, the largest STs were ST6 (8 isolates); ST9 and ST28, each comprising 7 isolates; and ST5 (6 isolates). These major STs included 35% of the clade A strains. Within the “human” genovar A7, 58% of the strains were grouped in 4 STs (ST3, ST5, ST6, and ST9). In each of these STs, strains appeared epidemiologically unrelated, i.e., they were isolated over a large period of time (from 5 to 10 years) and in some cases from geographically distant places. No obvious relationships between the STs and the types of clinical samples were observed, except for ST9, which comprised 6 of the 7 isolates from the human respiratory tract. Outside of genovar A7, only ST28 in genovar A3 comprised several strains (n = 7) with different lifestyles; these isolates were sampled over a 43-year period in three different countries (Tables 1 and 2).
Fifty-six strains sharing 15 identical STs, i.e., 37 clinical and 19 environmental isolates of clade A, were further analyzed by PFGE in order to detect the occurrence of genomically identical strains in the corresponding population. Genomic macrorestriction with the endonuclease SwaI produced PFGE patterns suitable for strain comparison and comprising an average of 10 bands (data not shown). A high level of genomic diversity was observed: the 56 strains studied showed 51 pulsotypes, and 74 distinguishable isolates were demonstrated among the 79 strains of clade A based on both multilocus and PFGE analysis results. However, the levels of genomic diversity in each ST varied greatly, with from 9% to 100% of shared bands obtained from the patterns of the same ST (Table 4). A relationship between the diversity of PFGE patterns in an ST and the lifestyle of the strains could be observed (Table 4). Identical or very similar patterns with more than 95% shared bands were observed for pairs of phytopathogenic strains that in other respects were unrelated on the basis of date and/or site of isolation, such as T37 and H100 (ST25), LMG 175 and LMG 197 (ST35), and LMG 167 and LMG 303 (ST45) (Table 4). Identical or closely related patterns were also observed for three clinical strains isolated from blood cultures from the same patient within a 5-day period (strains AGR28, AGR29, and AGR30 in ST14), between strains from two patients attending the same clinical ward 1 year apart (AGR10 and AGR18 in ST8), and between strains from two patients hospitalized in two different hospitals in Montpellier on the same date (AGR26 and AGR27 in ST9). The other clinical strains studied show no more than 40% shared bands (Table 4). An intermediate genomic-diversity level was observed for nonphytopathogenic environmental strains, except for strains RTP1 and RTP2 (ST28), originating from the same experimental field, which appeared identical after PFGE analysis.
Table 4.
PFGE analysis of Agrobacterium strains belonging to the same ST
| Parameter | Value |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST2 (n = 3)c | ST3 (n = 4) | ST5 (n = 6) | ST6 (n = 8) | ST8 (n = 2) | ST9 (n = 7) | ST11 (n = 3) | ST13 (n = 2) | ST14 (n = 3) | ST25 (n = 2) | ST27 (n = 3) | ST28 (n = 7) | ST35 (n = 2) | ST41 (n = 2) | ST45 (n = 2) | |
| Clinical strains (%)a | 33 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 17 | 0 | 0 | 0 |
| Phytopathogen strains (%)a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 100 | 100 |
| Other strains (%)a | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 83 | 0 | 0 | 0 |
| Shared bands (%)b | 42 | 22 | 40 | 10 | 75 | 31 | 21 | 9 | 93 | 95 | 35 | 29 | 100 | 86 | 100 |
Percentage among all strains of an ST.
Percentage of shared bands obtained from the PFGE patterns of strains belonging to the same ST.
n, number of strains.
Phenotypic traits in genovars and sequence types.
Among the 88 strains tested, including 49 clinical and 39 environmental isolates, all grew at 35°C but only 41 strains grew at 42°C (Tables 1 and 2). Among the latter, 40 isolates were of clinical origin and belonged to genovar A7; the remaining strain was A. radiobacter LMG 90 (genovar A3) from Rosa sp. galls. In other words, only 3 out of the 43 clinical strains of genovar A7 (AGR33, AGR40, and LMG 355) did not grow at 42°C. Clinical strains belonging to other genovars than A7 did not grow at 42°C, and only one strain of environmental origin showed significant growth at this temperature.
