Abstract
Ralstonia solanacearum is a soil-borne plant pathogen that causes bacterial wilt disease on many plant species. We previously showed that swimming motility contributes to virulence of this bacterium in the early stages of host invasion and colonization. In this study we identified a new negative regulator of motility, named motN, that is located in a cluster of motility-related genes. A motN mutant was hypermotile both on 0.3% agar motility plates and in rich and minimal medium broth. However, like its wild-type parent, it was largely nonmotile inside plants. The motN mutant cells appeared hyperflagellated, and sheared cell protein preparations from motN contained more flagellin than preparations from wild-type cells. The motN strain was significantly reduced in virulence in a naturalistic soil soak assay on tomato plants. However, the motN mutant had wild-type virulence when it was inoculated directly into the plant vascular system. This suggests that motN makes its contribution to virulence early in disease development. The motN mutant formed weaker biofilms than the wild type, but it attached normally to tomato roots and colonized tomato stems as well as its wild-type parent. Phenotypic analysis and gene expression studies indicated that MotN directly or indirectly represses transcription of the major motility regulator FlhDC. MotN was also connected with other known motility and virulence regulators, PehSR, VsrBC, and VsrAD, via uncertain mechanisms. Together, these results demonstrate the importance of precise regulation of flagellum-mediated motility in R. solanacearum.
INTRODUCTION
Ralstonia solanacearum, a soil-borne bacterium, causes lethal bacterial wilt disease in more than 200 plant species around the world, including such important crops as potato, tomato, tobacco, banana, and peanut (29). Due to its aggressiveness, wide geographic distribution, and unusually broad host range, bacterial wilt is considered one of the most destructive plant diseases (52). The bacterium normally invades plant roots from the soil through wounds or natural openings where secondary roots emerge, colonizes the intercellular space of the root cortex and vascular parenchyma, and eventually enters the xylem vessels and spreads up into the stem and leaves. The pathogen cell density in stems commonly surpasses 109 CFU/g of host tissue (4, 64). After R. solanacearum has colonized the xylem, large numbers of bacterial cells are shed from roots, providing a pathway for bacteria to return to the soil and initiate new infections (29). Affected plants suffer chlorosis, stunting, and wilting and usually die rapidly. Many factors contribute to the virulence of R. solanacearum, including bacterial extracellular polysaccharide (EPS) (17), a set of effector proteins secreted through the type 3 secretion system (5), and a consortium of plant cell wall-degrading enzymes (26, 33). Expression of bacterial wilt virulence genes is controlled by a complex regulatory network (18, 25, 56).
R. solanacearum has swimming motility mediated by one to four polar flagella (12); motility is a quantitative trait within populations, and its expression is affected by both cell density and the environment (14, 15). In culture, the bacteria are essentially nonmotile when the cell density is low (<106 CFU/ml) or high (>109 CFU/ml). During exponential growth, the percentage of motile cells increases with cell density, peaking at about 108 CFU/ml, when 65% of cells are motile, and then declining with higher cell densities (60). However, when R. solanacearum grows in the xylem of infected tomato plants, almost no motile bacteria are present until the pathogen reaches a density of >109 CFU/g of tissue, at which point fewer than 5% of cells are motile (60).
Motility is a virulence factor for R. solanacearum. A nonmotile flagellin (fliC) mutant of R. solanacearum was significantly reduced in virulence on unwounded tomato plants inoculated by a naturalistic soil soak assay (60). Motile but nontactic mutants lacking either CheA or CheW, which are core chemotaxis signal transduction proteins, also had reduced virulence indistinguishable from that of a nonmotile mutant (66). Together, these results demonstrate that directed motility is required for full virulence. In contrast, both nonmotile and nontactic strains were as virulent as the wild type when they were introduced directly into the plant stem through a cut petiole, indicating that motility and taxis make their contribution to virulence in the early stages of host invasion and colonization (60, 66).
In R. solanacearum, motility is coregulated with several other known virulence factors by a complex system. The central component of this network is PhcA, a cell density-responsive LysR-type global regulator (10, 57) that specifically responds to a quorum-sensing molecule, 3-hydroxypalmitic acid methyl ester (3-OH PAME) (15, 23). At a high cell density, accumulation of 3-OH PAME activates PhcA, which then induces expression of the virulence factors EPS, endoglucanase, and others (56). PhcA also represses expression of the two-component regulator PehSR (56), which positively regulates bacterial swimming and twitching motility as well as plant cell wall-degrading polygalacturonases (1, 35). At low cell densities, PhcA is not expressed, allowing PehSR to activate expression of early disease virulence factors like polygalacturonases and motility (56). The heterotetramer FlhDC is the master regulator of flagellar biosynthesis and motility, and it activates expression of class II flagellar genes (1, 59). Other regulators also affect motility in R. solanacearum, including the two-component regulators VsrBC, which positively regulates motility, and VsrAD, which negatively regulates motility (65).
In this study we characterize a new negative regulator of swimming motility in R. solanacearum, MotN. Since nonmotile R. solanacearum strains have reduced virulence, we hypothesized that the hypermotile motN mutant would have increased virulence. We used reporter gene fusions and regulatory mutants to determine how MotN fits into the network regulating motility in this pathogen. We found that being hypermotile reduces bacterial wilt virulence as much as being nonmotile, suggesting that precise regulation of this behavior is critical for pathogen success. The regulation of motility in R. solanacearum is even more complex than previously understood, involving not only many known regulators but also unknown components that interact transcriptionally and possibly also posttranscriptionally.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. solanacearum strains were grown at 28°C either in Casamino Acids-peptone-glucose (CPG) rich broth (30) or on tetrazolium chloride (TZC) plates containing CPG plus 1.8% (wt/vol) agar and 0.05% (wt/vol) 2,3,5-triphenyltetrazolium chloride (36). Boucher's minimal medium (BMM) (8) amended with 0.2% (wt/vol) glucose was used when minimal medium was required. Escherichia coli strains were grown in Luria-Bertani medium (40) at 37°C. Antibiotics were added to cultures when needed at the following concentrations: ampicillin, 50 mg/liter; kanamycin, 25 mg/liter; tetracycline, 15 mg/liter; gentamicin, 12.5 mg/liter, and rifampin, 25 mg/liter. Growth rates of wild-type and mutant strains were compared in CPG medium, in BMM with 0.2% glucose, and in tobacco leaf tissue (Nicotiana tabacum cv. Bottom Special) as previously described (60). Unless otherwise noted, medium components were purchased from Difco Laboratories (Detroit, MI), and other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169recA1endA1 hsdR17(rK− mK−) supE44thi-1gyrArelA1 | Invitrogen |
| R. solanacearum | ||
| K60 | Wild-type tomato isolate; phylotype II, sequevar 7 (historically known as race 1 biovar 1) | 36 |
| K713 | K60 ΔmotN::aacC1; Gmr | This study |
| K714 | K60 ΔmotN | This study |
| K701 | K60 fliC::aacC1; aflagellate and nonmotile; Gmr | 60 |
| K702 | K60 flhDC::aacC1; Gmr | 59 |
| K60 Rifr | K60 spontaneous Rifr mutant; motile and tactic, fully virulent | 66 |
| K716 | pJYTn7motNgus transposed in K714; Smr | This study |
| K710 | pJYTn7motNgus transposed in K60; Smr | This study |
| KB5 | K60 pehR::Tn5; Kmr | 1 |
| K791 | K60 ΔvsrAD::aacC1; Gmr | This study |
| K796 | K60 vsrBC::aacC1; Gmr | This study |
| K800 | K60 phcA::aacC1; Gmr | This study |
| K721 | KB5 Tn7-motN::gus; Kmr Smr | This study |
| K722 | K791 Tn7-motN::gus; Gmr Smr | This study |
| K723 | K796 Tn7-motN::gus; Gmr Smr | This study |
| K724 | K800 Tn7-motN::gus; Gmr Smr | This study |
| K725 | K60 flhDC::aacC1 ΔmotN; Gmr | This study |
| K726 | K60 Tn7-PflhDC::gus ΔmotNpehR::Tn5; Gmr | This study |
| K727 | K60 Tn7-PflhDC::gus ΔmotNvsrAD::aacC1; Smr Gmr | This study |
| K728 | K60 Tn7-PflhDC::gus ΔmotNvsrBC::aacC1; Smr Gmr | This study |
| K821 | pJYTn7PflhDCgus transposed in K60; Smr | This study |
| K822 | pJYTn7PflhDCgus transposed in K791; Smr Gmr | This study |
| K824 | pJYTn7PflhDCgus transposed in K796; Smr Gmr | This study |
| K826 | pJYTn7PflhDCgus transposed in K714; Smr | This study |
| K827 | pJYTn7PflhDCgus transposed in KB5; Smr Kmr | This study |
| Plasmids | ||
| pSTBlue-1 | Cloning vector; Apr Kmr | EMD Bioscience |
| pUFR80 | sacB+; gene replacement vector; Apr Kmr | 58 |
| pVO155 | Apr Kmr | 43 |
| pHP45Ω | Smr Ω cassette; Apr Smr | 51 |
| pUC18-mini-Tn7 | Mini-Tn7 delivery plasmid; Apr | 13 |
| pJYminiTn7gus | Mini-Tn7gus reporter; Ampr Smr | This study |
| pJYTn7motNgus | Mini-Tn7motN::gus; Ampr Smr | This study |
| pJYTn7PflhDCgus | Mini-Tn7 PflhDC::gus; Ampr Smr | This study |
Gmr, gentamicin resistance; Tcr, tetracycline resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance; Rifr, rifampin resistance.
