Abstract
The Rhodopseudomonas palustris transcriptional regulator RpaR responds to the RpaI-synthesized quorum-sensing signal p-coumaroyl-homoserine lactone (pC-HSL). Other characterized RpaR homologs respond to fatty acyl-HSLs. We show here that RpaR functions as a transcriptional activator, which binds directly to the rpaI promoter. We developed an RNAseq method that does not require a ribosome depletion step to define a set of transcripts regulated by pC-HSL and RpaR. The transcripts include several noncoding RNAs. A footprint analysis showed that purified His-tagged RpaR (His6-RpaR) binds to an inverted repeat element centered 48.5 bp upstream of the rpaI transcript start site, which we mapped by S1 nuclease protection and primer extension analyses. Although pC-HSL-RpaR bound to rpaI promoter DNA, it did not bind to the promoter regions of a number of RpaR-regulated genes not in the rpaI operon. This indicates that RpaR control of these other genes is indirect. Because the RNAseq analysis allowed us to track transcript strand specificity, we discovered that there is pC-HSL-RpaR-activated antisense transcription of rpaR. These data raise the possibility that this antisense RNA or other RpaR-activated noncoding RNAs mediate the indirect activation of genes in the RpaR-controlled regulon.
INTRODUCTION
Many bacteria control subsets of genes in a cell density-dependent manner. This coordinated group behavior is known as quorum sensing and response. More than 100 species of Proteobacteria contain acyl-homoserine lactone (acyl-HSL) quorum-sensing (QS) circuits (12, 45). Acyl-HSLs can diffuse into and out of cells, and once a threshold concentration is reached, acyl-HSLs bind specific transcriptional regulators that control target genes. A variety of genes are controlled by QS depending on the bacterial species, including protease genes, conjugal transfer genes, antibiotic synthesis genes, and bioluminescence genes (12, 45). Many QS-regulated gene products are “public goods,” exoproducts that can be shared by all of the individuals in a group.
Two types of genes are involved in most acyl-HSL-type QS systems: luxI- and luxR-type genes. LuxI proteins are QS signal synthases that catalyze amide bond formation between an acyl group on an appropriate side chain donor (most often acyl-acyl carrier protein) and S-adenosylmethionine (SAM) resulting in the final acyl-HSL product (22, 26, 34, 35). For fatty acyl-HSLs, signal specificity is conferred by the length (4 to 18 carbons) and side chain modifications of the fatty acyl group. LuxR homologs are homodimeric transcription factors, with each monomer consisting of two domains: an N-terminal acyl-HSL binding domain and a C-terminal DNA-binding domain that contains a helix-turn-helix motif (5, 6, 13, 50). Genes controlled by LuxR homologs often have specific inverted repeat DNA sequences in their promoter regions. These elements are known as lux box-like sequences. The lux box is a 20-bp palindromic sequence centered at bp −42.5 from the transcription start of the Vibrio fischeri lux operon, which encodes the luminescence functions (7, 9). With its cognate acyl-HSL, LuxR binds to the lux box and facilitates RNA polymerase binding (43). The number of genes controlled by LuxR-LuxI type proteins varies among systems. For example in V. fischeri only 25 (0.6% of total) genes are LuxR-3-oxo-hexanoyl-HSL-controlled (3), whereas about 350 genes (6% of total) are QS-controlled in the opportunistic pathogen, Pseudomonas aeruginosa (37, 44).
Recently, we identified a novel HSL-type QS signal in Rhodopseudomonas palustris (34). We found that this phototrophic purple nonsulfur bacterium uses a LuxI-type acyl-HSL synthase, RpaI, to produce p-coumaroyl-homoserine lactone (pC-HSL) rather than a fatty acyl-homoserine lactone. The p-coumaroyl side chain is derived from an exogenously provided plant metabolite, p-coumarate, rather than from endogenous fatty acid synthesis intermediates, as is the case for other acyl-HSL signaling systems. Like fatty acyl-HSL QS systems, pC-HSL gene control appears to depend on a LuxR homolog, RpaR. Microarray experiments showed that in R. palustris, pC-HSL influenced the expression of at least 17 genes, including the rpaI gene.