The production of 3-ketolactose from lactose and differential acid production, which are major characteristics commonly used for determining biovar affiliation, were studied for the 74 genomically distinguishable isolates belonging to clade A. The assays gave results congruent with biovar identifications for the LMG control strains tested. Forty-four strains out of 74 (59%) were positive for production of 3-ketolactose (Tables 1 and 2). Genovars A1, A2, A5, and A6 contained only 3-ketolactose-producing strains, while in other genovars, the ability to produce 3-ketolactose varied among strains. For example, genovar A7 contained 36% (15 out of the 42 strains tested) 3-ketolactose-producing strains, and production was also found to be variable among strains sharing the same ST (ST5, ST6, ST8, and ST13). In clade A, all the 3-ketolactose nonproducers were isolated from animal hosts (human or nematode), except for strain 181 from Populus sp.
Whatever their clinical or environmental origins, all clade A isolates are unable to produce large amounts of acid from glucose and, consequently, to dissolve CaCO3 from a glucose-containing medium: they were negative in the differential acid production assay.
Genetic statistics and recombination.
Genetic analysis of the population was performed on the 74 genomically distinguishable isolates of clade A. Genetic statistics were also calculated for the two main genovars, A3 and A7 (Table 5). Within clade A, a total of 544 single-nucleotide polymorphisms (SNPs) in the 7 loci were observed. This corresponded to a range of 13.1% to 23.4% polymorphic sites, depending on the gene (Table 5). The number of different alleles for the seven loci ranged from 22 ( glnA and groEL) to 28 (atpD) (Table 5) and did not depend on the size of the sequence studied. All loci had equivalent mol% G+C contents, from 59.6% to 61.1%, with a mean value of 60.5%, which was similar to the mean mol% G+C contents of the A. tumefaciens C58 chromosomes (59%) (18). The mean genetic diversity (H) and the genetic diversity at each locus (h) for clade A and genovars A3 and A7 indicated a high level of genetic diversity both inside clade A and inside the main genovars, A3 and A7 (Table 5). The locus trpE displayed the highest percentage of polymorphic sites, while groEL appeared to be the most conserved in both genovars A3 and A7. For instance, groEL displayed only 6 polymorphic sites among the 18 strains of genovar A3 tested. In contrast, the groEL gene was the locus for which a higher rate of nonsynonymous SNPs versus synonymous SNPs (dN/dS ratio) was observed at the clade level. Nevertheless, inside each of the genovars A3 and A7, no nonsynonymous mutations were observed at this locus. The dN/dS ratio for the other six loci was found to be weak (Table 5), indicating that these loci were not subjected to strong positive selective pressure. The nonsynonymous mutations did not correspond to any premature stop codon.
Table 5.
Sequence analysis of the seven loci
| Locus | Clade or genovar | No. of alleles | No. (%) of polymorphic sites | Genetic diversity (h) | No. of nonsynonymous codons | dN/dSa ratio |
|---|---|---|---|---|---|---|
| atpD | Clade A | 28 | 68 (14.6) | 0.9716 | 3 | 0.02 |
| Genovar A7 | 10 | 13 (2.8) | 0.8824 | 0 | ||
| Genovar A3 | 9 | 17 (3.7) | 0.9778 | 1 | 0.08 | |
| dnaK | Clade A | 26 | 67 (14) | 0.9730 | 2 | 0.02 |
| Genovar A7 | 11 | 15 (3.1) | 0.9085 | 0 | ||
| Genovar A3 | 6 | 14 (2.9) | 0.9111 | 0 | ||
| glnA | Clade A | 22 | 81 (17.1) | 0.9673 | 6 | 0.03 |
| Genovar A7 | 9 | 13 (2.7) | 0.9085 | 1 | 0.06 | |
| Genovar A3 | 6 | 16 (3.4) | 0.8889 | 0 | ||
| groEL | Clade A | 22 | 67 (13.3) | 0.9403 | 10 | 0.107 |
| Genovar A7 | 9 | 9 (1.8) | 0.9150 | 0 | ||
| Genovar A3 | 3 | 6 (1.2) | 0.3778 | 0 | ||
| rpoB | Clade A | 27 | 70 (13.1) | 0.9744 | 5 | 0.03 |
| Genovar A7 | 13 | 33 (6.2) | 0.9608 | 1 | 0.015 | |
| Genovar A3 | 5 | 8 (1.5) | 0.7556 | 0 | ||
| trpE | Clade A | 25 | 101 (19.8) | 0.9787 | 1 | 0.004 |
| Genovar A7 | 10 | 52 (10.2) | 0.9346 | 0 | ||
| Genovar A3 | 7 | 20 (3.9) | 0.9333 | 0 | ||
| zwf | Clade A | 26 | 90 (23.4) | 0.9730 | 6 | 0.02 |
| Genovar A7 | 9 | 15 (3.9) | 0.8954 | 0 | ||
| Genovar A3 | 8 | 13 (3.4) | 0.9556 | 1 | 0.08 |
dN, number of nonsynonymous substitutions per nonsynonymous site; dS, number of synonymous substitutions per synonymous site.
