Abstract
Mutants with deletion mutations in the glg and mal gene clusters of Escherichia coli MC4100 were used to gain insight into glycogen and maltodextrin metabolism. Glycogen content, molecular mass, and branch chain distribution were analyzed in the wild type and in ΔmalP (encoding maltodextrin phosphorylase), ΔmalQ (encoding amylomaltase), ΔglgA (encoding glycogen synthase), and ΔglgA ΔmalP derivatives. The wild type showed increasing amounts of glycogen when grown on glucose, maltose, or maltodextrin. When strains were grown on maltose, the glycogen content was 20 times higher in the ΔmalP strain (0.97 mg/mg protein) than in the wild type (0.05 mg/mg protein). When strains were grown on glucose, the ΔmalP strain and the wild type had similar glycogen contents (0.04 mg/mg and 0.03 mg/mg protein, respectively). The ΔmalQ mutant did not grow on maltose but showed wild-type amounts of glycogen when grown on glucose, demonstrating the exclusive function of GlgA for glycogen synthesis in the absence of maltose metabolism. No glycogen was found in the ΔglgA and ΔglgA ΔmalP strains grown on glucose, but substantial amounts (0.18 and 1.0 mg/mg protein, respectively) were found when they were grown on maltodextrin. This demonstrates that the action of MalQ on maltose or maltodextrin can lead to the formation of glycogen and that MalP controls (inhibits) this pathway. In vitro, MalQ in the presence of GlgB (a branching enzyme) was able to form glycogen from maltose or linear maltodextrins. We propose a model of maltodextrin utilization for the formation of glycogen in the absence of glycogen synthase.
INTRODUCTION
The synthesis of glycogen in bacteria occurs when they are grown with limited nutrients but an abundance of a carbon source (33, 34). Escherichia coli accumulates glycogen at levels of more than half of its cell mass under optimal conditions. The glycogen gene cluster in E. coli consists of two operons oriented in tandem, glgBX and glgCAP, encoding enzymes that synthesize and degrade glycogen (12). The encoded enzymes are a branching enzyme (glgB), a debranching enzyme (glgX), an ADP-glucose pyrophosphorylase (glgC), a glycogen synthase (glgA), and a glycogen phosphorylase (glgP). The polymerization of the α-1,4-linked glucosyl chain is mediated via the transfer of glucose from ADP-glucose by GlgA, the glycogen synthase, onto the nonreducing ends of linear dextrins that are subsequently branched (formation of α-1,6-glycosyl linkage) by GlgB, the branching enzyme. The expression of the glg gene cluster is complicated. It involves the global carbon storage regulator CsrA (2, 53), the cyclic AMP (cAMP)/catabolite gene activator protein (CAP) system (39), and the stringent response (38). In addition, the two-component regulatory system PhoP-PhoQ (29) connects the system to Mg2+ levels, and even the phosphotransferase system appears to affect the glycogen phosphorylase involved in the degradation of glycogen (42, 43). glgS, an additional gene involved in glycogen synthesis, is not part of the glg gene cluster. It is not essential for glycogen synthesis. glgS mutants show reduced glycogen contents and reveal RpoS dependency of glycogen synthesis (20). The role of GlgS in glycogen synthesis remains unclear, but a role in protein-protein interaction has been suggested (24). The mode of glycogen synthesis, in particular the initial priming reaction, is not well established in bacteria, but the mode of degradation is well established. GlgP, by forming glucose-1-phosphate (glucose-1-P), reduces the lengths of the α-1,6-branched chains to the sizes of maltotetraosyl and maltotriosyl residues, which then become substrates of GlgX, the debranching enzyme that releases maltotetrose and maltotriose (12, 46). It is the glycogen-dependent formation of maltotetraose and maltotriose that establishes the link to the maltose/maltodextrin-utilizing system.
The maltose/maltodextrin system of E. coli consists of 10 genes located in five operons that encode proteins involved in the uptake of maltose and short maltodextrins via a binding protein-dependent ABC transporter and four maltodextrin-utilizing enzymes (5). The key enzyme in maltose utilization is MalQ (amylomaltase) (28, 35, 51), a 4-α-glucanotransferase. It recognizes maltose and longer maltodextrins; it preferentially removes glucose from the reducing ends of maltose and small maltodextrins and transfers the remaining enzyme-bound dextrinyl residue to the nonreducing ends of other maltodextrins, thus forming longer maltodextrin chains in the process (31). When glucose is removed (in vivo, for instance, by glucokinase) and no other dextrin-utilizing enzymes are present, rather long maltodextrin chains are formed (16). With longer maltodextrins (for instance, maltoheptaose), very little glucose is formed in the disproportionation reaction, but the increase in chain length still occurs (23). MalP (maltodextrin phosphorylase) (41, 50) recognizes maltotetraose and longer maltodextrins and removes glucose from the nonreducing ends of the dextrins by phosphorolysis, forming glucose-1-P. Thus, the combination of MalQ and MalP yields via maltodextrin intermediates glucose and glucose-1-P that enter glycolysis. Mutants lacking MalQ cannot grow on maltose but can grow on maltotetraose and larger maltodextrins due to the action of MalP releasing glucose-1-P. Mutants lacking MalP can grow on maltose and short maltodextrins, but in the process they accumulate large amounts of long-chain maltodextrins, resulting in an expanded cell shape (40). Of the supplied maltose, they can utilize only one glucose moiety that is released by the action of MalQ. Thus, the combination of MalQ and MalP provides the means to efficiently utilize maltose or maltodextrins. However, E. coli has two additional enzymes that act on maltodextrins. MalS is an enzyme located in the periplasm that recognizes long dextrins and preferentially releases maltohexaose from the nonreducing ends of linear maltodextrins (18). This is in line with the specificity of the maltodextrin ABC transporter, which can accommodate maltodextrins of up to 7 glucosyl units (13, 49). MalZ (maltodextrin glucosidase) is located in the cytoplasm and cleaves glucose sequentially from the reducing ends of maltodextrins, with maltotriose as the smallest substrate (48). MalZ is also able to hydrolyze γ-cyclodextrin (32) and larger cyclodextrins. It exhibits transglycosylation activity and is able to produce nigerose-containing dextrins (47). Like all mal genes, malZ is under the control of MalT, the central activator, with maltotriose being the inducer (37). However, malZ is also induced at high osmolarity independent of MalT (15). In addition, E. coli contains a cytoplasmic α-amylase that is not part of the maltose system or of the glycogen system (36). The function of the enzyme is unclear; mutants lacking the enzyme have no apparent phenotype.
There is the phenomenon of endogenous induction of the maltose system that is caused by the formation of internal maltotriose derived from the degradation of glycogen (15). Two Glg enzymes are essential for endogenous maltotriose formation: GlgP, which shortens the branch chains of glycogen to the lengths of maltotetraosyl and maltotriosyl units, and GlgX, which releases the branch chains (12). On the other hand, of the mal enzymes, MalQ, MalP, and MalZ greatly affect glycogen-dependent endogenous induction (15). In particular, the osmo-dependent increase in the amount of MalZ strongly reduces endogenous induction by degrading maltodextrins, including the inducer maltotriose (37) to maltose (15). The expression of the maltose system is subject to strong catabolite repression by glucose. Not only are the operons encoding the ABC transporter dependent on the cAMP/CAP complex, but so is the expression of malT, encoding the activator of all mal genes (9, 10).