To further understand pC-HSL regulation, we studied RpaR activity. We show here that RpaR functions as an activator and that purified RpaR-pC-HSL can bind the promoter of the rpaI operon at a specific lux box-like sequence. However, RpaR does not appear to bind promoter regions of other genes influenced by pC-HSL and RpaR. Thus, we also used an RNAseq approach to identify transcripts controlled not only by pC-HSL but also by RpaR specifically. This enabled a confirmation and extension of previous results, demonstrated that pC-HSL-regulated genes are also RpaR regulated, and demonstrated that there is novel rpaR antisense transcript activated by pC-HSL.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacteria used are described in Table 1. All R. palustris strains were grown photoheterotrophically in photosynthetic medium with 10 mM succinate (PM-succinate) as described elsewhere (16). p-Coumarate was added at a concentration of either 0.5 or 1 mM, as indicated. For growth of P. aeruginosa and Escherichia coli, we used Luria-Bertani broth containing 50 mM morpholinepropanesulfonic acid (MOPS; pH 7.0). Antibiotics were added to growth medium at the following concentrations (per ml): 100 μg of gentamicin for R. palustris and 200 μg of carbenicillin and 50 μg of gentamicin for P. aeruginosa. pC-HSL (Syntech Solutions, San Diego, CA) was added as indicated.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| R. palustris | ||
| CGA009 | Wild type | 18 |
| CGA850 | rpaR mutant | This study |
| CGA814 | rpaI::lacZ chromosomal fusion; Kmr | 34 |
| P. aeruginosa PAO-T7-MW1 | T7 expression strain; lasI rhlI | 14 |
| E. coli S17-1 | recAthipro | 40 |
| Plasmids | ||
| pJQ200KS | Suicide vector, sacB; Gmr | 27 |
| pJLQhis | N-terminal His-QscR overexpression vector; Apr | 19 |
| pQF5016b.rpaR | N-terminal His-RpaR overexpression vector; Apr | This study |
| pQF5016b.rpaRD82N | N-terminal His-RpaR D82N mutant overexpression vector; Apr | This study |
| pBBR1MCS-5 | Broad-host-range vector; Gmr | 17 |
| pBBR1MCS-5lacZ | pBBR1MCS-5 with promoterless lacZ; Gmr | 34 |
| pBBR-PrpaI-lacZ | rpaI promoter reporter; Gmr | 34 |
| pBBR-RpaR-PrpaI-lacZ | rpaI promoter reporter plus rpaR; Gmr | 34 |
| pBBR-LB1mut | rpaI promoter w/mutated lux box-1 in pBBR1MCS5-lacZ; Gmr | This study |
| pBBR-LB2mut | rpaI promoter w/mutated lux box-2 in pBBR1MCS5-lacZ; Gmr | This study |
| pBBR-LB12mut | rpaI promoter w/mutated lux box-1 and -2 in pBBR1MCS5-lacZ; Gmr | This study |
Gmr, gentamicin resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance.
Construction of an rpaR deletion mutant.
The rpaR in-frame deletion mutant CGA850 was constructed by sequence overlap extension PCR. The DNA sequences of the primers used for this and all other primers used in the present study are provided in Table S1 in the supplemental material. The flanking DNA included about 500 bp upstream of rpaR, as well as the first three rpaR codons. The downstream DNA included the last three rpaR codons (including the stop codon) and about 500 bp of DNA downstream of rpaR. The product was ligated with the suicide vector pJQ200KS (27) and introduced into R. palustris CGA009 by conjugation with E. coli S17-1 as described previously (25, 29, 34). We confirmed the mutation in a sucrose-resistant, gentamicin-sensitive isolate by PCR analysis and DNA sequencing.
RNA extraction and RT-PCR.