Evidence in favor of clonal or recombining population structure can be obtained by assessing the levels of linkage between alleles at different loci around the chromosome. We assessed the linkage between alleles from the 7 loci by determination of the sIA value. The sIA value is expected to be zero when a population is at linkage equilibrium, i.e., when free recombination occurs. Analyses were carried out using one isolate from each ST in order to minimize any bias due to a possible epidemic population structure. The sIA values were calculated for clade A and for genovars A3 and A7. The sIA values ranged from 0.3425 to 0.3610 and were significantly different from 0 (P ≤ 1.00 × 10−3), suggesting that the recombination rates were low.
Linkage disequilibrium in clade A could be present in long-term recombining populations where adaptive clones have emerged over the short term. To explore this hypothesis, we performed decomposition analysis, which depicts all of the shortest pathways linking sequences, including those that produce an interconnected network. The split graph (obtained using the Neighbor-Net method) of all seven loci displayed parallel paths (Fig. 2) corresponding to recombination events. However, the number of events was low and confirmed the low rate of recombination deduced from the determination of the sIA. Recombination events occurred only inside genovars A3, A4, and A7. Finally, the recombination clusters generated by splitting trees were consistent with phylogenetic lineages. This suggested that genetic exchanges occurred inside each genovar but that each genovar formed a lineage genetically isolated from others.
Fig. 2.
SplitsTree decomposition analyses of MLSA data for A. radiobacter, A. tumefaciens, and A. vitis strains. The distance matrix was obtained from allelic profiles of strains. A network-like graph indicates recombination events. A starlike radiation from the central point indicates absence of recombination.
DISCUSSION
Agrobacterium spp. are described as environmental bacteria, and some strains are phytopathogenic by tumorigenesis. The phytopathogenic behavior is related to a particular genomic structure that includes a conjugative plasmid harboring genes involved in tumor formation in plants. Besides these highly specialized bacteria, nonphytopathogenic strains were found in diverse environments not always in association with plants. Agrobacterium is also a pathogen of human beings by a mechanism probably unrelated to plant tumorigenesis (29), but the characteristics of the pathogenicity of clinical strains remain mostly unknown. Virulence in humans is considered to be low; however, the frequent isolation of Agrobacterium in cases of nosocomial infections and in cystic fibrosis patients (3, 5, 23) suggests adaptation of the bacterium to humans, particularly in the context of hospitalization, immunosuppression, or underlying diseases. Typically, mild nosocomial and/or opportunistic pathogens of environmental origin, like Stenotrophomonas maltophilia or Ochrobactrum spp., displayed a high level of resistance to antibacterial compounds (27, 38). This resistance by itself could explain bacterial adaptation to nosocomial conditions. In the case of Agrobacterium, resistance to antibiotics is considered to be lower than that observed for other environmental opportunistic pathogens (3). On the other hand, genetic data suggested heterogeneity within the genus Agrobacterium and in the species A. tumefaciens and A. radiobacter (30, 31). This genetic diversity has been mainly demonstrated by DNA-DNA hybridization and amplified fragment length polymorphism (AFLP) approaches, because 16S rRNA genes displayed low polymorphism in the genus, as was also observed for the entire family Rhizobiaceae (42). Adaptation of some strains to human colonization and/or infection, taken together with the genetic heterogeneity in the genus, raises the question of the existence of a genetic subpopulation of Agrobacterium that has developed a particular relationship with humans.