In contrast, the role of mal enzymes in glycogen synthesis and metabolism is not known, although it was suggested briefly in an early review by Preiss (33). In this study we tested the amounts, the sizes, and the branch chain distributions of glycogen in isogenic mutants lacking MalP, MalQ, MalZ, and GlgA. We found that maltose/maltodextrin metabolism can lead to the formation of glycogen even in the absence of glycogen synthase. It is the activity of MalP (maltodextrin phosphorylase) that determines the strength of this pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and recombinant DNAs that were used or constructed in this study are listed in Table 1. Strains, all derivatives of MC4100, were constructed by P1 transduction (27). Deletions were introduced by the method of Datsenko and Wanner (11) in combination with use of the heat-inducible recombination system in strain DY330 (54). Some deletion alleles were obtained from the National BioResource Project (NIG, Japan; E. coli). When needed, the kanamycin resistance cassette was removed with plasmid pCP20 harboring the Saccharomyces cerevisiae FLP recombinase (11). For the expression of MalZ in MC4100 and the TBP38 (ΔglgA ΔmalP) deletion strain, plasmid p6×HismalZ (47) was transformed into the corresponding E. coli strains. The strains were cultured in M63 minimal salts [15 mM (NH4)2SO4, 22 mM KH2PO4, 40 mM K2HPO4, 1 mM MgSO4, 0.025 mM FeCl2] supplemented with a single carbon source at a concentration of 0.5% (wt/vol) at 37°C with shaking (250 rpm). The medium was supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (35 μg/ml) when necessary.
Table 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| K-12 MG1655 | F− λ−ilvGrfb = 50 rph = 1 | 4 |
| MC4100 | araD139 deoC1 flbB5301 ptsF25 rbsR relA1 rpsL150 Δ(argF-lac)U169 | 7 |
| MC1061 | Δ(araA-leu)7697 araD139 Δ(codB-lacI)3 galK16 galE15 mcrA0 relA1 rpsL50 spoT1 mcrB1 hsdR2 | 8 |
| TB05 | MC4100 ΔmalP::kan | T. Bergmiller, Konstanz, Germany |
| TB07 | MC4100 ΔmalZ::kan | T. Bergmiller, Konstanz, Germany |
| TB16 | MC4100 ΔmalQ::kan | T. Bergmiller, Konstanz, Germany |
| TB38 | MC4100 glgA::Tn10 | T. Bergmiller, Konstanz, Germany |
| TBP38 | MC4100 glgA::Tn10 ΔmalP::cat | This study |
| GW51 | MC4100 ΔmalP ΔglgA::kanglgS::Tn10 | G. Witz, Konstanz, Germany |
| Plasmids | ||
| pET29b | Protein expression vector | Novagen, Madison, WI |
| pET29b-malQ | MalQ expression plasmid | This study |
| p6×His119 | Protein expression vector | 22 |
| p6×H119-EBE | GlgB expression plasmid | This study |
| p6×HmalZ | MalZ overexpression plasmid | 47 |
Cloning and purification of MalQ and GlgB from E. coli K-12.
The malQ gene, encoding amylomaltase, was amplified from chromosomal DNA of E. coli K-12 strain MG1655 (4) by PCR with forward primer 5′-AAC CGC ACT CTC GAG CTT CTT CGC TGC AGC-3′ (an XhoI site is underlined) and reverse primer 5′-AAA CGC TAA GGA AGC CAT ATG GAA AGC-3′ (an NdeI site is underlined). The glgB gene, encoding branching enzyme, was amplified by PCR using forward primer 5′-AAT GGC AAG CTT TGT CAT TCT GCC TC-3′) (an HindIII site is underlined) and reverse primer 5′-CAG GAA GAC ACA TAT GTC CGA TCG TAT C-3′ (an NdeI site is underlined) from genomic DNA of E. coli K-12. The PCR products were treated with the restriction enzymes indicated above and isolated using agarose gel electrophoresis. Protein expression vectors pET29b and p6×H119 were used for ligation with the isolated malQ and glgB genes, respectively.
The recombinant plasmid harboring malQ was designated pET29b-malQ and transformed into E. coli BL21(DE3) for protein expression. In the case of GlgB, the recombinant plasmid was named p6×H119-EBE and transformed into E. coli MC1061. The transformants were cultured using 1 liter of Luria-Bertani medium (10 g tryptone, 5 g yeast extract, 5 g sodium chloride) at 37°C with shaking (250 rpm). For the induction of malQ, 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added when the absorbance of the culture medium at 600 nm reached 0.6 to 0.8. Cells were harvested using centrifugation (8,000 × g, 20 min), and the pellet was sonicated (VC-600; Sonics & Materials Inc.) (output 6, three times for 5 min each, 60% duty) in an ice-cold water bath. Cell debris was removed by centrifugation (12,000 × g, 20 min) at 4°C. The proteins were purified from the cell extracts using Ni-nitrilotriacetic acid (NTA) affinity chromatography.
Isolation of glycogen from E. coli.
Each E. coli strain was cultured in M63 minimal salts (500 ml) supplemented with a specific carbon source at 37°C for 20 to 40 h with shaking (250 rpm). The cells were harvested by centrifugation (6,000 × g) at 4°C for 20 min, washed with ice-cold water, and resuspended in 5 ml of 50 mM sodium acetate (pH 4.5). The resuspension was boiled for 10 min and sonicated (VC-600; Sonics & Materials Inc.) (output 6, three times for 5 min each, 60% duty) at room temperature. The crude cell extract was centrifuged (10,000 × g) at 25°C for 20 min, and glycogen in the supernatant was precipitated by adding 2 volumes of ethanol. The precipitated glycogen was collected by centrifugation (10,000 × g) at 4°C for 20 min and air dried. The dried glycogen was resuspended in 1 ml of sterile distilled water and stored at −18°C until further analysis. The quantification of the glycogen was carried out as described previously by Shim et al. (45).
TLC.
A Whatman K5F silica gel plate (Whatman, Maidstone, United Kingdom) was activated at 110°C for 1 h. Prepared samples were spotted on a plate using a pipette; the plate was placed in a thin-layer chromatography (TLC) chamber containing a solvent mixture of n-butanol-ethanol-water (5:5:3, vol/vol/vol) and developed twice at room temperature. The plate was dried, visualized by being dipped into a solution containing 0.3% N-(1-naphthyl)-ethylenediamine and 5% H2SO4 in methanol, dried, and then heated for 10 min at 110°C.
Spectrophotometric analysis of iodine-glycogen complexes.
