Abstract
Clostridium acetobutylicum is both a model organism for the understanding of sporulation in solventogenic clostridia and its relationship to solvent formation and an industrial organism for anaerobic acetone-butanol-ethanol (ABE) fermentation. How solvent production is coupled to endospore formation—both stationary-phase events—remains incompletely understood at the molecular level. Specifically, it is unclear how sporulation-specific sigma factors affect solvent formation. Here the sigF gene in C. acetobutylicum was successfully disrupted and silenced. Not only σF but also the sigma factors σE and σG were not detected in the sigF mutant (FKO1), and differentiation was stopped prior to asymmetric division. Since plasmid expression of the spoIIA operon (spoIIAA-spoIIAB-sigF) failed to complement FKO1, the operon was integrated into the FKO1 chromosome to generate strain FKO1-C. In FKO1-C, σF expression was restored along with sporulation and σE and σG protein expression. Quantitative reverse transcription-PCR (RT-PCR) analysis of a select set of genes (csfB, gpr, spoIIP, sigG, lonB, and spoIIR) that could be controlled by σF, based on the Bacillus subtilis model, indicated that sigG may be under the control of σF, but spoIIR, an important activator of σE in B. subtilis, is not, and neither are the rest of the genes investigated. FKO1 produced solvents at a level similar to that of the parent strain, but solvent levels were dependent on the physiological state of the inoculum. Finally, the complementation strain FKO1-C is the first reported instance of purposeful integration of multiple functional genes into a clostridial chromosome—here, the C. acetobutylicum chromosome—with the aim of altering cell metabolism and differentiation.
INTRODUCTION
The endospore-forming obligate anaerobe Clostridium acetobutylicum is best known for its acetone, butanol, and ethanol (ABE) fermentation and has recently received renewed attention for of its industrial potential, specifically as a biofuel producer (26, 37). Despite this increased attention and potential, many fundamental questions remain about clostridial physiology, differentiation, and metabolism, and the lack of this basic knowledge has limited the success of engineering of C. acetobutylicum for industrial processes (26, 37). Two of these key questions are how clostridial cells regulate differentiation and how differentiation is related to solvent formation (26, 37, 38). It has been well established that Spo0A is the master regulator of both solventogenesis and sporulation (8, 14, 41), but the regulation of sporulation downstream of Spo0A and any effect the regulation has on solventogenesis are not yet well understood (37, 38).
In contrast, sporulation in Bacillus subtilis has been studied extensively, and its regulation is well understood (9, 16, 48). To initiate sporulation, B. subtilis employs a multicomponent phosphorelay to phosphorylate Spo0A (4, 40), which then stimulates the expression of sigF, the prespore-specific sigma factor, and sigE, the mother cell-specific sigma factor (9, 16), in addition to many other genes (10, 34). σF is then followed by σG in the developing endospore, and σE is followed by σK in the mother cell (9, 16). Though translated, neither σF nor σE becomes active until after the cell completes asymmetric division. σF is held inactive by the anti-sigma factor SpoIIAB until the membrane-bound phosphatase SpoIIE can dephosphorylate the anti-anti-sigma factor SpoIIAA, which then binds to SpoIIAB to release σF from repression (23, 45, 58). Unbound σF then promotes the expression of about 48 genes in the prespore cell (56), including spoIIR, which is required to activate the processing of pro-σE into active σE (22, 29). Thus, σF is the first sigma factor to become active in the sporulation cascade, followed by σE. Although sigG is under the transcriptional control of σF, it does not become active until after σE is processed into an active form through a process still not fully understood (5, 9, 16, 31). In turn, σG plays a role in the processing and activation of σK, the last sporulation-related sigma factor in the regulation cascade (9, 16).
Upon sequencing of the C. acetobutylicum genome (35), genomic analysis identified homologs for much of the B. subtilis regulatory network in C. acetobutylicum, except for the multicomponent phosphorelay to phosphorylate Spo0A (38). In the first reported microarray analysis of any clostridial organism (50), sigF was observed to be severely downregulated in the C. acetobutylicum Spo0A inactivation strain SKO1 (14), demonstrating that sigF is under the control of Spo0A. This was further supported by the finding that upon butanol stress of a groESL-overexpressing strain of C. acetobutylicum, sigF had a gene expression pattern very similar to that of spo0A (51). Subsequent work using microarray analysis suggested that the regulatory network in C. acetobutylicum is similar to that in B. subtilis (1, 20). According to deduced activity plots in reference 20, Spo0A activity spiked first during early transitional phase, followed by σF, σE, and σG in close succession during mid-stationary phase. Only recently, though, have genetic mutants to specifically interrogate the roles of these sigma factors been generated. Inactivation mutants for all four sigma factors (σF, σE, σG, and σK) have been generated in Clostridium perfringens (15, 28), and although the regulation and roles of σE, σG, and σK appear to be different from those in B. subtilis (15, 28), σF was still the first sigma factor to become active and still seemed to be regulated by SpoIIAA and SpoIIAB (28). C. acetobutylicum σE and σG inactivation mutants have been reported recently, and although differences from the B. subtilis model have been identified (54), σF expression was not significantly altered in these mutants, suggesting that σF is still the first sigma factor to become active in C. acetobutylicum.
In B. subtilis, mutations in sigF or sigE blocked sporulation at stage II (asymmetric division) (18). Initially, cells developed single, normal-looking asymmetric septa, but instead of starting engulfment (stage III), cells formed an additional septum at the opposite end of the cell (disporic cells), and some even formed multiple septa (18). A separate sigF mutant formed a single asymmetric septum and started engulfment but did not complete it (18). When sigE is disrupted in C. acetobutylicum, sporulation is blocked before asymmetric division (stage II), and the cell exhibits longitudinal internal membranes (54). If σF is still the first sigma factor to become active in C. acetobutylicum, a sigF mutant would be expected to have a phenotype similar to that of the sigE mutant, in that sporulation should be blocked before stage II. This hypothesis was tested in the work reported here. Our goal was to examine the role of σF in differentiation and solventogenesis in C. acetobutylicum by silencing its expression. The sigF gene was successfully disrupted, and sporulation was blocked before asymmetric division. In addition, expression of σE and σG was not detected in the inactivation mutant, confirming the position of σF at the top of the regulatory cascade. Upon complementation, sporulation was restored, along with expression of σE and σG.
MATERIALS AND METHODS
Bacterial growth and maintenance conditions.
C. acetobutylicum ATCC 824 was grown anaerobically at 37°C in liquid CGM (80 g/liter glucose, buffered with 30 mM sodium acetate) (57) or on solid 2× YTG (pH 5.8) agar plates (36) supplemented with the appropriate antibiotic (erythromycin at 40 μg/ml for solid-medium plates and 100 μg/ml for liquid medium, thiamphenicol at 5 μg/ml, or tetracycline at 10 μg/ml). C. acetobutylicum strains were stored at −85°C in CGM supplemented with 15% glycerol and were revived by plating onto 2× YTG (pH 5.8) agar plates. For spore-forming strains, individual colonies at least 5 days old were transferred to 10-ml tubes of liquid CGM and were heat shocked at 75 to 80°C for 10 min to kill all vegetative cells and to induce germination. For non-spore-forming strains, individual colonies less than 3 days old were transferred to 10-ml tubes of liquid CGM and were allowed to grow. Escherichia coli strains were grown aerobically at 37°C in liquid LB medium or on solid LB agar plates supplemented with the appropriate antibiotic (50 μg/ml ampicillin, 35 μg/ml chloramphenicol, 100 μg/ml spectinomycin, or 10 μg/ml tetracycline). E. coli strains were stored at −85°C in LB medium supplemented with 15% glycerol.
Analytical methods.