In this study, we investigated genetic diversity and some phenotypic traits in the largest collection of clinical isolates of A. radiobacter and A. tumefaciens reported so far. The population of strains represents diverse clinical and geographic sources, as well as diverse dates of isolation. Environmental strains were also included in the population studied here. Finally, the overall collection represented different lifestyles encountered in the genus: free living and association with plants (phytopathogenic or not), with nematodes, and with humans. The collection was studied by multilocus analysis and PFGE, two methods applied for the first time to a large population of genomically and biologically diverse Agrobacterium isolates.
The multilocus-based analysis proposed here succeeded in analyzing the four represented genomic species defined by DNA-DNA hybridization in biovar 1 of A. tumefaciens and A. radiobacter, the K84 group (biovar 2), and A. vitis (biovar 3). Moreover, MLSA displayed good discriminatory power, defining seven genovars in clade A that roughly corresponded to biovar 1 of A. tumefaciens and A. radiobacter. The large majority of the clinical strains and isolates (88%) belonged to genovar A7, the only genovar encompassing only clinical strains. The few remaining clinical strains were scattered in three other genovars, either alone or associated with environmental, nonphytopathogenic strains. At first glance, genovar A7 could be considered a “clinical genovar” or a “human-associated” genovar. However, a sampling bias could be suspected in the population studied, since a number of the strains analyzed came from the same region of France (Teaching Hospitals of Nîmes and Montpellier, 60 kilometers apart in the south of France). For example, ST3 and ST5 in genovar A7 contained only strains from this geographic origin, although they were isolated 5 or 6 years apart. However, PFGE analysis of ST3 and ST5 strains showed genomic patterns differing by at least 8 or 6 bands out of approximately 10, respectively. This suggested that the grouping of strains in STs and then in genovars did not result from the spread of the same clone in a limited geographic zone. This was confirmed for some STs that included strains of remote origin, such as France and Sweden (ST6 and ST9) or France and the United States (ST13). Moreover, in the same ST, the dates of isolation could differ by up to 31 years. For instance, ST13 included strains isolated in 1974 in Texas and in 2008 in Montpellier (France). For these reasons, we considered that genovar A7 was actually a “human-associated” genovar and not a “geographic” genovar. A major phenotypic trait further supported this conclusion, since 91% of the strains belonging to genovar A7 grew at 42°C, whereas the optimal temperature for the genus Agrobacterium was defined as between 25°C and 28°C (34). The heat shock protein HSP60 is a chaperone protein involved in stress adaptation, including thermal stresses. The groEL gene was included in our MLSA scheme and displayed a higher frequency of nonsynonymous mutations than other loci. This suggested that groEL is subjected to particular selective pressure in the population tested. This selective pressure could be the host temperature, which clearly differs between the human body and plants or other environmental sources. The ability to grow at high temperature should be considered a major trait for adaptation to the human body or to other thermoregulated animals. Agrobacterium strains isolated from other warm-blooded animals have recently been described (36), and they should be analyzed in order to decide whether genovar A7 is a real “human-associated” genovar or a more generally “thermophilic” genovar that consequently succeeded in its relationship with the human body.