A complex of glycogen with iodine was formed in saturated CaCl2 solution by the method of Krisman (25) and Yamaguchi et al. (52). A 0.5-ml portion of iodine-iodide solution (0.26 g I2 and 2.6 g KI in 10 ml H2O) was added to 130 ml of saturated CaCl2 solution. A 0.78-ml portion of the iodine reagent was mixed with 0.12 ml of glycogen sample, and the absorption spectra were measured on a Shimadzu spectrophotometer (UV-160A; Japan) against a reference solution.
HPAEC.
The reaction mixtures (or cell lysates) were boiled for 10 min before centrifugation at 12,000 × g for 10 min, and the supernatants were filtered using a membrane filter kit (0.2-μm pore diameter; Gelman Sciences, Ann Arbor, MI). High-performance anion-exchange chromatography (HPAEC) was performed using a Dionex DX-500 system (Sunnyvale, CA) equipped with a pulsed amperometric detector (ED40; Dionex) and a CarboPac PA-1 column (4 by 250 mm; Dionex). Samples were eluted with a gradient of 0.6 M sodium acetate in 0.15 M NaOH at a flow rate of 1.0 ml/min. Gradients of 0.6 M sodium acetate were applied as follows: a linear gradient of 10 to 30% for min 0 to 10, 30 to 40% for min 10 to 16, 40 to 50% for min 16 to 27, 50 to 60% for min 27 to 44, and 60 to 64% for min 44 to 60.
SEC-MALLS/RI analysis.
Glycogen obtained from each E. coli strain was filtered through a disposable syringe filter (0.5-μm pore diameter; Millipore, Bedford, MA) and injected into the size exclusion chromatography (SEC)-multiangle laser light scattering (MALLS)/refractive index (RI) system (632.8 nm, DAWN DSP-F; Wyatt Technology, Santa Barbara, CA). The weight-average molecular weight (Mw) of the sample was calculated using ASTRA V4.90.07 software (Wyatt Technology), using the Berry extrapolation method for curve fitting and a dn/dc value of 0.146 ml/g. The details of the method were described by Shim et al. (45).
In vitro synthesis of glycogen using MalQ and GlgB with simultaneous removal of glucose.
One percent maltose (G2) or maltoheptaose (G7) was incubated with MalQ (0.04 U/mg substrate) at 30°C and pH 7.0 (50 mM Tris-HCl buffer) for 15 h. To compare the effect of simultaneous removal of glucose, glucose oxidase (10 U/mg substrate; Youngdong Pharmacy Co., Republic of Korea) was added to the reaction mixture, followed by treatment with branching enzyme (GlgB; 15 U/mg substrate) at 30°C and pH 7.0 (50 mM Tris-HCl buffer) for 4 h. Polysaccharides were precipitated by adding 2 volumes of ethanol and collected using centrifugation (10,000 × g) at 4°C for 20 min. The precipitates were washed twice with 70% ethanol and dried at 37°C overnight. To analyze the side chain distribution of the polysaccharides synthesized by the branching enzymes, the sample solution was treated with isoamylase (20 U/mg substrate) at pH 4.3 (50 mM sodium acetate buffer) and 55°C for 48 h, and then the debranched sample was analyzed using HPAEC.
RESULTS
Analysis of glycogen in different mutants.
Cellular glycogen was extracted by boiling and sonicating cells, followed by separation from insoluble material by centrifugation. The soluble material contained linear dextrins and soluble glycogen. The addition of ethanol precipitated glycogen as well as long linear dextrins with a degree of polymerization of greater than 7 to 8 glucosyl units (14). The precipitate was solubilized in water, and size exclusion chromatography (SEC) and multiple laser light scattering (MALLS)/refractive index analysis were performed with this material. This method gave a value for the average molecular weight of glycogen. To test the branch chain distribution, the glycogen sample was first treated with isoamylase, which cleaves the α-1,6-linked branches, releasing linear dextrins. The dextrin content of the incubation mixture was then analyzed by high-performance anion-exchange chromatography as well as by thin-layer chromatography (TLC). The difference in the amounts and lengths of dextrins before and after treatment with isoamylase revealed the chain length and side chain distribution of glycogen, as shown in Table 2. Figure 1 shows the analysis of glycogen isolated from the wild-type strain (MC4100) grown on glucose, maltose, and maltodextrins.
Table 2.
Characteristics of the glycogen accumulated in E. coli mutants
| Strain | Carbon sourcea | Glycogen characteristics (mean ± SD)b |
||
|---|---|---|---|---|
| Mol wt (× 106) | Total amt (mg/mg protein) | Chain length (DP)c | ||
| MC4100 | G1 | 2.0 ± 0.5 | 0.032 ± 0.002 | 11.0 ± 0.2 |
| G2 | 13.7 ± 0.4 | 0.049 ± 0.005 | 12.7 ± 0.3 | |
| MD | 2.7 ± 0.6 | 0.074 ± 0.03 | 12.4 ± 0.1 | |
| MalP− | G1 | 9.0 ± 3.2 | 0.043 ± 0.004 | 10.8 ± 0.4 |
| G2 | 11.1 ± 0.1 | 0.97 ± 0.29 | 14.0 ± 1.6 | |
| MD | 6.0 ± 0.1 | 1.31 ± 0.11 | 12.9 ± 0.2 | |
| MalZ− | G1 | 5.0 ± 0.7 | 0.013 ± 0.001 | 9.6 ± 0.3 |
| G2 | 21.2 ± 0.2 | 0.112 ± 0.028 | 13.3 ± 0.5 | |
| MD | 4.2 ± 0.2 | 0.129 ± 0.009 | 9.5 ± 0.2 | |
| GlgA− | G1 | — | 0 | — |
| G2 | — | 0 | — | |
| MD | 0.52 ± 0.13 | 0.176 ± 0.018 | 11.0 ± 0.1 | |
| GlgA−/MalP− (GlgS+) | G1 | — | 0 | — |
| G2 | 2.7 ± 0.76 | 1.19 ± 0.101 | 16.6 ± 1.1 | |
| MD | 5.0 ± 0.92 | 1.0 ± 0.083 | 11.2 ± 0.4 | |
| GlgA−/MalP− (GlgS−) | G1 | — | — | — |
| G2 | — | 1.21 ± 0.089 | 16 ± 0.5 | |
| MD | — | — | — | |
| MalQ− | G1 | 1.7 ± 0.4 | 0.073 ± 0.006 | 9.1 ± 0.8 |
| G2 | × | × | × | |
| MD | 8.5 ± 1.3 | 0.096 ± 0.012 | 8.2 ± 0.9 | |
| MalQ−/MalP− | G1 | — | 0.03 ± 0.001 | 10.1 ± 0.9 |
| G2 | × | × | × | |
| MD | × | × | × | |
| MC4100/p6×HmalZ+d | G1 | — | 0.02 ± 0.002 | 11 ± 0.3 |
| G2 | — | — | — | |
| MD | — | — | — | |
| MalP−/p6×HmalZ+d | G1 | — | — | — |
| G2 | — | 0.19 ± 0.028 | 13.7 ± 0.7 | |
| MD | — | — | — | |
| GlgA−/MalP−/p6×HmalZ+d | G1 | — | — | — |
| G2 | — | 0.12 ± 0.038 | 15.5 ± 0.9 | |
| MD | — | — | — | |
G1, glucose; G2, maltose; MD, maltodextrin.