RNA extractions and reverse transcription-PCR (RT-PCR) analyses were performed according to previously published protocols (25, 29, 34). Briefly, R. palustris cultures were grown to an optical density at 600 nm (OD660) of 0.8 in PM-succinate and diluted to an OD660 of 0.06 in fresh PM-succinate medium containing 0.5 mM p-coumarate. After 24 h (OD660 of 0.25 to 0.35), the cells were harvested, and the total RNA was extracted and purified by using an RNeasy minikit (Qiagen). Real-time PCRs included 1 ng of cDNA and 200 nM primers in 25 μl of SYBR green PCR amplification master mix (Applied Biosystems). Genomic DNA was used as a standard, and the constitutive fixJ (rpa4248) transcript was used as an internal control.
Plasmid construction.
To construct the His6-tagged rpaR expression vector pQF5016b.rpaR, the rpaR open reading frame (ORF) was PCR amplified from genomic DNA with primers containing NcoI and BglII restriction sites. After digestion with NcoI and BglII, the PCR product was ligated with NcoI- and BamHI-digested pJLQhis. The D82N mutant plasmid was constructed by a site-directed mutagenesis of pQF5016b.rpaR. The mutation was generated by using Pfu polymerase PCR and a QuikChange site-directed mutagenesis kit (Stratagene). To construct the mutant rpaI promoter plasmids, promoter fragments were PCR amplified and ligated with ApaI-NheI-digested pBBR1MCS-5lacZ (34). Constructs were confirmed by DNA sequencing.
Assessing RpaR solubility.
P. aeruginosa PAO-T7MW1 containing pQF5016b.rpaR was grown at 37°C to an OD600 of 0.5 in LB broth containing 50 mM MOPS and 20 μM pC-HSL and then chilled to 16°C. IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and culture growth was continued at 16°C for 16 h. Cells were harvested by centrifugation, and the pellet was suspended in buffer containing 20 mM Tris (pH 7.9), 500 mM NaCl, and 20 μM pC-HSL, and the cells were broken by sonication. The cell extract was clarified by centrifugation (18,000 × g), and the resulting pellet and supernatant fractions were separated by SDS-12% PAGE. The gels were stained with Coomassie brilliant blue dye. The histidine-tagged portion of His6-RpaR was detected by Western blot analysis with a SuperSignal West HisProbe kit (Pierce Protein Research Products).
Purification of His6-RpaR.
Clarified cell extracts of P. aeruginosa PAO-T7-MW1 (pQF5016b.rpaR) were prepared as described above. We used P. aeruginosa as an expression host because it has a high genomic GC content similar to that of R. palustris. The cell extract was mixed with Ni-NTA agarose (Qiagen) for 1 h. The agarose was pelleted (1,000 rpm) and washed with increasing amounts of imidazole (3 volumes each of 0, 20, 50, and 100 mM imidazole), and then His6-RpaR was eluted with 500 mM imidazole. Purified His6-RpaR was dialyzed (1-kDa cutoff) against buffer consisting of 20 mM Tris (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 10% glycerol, and 20 μM pC-HSL. The final yield of His6-RpaR obtained from a 150-ml culture was 44 μg.
Measurement of pC-HSL.
To compare pC-HSL levels in the wild-type and rpaR mutant strains, cells were grown to late logarithmic phase, diluted to an OD660 of 0.04 in fresh PM-succinate medium containing 1 mM p-coumarate, and grown photoheterotrophically for 20 h (OD660 of 0.3 to 0.4). Whole-cell cultures were extracted twice with ethyl acetate containing 0.1 ml of glacial acetic acid per liter, the solvent was removed by evaporation under a stream of nitrogen gas, and pC-HSL levels were measured by using a bioassay that specifically responds to pC-HSL (34). To measure pC-HSL levels associated with His6-RpaR (purified as described above except that pC-HSL was omitted from the buffer), 500 pmol of protein (estimated by Bradford assay) was digested with 6 μg of proteinase K in 500 μl of buffer for 1 h at 37°C. pC-HSL was extracted three times with equal volumes of acidified ethyl acetate and measured by using the pC-HSL bioassay described above.
EMSAs.