DNA manipulations.
DNA manipulations, including cloning, Southern blotting, restriction mapping, sequencing, and PCR, were performed using standard methods (7). R. solanacearum and E. coli were transformed by electroporation as previously described (2). DNA sequencing and oligonucleotide synthesis were performed at the University of Wisconsin—Madison Biotechnology Center, Madison, WI. DNA sequence was analyzed using the DNASTAR software package (DNASTAR, Inc., Madison, WI). Unless otherwise noted, molecular biology reagents and kits were purchased from Promega (Madison, WI).
Strain construction, mutagenesis, and complementation.
Splicing by overlap extension PCR (31) (SOE-PCR) was used to create a motN deletion construct by using primers motN-F (5′-CTTGTCCAGGTTGGTCTGGT-3′), motN-soe2 (5′-CTCCGACTGCAACACCCGCCAGATCTCCAGCTGTAGGGATCGCTA-3′; BglII site underlined), motN-soe3 (5′-AGATCTCCAGCTGTAGGGATCGCTA-3′; BglII site underlined) and motN-R (5′-TTGCCCTCGTTGGAGTACAGCTC-3′). The resulting 1,552-bp motN deletion construct containing an introduced BglII site was cloned into pSTBlue and pUFR80 to create pSJYΔmotN and pUJYΔmotN, respectively. A gentamicin resistant cassette from pUCGM was inserted into the BglII site of pSJYΔmotNGm. The deletion constructs were introduced into the chromosome of wild-type R. solanacearum strain K60 by double homologous recombination as previously described (66) or by sacB-assisted deletion mutagenesis to create K713 and K714, respectively. The correct allelic replacement in each mutant was confirmed by PCR, by sequencing, and by Southern blot analysis.
Transposon Tn7, which specifically inserts into the selectively neutral attTn7 site 25 bp downstream of the R. solanacearum glmS gene, was used to create cis-merodiploid transcriptional fusions to study gene expression. A mini-Tn7 delivery vector, pJYminiTn7gusSmΩ, was constructed from pUC18-mini-Tn7T-oriT, pVO155 (43), and pHP45Ω (51) as follows. A 2.1-kb SpeI/PvuII promoterless β-glucuronidase (gus) cassette with the ribosomal binding site from pVO155 (43) was moved into the SpeI/EcoRV sites of pBluscript II to create pBJYgus; a 2.0-kb HindIII SmrΩ cassette from pHP45Ω was cloned into the EcoRI site of pBJYgus in the same orientation as gus to create pBJYgusSmΩ. The 3.9-kb BamHI/HindIII gus-SmrΩ fragment from pBJYgusSmΩ was cloned into the StuI site of pUC18-mini-Tn7T-oriT, resulting in pJYminiTn7gusSmΩ, which was used to create transcriptional fusions for gene expression studies. The motN open reading frame (ORF) with its putative native promoter was amplified from K60 by using primers motN-F1 (5′-TTCTAGAGACGGCTCCTATATTCCCGT-3′) and motN-R1 (5′-TCTCGAGCTAGTGATGGTGATGGTGATGCAACTGGAGCCCGTTGCTGAC-3′; restriction sites are underlined) and cloned into SpeI and XhoI sites of pJYminiTn7gusSmΩ to create pJYTn7motNgus. The Tn7-motN::gus construct was transposed into various R. solanacearum strains for gene expression and complementation studies. To study flhDC expression, the 921-bp promoter region of flhDC was cloned to pJYminiTn7gusSmΩ to create pJYTn7PflhDCgus. The pJYTn7PflhDCgus construct was transposed into various R. solanacearum strains for gene expression studies.
Motility assays.
Motility of R. solanacearum strains was assayed on semisolid motility medium plates containing 1% (wt/vol) tryptone and 0.3% (wt/vol) noble agar as previously described (66, 67). We also directly observed bacterial motility by microscopy after growth in CPG broth, BMM broth, or xylem fluid collected from symptomatic plants, as previously described (60). Percentage of motile cells was quantified by ImagePro image tracking software.
Electron microscopy.
Bacteria were harvested from the edge of the motility halo on 48-h-old cultures growing on semisolid motility agar plates. Cells were placed gently on Pioloform-coated 300-mesh nickel grids and stained with methylamine tungstate negative stain (Nano-W; Polysciences, Inc.). At least 100 cells of each strain were analyzed. Images were taken on a Philips CM120 microscope at 80 kV with an SIS MegaView III digital camera.
Flagellin isolation and SDS-PAGE.
Bacterial cultures were grown in BMM broth supplemented with 0.2% (wt/vol) yeast extract to a cell density of ∼4 × 108 to 5 × 108 CFU/ml, the cell density corresponding to the maximum proportion of motile cells (60). Cell densities were determined spectrophotometrically and confirmed by dilution plating. Crude flagellin was isolated from sheared bacterial cell extracts as described previously (61). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% gels and stained with Coomassie brilliant blue (7). The loading amount of each sample was adjusted according to the culture cell density before flagellin isolation such that all the samples contained protein isolated from same number of cells.
Virulence assays.
We assayed the virulence of mutant and wild-type R. solanacearum strains by means of naturalistic soil-soak and more direct petiole inoculation virulence assays of the wilt-susceptible heirloom tomato cultivar Bonny Best, as previously described (60). For the soil soak assay, a bacterial suspension was poured onto the soil of pots containing an unwounded 16-day-old tomato plant to a final density of approximately 5 × 107 CFU/g of soil. For the petiole inoculation, about 2,000 bacteria in a 2-μl volume were placed onto the freshly cut petiole stump after removal of the first true leaf of a 21-day-old tomato plant. Plants were monitored daily for disease progress by a rater blind to the treatment conditions, and symptoms were scored on a 0 to 4 disease index, where 0 indicates no disease, 1 indicates 1 to 25% of leaves wilted, 2 indicates 26 to 50% of leaves wilted, 3 indicates 51 to 75% of leaves wilted, and 4 indicates 76 to 100% of leaves wilted. Each experiment contained a minimum of 16 plants per strain, and experiments were repeated at least three times. The disease index for each day is the average of 48 plants from three experiments.