Cell growth was monitored by measuring the A600 using a DU 730 spectrophotometer (Beckman-Coulter, Brea, CA). Supernatant samples from cultures were analyzed for glucose, acetate, butyrate, acetoin, acetone, butanol, and ethanol using an Agilent (Santa Clara, CA) high-performance liquid chromatograph (HPLC) (52). DNA concentrations and purities were measured at 260 nm and 280 nm using a NanoDrop (Wilmington, DE) spectrophotometer.
Bacterial transformations.
DNA was introduced into E. coli and C. acetobutylicum strains using electroporation with the Gene Pulser Xcell electroporation system (Bio-Rad, Hercules, CA). Prior to transformation into C. acetobutylicum, plasmids were methylated in E. coli ER2275 carrying the methylating plasmid pAN1 or pAN2 (32). Clostridial transformations were performed as described in reference 33.
Plasmid construction.
To construct the pKOSIGF plasmid, a 391-bp internal fragment of sigF (CAC2306) was PCR amplified from C. acetobutylicum genomic DNA with AmpliTaq Gold polymerase (Applied Biosystems, Carlsbad, CA) and primers SigF-F/SigF-R (see Table S1 in the supplemental material). The PCR product was cloned into the pCR8/GW/TOPOTA cloning plasmid (2.82 kb) and One Shot TOP10 E. coli (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, yielding plasmid pCR8-SigF (3.21 kb). pCR8-SigF was then digested and linearized using ScaI (New England Biolabs [NEB], Ipswich, MA), which has a single digestion site in the middle of the sigF fragment. After digestion, the linearized pCR8-SigF plasmid was treated with Antarctic Phosphatase (NEB, Ipswich, MA) to dephosphorylate the ends. A chloramphenicol/thiamphenicol resistance (Cm/Thr) cassette (encoding chloramphenicol resistance in E. coli and thiamphenicol resistance in C. acetobutylicum), under the control of the clostridial phosphotransbutyrylase promoter (Pptb) and an optimized Shine-Dalgarno sequence for C. acetobutylicum and with a rho-independent terminator at the end (47, 55), was then ligated into the vector using NEB (Ipswich, MA) Quick Ligase and was cloned into One Shot TOP10 E. coli to yield pCR8-SigF/TH (4.26 kb). The Cm/Thr cassette in pCR8-SigF/TH is flanked by 188 bp of the sigF gene on the 5′ end and 203 bp of the sigF gene on the 3′ end. Finally, pCR8-SigF/TH was recombined into the pKOREC Destination plasmid (54) using the Invitrogen (Carlsbad, CA) Gateway LR recombination reaction to yield pKOSIGF (6.08 kb).
To construct the pSPOIIA plasmid, the entire spoIIA operon, including the upstream (171-bp) and downstream (94-bp) intergenic regions, was amplified from C. acetobutylicum genomic DNA using Phusion High-Fidelity DNA polymerase (Finnzymes Oy, Espoo, Finland) and primers SpoIIA-F/SpoIIA-R (see Table S1 in the supplemental material), and BamHI digestion sites were added to each end of the PCR product. Next, both pTLH1 (13) and the PCR product were digested with BamHI (NEB, Ipswich, MA), ligated together using NEB Quick Ligase (Ipswich, MA), and cloned into One Shot TOP10 E. coli to yield pSPOIIA (7.54 kb).
To construct the pKISPOIIA plasmid, a 222-bp internal fragment of the Cm/Thr cassette was first amplified from the full Cm/Thr cassette with AmpliTaq Gold polymerase and primers TH-frag-F/TH-frag-R (see Table S1 in the supplemental material). Next, a rho-independent terminator and a SalI digestion site were added to the 3′ end of the fragment using a modified TH-frag-R primer. The full 272-bp fragment was then cloned into the pCR8/GW/TOPOTA cloning plasmid (2.82 kb) and One Shot TOP10 E. coli according to the manufacturer's instructions, yielding plasmid pCR8-TH_frag (3.1 kb). pCR8-TH_frag was linearized by digestion with SalI (NEB, Ipswich, MA) and was then treated with the DNA polymerase I, Large (Klenow) fragment (NEB, Ipswich, MA) and Antarctic Phosphatase. Next, the entire spoIIA operon, including the upstream (171-bp) and downstream (94-bp) intergenic regions, was amplified from C. acetobutylicum genomic DNA using Phusion High-Fidelity DNA polymerase (Finnymes Oy, Espoo, Finland) and primers SpoIIA-F/SpoIIA-R (see Table S1 in the supplemental material). The spoIIA operon was then ligated into the linearized pCR8-TH_frag plasmid using NEB Quick Ligase and was cloned into One Shot TOP10 E. coli to yield pCR8-TH_frag/spoIIA (4.93 kb). Finally, pCR8-TH_frag/spoIIA was digested with EcoRI (NEB, Ipswich, MA), and the TH_frag/spoIIA fragment (2,128 bp) was excised from a gel and purified using the Wizard SV gel cleanup system (Promega, Madison, WI). This fragment was then ligated into a shortened pTLH1 vector. pTLH1 (13) was digested with BglI (NEB, Ipswich, MA), and a 1,118-bp fragment (containing most of the ampicillin resistance gene) was removed. This shortened pTLH1 vector (4.6 kb) was then digested with EcoRI; the TH_frag/spoIIA fragment was ligated into the digested vector using NEB Quick Ligase; and the resulting plasmid was cloned into One Shot TOP10 E. coli to yield pKISPOIIA (6.71 kb).
Integration of pKOSIGF to disrupt sigF.
To induce chromosomal integration, a protocol similar to that described in reference 54 was used. Of primary importance is the expression of the B. subtilis resolvase gene recU, which is intended to enhance recombination, because like all clostridia, C. acetobutylicum lacks an identifiable resolvase gene (43). recU on the pKOSIGF plasmid is under the control of the strong promoter Pthl (55) to ensure expression during exponential phase. Colonies were vegetatively transferred every 24 h via replica plating onto fresh 2× YTG plates supplemented with 5 μg/ml of thiamphenicol, for a total of 10 transfers. The colonies were then vegetatively transferred every 24 h via replica plating onto fresh 2× YTG plates without antibiotics for a total of 6 transfers to cure the cells of the disruption plasmid. After the 6 transfers, the colonies were again replica plated onto fresh 2× YTG plates supplemented with 5 μg/ml of thiamphenicol and were allowed to grow for 36 to 48 h. These colonies were then replica plated onto fresh 2× YTG plates supplemented with 40 μg/ml of erythromycin and were allowed to grow for 36 to 48 h. The disruption cassette confers thiamphenicol resistance, while the plasmid backbone confers erythromycin resistance. As discussed in reference 54, four different types of colonies can arise: (i) cells that lost the plasmid and did not undergo any integration events would not grow on either antibiotic; (ii) cells still harboring the plasmid would grow rapidly on both thiamphenicol and erythromycin because of the high copy number of the plasmid (25); (iii) cells that lost the plasmid and underwent a double-crossover event would grow on thiamphenicol but not on erythromycin; and (iv) cells that underwent a single-crossover event would grow on both antibiotics, because the entire plasmid had been incorporated into the genome, but would have delayed growth on erythromycin (due to the single copy of the gene on the chromosome versus multiple plasmid copies). Putative integrants (either double- or single-crossover integrants) were selected and confirmed using colony PCR (47) with primers SigF-KO-F, SigF-KO-R, TH-F, and TH-R (see Table S1 in the supplemental material).
Integration of pKISPOIIA to complement the mutant.
In order to complement the disruption mutant (FKO1), plasmid pKISPOIIA was integrated into the chromosome. Since FKO1 already had resistance to thiamphenicol and erythromycin, the strategy to screen for integration events was to have pKISPOIIA disrupt the Cm/Thr cassette, making the integration mutants sensitive to thiamphenicol again but resistant to tetracycline. To accomplish this, a ∼200-bp internal fragment of the Cm/Thr cassette was ligated upstream of the full spoIIA operon. To prevent readthrough from the strong Pptb promoter upstream of the Cm/Thr cassette, a rho-independent terminator was placed after the ∼200-bp fragment (see “Plasmid construction” above for details).