Estimated from three replicates. —, not determined; ×, no growth.
The average chain length of glycogen was calculated as weight-average degree of polymerization (DP) using the equation DP = Σ(peak area of each chain × chain length of the corresponding chain)/Σ(peak area of each chain).
MalZ was overexpressed using plasmid p6×HmalZ.
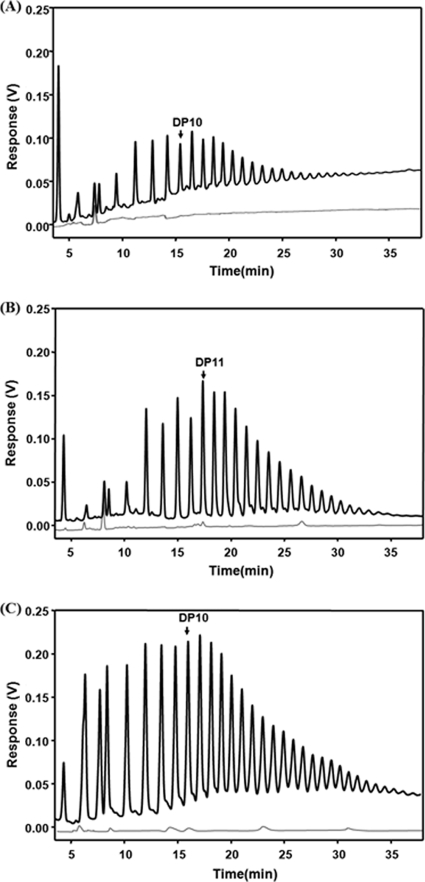
Fig. 1.
HPAEC analyses of the glycogen side chains isolated from E. coli MC4100 (laboratory wild-type strain) grown on glucose (A), maltose (B), and maltodextrins (C). The assays were carried out before (gray line) and after (black line) treatment with isoamylase. The glycogen content was calculated from the sum of the peak areas. Bovine glycogen was used as standard.
Growth of the wild type on maltose and maltodextrin increases the amount of glycogen in comparison to that with growth on glucose.
When the wild type was grown on glucose, maltose, or maltodextrin, the amount of glycogen increased from 0.03 mg/mg protein on glucose to 0.05 and 0.07 mg/mg protein on maltose and maltodextrin, respectively. The average chain length increased from 11 to 13, while the molecular weights were 2.0 × 106, 13.7 × 106, and 2.7 × 106 with glucose, maltose, and maltodextrin as the carbon source, respectively (Table 2). Figure 1 shows the analysis of the released linear branch chains from glycogen. This indicated that the induction of the maltose system by maltose and maltodextrin contributed to the amount of glycogen. When the strain is grown on glucose, the maltose system is turned off due to catabolite repression.
The lack of MalP strongly increases the amount of glycogen when growth is on maltose or maltodextrin.
When TB05, the ΔmalP derivative of MC4100 was grown on glucose, the amount of glycogen and its average chain length were similar to those of the wild type (Table 2). However, when this strain was grown on maltose or maltodextrin, the amount of glycogen dramatically increased, reaching 0.97 and 1.31 mg per mg protein, respectively. Figure 2A shows the analysis of the released branch chains with glucose or maltose as the carbon source. The molecular weight of glycogen when grown on all three carbon sources was in the range of 6 × 106 to 11 × 106, while the average chain length increased to 14 with maltose as the carbon source. This indicated that the long linear dextrins were formed from maltose by MalQ. They were not reduced in size due to the absence of MalP and thus were transformed into glycogen.
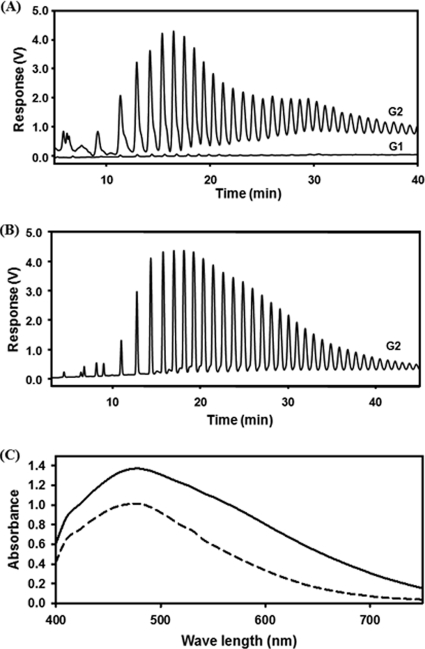
Fig. 2.
(A and B) HPAEC analyses of the glycogen side chains isolated from ΔmalP (A) and ΔglgA ΔmalP (B) derivatives of MC4100 after growth on maltose (G2) (A and B) or glucose (G1) (A). Note that the amount of glycogen in a ΔmalP strain grown on maltose is dramatically increased in comparison to that for growth on glucose. Under these conditions, the presence or absence of GlgA does not contribute to the amount of glycogen. (C) Spectral analysis of iodine-stained glycogen obtained from a ΔglgA ΔmalP strain, TBP38, grown on maltose (solid line) and of glycogen obtained from wild-type strain MC4100 grown on glucose (dashed line). Note that the sample of TBP38 had to be diluted to fall into the same absorption range as the sample of MC4100.
The lack of MalZ also results in an increase of glycogen when the strain is grown on maltose or maltodextrin but to a lesser extent than in a strain lacking MalP.
When TB07, the ΔmalZ derivative of MC4100, was grown on glucose, the amount of glycogen was, surprisingly, lower than that in the wild type (0.01 mg/mg protein). When growth was on maltose or maltodextrin, the amount of glycogen increased considerably (to 0.11 and 0.13 mg/mg protein, respectively), although not as much as in the otherwise isogenic ΔmalP strain (Table 2). Both MalP and MalZ reduce the size of linear maltodextrins, MalP by phosphorolysis of the nonreducing glucosyl residue and MalZ by cleaving glucose from the reducing ends of maltodextrins. This indicates that linear dextrins formed from maltose (and maltodextrins) by the action of MalQ will contribute to the formation of glycogen. MalZ, and to a higher degree MalP, will counteract the formation of glycogen by reducing the size of the linear maltodextrins.
Glycogen formed in the ΔmalP strain grown on maltose is not due to GlgA.
When TB38, a ΔglgA derivative of MC4100 (containing MalP and MalZ), was grown on glucose or maltose, no glycogen was formed. However, when it was grown on maltodextrin, a substantial amount of glycogen was produced (Table 2). Thus, glycogen can be formed from maltodextrins in the absence of GlgA. That no glycogen was produced with maltose as the carbon source must be due to the presence of MalP and MalZ, which effectively reduce the size of the maltodextrins formed from maltose by MalQ.