Hydrogen peroxide detection in tomato leaves.
Tomato leaves from 25-day-old tomato plants were infused with R. solanacearum cells or water as a control. The leaves were sampled 24 h after infusion, and hydrogen peroxide production in plant tissue was detected with 3,3′-diaminobenzidine (DAB; Sigma) stain as previously described (24). Oxidation of DAB by hydrogen peroxide creates a visible brown precipitate in the host tissue.
Biofilm assay.
To quantify biofilm formation, we used the polyvinylchloride (PVC) microtiter plate assay with minor modification (45). Briefly, 5-μl overnight cultures of R. solanacearum adjusted to an optical density at 600 nm (OD600) of 0.1 were used to inoculate 95 μl of CPG broth in wells of a PVC microtiter plate and incubated without shaking for 24 h at 28°C. Crystal violet staining and biofilm quantification by absorbance at 530 nm were performed as described previously (45).
GUS activity assays.
β-Glucuronidase (GUS) activity was measured over a 45-min time course as described previously (9). Briefly, to measure GUS activity in culture, bacteria were grown in either CPG broth or BMM with glucose at 28°C, and both log- and stationary-phase samples were harvested by centrifugation at 8,000 × g for 3 min. To measure gus gene fusion expression in planta, tobacco leaves (Nicotiana tabacum cv. Bottom Special) were infiltrated with a 1 × 108 CFU/ml bacterial suspension with a syringe, and 1-cm2 disks of plant tissue were harvested with a cork borer at 0, 8, and 24 h after inoculation, homogenized in sterile distilled water, and pelleted by centrifugation. Methylumbelliferone glucuronide (MUG) was used as a substrate. Activity was normalized to the number of CFU and expressed as pmol of 4-methylumbelliferone (4-MU) produced per minute per CFU.
Chemotaxis assays.
A slightly modified agarose plug method (68) was used to quantify R. solanacearum chemotaxis. After 30 min of incubation at room temperature in a chemotaxis chamber holding an agarose plug containing 0.1% of the known chemoattractant yeast extract, the chemotaxis ability of R. solanacearum cells was observed under an Olympus BX60 phase-contrast microscope (Olympus America Inc., Melville, NY) and recorded by a charge-coupled-device camera (66). All agarose plug assays were repeated at least three times, and at least two agarose plugs were used for each replicate.
Noncompetitive and competitive attachment to tomato roots.
R. solanacearum cell suspensions were prepared as for the chemotaxis assay above and resuspended in sterile chemotaxis buffer (10 mM potassium phosphate, pH 7.0, 0.1 mM EDTA, 1 mM MgSO4) to an OD600 of 0.1 for a noncompetitive attachment assay. For competitive studies, K60Rif (a spontaneous rifampin-resistant variant of the wild type) and motN mutant cell suspensions were combined in a 1:1 ratio. The attachment of R. solanacearum cells to tomato seedling roots was quantified as described previously (45). Each experiment contained at least 10 plants per treatment; all experiments were repeated three times. We also directly observed the attachment of the green fluorescent protein (GFP)-labeled wild-type and motN mutant strains to tomato roots under a Zeiss LSM510 Meta laser scanning confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY).
Colonization of tomato stems by R. solanacearum strains.
To measure bacterial competitive fitness during invasion and colonization, tomato plants were soil-soak inoculated with either the marked wild-type K60Rif strain or the motN mutant alone (single inoculation test) or with the wild-type and motN mutant strains together in a 1:1 mixture (competition assay). Population sizes of each strain were determined in tomato plants as soon as the first wilt symptoms appeared (disease index, 1) as described previously (66). R. solanacearum population sizes were determined as the number of CFU/g of plant tissue. Each experiment contained at least 16 plants per treatment; all experiments were repeated three times.
Data analysis.
For disease assays, the mean disease index for each treatment for each day was analyzed by analysis of variance (ANOVA) at the 95% level. For the motility assay and biofilm assay data, Fisher least significant differences were calculated and applied at the 95% confidence level. All statistical analyses were performed with Minitab statistical software (Minitab, Inc., State College, PA).
RESULTS
Analysis and mutagenesis of motN.
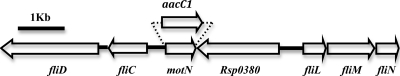
An apparent transcriptional regulator was identified in a cluster of motility genes between fliC and fliL in the R. solanacearum genome (Fig. 1). This 633-bp ORF encodes a putative protein of 210 amino acids. The gene, which we named motN (for motility negative regulator), is conserved among all the R. solanacearum strains sequenced to date (27, 53). The strain K60 MotN amino acid sequence is 100% identical to the sequences from the closely related R. solanacearum strains IPO1609 and UW551, 99% identical to banana strain MolK2, and 95% identical to GMI1000. The K60 MotN amino acid sequence is also 93% identical to a two-component transcriptional regulator in Ralstonia pickettii strain 12J/12D. No other sequence in the databases had more than 45% identity to MotN, suggesting that MotN has no direct homologs in other sequenced organisms. MotN has the characteristic structure of a cytoplasmic response regulator in a two-component system, with an N-terminal CheY-like signal receiver domain (REC domain) and a C-terminal helix-turn-helix DNA-binding domain. There is no apparent histidine protein kinase sensor gene nearby. The ORF adjacent to motN (RSp0380) encodes a hypothetical protein containing a membrane domain, an EAL domain (also known as DUF2, for domain of unknown function 2), and a diguanylate cyclase (GGDEF) domain. Deletion of this gene had no detectable effect on either motility or virulence (data not shown).
Fig. 1.
Cloning and mutagenesis of the motN region of R. solanacearum. Arrows represent open reading frames. motN is located within a cluster of motility genes between fliC and fliL. The motN ORF was either deleted to make the markerless motN deletion mutant K714 or replaced with a gentamicin resistance cassette (aacC1) to make strain K713.
SOE-PCR (31) was used to create an unmarked motN deletion mutant (K714) or to replace the motN region with a gentamicin resistance gene cassette (K713). Strains K713 and K714 had normal mucoid colony morphology on solid medium and grew as well as the wild-type parent strain K60 in rich medium, in minimal medium supplemented with 0.2% glucose, and in leaves of tobacco plants (data not shown). The two mutants behaved indistinguishably in virulence and motility assays (data not shown). Markerless motN mutant K714 was used for all assays except the competition and attachment assays that required a selectable marker; these were conducted with K713.
R. solanacearum motN mutants were hypermotile.
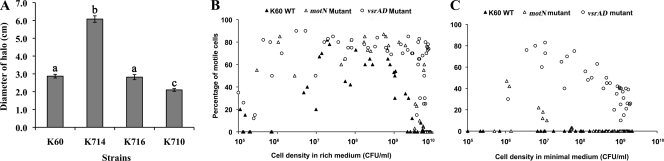
The location of motN within a cluster of motility genes suggested it might be involved in bacterial motility. To test this hypothesis, we evaluated swimming motility of R. solanacearum strains on semisolid motility agar. The motN mutant was hypermotile, producing swimming haloes at least 2-fold larger than those of K60 (Fig. 2 A). When complemented with a Tn7-motN construct transposed into the chromosomal attTn7 site, the motN mutant was restored to wild-type motility, while a strain that had two copies of motN (K710) formed a smaller halo than K60 (Fig. 2A).
Fig. 2.