The pKISPOIIA plasmid was integrated by using a protocol similar to that used to integrate pKOSIGF into the chromosome. Colonies were vegetatively transferred every 24 h via replica plating onto fresh 2× YTG plates supplemented with 10 μg/ml of tetracycline for a total of 6 transfers. The colonies were then vegetatively transferred every 24 h via replica plating onto fresh 2× YTG plates without antibiotics for a total of 5 transfers. After the 5 transfers, the colonies were replica plated onto fresh 2× YTG plates supplemented with 10 μg/ml of tetracycline and were allowed to grow 36 to 48 h. These colonies were then replica plated onto fresh 2× YTG plates supplemented with 5 μg/ml of thiamphenicol and were allowed to grow 36 to 48 h. Ideally, colonies with resistance to tetracycline but sensitivity to thiamphenicol, because pKISPOIIA disrupted the Cm/Thr cassette, would be selected. However, all colonies showed resistance to both antibiotics. Since pKISPOIIA could still have integrated into the chromosome through another region of homology (e.g., a copy of spoIIAA is located on both pKISPOIIA and the chromosome of FKO1), an alternative selection strategy was used. This was to select for spore formation, since σF expression should be restored along with sporulation. After 7 days, 12 colonies were selected at random, heat shocked, and grown with 5 μg/ml of thiamphenicol. Tetracycline was never added to liquid cultures because of its negative effect on protein synthesis, cell growth, and solvent production (13). Three colonies grew up within 24 h, and single-crossover integration was confirmed using PCR (see Fig. 3C and D) and sequencing.
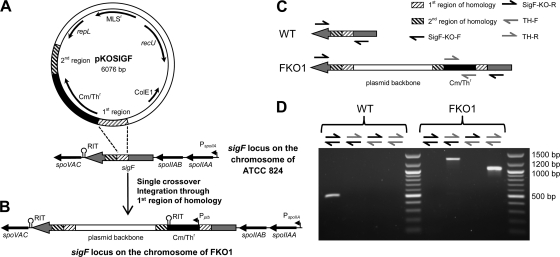
Fig. 3.
Integration of pKISPOIIA into the genome of FKO1. (A) The pKISPOIIA vector with the entire spoIIA operon under the control of its natural promoter and a ∼200-bp fragment (frag) of the Cm/Thr cassette upstream of the operon. (B) Integration of pKISPOIIA into the genome of FKO1 occurred through the repL locus. The Pptb promoter and rho-independent terminators (RIT) are indicated. The presumable PspoIIA promoters are also indicated. (C) Primers used to confirm the integration of pKISPOIIA. (D) PCR results from the three strains confirming integration and the presence of an intact sigF gene.
Southern blot analysis.
Intact, unsheared genomic DNA from wild-type (WT) C. acetobutylicum and the sigF mutant (FKO1) was prepared as described in reference 54. Biotinylated probes were created using the NEBlot Phototope kit (NEB, Ipswich, MA) according to the manufacturer's instructions. For a probe template, a PCR product was prepared from C. acetobutylicum genomic DNA with primers SigF-probe-F/SigF-probe-R (see Table S1 in the supplemental material) and AmpliTaq Gold (Applied Biosystems, Carlsbad, CA). Blots were prepared, probed, and imaged as described in reference 54.
Western blot analysis.
Crude cell extracts and Western blots were prepared as described in reference 54. Sixty micrograms of total protein was loaded onto the gels, and the antibodies against Spo0A, σF, σE, and σG were diluted 1:5,000, 1:1,000, 1:250, and 1:500, respectively. Membranes were incubated with the antibodies against Spo0A, σF, and σG for 1 h at room temperature and with the antibody against σE overnight at 4°C. Secondary antibodies were diluted 1:3,000 and were incubated for 1 h at room temperature.
Sporulation assays.
Cells were heat shocked as described under “Bacterial growth and maintenance conditions” above and were treated with chloroform as described in reference 54.
Flow cytometry and microscopy.
Samples for flow cytometric (FC) analysis and light microscopy were prepared and imaged as described in reference 53. Phase-contrast images were taken on a Zeiss (Thornwood, NY) Axioskop 2 microscope, and a Becton Dickinson (BD; Franklin Lakes, NJ) FACSAria system was used for all flow cytometric analyses. Cells stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Invitrogen, Carlsbad, CA) were treated with SlowFade Gold antifade reagent (Invitrogen, Carlsbad, CA) and were imaged on a Zeiss (Thornwood, NY) 5 Live Duo microscope. Transmission electron microscopy (TEM) samples were prepared and imaged as described in reference 20.
RNA isolation and Q-RT-PCR.
Cells were sampled and RNA isolated as described in reference 20. RNA was reverse transcribed into cDNA, and quantitative reverse transcription-PCR (Q-RT-PCR) analysis was performed as described in reference 3. As before, CAC3571 was used as the housekeeping gene, and its expression was not affected in the sigF disruption strain (FKO1) or the complementation strain (FKO1-C) (data not shown). Two biological replicates were prepared for each strain (WT, FKO1, and FKO1-C), and three technical replicate reactions per biological replicate were performed for each primer set. Cycle threshold (CT) values were then averaged together (n = 6) for subsequent analyses. Primer sequences are listed in Table S1 in the supplemental material.
RESULTS
Disruption of sigF.
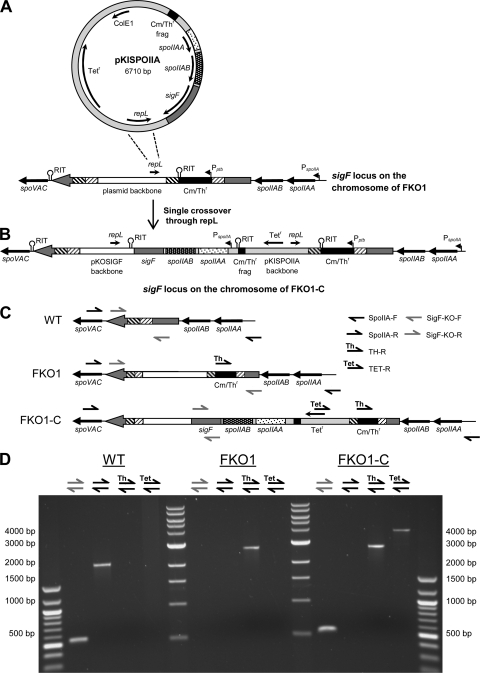
In order to disrupt the sigF gene (CAC2306), the replicating vector pKOSIGF was constructed such that two regions of homology (each ∼200 bp) flank a chloramphenicol/thiamphenicol resistance (Cm/Thr) cassette optimized for C. acetobutylicum (47) (Fig. 1 A). The plasmid was integrated using a method similar to previous clostridial disruption methods (14, 54), and a single integration through the first region of homology was achieved (Fig. 1B). Integration was stimulated by successive vegetative transfers coupled with the expression of the B. subtilis resolvase recU (see Materials and Methods for details). To confirm the integration, putative integrants were selected for colony PCR using four different primer sets (Fig. 1C and D). In wild-type (WT) cells, only SigF-KO-F/SigF-KO-R generate a PCR product (554 bp), since WT cells lack the Cm/Thr cassette (Fig. 1D). For the disruption mutant (referred to below as the FKO1 strain), three outcomes were possible: a single integration through the first region of homology, a single integration through the second region of homology, or a double crossover. For a single integration through the first region of homology, primers SigF-KO-F/TH-R would generate a PCR product of 1,342 bp (Fig. 1D). Although TH-F/SigF-KO-R could also generate a product (6,338 bp), the extension time for the PCR was not long enough to generate it. Alternatively, had a single integration occurred through the second region of homology, TH-F/SigF-KO-R would have generated a PCR product of 1,312 bp, which would have been visible on the gel (Fig. 1D). Additionally, had a double crossover occurred, both the 1,342-bp and the 1,312-bp product would have been generated in addition to a product for SigF-KO-F/SigF-KO-R (1,604 bp). Since the sole product generated was the 1,342-bp band, pKOSIGF could have been incorporated only via a single integration through the first region of homology (Fig. 1D). To further confirm this orientation, the entire integration region was amplified from FKO1 with primers SigF-KO-F/SigF-KO-R, which lie outside the disrupted region, and was sequenced. The sequenced product matched the expected sequence exactly (Fig. 1B). In addition to confirming the orientation, this sequencing also demonstrates that only a single copy of pKOSIGF integrated within the sigF locus. Had more than one copy of pKOSIGF integrated within sigF, the product produced using primers SigF-KO-F/SigF-KO-R would have been much larger.