When TBP38, a ΔmalP ΔglgA derivative of MC4100, was grown on glucose, again no glycogen was formed. Under these growth conditions, the maltose system is turned off due to catabolite repression by glucose. However, when TBP38 was grown on maltose (or maltodextrin), large amounts of glycogen were found (1.2 or 1.0 mg/mg protein, respectively). The analysis of the released branch chains is shown in Fig. 2B. The molecular weight was 2.7 × 106 and the average chain length was 17, significantly higher than for glycogen from the wild type when grown on glucose (Table 2). This demonstrates that glycogen can be formed when growth is with maltose as the carbon source in the absence of GlgA provided that the long linear dextrins originating from the action of MalQ on maltose or maltodextrin are not degraded by MalP. Glycogen produced in TBP38 after growth on maltose was also tested by iodine staining followed by spectrophotometric analysis and compared with glycogen formed in MC4100 grown on glucose and formed exclusively via the GlgA-dependent pathway (Fig. 2C). The λmax of glycogen from both strains was 480 nm. This qualitative analysis showed no difference in glycogen derived by the MalQ-dependent pathway and the GlgA-dependent pathway.
Dual role of MalP in the synthesis of glycogen.
As shown above, the lack of MalP strongly increases the synthesis of glycogen when maltose is the carbon source in a ΔglgA strain. This is due to the action of MalQ on maltose (as well as on maltodextrin) to polymerize maltose to long maltodextrins that subsequently become substrates for GlgB, the branching enzyme. The presence of MalP in such a strain would reduce glycogen synthesis by degrading the dextrins produced by MalQ. In contrast, in a strain that lacks MalQ, the presence of MalP increases the amount of glycogen when grown on maltodextrin. This is shown in Table 2. In this strain, growth on glucose produces glycogen in a GlgA-dependent fashion. Growth on maltose is not possible, since MalQ is essential for maltose metabolism. However, growth on maltodextrin is possible. The dextrins (up to a chain length of seven glucosyl units) are transported by the ABC transporter, and MalP releases glucose-1-P by phosphorolysis from these dextrins, which enter glycolysis for growth and energy production. We interpret this glycogen synthesis as a result of the surplus of glucose-1-P formed by the action of MalP, which can fuel the formation of ADP-glucose and subsequently the formation of glycogen by GlgA. Indeed, when a double mutant lacking MalQ as well as MalP is used (Table 2), the strain is unable to grow on maltodextrin, whereas growth on glucose still produces glycogen. Thus, MalP-dependent phosphorolysis of maltodextrins increases GlgA-dependent glycogen synthesis.
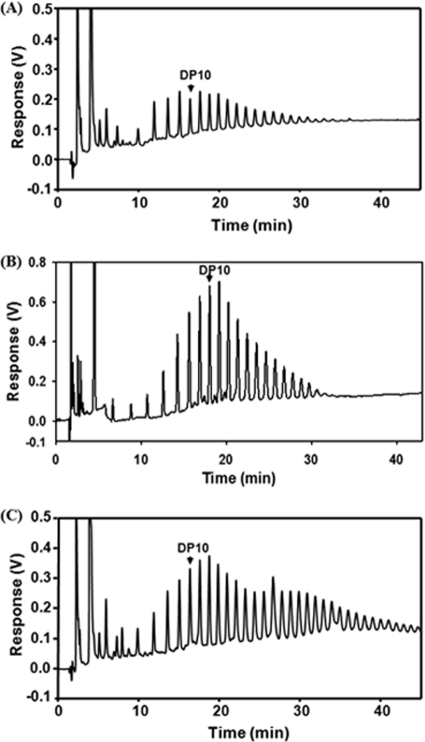
Overexpression of MalZ reduces the amount of glycogen in a ΔmalP ΔglgA strain grown on maltose but not in a wild-type (glgA+) strain grown on glucose.
When grown on maltose, GW51, a ΔmalP ΔglgA ΔglgS derivative of MC4100, produced large amounts of glycogen (1.21 mg/mg protein), as did strain TBP38 (1.19 mg/mg protein), the glgS+ variant of GW51, as shown above. This showed that the lack of GlgS did not affect the GlgA-independent synthesis of glycogen. However, when the ΔmalP ΔglgA strain was transformed with a plasmid harboring the malZ gene, the amount of glycogen was strongly reduced, to 0.1 mg/mg protein (Fig. 3C and Table 2). In contrast, when the wild-type MC4100 was grown on glucose, the amount of glycogen was reduced by only 33% in the presence of overexpressed MalZ (Fig. 3A and Table 2). This demonstrates that GlgA-dependent formation of glycogen was largely resistant to MalZ (two-thirds remaining), while the formation of glycogen via the maltose system was highly sensitive to MalZ (more than a 10-fold reduction). We conclude that maltodextrins formed via the GlgA pathway may not be readily accessible to MalZ. The side chains in glycogen, once they are formed by the branching enzyme, are resistant to MalZ, whereas the linear maltodextrins, the precursors of maltose-dependent glycogen produced by the activity of MalQ, are degraded by MalZ, and therefore much less glycogen was formed. This is in line with the observation that MalZ hydrolyzes glucosyl residues from the reducing end (48), which is not present in the branches of glycogen. When MalZ was overproduced in TB05, the glgA+ derivative of TBP38 (ΔmalP ΔglgA), after growth in maltose, the reduction in glycogen content was 5-fold instead of the 10-fold in the absence of GlgA (Table 2 and Fig. 3B). This indicates that the presence of GlgA may rescue part of the maltodextrin precursors from the action of MalZ while being synthesized in the ADP-glucose-mediated polymerization reaction.
Fig. 3.
Comparison of glycogen branch chain distributions in wild-type MC4100 (A) and the ΔmalP (B) and ΔglgA ΔmalP (C) derivatives in the presence of a plasmid encoding and overproducing MalZ after growth on glucose (A) and maltose (B and C). Compare panel A with Fig. 1A, panel B with Fig. 2A, and panel C with Fig. 2B. Note that there is more than a 10-fold reduction in glycogen content caused by MalZ in the ΔglgA ΔmalP strain grown on maltose but not in the wild-type MC4100 grown on glucose.
In vitro, maltose can be transformed into glycogen by MalQ and GlgB (branching enzyme) provided that glucose is removed from the reaction mixture.
The incubation of maltose with homogeneous MalQ yields glucose and linear maltodextrins with a chain length of at least 10 glucosyl residues, in decreasing amounts as the dextrins become longer (16). The mixture of dextrins represents an equilibrium where the dextrins are substrates as well as acceptors in the dextrinyl transfer reaction of MalQ. Glucose also acts as an acceptor limiting the extent of polymerization and in increasing concentration becomes a feedback inhibitor of MalQ. The removal of glucose from the reaction mixture, for instance, by ATP and glucokinase (to form glucose-6-P that is not part of the equilibrium), allows the formation of maltodextrins of continually increasing chain length (26). This situation mimics the cytosol of a ΔmalP strain grown on maltose, where the maltodextrins formed by the MalQ activity are not reduced in size by the missing MalP enzyme. The removal of glucose by glucokinase on the one hand allows growth on glucose-6-P and on the other hand ensures the uncurbed activity of MalQ. We used a similar setup to produce glycogen from maltose and MalQ in vitro by removing liberated glucose using glucose oxidase. When the reaction was carried out for 15 h, GlgB, the branching enzyme, was added. The glycogen formed had a molecular weight of 1.0 × 106 and a peak branch chain length of 10 (Fig. 4A).