The motN mutant was hypermotile on 0.3% agar plates, in rich medium broth, and in minimal medium with 0.2% (wt/vol) glucose. (A) Diameters of motility halos formed on semisolid 0.3% agar plates were measured from two directions for each plate for four strains: K60 (wild type), K714 (motN mutant), K716 (complemented motN strain), and K710 (motN overexpressed strain). The experiment was repeated three times, with at least three plates for each strain per replicate. Bars indicate standard error. Columns with different letters above differ significantly according to the Fisher least significant difference test (P < 0.05). (B and C) Each point shows the percentage of R. solanacearum cells exhibiting swimming motility at different cell densities during growth in culture. Closed triangles, K60 wild type (WT); open triangles, K714 motN; open circles, K791 vsrAD. The initial density of each strain was 1 × 105 CFU/ml. The percentage of motile cells was determined microscopically by motility track photography, and combined results are shown from three time course experiments for each strain.
To directly visualize bacterial motility, we used microscopy to compare motN mutant cells grown in either CPG broth or BMM-glucose broth to wild-type cells. In rich broth, wild-type strain K60 is rarely motile at very low (<106 CFU/ml) or high (>109) cell density, but the motN mutant had both a wider motile cell density range and an overall higher percentage of motile cells (Fig. 2B). However, the motN mutant was not as hypermotile as a mutant lacking global regulator VsrAD (32, 55, 56). In minimal medium with glucose, wild-type strain K60 is completely nonmotile at all cell densities, but the motN mutant was still motile, albeit over a narrower cell density range, while the vsrAD mutant was motile across a wide range of cell densities (Fig. 2C). These results suggested that motN encodes a negative regulator of motility. We also examined the motility of bacteria in xylem fluid from tomato plants inoculated with various GFP-labeled R. solanacearum strains. In the xylem, both wild-type and motN strains were essentially nonmotile at low cell density, and fewer than 5% of the bacterial cells were motile at 109 CFU/ml (data not shown), indicating that conditions or signals in planta suppress the hypermotility of a motN mutant. This is consistent with our previous finding that when R. solanacearum grows in tomato xylem, the bacteria become almost entirely nonmotile, and the motility regulon appears to be shut down (60).
The motN mutant was hyperflagellated and overproduced flagellin.
The swimming motility of R. solanacearum is mediated by flagella. We used electron microscopy to determine if the motN mutant has normal flagella. The abundant EPS produced by R. solanacearum in broth culture interferes with electron microscopy, so we collected cells from the edges of the halo on motility plates. Under these conditions, approximately 30% of wild-type strain K60 cells had at least one and occasionally two or three helical polar flagella (data not shown), while the rest were aflagellate. In contrast, more than 50% of the motN mutant cells had multiple (three to five) flagella.
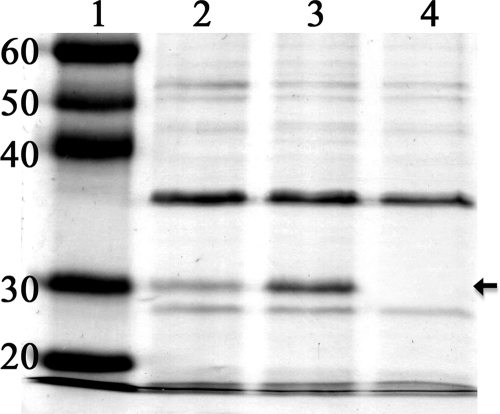
To confirm that the motN mutant overproduced flagella, we compared the production of the flagellar filament protein flagellin by similar numbers of wild-type and motN mutant cells. Sheared-cell protein extracts from both wild-type strain K60 and the motN mutant, normalized to cell number, contained a band with the 30-kDa molecular mass of R. solanacearum flagellin (Fig. 3). However, the flagellin band produced by a comparable number of motN mutant cells was much thicker, indicating that motN produced a larger amount of flagellin per cell than the wild-type strain. As expected, the flagellin band was absent from sheared cell extracts of fliC mutant K701 (Fig. 3, lane 4). These data suggest that MotN mutation directly or indirectly increases the synthesis of flagella.
Fig. 3.
Crude flagellin in sheared cell extracts from different R. solanacearum strains, normalized by loading total protein from equivalent numbers of cells of each strain. Proteins were separated by SDS-PAGE on 12% gels and stained with Coomassie brilliant blue. Lane 1, molecular size markers (masses are indicated in kilodaltons); lane 2, wild-type strain K60; lane 3, K714 (motN); lane 4, K701 (fliC). The arrow indicates R. solanacearum flagellin.
Loss of the motN mutant reduced R. solanacearum virulence.
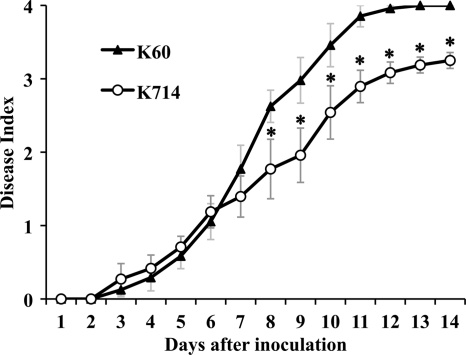
Hypermotility could provide a competitive advantage during the early stages of bacterial colonization and thus increase virulence. We tested this hypothesis using a biologically representative soil soak inoculation that requires bacteria to actively find and invade host plant roots from the soil. Unexpectedly, the hypermotile motN mutant was significantly reduced in virulence (P < 0.05, ANOVA). At the end of the assay, all plants inoculated with the wild-type parent strain were dead (disease index, 4), while plants inoculated with the motN mutant had an average disease index of 3.2 because about 20% of plants remained symptomless (Fig. 4). However, there were no differences between the virulence of the wild type and the motN mutant in a cut petiole inoculation assay that bypassed the normal infection route by directly inoculating bacteria into the vascular system of tomato plants (data not shown).
Fig. 4.
A motN mutant of R. solanacearum had reduced virulence. Virulence was measured by inoculating unwounded 16-day-old tomato plants (cultivar Bonny Best) with a suspension of R. solanacearum cells poured onto each pot to a final concentration of about 5 × 107 CFU/g of soil. Plants were incubated at 28°C and rated daily on a 0 to 4 disease index scale. Each point represents the mean of three independent assays with 16 plants per treatment inoculated with either wild-type strain K60 or the motN mutant K714. Bars indicate the standard error of the mean. Asterisks indicate days where the wild type and K714 were significantly different by ANOVA (P < 0.05).
R. solanacearum motN mutant induced wild-type levels of oxidative bursts.
Bacterial flagellin is an important elicitor of plant defenses (22). To test the possibility that the motN mutant had decreased virulence because its abundant flagellin triggered increased defense responses in the tomato host, we measured the accumulation of hydrogen peroxide, an indicator of plant defense activation, in tomato leaves infected with either motN or wild-type bacteria. The two strains induced indistinguishable oxidative bursts in tomato (data not shown), suggesting that the motN mutant was not reduced in virulence because its hyperflagellation elicited stronger host defense responses.
The motN mutant had normal chemotaxis ability.
Previous studies found that R. solanacearum needs directed motility (taxis) for full virulence; indeed, random motility is just as deleterious as none at all (66). It was possible that the hypermotile motN mutant had reduced virulence because of a chemotaxis defect, leading to random undirected movement. We tested this hypothesis by measuring the strain's chemotaxis to yeast extract, a known attractant, using an agarose plug assay. Wild-type and motN cells accumulated in similar numbers around attractant-containing plugs (data not shown), indicating that the motN mutant retained wild-type chemotactic behavior.
A motN strain produced weaker biofilms.