Fig. 1.
Integration of pKOSIGF into the C. acetobutylicum genome to disrupt the sigF gene. (A) The pKOSIGF vector with two regions of homology for sigF and the orientation of sigF in the genome. (B) A single-crossover event with pKOSIGF via the first region of homology resulted in the integration of the entire plasmid. The Pptb promoter and rho-independent terminators (RIT) are indicated. The presumable PspoIIA promoter is also indicated. (C) Primers used to confirm the integration and orientation of pKOSIGF. (D) PCR results for the WT and FKO1 strains demonstrating the integration of pKOSIGF through the first region of homology.
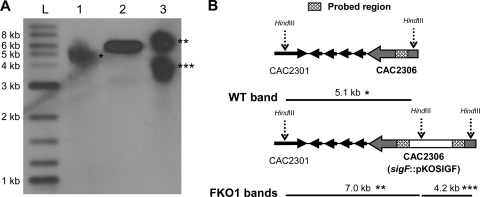
Finally, to demonstrate that pKOSIGF integrated into the chromosome only at the sigF locus, a Southern blot was prepared (Fig. 2). Genomic DNAs from the WT and FKO1 strains, along with pKOSIGF, were digested with HindIII, for which there are digestion sites flanking the sigF locus on the WT chromosome and a single digestion site within the recU gene on pKOSIGF (Fig. 2B). When this digested DNA is probed with a biotinylated probe prepared using the first region of homology as a template, a single band should be visible for WT DNA and pKOSIGF, and two bands should be visible for FKO1 DNA, since the fist region of homology is duplicated in the mutant. Following the digestion and hybridization of the probe, a single band is visible for the WT (∼5.0 kb) and pKOSIGF (∼6.0 kb), while two bands are visible for FKO1 (∼7.0 kb and ∼4.0 kb) (Fig. 2A). This demonstrates that pKOSIGF integrated only within the sigF locus.
Fig. 2.
(A) Southern blot of HindIII-digested genomic DNA from C. acetobutylicum WT and FKO1 strains. Lane 1, WT DNA; lane 2, pKOSIGF; lane 3, FKO1 DNA; lane L, 2-log ladder from NEB. The membrane was exposed for 10 min. (B) Diagrams of the bands expected from the WT and FKO1 strains and of the probed region. The four short arrows represent CAC2302, CAC2303, CAC2304, and CAC2305 and are not drawn to scale.
Complementation of sigF.
In order to complement FKO1, the entire spoIIA operon (spoIIAA-spoIIAB-sigF), including the upstream intergenic region (171 bp), presumably containing the natural promoter and ribosomal binding site, and the downstream intergenic region (94 bp), containing a rho-independent transcriptional terminator (as predicted by TransTermHP [24]), was ligated into a tetracycline-based replicating plasmid to produce pSPOIIA, and this plasmid was transformed into FKO1. The entire spoIIA operon was used, because in the WT, sigF is expressed only as part of this operon (20, 39), and because the entire operon was needed to restore σF expression in the C. perfringens sigF mutant (SM101::sigF) (28). However, in contrast to its effect on C. perfringens (28), plasmid expression of the spoIIA operon failed to restore sporulation to FKO1. FKO1 pSPOIIA colonies were tested for both chloroform resistance and heat resistance, but no CFU developed on solid agar plates following either stress. It was speculated that, as with the C. acetobutylicum EKO1 and GKO1 mutants (sigE and sigG disruption mutants, respectively), plasmid-based complementation would not restore sporulation (54). In B. subtilis, plasmid complementation of a sigE mutant failed to restore WT levels of sporulation, and only by integrating the operon back onto the chromosome could WT levels of sporulation be restored (2). Although no equivalent B. subtilis study with a sigF mutant could be identified in the literature, it is possible that this is the case for sigF mutants as well. Therefore, we decided to “knock in” a single copy of the full operon into the genome of FKO1. For this purpose, the tetracycline-based replicating plasmid pKISPOIIA was constructed (Fig. 3 A) and integrated into the chromosome (Fig. 3B).
To confirm the loss of the plasmid in the putative integration mutant, a minipreparation procedure was performed on the culture to isolate plasmid DNA, and the resulting eluate was used to transform One Shot TOP10 E. coli. When cells were plated onto solid LB agar plates supplemented with tetracycline, no tetracycline-resistant CFU developed. This result suggested that the cells had been cured of plasmid pKISPOIIA. To confirm plasmid integration, genomic DNA from the mutant colony was isolated and used in confirmation PCRs with four different PCR primer sets (Fig. 3C).
The first PCR primer set, SigF-KO-F/SigF-KO-R, tested for an intact sigF gene. As shown in Fig. 1D, the WT generates a PCR product, but FKO1 does not. The new integration mutant (referred to below as the FKO1-C strain) also generated a PCR product, indicating an intact sigF gene (Fig. 3D). The second PCR primer set, SpoIIA-F/SpoIIA-R, will generate a product only if the native spoIIA operon is intact; only the WT generates a product (Fig. 3D). The third PCR primer set, SpoIIA-F/TH-R, was used to determine where the Cm/Thr cassette was located. FKO1 and FKO1-C generated products of the same size, indicating that the integration occurred downstream of the original disruption site (Fig. 3D). The final PCR primer set, SpoIIA-F/TET-R, was used to determine where the Tetr cassette was located, and only FKO1-C generated a product (Fig. 3D). This product was sent for sequencing, which indicated that pKISPOIIA integrated through the repL locus (Fig. 3B). To confirm this, the entire region was PCR amplified and sequenced. The entire region (more than 14 kb) was too large to amplify in one PCR, so three overlapping products were generated using SpoIIA-F/TET-Seq-R, TET-Seq-F/RecU-Seq-R, and RecU-Seq-F/SpoIIA-R (see Table S1 in the supplemental material) and were sequenced. The sequenced product matched the expected sequence exactly, with integration occurring through the repL locus (Fig. 3B).
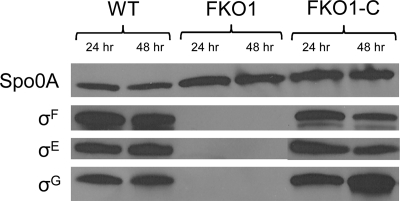
σF is needed for the expression of both σE and σG.
As discussed earlier, σF is the first sporulation-related sigma factor to become active in B. subtilis (9, 16). To test if this is also true for C. acetobutylicum, Western blotting was performed for σF, Spo0A, σE, and σG (Fig. 4). Spo0A expression was not affected by the disruption of sigF, but none of the three sporulation-related sigma factors examined were detected in FKO1 (Fig. 4). The lack of σF in FKO1 confirms that sigF was successfully disrupted and silenced, while the lack of σE and σG suggests that σF is needed for the expression of both σE and σG. In B. subtilis, sigE is initially translated into inactive pro-σE, which is processed into active, mature σE by the proteolytic removal of 27 residues from its N terminus, and this maturation is a σF-directed process (16). Thus, a lack of σF leads to a lack of mature σE. However, in the Western blot of C. acetobutylicum using the anti-σE antibody, no band corresponding to the approximate size of pro-σE was observed. The transcription of sigG in B. subtilis is controlled by σF, and thus, without σF, sigG is not transcribed (9, 16). The transcription pattern of sigG in FKO1 is discussed below. Expression of all three sigma factors is restored in the complemented strain FKO1-C (Fig. 4), confirming that a functional σF restores σE and σG expression.