To assess RpaR binding to DNA in electrophoretic mobility shift assays (EMSAs), we used a 103-bp DNA fragment (including the entire 86-bp rpaR-rpaI intergenic region) as the probe. The probe was generated by PCR amplification with pBBR-RpaR-PrpaI-lacZ (Table 1) as a template. The DNA fragments (0.30 pmol) were mixed with His6-RpaR (0 to 16 pmol) in a 10-μl reaction mixture containing 20 mM Tris (pH 7.5), 50 mM KCl, 10% glycerol, 1 mM dithiothreitol, and 20 μM pC-HSL. After 20 min at room temperature, the samples were separated by electrophoresis on a 5% nondenaturing acrylamide-Tris-glycine-EDTA gel in Tris-glycine-EDTA buffer at 4°C. Gels were soaked in 10,000-fold-diluted SYBR green I nucleic acid stain (Lonza Group, Ltd.), and DNA was visualized under UV light at 300 nm. The same protocol was used to determine which of the lux box-like elements was required for RpaR binding, except that the DNA probes contained nucleotide substitutions as indicated and were generated by PCR amplification with pBBR-LB1mut, pBBR-LB2mut, or pBBR-LB12mut (Table 1) as templates.
To test for RpaR binding to the other pC-HSL-regulated genes (34), we followed a similar protocol except that probes specific for the promoter of each gene were generated by PCR amplification with genomic R. palustris CGA009 DNA as a template. For each probe, the entire intergenic region was amplified except where indicated. The DNA probe sizes were as follows: rpa0905, 152 bp; rpa1096, 198 bp; rpa1098, 200 bp; rpa1674, 179 bp; rpa1845, 336 bp; rpa2519, 100 bp; rpa2883, 132 bp; rpa3185, 200 bp of the 905-bp intergenic region was used; rpa3306, 266 bp; rpa3599, 139 bp; rpa3892, 200 bp of the 480-bp intergenic region was used; rpa3929, 309 bp; rpa4296, 171 bp; rpa4684, 200 bp of the 559-bp intergenic region was used; and rpa4713, 246 bp. For pC-HSL-regulated genes newly identified by “not-so-random” (NSR) RNAseq analysis (rpa0745 and rpa1876), the DNA probes consisted of the 300 bp upstream of the start codon.
DNase I footprinting.
The DNase I footprint analysis was performed by using a previously described nonradiochemical capillary electrophoresis method (48). A 6-FAM-labeled (5′), 239-bp DNA fragment (starting 186 bp upstream of the rpaI ORF and ending 53 bp downstream of the ORF) was generated by PCR amplification. The DNA fragment (0.45 pmol) was mixed with purified His6-RpaR in a 50-μl reaction mixture containing the buffer used for gel shift assays. After 20 min at room temperature, reaction mixtures were treated for 1 min with DNase I (0.3 U; Promega). Samples were purified for GeneScan sequencing analysis and separated by using an ABI Prism genetic analyzer equipped with an ABI Prism GeneScan (PE Applied Biosystems). DNA fragment sizes were determined by using an ABI peak scanner software.
S1 nuclease protection assays.
The probe was generated by PCR amplification with genomic DNA as a template, a 6-FAM-labeled forward primer, and unlabeled reverse primer (455 bp upstream of the rpaI ORF). The probe was denatured by heating for 10 min at 95°C and then immediately chilled. RNA (30 μg) from logarithmic-phase (19-h) or stationary-phase (31-h) cells was mixed with 0.1 pmol of 6-FAM-labeled probe in hybridization buffer (38 mM HEPES [pH 7.0], 0.3 M NaCl, 1 mM EDTA, and 0.01% Triton X-100) for 16 h at 55°C. Hybridization products were digested for 30 min at 37°C by using S1 nuclease (100 U; Promega Corp.) and purified for GeneScan sequencing analysis as described above.
Primer extension analyses.