No differences were observed in epidemiological data, such as geographic origin, clinical site of isolation, medical unit, and date of isolation, between strains in genovar A7 and the few other human strains scattered in other genovars or between strains from different STs in genovar A7. The genomic structure and its dynamics have been related to the lifestyles of bacteria (20). Pathogenic bacteria, which have close relationships with their hosts, generally inhabit a narrow and stable niche and show a low level of genomic polymorphism. Comparison of macrorestriction profiles could provide a snapshot of the genome dynamics. In the alphaproteobacteria, the genome of Brucella, which lives in mammalian macrophages, appears highly conserved by diverse comparative methods (14, 15, 25), whereas the genome of its phylogenetic neighbor Ochrobactrum, a free-living bacterium and an opportunistic pathogen, is highly fluid and polymorphic (33, 39). We observed differences in the levels of genomic polymorphism assessed by PFGE between the phytopathogenic strains and the clinical strains. The genome of the phytopathogens appeared globally conserved, suggesting the existence of constraints limiting genomic variation and adaptation to a narrow niche, including a specialized virulence mechanism. In contrast, PFGE profiles were highly polymorphic among A. tumefaciens (A. radiobacter) clinical strains. Variation generated by genome rearrangements has been reported to provide advantages for invading or for merely inhabiting complex environments, a condition that corresponded to the opportunistic behavior of A. radiobacter. For epidemiological purposes, PFGE-RFLP appeared more discriminative than MLST, since it was able to differentiate strains belonging to the same multilocus sequence type and is probably a suitable method for the epidemiological follow-up of human outbreaks due to Agrobacterium. PFGE previously permitted detection of the nosocomial transmission of Agrobacterium isolates from intravenous catheters of two hospitalized patients (17).
Previous phylogenetic studies based on the 16S rRNA gene (42) suggested a high level of recombination in the genus Agrobacterium, making its utilization in taxonomy difficult. Our results showed, by the use of a multilocus approach, that the Agrobacterium population was structured in robust subpopulations and had a low rate of recombination. The clade A population displayed a basically clonal structure, with no recombination among clones and a low level of recombination within each clone (genovar). The genovars obtained by the multilocus approach seem to be in accordance with the genomic species determined in previous studies (7, 8, 30). Consequently, our multilocus scheme appeared to be a suitable tool for taxonomic studies in the genus and allowed us to suggest that the “human-associated” genovar A7, which contains a strain previously affiliated with genomospecies G2 by DNA-DNA hybridization (30) and presents particular phenotypic features, such as growth temperature and inconstant production of 3-ketolactose, may represent a new species in the genus Agrobacterium (Rhizobium). However, further investigations, such as DNA-DNA hybridization between diverse members of genovar A7 and a more complete phenotypic study, remain to be performed.
Agrobacterium spp. are generally mild pathogens, but the existence of a clone adapted to clinical conditions should stress its detection and epidemiological surveillance, as well as its evolution, in order to detect the emergence of virulent and/or resistant strains and to prevent their spread.
ACKNOWLEDGMENTS
We are particularly indebted to the microbiology laboratory team of the Nîmes, Montpellier, and Toulouse, France, academic hospitals for providing some clinical isolates. We also thank CNRS UPR2355 ISV at Gif-sur-Yvette and UMR1133 EMIP of the University Montpellier 2 for providing isolates from plants and nematodes, respectively.
Parts of this study were supported by grants from ADEREMPHA (Sauzet, France).