Fig. 4.
Western blot of sporulation-related proteins Spo0A, σF, σE, and σG in the WT, FKO1, and FKO1-C strains. Membranes were exposed for 30 s, 1 min, and 10 min for Spo0A, σF, and both σE and σG, respectively.
Disruption of sigF abolishes sporulation prior to asymmetric septum formation.
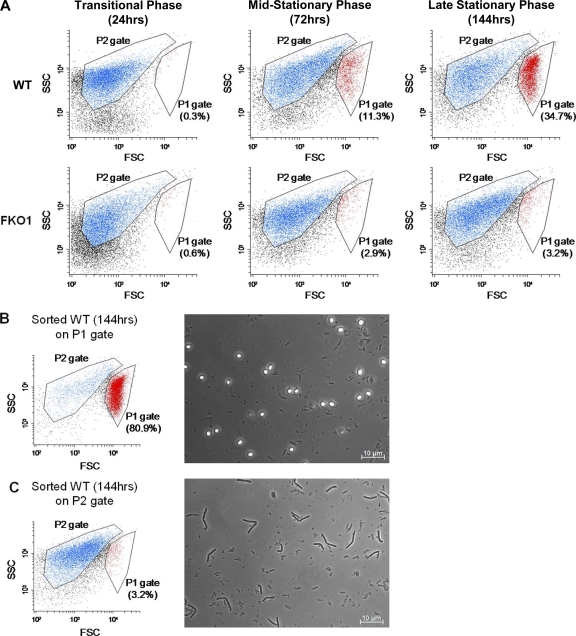
Since none of the three sigma factors σF, σE, and σG were detected by Western blotting, FKO1 should be unable to generate mature spores. To test for mature spores, both chloroform and heat shock assays were used. For the chloroform assay, aliquots of culture were treated with chloroform and were then plated onto 2× YTG plates with thiamphenicol. Typically, the WT produces at least 1 × 105 spores/ml by 120 h (5 days), but no FKO1 colonies formed after chloroform treatment, even after 168 h (7 days). In addition, when FKO1 colonies, at least 7 days old, were subjected to heat shock and were supplemented with thiamphenicol, no cultures grew up. These assays demonstrate that FKO1 does not produce mature spores. To investigate further the effect of the silencing of σF expression on differentiation, flow cytometric (FC) analysis was performed on both the WT and FKO1 strains (Fig. 5 A). Previously, FC was used to track differentiation using the forward-scatter (FSC) and side-scatter (SSC) characteristics of the various morphologies of C. acetobutylicum (53). When the WT was analyzed by FC, the typical shift to high FSC was observed, and a distinct, separate population developed by the late stage of the culture (Fig. 5A). This separate population with high-FSC characteristics represents primarily mature free spores (53). To confirm this, the 144-h WT sample was sorted on gates P1 and P2 (Fig. 5A). Following sorting, the samples were reanalyzed using the same gates and were examined by phase-contrast microscopy (Fig. 5B and C). After sorting on the P1 gate, the population within this gate was enriched from 34.7% to 80.9%, while the sample sorted on the P2 gate had only 3.2% of its population falling within the P1 gate. This 3.2% within the P1 gate can be attributed to scatter from the main population and does not make up a distinct, separate population. The sorted populations were also examined using phase-contrast microscopy. The population sorted on the P1 gate consists of many mature free spores (phase-bright cells in Fig. 5B). While some vegetative cells are still present, a large fraction of the population is made up of mature spores. In contrast, the population sorted on the P2 gate consists exclusively of vegetative cells, with no phase-bright spores visible (Fig. 5C). FC analysis of FKO1 shows that no population develops in the P1 gate (Fig. 5A). Although some events fall within the P1 gate (2.9% and 3.2% at 72 h and 144 h, respectively), these are scatter from the main population, as seen when the WT was sorted on the P2 gate (Fig. 5C). The lack of a population in the P1 gate further demonstrates that FKO1 does not develop mature spores.
Fig. 5.
Flow cytometric analysis of a time course of the C. acetobutylicum WT and FKO1 strains. (A) Abbreviated time course of the WT and FKO1 strains with two gates indicated. The WT sample at 144 h was sorted on the P1 and P2 gates. (B and C) Flow cytometric analysis of the WT sample at 144 h sorted on the P1 gate (B) and the P2 gate (C) with accompanying phase-contrast microscopic images of the sample.
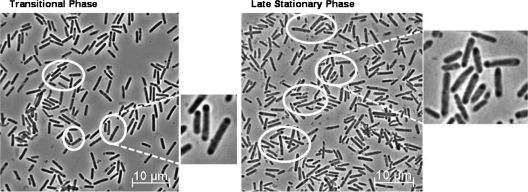
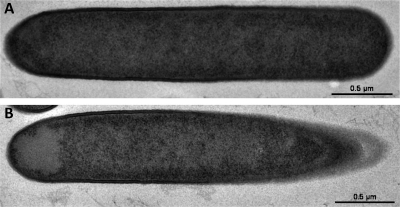
To determine whether FKO1 undergoes any morphological changes, a pH-controlled culture (where the pH was kept above 5.0) was analyzed by phase-contrast microscopy at several time points (Fig. 6). During the transitional phase, only rod-shaped vegetative cells were observed for FKO1, while typically during transitional phase, the clostridial form (characterized by swollen cells with phase-bright granulose) is visible (19, 30, 53). Even during late-stationary phase, only rod-shaped vegetative cells were present, with no visible clostridial forms or endospores (characterized by a phase-bright prespore within the mother cell) (19, 30, 53), much less mature free spores. The only discernible feature was a phase-dark area at one or both poles of the cell. To determine if an asymmetric septum or any membrane developed within the cell, transmission electron microscopy (TEM) was used. Two cell types were identified in the 70 cells analyzed: rod-shaped cells with no distinguishing features (67% of the population) (Fig. 7 A) and rod-shaped cells with a slightly electron translucent region at one pole (31% of the population) (Fig. 7B). For one cell, an electron-translucent region was located at both poles (1% of the population). Importantly, no membrane enclosed these regions, and in all the cells analyzed, no asymmetric septa were observed, in contrast with the B. subtilis σF inactivation strain (18). Although they are not directly correlated, we believe the phase-dark regions and the electron-translucent regions are the same regions, based on the similarity of their intracellular locations. Initially, we hypothesized that these phase-dark/electron-translucent regions were condensed DNA, potentially from the cell getting ready for asymmetric division. To test this, cells were stained with DAPI and were imaged. Instead of indicating DNA enrichment, the DAPI stain indicated a lack of DNA in these phase-dark regions (see Fig. S1 in the supplemental material). We are therefore unsure what these regions are composed of. In B. subtilis, it was found that slight phase-dark regions were located at each pole of the cell, indicating a lack of nucleoid DNA and an increased presence of ribosomal proteins (27), but it is not clear whether a similar phenomenon occurs in FKO1 cells.
Fig. 6.
Phase-contrast microscopic analysis of the C. acetobutylicum FKO1 strain from a pH-controlled fermentor during a typical time course. Cells displaying phase-dark regions are circled, and a representative area is enlarged to the right of each panel.
Fig. 7.
TEM of the C. acetobutylicum FKO1 strain. Rod-shaped cells with no noticeable features were observed for the majority of the culture (A), but a subpopulation of cells exhibited a slight electron-translucent region at one pole (B).
Complementation restores sporulation.