We annealed 20 μg of total RNA from logarithmic-phase (19-h) or stationary-phase (31-h) cultures with 10 nM a 6-FAM-labeled primer complementary to a region 63 to 83 bp downstream of the rpaI translation start site. Reverse transcription was performed with SuperScript III reverse transcriptase (Invitrogen). After RNase H digestion of RNA, cDNA was purified for GeneScan sequencing analysis as described above.
NSR RNAseq analysis.
RNA was purified from mid-logarithmic-phase cells, which were chilled in an ice-water bath, harvested by centrifugation, frozen in liquid nitrogen, and stored at −80°C. Thawed cells were resuspended in QIAzol lysis reagent and disrupted by bead beating. RNA was purified from the cell lysate by using a miRNeasy minikit (Qiagen) according to the manufacturer's instructions. RNA was treated with Turbo DNase (Ambion) and purified with a RNeasy MinElute cleanup kit (Qiagen).
First-strand and second-strand cDNA synthesis, NSR library construction, and sequencing with an Illumina GA2 were performed as described previously (4). The R. palustris-specific NSR primers were based on the genome sequences of six R. palustris strains (CGA009, TIE-1, Ha2, BisB5, BisB18, and BisA53) (24). We used a pool of 1,203 NSR hexamers with no perfect match to any rRNA transcripts. NSR hexamers were synthesized individually with a 5′ amplification-annealing site for first-strand (5′-TCCGATCTCTTAN-NSR hexamer reverse complement-3′) and second strand (5′-TCCGATCTGAN-NSR hexamer-3′) priming events and pooled prior to library construction. An additional set of 278 hexamers, which caused the majority of rRNA priming events in test NSR libraries, was further removed leaving a final set of 925 NSR hexamers. The NSR hexamer sequences of the final primer pools for the first and second strand priming events are given in Table S2 in the supplemental material. One limitation of our approach is that coverage biases introduced during cDNA synthesis make it difficult to define operon structures with precision. In particular, the 5′ universal sequences attached to each NSR hexamer can enhance the priming efficiency at complementary template sites and reduce the uniformity of read distribution across individual transcripts.
Sequence analysis.
Raw sequencing reads (31 nucleotides in length) were first filtered based on their PHRED quality scores. Reads having a base-call accuracy of ≥99% were selected for alignment with the R. palustris CGA009 genomic sequence. The alignment was performed by using the Burrows-Wheeler alignment tool (20). Reads that mapped to the R. palustris genome sequence were categorized as (i) uniquely mapped, (ii) partially mapped, or (iii) nonuniquely mapped as described in Table S3 in the supplemental material. Only the uniquely mapped reads, defined as those that map to a nonduplicated genome locations with ≤2 of 31 mismatches with the genome, were subjected to further analysis. The number of raw reads overlapping each gene was recorded and normalized based on reads per million uniquely mapped reads (RPM). Differentially expressed features were identified by using the DESeq statistical package (http://www-huber.embl.de/users/anders/DESeq/), which applied the Benjamini-Hochberg procedure for adjusting P values (Padj). Features with a Padj of ≤0.05 and with fold change ratios ≥ 2.5 were considered to be differentially expressed. All statistical tests were performed by using R project software version 2.10.1 (www.r-project.org) and Bioconductor version 2.6 (www.bioconductor.org). The DNA raw sequencing reads have been deposited in the NCBI Gene Expression Omnibus under the accession number GSE27365 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27365).
Visualization of mapped reads.