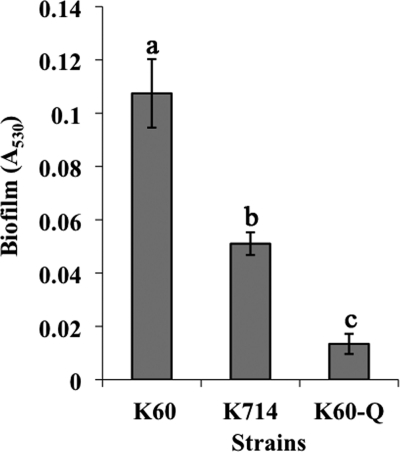
R. solanacearum produces biofilms on both abiotic and biotic surfaces (35, 66). Several Gram-negative bacteria need swimming motility to form biofilms (44). Could hypermotility lead to altered biofilms? To determine if the MotN regulator contributes to normal biofilm formation, we compared the biofilms formed by different strains following static culture in PVC plate wells stained with crystal violet. Under these conditions, the motN mutant produced significantly less biofilm than the wild-type K60, indicating that MotN plays a role in this multicellular behavior (Fig. 5).
Fig. 5.
A motN mutant was reduced in biofilm formation. R. solanacearum strains were incubated statically for 24 h in PVC microtiter plates, and biofilm formation was quantified by measuring the A530 of crystal violet-stained wells rinsed with ethanol. K60, wild-type strain; K714, motN mutant; K60-Q, pilQ mutant biofilm-defective negative control. Data shown are the means of four independent experiments with at least three replicates for each strain per experiment. Bars represent standard errors of the means. Columns with different letters were significantly different according to Fisher's least significant difference test (P < 0.05).
Competitive and noncompetitive attachment and colonization assay.
Biofilms begin with the attachment of bacteria to a surface (16), and attachment of soil bacteria to plant cells is a crucial early step in many plant-microbe interactions (54). We wondered if the hypermotile motN mutant formed less biofilm and had reduced virulence because it attached poorly to host plant roots. However, there was no significant difference in the numbers of wild-type and mutant cells that attached to tomato roots in either individual or 1:1 dual-strain competitive root attachment assays (see Table S1 in the supplemental material). There was also no discernible qualitative difference in the spatial patterns of attachment of GFP-labeled wild-type and motN mutant strains to tomato seedling roots, as assessed by laser scanning confocal microscopy (data not shown).
We also measured the competitive fitness of the motN mutant during invasion and colonization of tomato stems. We quantified pathogen population sizes in stems of wilting tomato plants that had been soil-soak inoculated with K60 Rifr (a spontaneous rifampin-resistant variant of the wild type) or strain K713 (motN Gmr) individually or in a 1:1 mixture. There was no significant difference in population sizes in planta between K713 and strain K60 Rifr in either the competitive or noncompetitive plant colonization assays (see Table S1 in the supplemental material). Thus, once they get into the plant stems, motN cells can colonize tomato as well as the wild-type strain.
Interactions between MotN and other regulators.
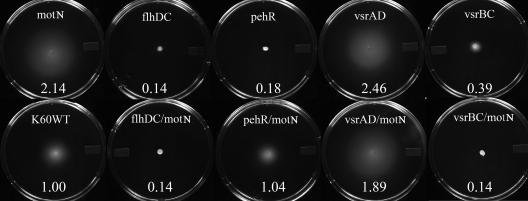
Motility behavior of R. solanacearum is controlled by a complex set of interacting factors, including both environmental signals and virulence regulators. Mutants lacking PhcA, a quorum-sensing global virulence regulator, retain abnormally high motility as cell densities increase (11). Mutants lacking FlhDC, a master regulator of swimming motility, are nonmotile (59). Mutants lacking PehSR, a two-component positive regulator of polygalacturonase production, swimming motility, and twitching motility, are also nonmotile (1, 59). Mutants lacking the global virulence and stress regulator VsrAD are hypermotile, and mutants lacking VsrBC, a two-component positive regulator of swimming motility and EPS production, are nonmotile (65). To directly compare the phenotypic effects of these regulators on R. solanacearum motility, we assessed the swimming motility of single and double regulatory mutant strains on 0.3% agar plates. FlhDC and VsrBC appear to be phenotypically dominant over MotN since both flhDC motN (K725) and vsrBC motN (K728) double mutants were nonmotile, like flhDC (K702) and vsrBC (K796) single mutants. Interestingly, while a pehR (KB5) mutant was nonmotile and a motN mutant (K714) was hypermotile, a pehR motN (K726) double mutant had wild-type motility. The vsrAD motN (K727) double mutant had motility similar to that of the single mutants vsrAD (K791) and motN (K714), indicating that there was no addictive effect of vsrAD and motN on motility (Fig. 6).
Fig. 6.
Motility phenotypes of single and double regulatory mutants. Typical bacterial halos formed after 48 h of incubation at 28°C on semisolid 0.3% agar plates are shown. The number in each plate is the motility ratio, calculated as the halo diameter of the mutant divided by the halo diameter of wild-type strain K60. Experiments to determine the motility ratio were repeated three times, with each replicate containing at least three plates for each strain.
To determine how motN fits into the motility regulatory network, we used a gus transcriptional fusion to measure motN expression in strains lacking known motility regulators (Table 2). motN expression decreased about 2-fold when VsrAD was deleted (strain K721) at stationary phase or in minimal medium, indicating that under these conditions the global virulence and stress regulator VsrAD induced motN expression. The quorum-sensing global virulence regulator PhcA regulated motN expression in a cell density-dependent manner, inducing motN expression in log phase but inhibiting motN expression in stationary phase when cell density was high. When vsrBC was deleted (strain K723), motN expression decreased slightly, suggesting that VsrBC has a minor effect on MotN. Deletion of PehR (strain K721) did not affect motN expression at the transcriptional level (Table 2). We also measured expression of the motN::gus fusion in the wild-type and the above-mentioned mutant backgrounds when the bacteria were growing in tobacco leaves, but we did not detect motN::gus expression in planta in any background (data not shown). This indicates that once the pathogen is inside the plant, motN is no longer expressed.
Table 2.
Effect of various regulatory mutations on motN::gus expression in R. solanacearum
| Strain name | Genotype | Fold change in motN::gus expression (GUS activity) in the indicated medium (growth phase)d |
||
|---|---|---|---|---|
| CPG (log phase)a | CPG (S phase)b | BMM-0.2% glucose (log phase)c | ||
| K716 | K60 wild type | 1 (166.82 ± 5.54) | 1 (183.37 ± 13.39) | 1 (175.06 ± 4.29) |
| K721 | pehR | 0.89 (148.10 ± 3.22) | 0.95 (173.98 ± 10.28) | 1.10 (191.98 ± 15.91) |
| K722 | vsrAD | 0.96 (159.58 ± 19.61) | 0.38 (69.75 ± 5.90) | 0.51 (89.62 ± 14.62) |
| K723 | vsrBC | 0.58 (97.36 ± 18.74) | 0.62 (113.74 ± 22.17) | 0.84 (146.36 ± 8.90) |
| K724 | phcA | 0.45 (75.41 ± 20.02) | 1.80 (330.42 ± 33.14) | 0.86 (150.21 ± 29.76) |
Cells were grown in CPG rich broth to log phase (5 × 108 CFU/ml).
Cells were grown in CPG rich broth or to stationary phase (7.5 × 109 CFU/ml).
Cells were grown in BMM minimal broth to log phase (3 × 108 CFU/ml).
Fold change was determined relative to the wild-type strain under the same conditions. β-Glucuronidase (GUS) activity is expressed as 10−9 pmol of 4-methylumbelliferone produced per min per CFU by extracts from cultures of cis-merodiploid strains carrying a Tn7-motN::gus fusion. Data represent means ± standard errors from three biological experiments, each containing two replicates per strain.