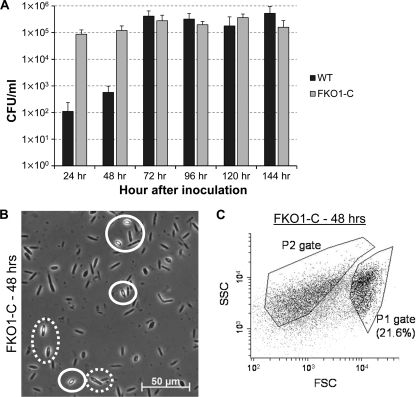
The Western blot confirms that FKO1-C expresses all sporulation-related sigma factors tested, in a manner similar to that of the WT (Fig. 4). To test for functional spores, chloroform assays were used. WT and FKO1-C cultures were grown in flasks, and chloroform assays were performed every 24 h (Fig. 8 A). Both the WT and FKO1-C strains developed typical numbers of spores (3 × 105 to 5 × 105 CFU/ml), but FKO1-C developed these spores faster than the WT (Fig. 8A). FKO1-C produced more than 1 × 105 spores/ml by 48 h, while the WT did not cross this threshold until 72 h (Fig. 8A). In FKO1-C cultures, developing endospore forms and mature free spores are visible by 48 h (Fig. 8B), and a significant spore population is evident by flow cytometry at 48 h (Fig. 8C). Therefore, integration of a complete spoIIA operon onto the chromosome reinstated both expression of all sporulation-related sigma factors and sporulation, though at a higher rate than that for the WT.
Fig. 8.
FKO1-C sporulates faster than the C. acetobutylicum WT strain. (A) Spores produced by the WT and FKO1-C strains over time. (B) Phase-contrast microscopy of FKO1-C after 48 h. Dashed white circles indicate developing endospore forms, and solid white circles indicate mature free spores. (C) Flow cytometric analysis of an FKO1-C culture at 48 h. A spore population constituting a percentage of the total population similar to that for the WT at 144 h (Fig. 5A) has evolved by 48 h.
A side effect of integrating a new, full spoIIA operon into the chromosome is the duplication of both spoIIAA and spoIIAB (Fig. 3B). The expression of these two genes is slightly higher in FKO1-C than in the WT (1.12- and 1.10-fold higher for spoIIAA and 1.17- and 1.11-fold higher for spoIIAB at 12 h and 24 h, respectively) (see Table S2 in the supplemental material) but not significantly higher. The activation of σF has been studied in detail in B. subtilis, and the concentrations of SpoIIAA and SpoIIE have been found to be crucial for the release of σF from SpoIIAB (17). Although spoIIAA and spoIIAB have slightly higher expression in FKO1-C, spoIIE actually has slightly lower expression than that in the WT (0.70- and 0.38-fold less at 12 h and 24 h, respectively) (see Table S2 in the supplemental material). From these data, it is unclear whether the duplication of spoIIAA and spoIIAB is responsible for the accelerated sporulation in FKO1-C or not. Importantly, all three of these genes (spoIIAA, spoIIAB, and spoIIE) are expressed in FKO1 (see Table S2). Both spoIIAA and spoIIAB reach about half the expression level of that in the WT, and spoIIE actually reaches a slightly higher expression level than that in the WT. The decreased expression of spoIIAA and spoIIAB in FKO1 may be explained by the altered length and structure of the spoIIA operon due to the integration of pKOSIGF (Fig. 1B).
Identification of σF-controlled genes.
With sigF successfully silenced in FKO1, the genes controlled by σF could be investigated. For a first assessment, we were interested in investigating whether the genes of the σF regulon in B. subtilis belonged to the σF regulon in C. acetobutylicum. Six genes were chosen for analysis by Q-RT-PCR: csfB (CAC0296), gpr (CAC1275), spoIIP (CAC1276), sigG (CAC1696), lonB (CAC2638), and spoIIR (CAC2898). These six are well-characterized genes of the σF regulon of B. subtilis (56) for which strong homologs can be identified on the C. acetobutylicum genome, and of the six, csfB and gpr-spoIIP (which are predicted to be transcribed as an operon [39]) have a predicted σF/G-binding motif, a binding motif for either σF or σG (39). In B. subtilis, CsfB (also called Gin) is believed to bind σG to prevent premature transcriptional activity (6, 21, 42); gpr encodes a protease responsible for the degradation of small acid-soluble proteins (SASPs) during germination (44, 49); SpoIIP is involved in the dissolution of the septal cell wall and is required for engulfment (16); lonB encodes an ATP-dependent protease whose precise function is unknown (46); SpoIIR is necessary for triggering the processing of pro-σE into σE (22, 29); and sigG encodes σG. The expression of these six genes was examined in both the WT and FKO1 at 12, 24, and 36 h, and expression ratios were calculated (Table 1). Of the six genes, only sigG exhibited a potential σF-dependent expression pattern. At all three time points (when sigG is typically expressed [20, 54]), the expression ratio for sigG was less than 1.0, indicating possible σF dependence (Table 1). This pattern was replicated when FKO1 was compared to FKO1-C (Table 1). Interestingly, sigG expression was only about 2- to 2.5-fold lower in FKO1 than in the WT or FKO1-C at 24 and 36 h (Table 1), but absolutely no protein was detected at either 24 or 48 h in FKO1 (Fig. 4). In contrast, for gpr, spoIIP, lonB, and spoIIR, the expression ratios at all three time points were greater than 1.0, indicating that expression was actually greater in FKO1 than in the WT. For csfB, the ratios were 0.94, 2.04, and 0.84, respectively, for the three time point, but this pattern still does not agree with a σF-dependent expression pattern.
Table 1.
Expression ratios of possible σF-regulated genes in C. acetobutylicum WT, FKO1, and FKO1-C strains
| Strains compared and time | Expression ratioa of: |
|||||
|---|---|---|---|---|---|---|
| csfB (CAC0296) | gpr (CAC1275) | spoIIP (CAC1276) | sigG (CAC1696) | lonB (CAC2638) | spoIIR (CAC2898) | |
| FKO1/WT | ||||||
| 12 h | 0.94 | 1.53 | 1.56 | 0.07 | 1.59 | 1.94 |
| 24 h | 2.04 | 2.10 | 1.55 | 0.49 | 1.86 | 2.57 |
| 36 h | 0.84 | 3.71 | 1.38 | 0.40 | 1.67 | 3.17 |
| FKO1/FKO1-C | ||||||
| 12 h | 5.21 | 4.79 | 2.24 | 0.10 | 3.75 | 2.13 |
| 24 h | 1.13 | 5.31 | 1.89 | 0.32 | 2.75 | 17.21 |
| 36 h | 1.50 | 8.77 | 4.32 | 0.59 | 2.42 | 16.43 |
Cycle threshold (CT) values from two biological replicates (with three technical replicates each) were averaged together (n = 6). Fold differences were calculated using the housekeeping gene CAC3571.
FKO1 produces normal levels of solvents in an inoculum-dependent manner.