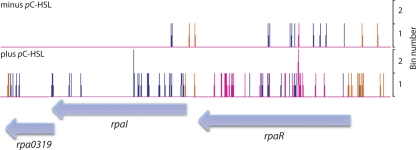
In order to visualize the location, number, and type of DNA sequence reads, we developed a program for generating customized plots, which can be opened with Artemis sequence visualization software as described at ftp.sanger.ac.uk/pub4/resources/software/artemis/artmis.pdf (33). The plots displayed the number and category (sense, antisense, intergenic positive DNA strand, or intergenic negative DNA strand) of DNA sequence reads at the R. palustris CGA009 genome location where the DNA sequence read begins (for an example, see Fig. 5). Because it is difficult to display widely varying numbers in a single graph, the normalized number of DNA sequence reads were first placed into six bins: (bin 1) 0 to 10, (bin 2) 11 to 100, (bin 3) 101 to 1,000, (bin 4) 1,001 to 10,000, (bin 5) 10,001 to 100,000, and (bin 6) 100,001 to 1,000,000. The bin number was then used by Artemis to draw a vertical line representing the actual number of reads at each genomic position. For instance, if there were 3,024 uniquely aligned DNA reads that begin at genome bp 85, it would be placed in bin 4, and Artemis would draw a vertical line of length 4 at position 85. The type of DNA sequence read is indicated by color as follows: sense reads, blue; antisense reads, pink; intergenic regions on the positive DNA strand, orange; and intergenic regions on the negative DNA strand, green.
Fig. 5.
Comparison of RNAseq profiles of the rpaI-rpaR genomic region from cells grown in the absence (top) or presence (bottom) of pC-HSL. Numbers of DNA sequence reads were binned by their normalized values and their start location was overlaid on the R. palustris CGA009 genome using Artemis software as described in Materials and Methods. The type of DNA sequence read is indicated by line color: sense reads, blue; antisense reads, pink; IG regions on the positive DNA strand, orange; and IG regions on the negative DNA strand, green. The normalized read values (reported as reads per million uniquely mapped reads [RPM]) for sense, antisense, IG-positive, and IG-negative reads were, respectively, as follows: for rpa0319 without pC-HSL, 0, 0, 0, and 0 RPM; for rpa0319 with pC-HSL, 8.0, 0, 7.7, and 0; for rpaI without pC-HSL, 1.6, 0, 0, and 0 RPM; for rpaI with pC-HSL 91.3, 0, 0, and 0 RPM; for rpaR without pC-HSL, 6.1, 0.4, 0, and 4.9 RPM; and for rpaR with pC-HSL, 7.6, 67.7, 8.0, and 0 RPM.
RESULTS
An R. palustris rpaR mutant has reduced rpaI expression and pC-HSL production.
Our previous work (34) suggested that RpaR might function as a repressor of the pC-HSL-controlled rpaI promoter and that pC-HSL served as a derepressor. To further understand RpaR regulation of gene expression, we constructed an R. palustris rpaR mutant (see Materials and Methods) and examined rpaI expression in this strain by using real-time PCR (Fig. 1A). Unlike the wild-type strain, the rpaR mutant did not show elevated rpaI expression when grown in the presence of p-coumarate or pC-HSL (Fig. 1A). The rpaR mutant phenotype was complemented with a wild-type rpaR allele. These results provide genetic evidence supporting an activator role rather than the suspected repressor function for RpaR (34). Consistent with the lack of rpaI induction in the rpaR mutant, pC-HSL levels were ca. 5% of wild-type levels (Fig. 1B).
Fig. 1.
An rpaR mutant is impaired in rpaI transcription and pC-HSL synthesis. (A) Real-time PCR analysis of rpaI transcript levels (normalized to the fixJ housekeeping gene) of the wild type (wt), rpaR mutant, or rpaR mutant complemented with pBBR-RpaR-PrpaI-lacZ. Cells were grown in PM-succinate with no addition (□), with 0.5 mM p-coumarate (▩), or with 250 nM pC-HSL (▪). Wild-type and mutant strains grew similarly. The data are the means of two biological replicates, and error bars indicate the range. (B) pC-HSL production by wild-type (wt) and rpaR mutant strains grown in the presence of 1 mM p-coumarate. The data are the means of three biological replicates. Error bars indicate the standard deviation.
Purification of His-tagged RpaR.
For in vitro experiments (see below), we used His-tagged RpaR purified from P. aeruginosa PAO-T7-MW1 containing pQFhis5016.rpaR (see Materials and Methods). As is the case for many LuxR family members (8, 38, 43, 51), the production of soluble RpaR is enhanced by the growth of recombinant bacteria in the presence of cognate acyl-HSL (Fig. 2 A and B). When cells were grown with the fatty acyl-HSLs hexanoyl-HSL or 3-oxo-dodecanoyl-HSL, His6-RpaR was detected only in the insoluble fraction of cell lysates, whereas we detected some soluble His6-RpaR in lysates of pC-HSL-grown cells (Fig. 2A and B).