Footnotes
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Amaya R. A., Edwards M. S. 2003. Agrobacterium radiobacter bacteremia in pediatric patients: case report and review. Pediatr. Infect. Dis. J. 22:183–186 [DOI] [PubMed] [Google Scholar]
- 2. Bouzar H., Jones J. B., Bishop A. L. 1995. Simple cultural tests for identification of Agrobacterium biovars. Methods Mol. Biol. 44:9–13 [DOI] [PubMed] [Google Scholar]
- 3. Chen C.-Y., Hansen K. S., Hansen L. K. 2008. Rhizobium radiobacter as an opportunistic pathogen in central venous catheter associated bloodstream infection: case report and review. J. Hosp. Infect. 68:203–207 [DOI] [PubMed] [Google Scholar]
- 4. Chilton M. D., et al. 1982. Agrobacterium rhizogenes inserts T-DNA into the genomes of the host plant root cells. Nature 295:432–434 [Google Scholar]
- 5. Coenye T., Goran J., Spilker T., Vandamme P., LiPuma J. J. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov. sp. nov. J. Clin. Microbiol. 40:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeLey J., Bernaerts M., Rassel A., Guilmot J. 1966. Approach to an improved taxonomy of the genus Agrobacterium. J. Gen. Microbiol. 43:7–17 [DOI] [PubMed] [Google Scholar]
- 7. DeLey J., Tytgat R., De Smedt J., Michels M. 1973. Thermal stability of DNA:DNA hybrids within the genus Agrobacterium. J. Gen. Microbiol. 78:241–252 [Google Scholar]
- 8. DeLey J. 1974. Phylogeny of prokaryotes. Taxon 23:291–300 [Google Scholar]
- 9. Dereeper A., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edmond M. B., Riddler S. A., Baxter C. M., Wicklund B. M., Pascuile A. W. 1993. Agrobacterium radiobacter a recently recognized opportunistic pathogen. Clin. Infect. Dis. 16:388–391 [DOI] [PubMed] [Google Scholar]
- 11. Erol Cipe F., Dogu F., Sucuoglu D., Aysev D., Ikinciogullari A. 2010. Asymptomatic catheter related Rhizobium radiobacter infection in a haploidentical hemapoetic stem cell recipient. J. Infect. Dev. Ctries. 4:530–532 [DOI] [PubMed] [Google Scholar]
- 12. Farrand S. K., van Berkum P. B., Oger P. 2003. Agrobacterium is a definable genus of the family Rhizobiaceae. Int. J. Syst. Evol. Microbiol. 53:1681–1687 [DOI] [PubMed] [Google Scholar]
- 13. Felsenstein J. 1984. Distance methods for inferring phylogenies: a justification. Evolution 38:16–24 [DOI] [PubMed] [Google Scholar]
- 14. Foster J. T., et al. 2009. Whole-genome-based phylogeny and divergence of the genus Brucella. J. Bacteriol. 191:2864–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gándara B., Merino A. L., Rogel M. A., Martinez-Romero E. 2001. Limited genetic diversity of Brucella spp. J. Clin. Microbiol. 39:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garrity G. M., et al. 2007. Part 3, The Bacteria: phylum “Proteobacteria”, class Alphaproteobacteria, p. 52–111 In Cole J. R., et al. (ed.), The taxonomic outline of Bacteria and Archaea, release 7.7. Michigan State University, East Lansing, MI: http://www.taxonomicoutline.org/index.php/toba/article/view/179/212 [Google Scholar]
- 17. Giammanco G. M., et al. 2004. Molecular typing of Agrobacterium species isolates from catheter-related bloodstream infections. Infect. Control Hosp. Epidemiol. 25:885–887 [DOI] [PubMed] [Google Scholar]
- 18. Goodner B., et al. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328 [DOI] [PubMed] [Google Scholar]
- 19. Haubold B., Hudson R. R. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage analysis. Bioinformatics 16:847–848 [DOI] [PubMed] [Google Scholar]
- 20. Hughes D. 8 December 2000. Evaluating genome dynamics: the constraints on rearrangements within bacterial genomes. Genome Biol. 1:reviews0006-reviews0006.8. doi: 10.1186/gb-2000-1-6-reviews0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 22. Kerters K., DeLey J., Sneath P. H. A., Sackin M. 1973. Numerical taxonomic analysis of Agrobacterium. J. Gen. Microbiol. 78:227–239 [Google Scholar]
- 23. Lai C.-C., et al. 2004. Clinical and microbiological characteristics of Rhizobium radiobacter infections. Clin. Infect. Dis. 38:149–153 [DOI] [PubMed] [Google Scholar]
- 24. Lippincott J. A., Lippincott B. B. 1969. Tumour-initiating ability and nutrition in the genus Agrobacterium. J. Gen. Microbiol. 59:57–75 [Google Scholar]