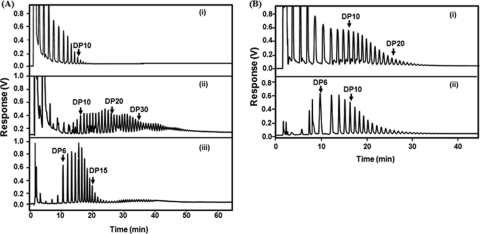
Fig. 4.
In vitro synthesis of glycogen by MalQ and GlgB. (A) Maltodextrins were analyzed by HPAEC after incubation of maltose with MalQ only (i), MalQ and glucose oxidase (ii), or MalQ, glucose oxidase, and GlgB (iii), followed by ethanol precipitation and treatment of the precipitate with isoamylase. Note that the maltodextrins seen in panel iii represent the branch chains of in vitro-formed glycogen. (B) Maltoheptaose was incubated with MalQ only (i) or MalQ and GlgB (ii), followed by ethanol precipitation and treatment of the precipitate with isoamylase. Note that the incubation of maltoheptaose with MalQ leads to maltodextrins long enough (larger than 10 glucosyl units) to allow glycogen formation with GlgB.
When instead of maltose maltoheptaose was used as substrate for MalQ, only small amounts of glucose were released and the addition of glucose oxidase could be omitted. The maltodextrins formed under these conditions were long enough to form glycogen in the presence of GlgB (Fig. 4B). These in vitro tests demonstrate that MalQ alone in the absence of GlgA is able to form from maltose (or small maltodextrins) maltodextrins that are long enough for the branching enzyme to proceed in the direction of glycogen synthesis.
DISCUSSION
Here we describe a novel mode of glycogen formation in E. coli that is based on the metabolism of maltose and maltodextrins and that is independent of the classical glycogen-synthesizing enzyme glycogen synthase (GlgA). There are four enzymes that participate in maltose/maltodextrin metabolism. Three are cytoplasmic: MalQ (amylomaltase), MalP (maltodextrin phosphorylase), and MalZ (maltodextrin glucosidase). The fourth is MalS, a periplasmic amylase. In contrast to the classical synthesis pathway involving GlgA, neither sugar phosphorylation nor ADP-dependent sugar activation is necessary for the polymerization of the α-1,4-linked glucosyl backbone. Instead, maltose and small maltodextrins (up to a chain length of seven glucosyl units) that are taken up by the binding protein-dependent ABC transporter are polymerized by MalQ to long dextrins that can function as substrates for the branching enzyme GlgB to form glycogen. Since glucose released by the action of MalQ is continually removed by glucokinase, it is no longer an acceptor for the transfer reaction of MalQ, nor does it inhibit MalQ activity. Thus, the chain lengths of the maltodextrins increase until they reach the minimal length to become substrates for the branching enzyme GlgB (19). The molecular weight and side chain distribution of the product formed are very similar to those of the glycogen formed by the GlgA-dependent mechanism.
In a wild-type E. coli strain (containing GlgA), the contribution of the maltose system to the synthesis of glycogen when growth is on glucose, maltose, or maltodextrins can be seen only by the increase in the total amount of glycogen (from 0.03 to 0.05 and 0.07 mg/mg protein) (Table 2). This increase is indeed due to maltodextrin metabolism and not due to a stimulation of the GlgA-dependent pathway, as can be seen by analyzing the effect that deletions in the maltose enzymes have on glycogen synthesis.
The most dramatic effect is seen when MalP (maltodextrin phosphorylase) is deleted. Growth on glucose, a condition that represses the maltose system, allows only the GlgA pathway to function. The glycogen content under these conditions is comparable to that of the wild type (Table 2). When maltose or maltodextrins are the carbon sources, the glycogen content increases more than 20-fold. We interpret this phenomenon as the result of the lack of MalP, which would reduce the sizes of maltodextrins by consecutively removing glucosyl residues from the nonreducing ends of maltodextrins. Thus, the amount of maltodextrins that can become substrates of GlgB is being controlled (reduced) by MalP. However, there might also be another aspect to consider. The active sites in the structures of MalP and GlgA with bound maltohexaose and pentose are superimposable with respect to the nonreducing end of the maltodextrin (44, 50). Thus, MalP and GlgA compete for the same residue in maltodextrin. Therefore, an unlikely alternative explanation might be that in the ΔmalP strain, GlgA is free of a competitor and can act faster.
Similarly, a deletion of MalZ also affects the amount of glycogen but much less dramatically than that of MalP. MalZ cleaves glucosyl residues from the reducing ends of maltodextrins (48), curbing their length and thus interfering with the substrate formation for GlgB.
To demonstrate that maltose/maltodextrin metabolism is leading to glycogen formation per se and not just by a stimulating effect on GlgA, the amount of glycogen was assayed in a series of strains lacking GlgA. The glgA malQ+ malP+ strain produced no glycogen when grown on glucose or on maltose (Table 2). However, when it was grown on maltodextrin, substantial amounts of glycogen were found. Apparently the action of MalQ to produce maltodextrins from maltose was efficiently counteracted by MalP and MalZ. However, with maltodextrins as the substrate the formation of sufficiently long dextrins by MalQ (under conditions where only small amounts of glucose were produced) was possible; these were long enough to be substrates for GlgB despite the presence of MalP and MalZ.
The glgA malP malQ+ strain showed a very high content of glycogen when grown on maltose. Here the role of MalP becomes obvious. The linear dextrins produced from maltose by MalQ are not reduced in size by MalP, and a large amount of glycogen was formed in the absence of GlgA.
The essential role of MalQ in the synthesis of maltodextrins as glycogen precursors can be deduced only indirectly. A deletion in MalQ in a glgA+ strain no longer allows growth on maltose. When this strain is grown on glucose, it produces normal amounts of glycogen in a GlgA-dependent manner. When it is grown on maltodextrins, growth and energy production proceed via the degradation of the linear dextrins by MalP, forming glucose-1-P. Presumably, it is the surplus of glucose-1-P that stimulates glycogen synthesis via GlgA (Table 2). In this situation, glycogen formation could not have been achieved directly by the dextrins taken up via the maltose/maltodextrin ABC transporter followed by the action of GlgB. The ABC transporter has a size limit of seven glucosyl units (13, 21, 49), which is smaller than the substrate size for GlgB (3, 19). In addition, larger dextrins entering the periplasm would be curbed in size by MalS, an amylase (the fourth maltose enzyme) preferentially forming maltohexaose units from larger dextrins (18). Therefore, maltodextrins entering from the outside into the malQ strain are prone only to degradation via MalP and MalZ and cannot be directly transformed into glycogen by GlgB.