FlhDC is a major regulator of flagellar biosynthesis and bacterial motility (59). We therefore determined the effect of various regulatory mutations on flhDC expression by measuring Gus activity levels from PflhDC::gus reporter gene fusions in diverse mutant backgrounds. Expression of flhDC was between 8- and 16-fold higher in the motN mutant (strain K826) than in the wild type (strain K821), depending on culture conditions (Table 3). flhDC::gus expression also increased 8- to 125-fold in a vsrAD strain (K822) and 6- to 88-fold in a vsrAD motN double mutant (strain K727) (Table 3). These data suggest that both motN and vsrAD negatively regulate transcription of flhDC, but they do not have additive effects, which is consistent with the phenotype on motility plates (Fig. 6). In contrast, flhDC expression was reduced 2- to 9-fold in a vsrBC mutant (strain K824) background, indicating that vsrBC positively regulates flhDC transcription. However, flhDC::gus expression was unchanged relative to the wild type in a vsrBC motN double mutant (strain K728) background. Interestingly, flhDC::gus expression was significantly higher in a pehR motN double mutant (strain K726) background than in the wild type (strain K821) or pehR single mutant (strain K827) background (Table 3) even though the pehR motN double mutant strain had wild-type motility on motility agar plates (Fig. 6).
Table 3.
Effect of various regulatory mutations on PflhDC::gus expression in R. solanacearum
| Strain name | Genotype | Fold change in PflhDC::gus expression by medium and growth phase (GUS activity)c |
|||
|---|---|---|---|---|---|
| CPGa |
BMM-0.2% glucoseb |
||||
| Log phase | S phase | Log phase | S phase | ||
| K821 | K60 wild type | 1 (154.27 ± 36.31) | 1 (214.20 ± 17.85) | 1 (52.91 ± 11.15) | 1 (115.52 ± 78.21) |
| K826 | motN | 8.57 (1,322.79 ± 54.44) | 8.13 (1,741.15 ± 104.87) | 16.71 (884.17 ± 372.72) | 11.49 (1,327.42 ± 492.68) |
| K827 | pehR | 1.41 (216.79 ± 42.06) | 0.73 (156.89 ± 11.02) | 0.92 (48.51 ± 13.58) | 0.35 (40.52 ± 14.19) |
| K726 | pehR motN | 14.68 (2,265.14 ± 509.49) | 8.96 (1,919.04 ± 66.76) | 35.95 (1,901.88 ± 333.10) | 12.16 (1,404.23 ± 169.71) |
| K822 | vsrAD | 8.18 (1,261.46 ± 392.91) | 12.97 (2,777.91 ± 489.20) | 73.19 (3,872.61 ± 1,058.06) | 124.74 (14,409.60 ± 1,930.99) |
| K727 | vsrAD motN | 6.69 (1,031.82 ± 167.81) | 8.26 (1,768.29 ± 462.08) | 60.08 (3,178.74 ± 919.83) | 88.28 (10,197.71 ± 1,462.11) |
| K824 | vsrBC | 0.25 (38.01 ± 8.59) | 0.12 (26.64 ± 11.41) | 0.49 (25.74 ± 4.96) | 0.25 (29.11 ± 7.91) |
| K728 | vsrBC motN | 1.10 (170.04 ± 64.64) | 0.98 (210.07 ± 25.53) | 0.68 (36.09 ± 7.31) | 0.79 (90.86 ± 11.41) |
Cells were grown in CPG rich broth to log phase(5 × 108 CFU/ml) or stationary(S) phase(7.5 × 109 CFU/ml).
Cells were grown in BMM minimal broth to log phase(3 × 108 CFU/ml) or to stationary(S) phase(3.5 × 109 CFU/ml).
Fold change was determined relative to the wild-type strain under the same conditions. β-Glucuronidase(GUS) activity is expressed as 10−9 pmol 4-methylumbelliferone produced per min per CFU by extracts from cultures of cis-merodiploid strains carrying a Tn7-motN::gus fusion. Data represent means ± standard errors from three biological experiments, each containing two replicates per strain.
DISCUSSION
The ability to move can be advantageous to bacteria. Motile cells may obtain more or better nutrients, avoid toxic or unfavorable environments, move to preferred hosts, and disperse more effectively (47). It is thus unsurprising that swimming motility is a virulence factor for many animal- and plant-pathogenic bacteria, such as Salmonella spp. (3), Vibrio spp. (41, 46), and Pseudomonas spp. (28, 34, 48). The bacterial wilt pathogen R. solanacearum needs motility for full virulence, apparently because motility facilitates the early stages of host plant invasion and colonization (60). A previously uncharacterized negative regulator of motility in R. solanacearum, named motN, is present and highly conserved in all R. solanacearum species complex strains sequenced to date. This suggests that motN contributes to adaptive fitness of this pathogen. Further, a motN deletion conferred an identical hypermotile phenotype on three phylogenetically diverse R. solanacearum strains: K60 (phylotype II, sequevar 7; historically known as race 1 biovar 1), UW551 (phylotype II, sequevar 1; historically known as race 3 biovar 2), and GMI1000 (phylotype I, sequevar 18; historically known as race 1 biovar 3) (data not shown). We predicted that the motN mutant would have increased virulence because a larger proportion of cells could move to and infect optimal sites of plant roots. This proved incorrect: the motN mutant was significantly reduced in virulence in a naturalistic soil soak assay. However, it caused wild-type disease levels when introduced directly into tomato stems, suggesting that motN, like motility in general, makes its contribution to virulence early in disease development. But what specific mechanism underlies this phenotype?
The motN mutant had more flagella per cell than the wild-type parent and also produced noticeably more flagellin protein, suggesting that the physical basis of the motN hypermotile phenotype is overproduction of flagella. This finding raised the possibility that the motN mutant had decreased virulence because its abundant flagellin triggered increased defense responses in the tomato host. We previously found that R. solanacearum flagellin did not elicit defense responses in Arabidopsis and tobacco, likely because it does not contain the typically conserved N-terminal flg22 motif, but the response of tomato to R. solanacearum flagellin was unknown (49). The similar levels of hydrogen peroxide in susceptible tomato leaves in response to R. solanacearum wild type and motN mutant infection suggest that the reduced virulence of motN is not a result of enhanced host defense response.
We also considered the possibility that the hypermotile motN mutant was reduced in virulence because of defective taxis behavior that caused it to move randomly rather than toward an optimal infection or attachment site. The chemotaxis ability of a motN mutant was indistinguishable from that of its wild-type parent in an in vitro assay using yeast extract, a known chemoattractant of R. solanacearum. However, we cannot rule out the possibility that MotN negatively regulates R. solanacearum motility in response to sensing a specific but unknown plant or environmental signal. This hypothesis could be tested by comparing the swimming behavior of marked wild-type and hypermotile cells interacting with tomato roots in soil.
Motility allows bacteria to efficiently migrate to and contact host cells. Attachment of bacteria to the plant cell surface is a critical early step for both host invasion and biofilm formation (16, 54). For example, motile strains of the rhizosphere bacteria Pseudomonas fluorescens and Pseudomonas putida PaW8 had a significant advantage in attachment to wheat roots over nonmotile strains (62, 63). It seemed possible that a highly motile bacterium would be less adherent, so we tested the hypothesis that poor attachment to host roots caused the reduced virulence of the hypermotile motN mutant. However, the motN mutant attached to tomato seedling roots as well as the wild type in both competitive and noncompetitive attachment assays, indicating that the motN mutant does not have an attachment defect.
In a naturalistic soil soak assay where the bacteria must locate and invade host plant roots from the soil, the motN mutant could not kill all the inoculated plants, unlike the wild type. However, the motN mutant had wild-type virulence when it was inoculated directly into the plant vascular system, and it had no colonization defect once it was inside host plants. These data, together with our observation that the motN mutant is nonmotile in planta and that motN is not expressed once the bacteria get inside the plant, suggest that motN makes its contribution to virulence early in disease development.