- 25. Michaux-Charachon S., et al. 1997. Genome structure and phylogeny in the genus Brucella. J. Bacteriol. 179:3244–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mougel C., Thioulouse J., Perriere G., Nesme X. 2002. A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. Evol. Microbiol. 52:573–586 [DOI] [PubMed] [Google Scholar]
- 27. Nicodemo A. C., Garcia Paez J. I. 2007. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J. Clin. Microbiol. Infect. Dis. 26:229–237 [DOI] [PubMed] [Google Scholar]
- 28. Paphitou N. I., Rolston K. V. 2003. Catheter related bacteremia caused by Agrobacterium radiobacter in a cancer patient: case report and literature review. Infection 31:421–424 [DOI] [PubMed] [Google Scholar]
- 29. Petrunia I. V., et al. 2008. Agrobacterium tumefaciens-induced bacteraemia does not lead to reporter gene expression in mouse organs. PLoS One 3:e2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Popoff M. Y., Kersters K., Kiredjian M., Miras I., Coynault C. 1984. Position taxonomique de souches d'Agrobacterium d'origine hospitalière. Ann. Microbiol. 135:427–442 [PubMed] [Google Scholar]
- 31. Portier P., et al. 2006. Identification of genomic species in Agrobacterium biovar 1 by AFLP genomic markers. Appl. Environ. Microbiol. 72:7123–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ream L. W., Gordon M. P. 1982. Crown gall disease and prospects for genetic manipulation of plants. Science 218:854–859 [DOI] [PubMed] [Google Scholar]
- 33. Romano S., et al. 2009. Multilocus sequence typing supports the hypothesis that Ochrobactrum anthropi displays a human-associated subpopulation. BMC Microbiol. 9:267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sawada H., Ieki H., Oyaizu H., Matsumoto S. 1993. Proposal for rejection of Agrobacterium tumefaciens and revised descriptions for the genus Agrobacterium and for Agrobacterium radiobacter and Agrobacterium rhizogenes. Int. J. Syst. Bacteriol. 43:694–702 [DOI] [PubMed] [Google Scholar]
- 35. Slater S. C., et al. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 191:2501–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tabar M. D., et al. 2010. Presence of opportunistic bacteria (Rhizobium spp.) with potential for molecular misdiagnosis among canine and feline clinical samples. Can. Vet. J. 51:895–897 [PMC free article] [PubMed] [Google Scholar]
- 37. Teyssier C., et al. 2003. Species identification and molecular epidemiology of bacteria belonging to Ochrobactrum genus. Pathol. Biol. 51:5–12 [DOI] [PubMed] [Google Scholar]
- 38. Teyssier C., et al. 2005. Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J. Med. Microbiol. 54:945–953 [DOI] [PubMed] [Google Scholar]
- 39. Teyssier C., et al. 2005. Pulsed-field gel electrophoresis to study the diversity of whole genome organization in the genus Ochrobactrum. Electrophoresis 26:2898–2907 [DOI] [PubMed] [Google Scholar]
- 40. Tzfira T., Citovsky V. 2006. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 17:147–154 [DOI] [PubMed] [Google Scholar]
- 41. Velázquez E., et al. 2010. Analysis of core genes supports the reclassification of strains Agrobacterium radiobacter K84 and Agrobacterium tumefaciens AKE10 into the species Rhizobium rhizogenes. Syst. Appl. Microbiol. 33:247–251 [DOI] [PubMed] [Google Scholar]
- 42. Young J. M., Kuykendall L. D., Martinez-Romero E., Kerr A., Sawada H. 2001. A revision of Rhizobium Frank 1889 with an emended description of the genus and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola deLajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 51:89–103 [DOI] [PubMed] [Google Scholar]
- 43. Young J. M., Kuykendall L. D., Martinez-Romero E., Kerr A., Sawada H. 2003. Classification and nomenclature of Agrobacterium and Rhizobium—a reply to Farrand et al. Int. J. Syst. Evol. Microbiol. 53:1689–1695 [DOI] [PubMed] [Google Scholar]
- 44. Young J. M., Pennycook S. R., Watson D. R. W. 2006. Proposal that Agrobacterium radiobacter has priority over Agrobacterium tumefaciens. Request for an opinion. Int. J. Syst. Evol. Microbiol. 56:491–493 [DOI] [PubMed] [Google Scholar]
- 45. Zhu J., et al. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]