Fig. 2.
Solubility and purification of RpaR from cell extracts of recombinant P. aeruginosa grown in the presence of pC-HSL. (A) SDS-PAGE analysis of soluble and insoluble polypeptides from P. aeruginosa PAO-T7-MW1 containing the His6-RpaR expression plasmid pQF5016b.rpaR. A control with no IPTG induction is shown in lane 1. Cells were grown in the absence of added acyl-HSLs (lane 2), the presence of pC-HSL at 10 μM (lane 3) or 20 μM (lane 4), or the presence of 20 μM C6-HSL (lane 5). The predicted molecular weight of His6-RpaR is 27,800. Molecular mass markers are indicated in the left lane (M) in kilodaltons. (B) The His tag was visualized by using a SuperSignal West HisProbe kit. Lanes are as described for panel A. (C) SDS-PAGE of clarified extract (lane 1) and His6-RpaR purified from the cell extract (lane 2). The left lane (M) shows the molecular mass standards.
To provide evidence that pC-HSL binding to RpaR is important for the generation of soluble RpaR, we constructed a gene coding for RpaR with a D-to-N substitution at amino acid residue 82. This residue is conserved among LuxR homologs (D79 in LuxR, D70 in TraR) and is important for hydrogen bonding with the imino group of the acyl-HSL ligand (21, 50). We did not detect soluble RpaR-D82N in lysates of cells grown in the presence of pC-HSL. RpaR-D82N was entirely in the insoluble fraction (data not shown). Although there are other interpretations, this is consistent with the idea that D82 plays a role in pC-HSL-binding, and without pC-HSL binding soluble RpaR exists at very low levels.
We purified His6-RpaR from clarified cell extracts of recombinant P. aeruginosa (Fig. 2C) and then measured the pC-HSL retained by the purified RpaR. RpaR monomers and pC-HSL were equimolar (1:1.2 [RpaR/pC-HSL]). Even after 16 h of dialysis against pC-HSL-free buffer, the protein-to-ligand ratio remained equimolar (1:1.3).
RpaR behaves as an activator at the luxI promoter by binding a lux box-like element centered at bp −48.5 upstream of the transcript start site.
To investigate the DNA-binding properties of RpaR by EMSA, we used a 103-bp DNA fragment encompassing the 86-bp rpaR-rpaI intergenic region as a specific probe. We previously identified two sequences with similarity to the V. fischeri lux box (boxes 1 and 2). Box 1 shows lux box identity in 15 of 20 base positions, and box 2 is identical to the lux box in 9 of 20 bases (Fig. 3 A), and we know rpaI expression is activated by RpaR (Fig. 1). RpaR bound to the specific probe but not to a similar-sized fragment from pUC19 (Fig. 3B).
Fig. 3.
RpaR binds DNA containing a lux box-like element. (A) Diagram of the two lux box-like sequences in the rpaR-rpaI intergenic region (box 1 and box 2). Mutations were introduced into the boxes at positions indicated by the arrows. (B) Gel mobility shift assays with His6-RpaR binding (concentrations are indicated in pmol) with wild-type (wt) and mutant (mut) lux box-like elements. Reaction mixtures contained 20 μM pC-HSL and 0.3 pmol of probe DNA. DNA from pUC19 was used as a control. (C) DNase I footprinting of the rpaI promoter region. A 239-bp, 6-FAM-labeled DNA fragment was incubated in the presence of His6-RpaR (0, 6, and 12 pmol) and then subjected to DNase I digestion. The fluorescence intensities of the DNA fragments are plotted relative to their sizes. The fragments corresponding to the lux box-like elements are boxed. One region, corresponding to bp 85 to 120 relative to the 6-FAM probe, was protected from DNase I digestion in the presence of RpaR (indicated by gray shading). This region covers box 1. DNase I protection was not observed for the region corresponding to box 2.