The effect of overproduced MalZ on the amount of glycogen synthesized in the two different pathways was surprising. Glycogen synthesized exclusively via the MalQ-dependent pathway was highly sensitive to MalZ (there was more than a 10-fold reduction in glycogen content), indicating degradation of smaller maltodextrin precursors before they reach the minimal size required for the action of GlgB. In contrast, when growth was on glucose under conditions where the maltose system is turned off, glycogen produced exclusively via the GlgA pathway was reduced by only 33% in the presence of overproduced MalZ. Clearly, once synthesized, intact glycogen is resistant to MalZ (because of the lack of reducing glucosyl residues), but why are the putative linear precursors onto which GlgA transfers consecutive glucosyl residues not sensitive to MalZ? One can only speculate about the inaccessibility of MalZ to the nascent GlgA maltodextrin products or substrates. It is possible that during the GlgA-catalyzed transfer reaction onto the nonreducing end of the acceptor maltodextrin, the reducing end of the nascent chain (which is the target for MalZ-dependent cleavage) is occluded long enough from the attack by MalZ. It is also possible that GlgA processively adds glucose from ADP-glucose to the growing glycan chain. Once the length of the nascent chain exceeds seven glucosyl units, it is no longer a substrate for MalZ. When the effect of overproduced MalZ on the glycogen synthesized via the MalQ-dependent pathway was tested in the presence of GlgA, the reduction of the glycogen content was not as strong as in the absence of GlgA. This must mean that the GlgA-dependent pathway was strongly stimulated under these conditions. The MalZ action on the MalQ-derived linear small maltodextrins will produce a large amount of glucose that will subsequently be transformed into glucose-6-P, glucose-1-P, and finally ADP-glucose. As reported previously (30), the level of ADP-glucose controls the amount of glycogen by controlling the activity of GlgA. Also, as discussed above, glycogen synthesized exclusively via the GlgA pathway is largely resistant to MalZ. In any case, the effect of overproduced MalZ clearly demonstrates the difference in the mechanisms of the two pathways.
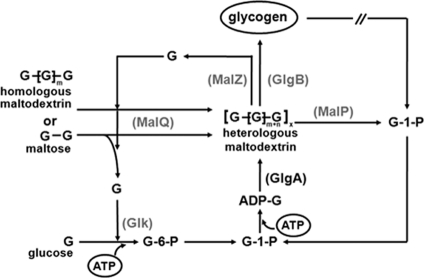
The data obtained for the series of strains lacking GlgA reveal a novel pathway of glycogen synthesis based on the metabolism of the maltose system. The roles of the different enzymes are summarized in Fig. 5.
Fig. 5.
Model for the synthesis of glycogen by the enzymes of the maltose system. Maltose or maltodextrins entering the cell via the ABC maltose/maltodextrin transporter (not shown) are polymerized by MalQ to maltodextrins long enough to become substrates of GlgB, the branching enzyme, to form glycogen. The presence of glucokinase (Glk) supports the formation of long maltodextrins by removing free glucose formed in the MalQ-catalyzed reaction. MalP and MalZ counteract the formation of long dextrins by removing glucose-1-P from the nonreducing ends (MalP) or by removing glucose from the reducing ends (MalZ) of maltodextrins. Thus, mutants lacking MalP or MalZ produce large amounts of glycogen with maltose/maltodextrin as the carbon source in the absence of GlgA.
Considering the different aspects of maltose and glycogen metabolism, it is interesting to identify the key enzymes on the crossroads of their interaction. The discovery of endogenous induction of the maltose system by maltotriose derived from the degradation of glycogen has become apparent by the use of mutants lacking MalQ. These mutants are nearly constitutive for the synthesis of the maltose enzymes (6, 15, 17). The explanation for this phenomenon is that maltotetraose and maltotriose which are released from phosphorylase-limited glycogen by GlgX, the debranching enzyme, are no longer repolymerized by the missing MalQ activity and result in the activation of MalT by the endogenously accumulating inducer maltotriose. In contrast, the polymerizing activity of MalQ to form long maltodextrins from maltose for the GlgA-independent synthesis of glycogen became apparent only in mutants lacking MalP.
The present publication cannot make any contribution to the understanding of carbon or energy flux from glycogen into glycolysis or gluconeogenesis. We determined the steady-state concentration of glycogen under standardized growth conditions by varying the composition of the maltose enzymes with the implicit assumption that the degradation of glycogen remained unaltered. Clearly, the steady-state amount of glycogen represents a balance between synthesis and degradation. Continuous degradation of glycogen must occur. For instance, deletion of glgP, encoding glycogen phosphorylase, strongly increased the amount of glycogen (1). Also, the phenomenon of endogenous induction of the maltose system by maltotriose formed by the degradation of glycogen, as discussed above, is evidence of continuous degradation of glycogen. Therefore, we conclude that the observed changes in the amount of glycogen upon changing maltose enzymes were due to changes in the rate of glycogen synthesis. Alterations in enzymes other than the maltose enzymes have been shown to affect the amount of glycogen. For instance, the overproduction of an ADP sugar pyrophosphatase dramatically reduced the amount of glycogen by degrading ADP-glucose, the donor substrate for GlgA (30).
ACKNOWLEDGMENTS
We gratefully acknowledge the construction of strains by Tobias Bergmiller and Gabi Witz.