Finally, we wanted to determine what, if any, effect disruption of sigF would have on solventogenesis. Playing a key role in the switch from acidogenesis to solventogenesis, Spo0A has been found to upregulate the expression of the key genes (the sol operon and the adc gene, all located on the pSOL1 megaplasmid [7]) involved in solventogenesis (8, 14, 41), in addition to its previously discussed role in sporulation. Since σF acts downstream of Spo0A, solvent production in FKO1 was expected to be similar to that in the WT, although the possibility of additional control of the solventogenic genes by σF could not be excluded. To test solvent production, cultures of both the WT and FKO1 were started from mid- or late-exponential-phase cells, as is typical for a batch culture, and were allowed to grow for 5 days. The supernatants were then analyzed for residual glucose, butyrate, acetate, ethanol, acetone, and butanol (Table 2). The WT performed as expected, consuming more than 300 mM glucose, reducing the butyric acid concentration to ∼15 mM, and producing ∼80 mM acetone and ∼150 mM butanol. However, FKO1 greatly underperformed by consuming only ∼100 mM glucose and producing only ∼17 mM acetone and ∼34 mM butanol (Table 2). When both the WT and FKO1 cultures are inoculated with stationary-phase cells, they perform similarly, both consuming ∼330 mM glucose and producing ∼80 mM acetone and more than 150 mM butanol (Table 2). Interestingly, FKO1-C exhibits the same inoculum-dependent behavior as FKO1. When inoculated with an exponentially growing culture, FKO1-C consumes only ∼87 mM glucose and produces ∼10 mM butanol, but when inoculated from a stationary-phase culture, it consumes ∼407 mM glucose and produces ∼201 mM butanol, significantly outperforming the WT culture (Table 2). Importantly, whether inoculated from an exponentially growing culture or from a stationary-phase culture, FKO1-C still formed spores (at least 2 × 105 CFU/ml). It is not known what causes this behavior. The same inoculum-dependent phenomenon was also observed in the C. acetobutylicum sigE disruption mutant (but not the sigG disruption mutant) (54), and it was suggested that this trait may be tied to early disruption of differentiation (thus affecting the sigF and sigE mutants but not the sigG mutant). However, FKO1-C, which forms spores, displays the same inoculum dependency of solvent formation.
Table 2.
Metabolite concentrations after 120 h of growth
| Inoculum and growth phase | Metabolite concn (mM)a |
|||||
|---|---|---|---|---|---|---|
| Consumed glucose | Butyrate | Acetate | Ethanol | Acetone | Butanol | |
| WT | ||||||
| Exponential | 321.8 (±8.2) | 15.2 (±2.3) | 6.9 (±1.4) | 25.8 (±4.0) | 83.5 (±14.3) | 149.3 (±5.6) |
| Stationary | 330.1 (±19.3) | 15.2 (±2.3) | 5.2 (±1.6) | 27.9 (±2.2) | 81.1 (±8.5) | 152.1 (±8.4) |
| FKO1 | ||||||
| Exponential | 116.5 (±10.6) | 43.2 (±4.1) | 17.5 (±1.5) | 10.0 (±1.3) | 17.7 (±5.5) | 34.0 (±8.5) |
| Stationary | 330.6 (±13.6) | 11.0 (±2.4) | 5.6 (±0.7) | 28.6 (±2.4) | 81.8 (±7.0) | 167.7 (±5.0) |
| FKO1-C | ||||||
| Exponential | 87.5 (±8.2) | 47.4 (±3.5) | 15.9 (±3.1) | 3.1 (±0.8) | 2.6 (±1.0) | 10.3 (±1.3) |
| Stationary | 407.1 (±35.8) | 2.8 (±2.7) | 0.5 (±1.5) | 40.0 (±8.7) | 97.0 (±12.1) | 201.1 (±18.5) |
Concentrations are averages for at least two biological replicates and two technical replicates(n, 4 to 11). Standard deviations are given in parentheses.
DISCUSSION
In this study, sigF expression in C. acetobutylicum was successfully silenced by disrupting the gene in the WT strain with pKOSIGF, thereby generating strain FKO1, and sigF expression was restored by integrating pKISPOIIA into the chromosome of FKO1. By silencing σF expression, we are now able to fully assess its role in sporulation and solventogenesis and to compare it with that in B. subtilis and C. perfringens. Earlier studies revealed that sigF is under the control of Spo0A (50), and its deduced activity plot suggested that, as in the B. subtilis model, σF is the first sporulation-related sigma factor to become active (20). In the current study, we have confirmed that σF is the first sporulation-related sigma factor to become active in C. acetobutylicum and that it is needed for the expression of both σE and σG (Fig. 4). This is in agreement with the findings for C. perfringens, where the expression of σE, σG, and σK is completely or largely absent in the sigF disruption mutant but is restored upon complementation (28). Interestingly, expression of the full spoIIA operon (spoIIAA-spoIIAB-sigF) from a replicating plasmid was able to complement the C. perfringens sigF mutant but not C. acetobutylicum FKO1 in this study. Whether this is an artifact resulting from the way in which the mutants were generated (replicating plasmid insertion versus group II intron insertion) or the plasmid copy number or whether it is a real physiological difference is difficult to determine. Unfortunately, no comparable B. subtilis study could be identified in the literature in which plasmid complementation was attempted for a sigF inactivation mutant.
In B. subtilis, sigF mutants entered stage II of differentiation (asymmetric cell division) but did not complete stage III (engulfment) (18). In contrast, FKO1 does not enter stage II of differentiation (Fig. 6 and 7). It is not clear at which stage sporulation is blocked in the C. perfringens sigF mutant (28). This is a significant difference between differentiation in B. subtilis and that in C. acetobutylicum, though how or whether σF is involved in asymmetric septation is unknown. We also note the developmental differences, based on TEM analysis, between C. acetobutylicum σF (FKO1) and σE (EKO1 [54]) inactivation strains. While both of these inactivation strains block sporulation prior to asymmetric division and prior to granulose biosynthesis, EKO1, in contrast to FKO1, appears to consistently initiate the biosynthesis of a septum, though it is not complete and appears as a dysfunctional, ill-formed membrane in the vegetative cells (54). Interestingly, the complementation strain FKO1-C completed sporulation faster than the WT (Fig. 8). It was originally hypothesized that the duplication of spoIIAA and spoIIAB was responsible for this phenotype, but the expression of these two genes was not significantly greater in FKO1-C than in the WT (see Table S2 in the supplemental material). Another possible explanation is the use of the entire downstream region of sigF in the complementation. This region presumably contains the promoter for spoVAC, though no known motif is recognizable. However, if this promoter were to become active, it would only produce a transcript that is antisense to repL (Fig. 3B), and it is doubtful that this causes the accelerated sporulation phenotype.
Although a functional σF seems to be needed for the expression of σE, the exact regulatory relationship between the two sigma factors is unclear. As noted above, our anti-σE antibody does not detect a pro-σE band. Based on homology with the B. subtilis σE protein, there does appear to be a 23-amino-acid prosequence in C. acetobutylicum σE, but no band corresponding to this slightly larger protein is detected, though it is possible that this band is too faint to be detected. In B. subtilis, σF is responsible for triggering the processing of pro-σE into σE via SpoIIR (16). However, in C. acetobutylicum, spoIIR did not display a σF-dependent expression pattern and actually appeared to have higher expression in FKO1 than in the WT or FKO1-C (Table 1). This would suggest that a different mechanism is used in C. acetobutylicum to regulate the expression and activation of σE.
By use of Q-RT-PCR to investigate the expression of six homologs of well-known and well-characterized σF-controlled genes in B. subtilis, only sigG exhibited a potential σF-dependent expression pattern (Table 1). Interestingly, expression of gpr, spoIIP, lonB, and spoIIR was actually higher in FKO1 than in the WT and FKO1-C (Table 1). There are two possible explanations for this. First, a repressor of these genes may be under the control of σF, so that in the WT and FKO1-C, which express σF, these genes are repressed, and thus, their expression is higher in FKO1. Alternatively, since these genes are presumably involved in sporulation, their expression should be transient, and they should be expressed only at a certain time by an unknown transcription/sigma factor. Since both the WT and FKO1-C sporulate, these genes may be expressed at the time needed and then shut down, while in FKO1 they may be expressed constantly, since sporulation is stalled. This could also explain why the FKO1/FKO1-C ratios are greater than the FKO1/WT ratios, since FKO1-C sporulates faster than the WT (Fig. 8), and these genes would be shut down sooner. Regardless of the explanation, these genes are almost certainly not σF-controlled genes.