Another explanation for the virulence defect of the hyperflagellated and hypermotile motN mutant could be the metabolic cost associated with hyperflagellation and hypermotility. Synthesis and operation of swimming motility systems require a sizable investment of cellular energy reserves: many genes are expressed, some at very high levels. Moreover, powering flagellar rotation demands significant energy (38). In E. coli, about 2% of the cell's total energy is required to synthesize, regulate, and rotate flagellar organelles (37). Production of flagella by Photorhabdus luminescens also incurs a significant metabolic cost (21). Because flagellar synthesis and rotation are metabolically expensive, there are clearly conditions when the fitness advantage of motility is outweighed by the energetic cost of the behavior. However, hypermotility apparently does not impose a cost large enough to measurably reduce the competitive fitness of the motN mutant strain during either root attachment or stem colonization. This therefore does not seem the most likely reason for the strain's reduced virulence.
The hypermotile motN mutant formed weaker biofilms on PVC plates than its wild-type parent. Biofilms play important roles in many bacteria-host interactions, and this behavior may contribute to the persistence and pathogenicity of R. solanacearum (16, 42). R. solanacearum forms biofilm-like aggregations on tomato seedling roots (35, 66), but the factors underlying this behavior and the importance of biofilms later in wilt pathogenesis are still unknown. It has been proposed that flagella might act both as surface adhesins and as providers of force-generating motility during biofilm formation. Flagellum-mediated motility is required for initial attachment and/or subsequent biofilm formation in several Gram-negative bacteria (44, 45, 50, 66). The xylem-dwelling plant pathogen Xylella fastidiosa forms thick biofilms on host xylem vessel walls, occluding water transport and causing water stress (19, 20). Like X. fastidiosa, R. solanacearum also inhabits the xylem vessel, a highly turbulent, negative-pressure environment. Biofilms could increase pathogen success by anchoring cells to vessel walls, protecting bacteria from antimicrobial host defenses, and facilitating nutrient filtration from the dilute flow of xylem fluid. Thus, reduced biofilm production may explain the lower virulence of the hypermotile motN mutant.
Hypermotile lsr2 mutants of Mycobacterium smegmatis could not form biofilms (6), consistent with our finding that enhanced motility can lead to decreased biofilm formation. Further, lack of motility increases biofilm formation in R. solanacearum. We observed that mutants lacking flagella (fliC), taxis in general (cheW), and aerotaxis in particular (aer1 and aer2), all formed significantly more biofilm on static PVC wells than the wild-type parent strain (67). In contrast, nonmotile mutants of another soil-borne plant pathogen, Agrobacterium tumefaciens, were deficient in biofilm formation under static conditions. However, in a flow cell, an aflagellate mutant rapidly formed aberrantly dense, tall biofilms, while a flagellated but paralyzed mutant formed weak, thin biofilms (39). The complex and sometimes conflicting results observed in static and flow cell studies of other species suggest that biofilm formation is significantly affected by assay conditions. Thus, microscopy studies of R. solanacearum cells inside xylem vessels of naturally infected host plants are needed to understand the biological role of this behavior during mid- and end-stage bacterial wilt disease.
Given the metabolic expense of swimming motility, it is not surprising that bacteria regulate it tightly and redundantly. Collectively, the data presented here and previously indicate that expression of R. solanacearum swimming motility is finely tuned to optimize bacterial fitness. Reducing, increasing, or altering the timing of motility expression all decrease bacterial wilt virulence. In R. solanacearum, motility is controlled by a complex interlocking regulatory cascade. It is negatively regulated by PhcA and VsrAD and positively regulated by VsrBC, PehSR, and FlhDC (1, 11, 32, 59, 65). In this study we found that motN negatively regulates expression of the master motility regulator FlhDC at the transcriptional level. In turn, expression of motN is positively regulated by vsrAD at stationary phase or in minimal medium when the conditions are stressful. The PhcA quorum-sensing system increases motN expression at low cell density but negatively regulates it at high cell density; this correlates with the previously described effect of cell density on motility itself (15). The generally 2-fold or smaller effect on motN expression by the regulators tested suggests that these factors rather weakly influence motN at the transcriptional level. Reporter gene experiments indicated that mutating pehR had no effect on motN transcription (Table 2). However, a pehR motN double mutant had wild-type motility, an intermediate phenotype between that of the nonmotile pehR strain and the hypermotile motN strain (Fig. 6). Expression of flhDC was significantly increased in a pehR motN double mutant background (Table 3), suggesting that MotN-mediated overexpression of FlhDC may have compensated for the motility defect caused by the absence of PehR. We previously reported that PehSR is required for flhDC expression in minimal medium (59), but here we found that, except in minimal medium at stationary phase, flhDC expression was almost equal to that of the wild type in a pehR mutant background (Table 3). This discrepancy may result from a slightly different minimal medium or, more likely, from the use of a multicopy plasmid-borne flhDC::gus fusion in the previous study as opposed to the chromosomal single-copy fusion used here. Our data also suggest that there are additional unidentified motility regulators in R. solanacearum, and these may interact with MotN and other known regulators to coregulate motility in R. solanacearum. Experiments measuring motN-related gene expression during pathogen interactions with host roots and wilt disease are needed to fully understand the complex regulation of motility in this bacterium.
Hypermotility is the most likely explanation for the reduced virulence of motN mutants. Nonetheless, our data do not rule out the possibility that the motN mutation has pleiotropic effects that reduce virulence by affecting expression or production of one or more virulence factors not associated with swimming motility. However, the motN gene is located within a large cluster of motility genes, and hypermotility is the most apparent phenotype of motN mutants; R. solanacearum mutants lacking the phcA and vsrAD regulators are also hypermotile. But phcA and vsrAD mutants are affected in several traits, such as EPS and extracellular enzyme production, while the motN mutants are altered only in motility and in biofilm formation, a behavior consistently linked with motility. Additional genes that might be regulated by motR could be identified by comparing the transcriptome profiles of wild-type and motN strains.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Research Initiative of the USDA-CSREES (Plant Biosecurity project 2006-04560), by a fellowship from the Storkan-Hanes-MacCaslin Foundation to F. Meng, and by the University of Wisconsin—Madison College of Agricultural and Life Sciences.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Allen C., Gay J., Simon-Buela L. 1997. A regulatory locus, pehSR, controls polygalacturonase production and other virulence functions in Ralstonia solanacearum. Mol. Plant Microbe Interact. 10:1054–1064 [DOI] [PubMed] [Google Scholar]
- 2. Allen C., Huang Y., Sequeira L. 1991. Cloning of genes affecting polygalacturonase production in Pseudomonas solanacearum. Mol. Plant Microbe Interact. 4:147–154 [Google Scholar]
- 3. Allen-Vercoe E., Sayers A. R., Woodward M. J. 1999. Virulence of Salmonella enterica serotype Enteritidis aflagellate and afimbriate mutants in a day-old chick model. Epidemiol. Infect. 122:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Araud-Razou I., Vasse J., Montrozier H., Etchebar C., Trigalet A. 1998. Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur. J. Plant Pathol. 104:795–809 [Google Scholar]
- 5. Arlat M., et al. 1992. Transcriptional organization and expression of the large hrp gene cluster in Pseudomonas solanacearum. Mol. Plant Microbe Interact. 5:187–193 [DOI] [PubMed] [Google Scholar]
- 6. Arora K., Whiteford D. C., Lau-Bonilla D., Davitt C. M., Dahl J. L. 2008. Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. J. Bacteriol. 190:4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ausubel F., et al. 1995. Short protocols in molecular biology, 3rd ed. John Wiley and Sons, New York, NY [Google Scholar]