This study was supported in part by the Korea-Germany Common Fund Program and in part by the Basic Science Research Program (2009-0087146) through the National Research Foundation. J.-H. Shim acknowledges Hallym University for support of this work.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Alonso-Casajús N., et al. 2006. Glycogen phosphorylase, the product of the glgP gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J. Bacteriol. 188:5266–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker C. S., Morozov I., Suzuki K., Romeo T., Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44:1599–1610 [DOI] [PubMed] [Google Scholar]
- 3. Binderup K., Mikkelsen R., Preiss J. 2002. Truncation of the amino terminus of branching enzyme changes its chain transfer pattern. Arch. Biochem. Biophys. 397:279–285 [DOI] [PubMed] [Google Scholar]
- 4. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 5. Boos W., Shuman H. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bukau B., Ehrmann M., Boos W. 1986. Osmoregulation of the maltose regulon in Escherichia coli. J. Bacteriol. 166:884–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casadaban M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coil using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 8. Casadaban M. J., Cohen S. N. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207 [DOI] [PubMed] [Google Scholar]
- 9. Chapon C. 1982. Expression of malT, the regulator gene of the maltose region in Escherichia coli, is limited both at transcription and translation. EMBO J. 1:369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chapon C. 1982. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J. Bacteriol. 150:722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dauvillée D., et al. 2005. Role of the Escherichia coli glgX gene in glycogen metabolism. J. Bacteriol. 187:1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davidson A. L., Dassa E., Orelle C., Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Defloor I., Vandenreyken V., Grobet P. J., Delcour J. A. 1998. Fractionation of maltodextrins by ethanol. J. Chromatogr. A 803:103–109 [Google Scholar]
- 15. Dippel R., Bergmiller T., Bohm A., Boos W. 2005. The maltodextrin system of Escherichia coli: glycogen-derived endogenous induction and osmoregulation. J. Bacteriol. 187:8332–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dippel R., Boos W. 2005. The maltodextrin system of Escherichia coli: metabolism and transport. J. Bacteriol. 187:8322–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ehrmann M., Boos W. 1987. Identification of endogenous inducers of the mal regulon in Escherichia coli. J. Bacteriol. 169:3539–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freundlieb S., Boos W. 1986. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J. Biol. Chem. 261:2946–2953 [PubMed] [Google Scholar]
- 19. Guan H., Li P., Imparl-Radosevich J., Preiss J., Keeling P. 1997. Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch. Biochem. Biophys. 342:92–98 [DOI] [PubMed] [Google Scholar]
- 20. Hengge-Aronis R., Fischer D. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 6:1877–1886 [DOI] [PubMed] [Google Scholar]
- 21. Khare D., Oldham M. L., Orelle C., Davidson A. L., Chen J. 2009. Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell 33:528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J. S., et al. 1999. Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J. Biol. Chem. 274:26279–26286 [DOI] [PubMed] [Google Scholar]
- 23. Kitahata S. 1988. Microbial disproportionating enzymes, p. 169–172 In Amylase Research Society of Japan (ed.) Handbook of amylases and related enzymes. Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 24. Kozlov G., Elias D., Cygler M., Gehring K. 2004. Structure of GlgS from Escherichia coli suggests a role in protein protein interactions. BMC Biol. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krisman C. R. 1962. A method for the colorimetric estimation of glycogen with lodine. Anal. Biochem. 4:17–23 [DOI] [PubMed] [Google Scholar]
- 26. Lengsfeld C., Schonert S., Dippel R., Boos W. 2009. Glucose-and glucokinase-controlled mal gene expression in Escherichia coli. J. Bacteriol. 191:701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Habor Laboratory Press, New York, NY [Google Scholar]
- 28. Monod J., Torriani A. M. 1950. De l'amylomaltase d'Escherichia coli. Ann. Inst. Pasteur (Paris) 78:65–77 [PubMed] [Google Scholar]
- 29. Montero M., et al. 2009. Escherichia coli glycogen metabolism is controlled by the PhoP-PhoQ regulatory system at submillimolar environmental Mg2+ concentrations, and is highly interconnected with a wide variety of cellular processes. Biochem. J. 424:129–141 [DOI] [PubMed] [Google Scholar]
- 30. Moreno-Bruna B., et al. 2001. Adenosine diphosphate sugar pyrophosphatase prevents glycogen biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 98:8128–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmer T. N., Wober G., Whelan W. J. 1973. The pathway of exogenous and endogenous carbohydrate utilization in Escherichia coli: a dual function for the enzymes of the maltose operon. Eur. J. Biochem. 39:601–612 [DOI] [PubMed] [Google Scholar]
- 32. Peist R., Schneider-Fresenius C., Boos W. 1996. The MalT-dependent and malZ-encoded maltodextrin glucosidase of Escherichia coli can be converted into a dextrinyltransferase by a single mutation. J. Biol. Chem. 271:10681–10689 [DOI] [PubMed] [Google Scholar]
- 33. Preiss J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38:419–458 [DOI] [PubMed] [Google Scholar]
- 34. Preiss J., Romeo T. 1989. Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv. Microb. Physiol. 30:183–238 [DOI] [PubMed] [Google Scholar]
- 35. Pugsley A. P., Dubreuil C. 1988. Molecular characterization of malQ, the structural gene for the Escherichia coli enzyme amylomaltase. Mol. Microbiol. 2:473–479 [DOI] [PubMed] [Google Scholar]
- 36. Raha M., Kawagishi I., Muller V., Kihara M., Macnab R. M. 1992. Escherichia coli produces a cytoplasmic alpha-amylase, AmyA. J. Bacteriol. 174:6644–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raibaud O., Richet E. 1987. Maltotriose is the inducer of the maltose regulon of Escherichia coli. J. Bacteriol. 169:3059–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romeo T., Black J., Preiss J. 1990. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vivo effects of the catabolite repression and stringent response systems in glg gene expression. Curr. Microbiol. 21:131–137 [Google Scholar]
- 39. Romeo T., Preiss J. 1989. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5′-diphosphate 3′-diphosphate and analysis of in vivo transcripts. J. Bacteriol. 171:2773–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwartz M. 1967. Phenotypic expression and genetic localization of mutations affecting maltose metabolism in Escherichia coli K12. Ann. Inst. Pasteur (Paris) 112:673–698 [PubMed] [Google Scholar]
- 41. Schwartz M., Hofnung M. 1967. Maltodextrin phosphorylase of Escherichia coli. Eur. J. Biochem. 2:132–145 [DOI] [PubMed] [Google Scholar]
- 42. Seok Y. J., Koo B. M., Sondej M., Peterkofsky A. 2001. Regulation of E. coli glycogen phosphorylase activity by HPr. J. Mol. Microbiol. Biotechnol. 3:385–394 [PubMed] [Google Scholar]
- 43. Seok Y. J., et al. 1997. High affinity binding and allosteric regulation of Escherichia coli glycogen phosphorylase by the histidine phosphocarrier protein, HPr. J. Biol. Chem. 272:26511–26521 [DOI] [PubMed] [Google Scholar]
- 44. Sheng F., Jia X., Yep A., Preiss J., Geiger J. H. 2009. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J. Biol. Chem. 284:17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shim J. H., et al. 2009. Role of maltogenic amylase and pullulanase in maltodextrin and glycogen metabolism of Bacillus subtilis 168. J. Bacteriol. 191:4835–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song H. N., et al. 2010. Structural rationale for the short branched substrate specificity of the glycogen debranching enzyme GlgX. Proteins 78:1847–1855 [DOI] [PubMed] [Google Scholar]
- 47. Song K. M., et al. 2010. Transglycosylation properties of maltodextrin glucosidase (MalZ) from Escherichia coli and its application for synthesis of a nigerose-containing oligosaccharide. Biochem. Biophys. Res. Commun. 397:87–92 [DOI] [PubMed] [Google Scholar]
- 48. Tapio S., Yeh F., Shuman H. A., Boos W. 1991. The malZ gene of Escherichia coli, a member of the maltose regulon, encodes a maltodextrin glucosidase. J. Biol. Chem. 266:19450–19458 [PubMed] [Google Scholar]
- 49. Wandersman C., Schwartz M., Ferenci T. 1979. Escherichia coli mutants impaired in maltodextrin transport. J. Bacteriol. 140:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Watson K. A., Schinzel R., Palm D., Johnson L. N. 1997. The crystal structure of Escherichia coli maltodextrin phosphorylase provides an explanation for the activity without control in this basic archetype of a phosphorylase. EMBO J. 16:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiesmeyer H., Cohn M. 1960. The characterization of the pathway of maltose utilization by Escherichia coli. II. General properties and mechanism of action of amylomaltase. Biochim. Biophys. Acta 39:427–439 [DOI] [PubMed] [Google Scholar]
- 52. Yamaguchi H., Kanda Y., Iwata K. 1974. Macromolecular structure and morphology of native glycogen particles isolated from Candida albicans. J. Bacteriol. 120:441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang H., Liu M. Y., Romeo T. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178:1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu D., Ellis H. M., Lee E. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]