Finally, this is the first reported instance of purposeful integration of a plasmid carrying functional genes (other than antibiotic resistance genes) into the genome of C. acetobutylicum with the aim of altering cellular metabolism and differentiation. Previous work integrating plasmids into the chromosome of C. acetobutylicum had sought only to disrupt genes and had used antibiotic resistance genes to screen for integration (11, 12, 14, 54). Although the procedure to integrate pKISPOIIA did not differ significantly from that used to integrate pKOSIGF, this study proves that genes beyond those coding for antibiotic resistance can be expressed from a plasmid integrated into the genome. This technique can prove useful when there is a need to integrate genes into the chromosome for stable expression in the context of metabolic engineering or when only a single copy of a gene is required or needed, as is presumably the case for sigF. While expression of the spoIIA operon from a multicopy replicating plasmid failed to restore sporulation, expression from the integrated plasmid did restore sporulation.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the use of the Delaware Biotechnology Institute Bio-Imaging Facility for all microscopy images, and we thank Mohab Al-Hinai for help in the phase-contrast microscopy analysis of the FKO1-C strain.
This work was supported by National Science Foundation (NSF) grant 0853490.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Alsaker K. V., Papoutsakis E. T. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 187:7103–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arcuri E. F., Wiedmann M., Boor K. J. 2000. Phylogeny and functional conservation of σE in endospore-forming bacteria. Microbiology 146(Pt 7):1593–1603 [DOI] [PubMed] [Google Scholar]
- 3. Borden J. R., Jones S. W., Indurthi D., Chen Y., Papoutsakis E. T. 2010. A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab. Eng. 12:268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burbulys D., Trach K. A., Hoch J. A. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 5. Camp A. H., Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chary V. K., Xenopoulos P., Piggot P. J. 2007. Expression of the σF-directed csfB locus prevents premature appearance of σG activity during sporulation of Bacillus subtilis. J. Bacteriol. 189:8754–8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cornillot E., Nair R. V., Papoutsakis E. T., Soucaille P. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dürre P., et al. 2002. Transcriptional regulation of solventogenesis in Clostridium acetobutylicum. J. Mol. Microbiol. Biotechnol. 4:295–300 [PubMed] [Google Scholar]
- 9. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 10. Fawcett P., Eichenberger P., Losick R., Youngman P. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97:8063–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Green E. M., Bennett G. N. 1996. Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. Appl. Biochem. Biotechnol. 57–58:213–221 [DOI] [PubMed] [Google Scholar]
- 12. Green E. M., et al. 1996. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142(Pt. 8):2079–2086 [DOI] [PubMed] [Google Scholar]
- 13. Harris L. M., Desai R. P., Welker N. E., Papoutsakis E. T. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67:1–11 [PubMed] [Google Scholar]
- 14. Harris L. M., Welker N. E., Papoutsakis E. T. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harry K. H., Zhou R., Kroos L., Melville S. B. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hilbert D. W., Piggot P. J. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iber D., Clarkson J., Yudkin M. D., Campbell I. D. 2006. The mechanism of cell differentiation in Bacillus subtilis. Nature 441:371–374 [DOI] [PubMed] [Google Scholar]
- 18. Illing N., Errington J. 1991. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J. Bacteriol. 173:3159–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones D. T., et al. 1982. Solvent production and morphological changes in Clostridium acetobutylicum. Appl. Environ. Microbiol. 43:1434–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones S. W., et al. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 9:R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karmazyn-Campelli C., et al. 2008. How the early sporulation sigma factor σF delays the switch to late development in Bacillus subtilis. Mol. Microbiol. 67:1169–1180 [DOI] [PubMed] [Google Scholar]
- 22. Karow M. L., Glaser P., Piggot P. J. 1995. Identification of a gene, spoIIR, that links the activation of sigma E to the transcriptional activity of sigma F during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 92:2012–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. King N., Dreesen O., Stragier P., Pogliano K., Losick R. 1999. Septation, dephosphorylation, and the activation of σF during sporulation in Bacillus subtilis. Genes Dev. 13:1156–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kingsford C. L., Ayanbule K., Salzberg S. L. 2007. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S. Y., Mermelstein L. D., Bennett G. N., Papoutsakis E. T. 1992. Vector construction, transformation, and gene amplification in Clostridium acetobutylicum ATCC 824. Ann. N. Y. Acad. Sci. 665:39–51 [DOI] [PubMed] [Google Scholar]
- 26. Lee S. Y., et al. 2008. Fermentative butanol production by clostridia. Biotechnol. Bioeng. 101:209–228 [DOI] [PubMed] [Google Scholar]
- 27. Lewis P. J., Thaker S. D., Errington J. 2000. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19:710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J., McClane B. A. 2010. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect. Immun. 78:4286–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Londoño-Vallejo J. A., Stragier P. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503–508 [DOI] [PubMed] [Google Scholar]
- 30. Long S., Jones D. T., Woods D. R. 1983. Sporulation of Clostridium acetobutylicum P262 in a defined medium. Appl. Environ. Microbiol. 45:1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meisner J., Wang X., Serrano M., Henriques A. O., Moran C. P., Jr 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. U. S. A. 105:15100–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mermelstein L. D., Papoutsakis E. T. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mermelstein L. D., Welker N. E., Bennett G. N., Papoutsakis E. T. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology (NY) 10:190–195 [DOI] [PubMed] [Google Scholar]
- 34. Molle V., et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 35. Nölling J., et al. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oultram J. D., et al. 1988. Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol. Lett. 56:83–88 [Google Scholar]
- 37. Papoutsakis E. T. 2008. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 19:420–429 [DOI] [PubMed] [Google Scholar]
- 38. Paredes C. J., Alsaker K. V., Papoutsakis E. T. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 39. Paredes C. J., Rigoutsos I., Papoutsakis E. T. 2004. Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res. 32:1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piggot P. J., Hilbert D. W. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579–586 [DOI] [PubMed] [Google Scholar]
- 41. Ravagnani A., et al. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172–1185 [DOI] [PubMed] [Google Scholar]
- 42. Rhayat L., Duperrier S., Carballido-Lopez R., Pellegrini O., Stragier P. 2009. Genetic dissection of an inhibitor of the sporulation sigma factor σG. J. Mol. Biol. 390:835–844 [DOI] [PubMed] [Google Scholar]
- 43. Rocha E. P., Cornet E., Michel B. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanchez-Salas J. L., Setlow P. 1993. Proteolytic processing of the protease which initiates degradation of small, acid-soluble proteins during germination of Bacillus subtilis spores. J. Bacteriol. 175:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schmidt R., et al. 1990. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 87:9221–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Serrano M., Hovel S., Moran C. P., Jr., Henriques A. O., Volker U. 2001. Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis. J. Bacteriol. 183:2995–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sillers R., Chow A., Tracy B., Papoutsakis E. T. 2008. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab. Eng. 10:321–332 [DOI] [PubMed] [Google Scholar]
- 48. Stragier P., Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297–341 [DOI] [PubMed] [Google Scholar]
- 49. Sussman M. D., Setlow P. 1991. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J. Bacteriol. 173:291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tomas C. A., et al. 2003. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol. 185:4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tomas C. A., Beamish J., Papoutsakis E. T. 2004. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J. Bacteriol. 186:2006–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tomas C. A., Welker N. E., Papoutsakis E. T. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl. Environ. Microbiol. 69:4951–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tracy B. P., Gaida S. M., Papoutsakis E. T. 2008. Development and application of flow-cytometric techniques for analyzing and sorting endospore-forming clostridia. Appl. Environ. Microbiol. 74:7497–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tracy B. P., Jones S. W., Papoutsakis E. T. 2011. Inactivation of σE and σG in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J. Bacteriol. 193:1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tummala S. B., Welker N. E., Papoutsakis E. T. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang S. T., et al. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16–37 [DOI] [PubMed] [Google Scholar]
- 57. Wiesenborn D. P., Rudolph F. B., Papoutsakis E. T. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl. Environ. Microbiol. 55:323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu L. J., Feucht A., Errington J. 1998. Prespore-specific gene expression in Bacillus subtilis is driven by sequestration of SpoIIE phosphatase to the prespore side of the asymmetric septum. Genes Dev. 12:1371–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.