- 8. Boucher C., Barberis P., Trigalet A., Demery D. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449–2457 [Google Scholar]
- 9. Brown D., Allen C. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53:1641–1660 [DOI] [PubMed] [Google Scholar]
- 10. Brumbley S. M., Carney B. F., Denny T. P. 1993. Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of phcA, a putative lysR transcriptional regulator. J. Bacteriol. 175:5477–5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brumbley S. M., Denny T. P. 1990. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J. Bacteriol. 172:5677–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchanan R. E., Gibbons N. E.(ed.). 1974. Bergey's manual of determinative bacteriology, 8th ed. Williams and Wilkins Co., Baltimore, MD [Google Scholar]
- 13. Choi K.-H., et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 14. Clough S., Lee K.-E., Schell M., Denny T. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates the production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clough S. J., Flavier A. B., Schell M. A., Denny T. P. 1997. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl. Environ. Microbiol. 63:844–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danhorn T., Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61:401–422 [DOI] [PubMed] [Google Scholar]
- 17. Denny T., Baek S. 1991. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 4:198–206 [Google Scholar]
- 18. Denny T. P. 2006. Plant pathogenic Ralstonia species, p. 573–644In Gnanamanickam S. S. (ed.), Plant-associated bacteria. Springer Publishing, Dordrecht, Netherlands [Google Scholar]
- 19. de Souza A. A., et al. 2004. Gene expression profile of the plant pathogen Xylella fastidiosa during biofilm formation in vitro. FEMS Microbiol. Lett. 237:341–353 [DOI] [PubMed] [Google Scholar]
- 20. de Souza A. A., Takita M. A., Pereira E. O., Coletta H. D., Machado M. A. 2005. Expression of pathogenicity-related genes of Xylella fastidiosa in vitro and in planta. Curr. Microbiol. 50:223–228 [DOI] [PubMed] [Google Scholar]
- 21. Easom C. A., Clarke D. J. 2008. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol. 8:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Felix G., Duran J. D., Volko S., Boller T. 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18:265–276 [DOI] [PubMed] [Google Scholar]
- 23. Flavier A. B., Clough S. J., Schell M. A., Denny T. P. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26:251–259 [DOI] [PubMed] [Google Scholar]
- 24. Flores-Cruz Z., Allen C. 2009. Ralstonia solanacearum encounters an oxidative environment during tomato infection. Mol. Plant Microbe Interact. 22:773–782 [DOI] [PubMed] [Google Scholar]
- 25. Genin S., Boucher C. 2002. Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Mol. Plant Pathol. 3:111–118 [DOI] [PubMed] [Google Scholar]
- 26. Gonzalez E. T., Allen C. 2003. Characterization of a Ralstonia solanacearum operon required for polygalacturonate degradation and uptake of galacturonic acid. Mol. Plant Microbe Interact. 16:536–544 [DOI] [PubMed] [Google Scholar]
- 27. Guidot A., et al. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hatterman D. R., Ries S. M. 1989. Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology 79:284–289 [Google Scholar]
- 29. Hayward A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65–87 [DOI] [PubMed] [Google Scholar]
- 30. Hendrick C. A., Sequeira L. 1984. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horton R. M., Hund H. D., Ho S. N., Ullen J. K., Pease L. R. 1989. Engineering hybrid genes without the use of restriction enzymes; gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 32. Huang J., Carney B. F., Denny T. P., Weissinger A. K., Schell M. A. 1995. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J. Bacteriol. 177:1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang J., Schell M. 1990. Evidence that extracellular export of the endoglucanase encoded by egl of Pseudomonas solanacearum occurs by a two-step process involving a lipoprotein intermediate. J. Biol. Chem. 265:11628–11632 [PubMed] [Google Scholar]
- 34. Josenhans C., Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605–614 [DOI] [PubMed] [Google Scholar]
- 35. Kang Y., Liu H., Genin S., Schell M. A., Denny T. P. 2002. Ralstonia solanacearum requires type-4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427–437 [DOI] [PubMed] [Google Scholar]
- 36. Kelman A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in tetrazolium medium. Phytopathology 44:693–695 [Google Scholar]
- 37. Macnab R. M. 1996. Flagella and motility, p. 123–145In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC [Google Scholar]
- 38. McCarter L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180–186 [DOI] [PubMed] [Google Scholar]
- 39. Merritt P. M., Danhorn T., Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189:8005–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Milton D., O'Toole R., Horstedt P., Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morris C. E., Monier J.-M. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41:429–453 [DOI] [PubMed] [Google Scholar]
- 43. Oke V., Long S. R. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837–849 [DOI] [PubMed] [Google Scholar]
- 44. O'Toole G., Kaplan H. B., Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79 [DOI] [PubMed] [Google Scholar]
- 45. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 46. O'Toole R., Milton D. L., Wolf-Watz H. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625–637 [DOI] [PubMed] [Google Scholar]
- 47. Ottemann K. M., Miller J. F. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109–1117 [DOI] [PubMed] [Google Scholar]
- 48. Panapoulos N. J., Schroth M. N. 1974. Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 64:1389–1397 [Google Scholar]
- 49. Pfund C., et al. 2004. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17:696–706 [DOI] [PubMed] [Google Scholar]
- 50. Pratt L., Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type 1 pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 51. Prentki P., Krisch H. M. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313 [DOI] [PubMed] [Google Scholar]
- 52. Prior P., Allen C., Elphinstone J.(ed.). 1998. Bacterial wilt disease: molecular and ecological aspects. Springer Verlag, Berlin, Germany [Google Scholar]
- 53. Remenant B., et al. 2010. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rodriguez-Navarro D. N., Dardanelli M. S., Ruiz-Sainz J. E. 2007. Attachment of bacteria to the roots of higher plants. FEMS Microbiol. Lett. 272:127–136 [DOI] [PubMed] [Google Scholar]
- 55. Schell M. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597–626 [DOI] [PubMed] [Google Scholar]
- 56. Schell M. A. 2000. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38:263–292 [DOI] [PubMed] [Google Scholar]
- 57. Schell M. A. 1996. To be or not to be: how Pseudomonas solanacearum decides whether or not to express virulence genes. Eur. J. Plant Pathol. 102:459–469 [Google Scholar]
- 58. Sun W., Dunning F. M., Pfund C., Weingarten R., Bent A. F. 2006. Within-species flagellin polymorphism in Xanthomonas campestris pv. campestris and its impact on elicitation of Arabidopsis FLS2-dependent defenses. Plant Cell 18:764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tans-Kersten J., Brown D., Allen C. 2004. Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant Microbe Interact. 17:686–695 [DOI] [PubMed] [Google Scholar]
- 60. Tans-Kersten J., Huang H., Allen C. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Totten P. A., Lory S. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Turnbull G. A., Morgan J. A., Whipps J. M., Saunders J. R. 2001. The role of bacterial motility in the survival and spread of Pseudomonas fluorescens in soil and in the attachment and colonisation of wheat roots. FEMS Microbiol. Ecol. 36:21–31 [DOI] [PubMed] [Google Scholar]
- 63. Turnbull G. A., Morgan J. A., Whipps J. M., Saunders J. R. 2001. The role of motility in the in vitro attachment of Pseudomonas putida PaW8 to wheat roots. FEMS Microbiol. Ecol. 35:57–65 [DOI] [PubMed] [Google Scholar]
- 64. Vasse J., Frey P., Trigalet A. 1995. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant Microbe Interact. 8:241–251 [Google Scholar]
- 65. Yao J. 2007. The role of bacterial taxis in Ralstonia solanacearum-host interactions. Ph.D. thesis. University of Wisconsin—Madison, Madison, WI [Google Scholar]
- 66. Yao J., Allen C. 2006. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 188:3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yao J., Allen C. 2007. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189:6415–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu H. S., Alam M. 1997. An agarose-in-plug bridge method to study chemotaxis in the archaeon Halobacterium salinarum. FEMS Microbiol. Lett. 156:265–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.