To determine whether RpaR binds to one or both of the potential lux box-like elements, we created probes containing the nucleotide substitutions C3T, T4A, A17T, and G18T in each of the boxes individually, as well as both combined (Fig. 3). Nucleotides 3, 4, 17, and 18 are important for binding of other LuxR homologs to target DNA (1, 2, 32, 47). A probe containing nucleotide substitutions in box 2 bound RpaR in a manner indistinguishable from the native probe. However, a probe containing substitutions in box 1 did not bind RpaR (Fig. 3B), indicating that box 1 and not box 2 is in the RpaR binding site. We confirmed that box 1 is essential for gene expression by using a series of rpaI::lacZ reporter plasmids with native or mutant lux box-type sequences. Consistent with the EMSA results, cells containing plasmids with substitutions in box 1 showed low levels of rpaI::lacZ expression in the presence of pC-HSL, while box 2 mutant constructs showed high, wild-type levels of rpaI::lacZ expression (data not shown). The identification of box 1 as an RpaR binding site was further confirmed by DNase I footprinting, which showed that RpaR protected a region of about 25 bp that included the predicted box 1 (Fig. 3C). This indicates that RpaR covers a DNA region similar in size to the regions covered by other LuxR homologs (38, 46, 51).
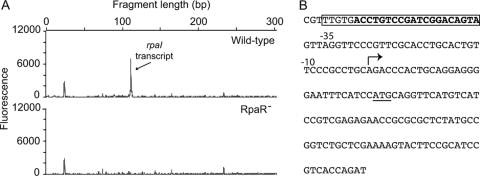
Activators generally bind upstream of the −35 region of a promoter, and repressor binding commonly overlaps the RNA polymerase binding region (11, 15, 30, 41). To gain additional insight about the function of RpaR, we mapped the rpaI transcript start site(s) by S1 nuclease protection analysis of RNA isolated from wild-type and rpaR mutant strains (Fig. 4 A). Regardless of the growth phase (i.e., the logarithmic or stationary phase [data not shown]), rpaI transcript was only detected in p-coumarate-grown, wild-type cells. The start of the transcript was 29 bp from the predicted rpaI translational start site. This centers the RpaR binding site (box 1) at bp −48.5 from the start of the transcript (Fig. 4B). A primer extension analysis showed the same transcript start site (data not shown). The first nucleotide in the transcript was an adenine, as is the case for at least two other LuxR homolog-regulated transcripts (9, 39, 47).
Fig. 4.
Identification of the rpaI transcript start site. (A) S1 nuclease protection analysis of the rpaI transcript from the R. palustris wild-type (top) and the RpaR-mutant (bottom) strains. The rpaI product was detected in the wild type but not the mutant. The rpaI fragment was 111 bp in length, thus mapping the rpaI start site to a location 29 bp upstream of the ATG start codon. (B) Sequence of the rpaR-rpaI intergenic region. The RpaR binding site, which includes the box 1 element (indicated in bold), is boxed. The arrow indicates the rpaI transcript start site and its translation start codon is underlined. The −35 and −10 regions are indicated but show little similarity to those defined for E. coli sigma-70 promoters. This is often the case for GC-rich alphaproteobacteria (R. palustris has a GC content of 65%), which have −35 and −10 features that differ from organisms with lower G+C contents (28, 31).
RpaR does not bind promoters of other pC-HSL-regulated genes.
A microarray analysis revealed that in addition to rpaI, 16 other R. palustris genes showed differential gene expression in the presence of pC-HSL (34). One of the genes, rpa0319, is downstream of rpaI (rpa0320) and cotranscribed with it. The other 15 genes (rpa0905, rpa1096, rpa1098, rpa1674, rpa1845, rpa2519, rpa2883, rpa3185, rpa3306, rpa3599, rpa3892, rpa3929, rpa4296, rpa4684, and rpa4713) are predicted to be monocistronic (23). A search of the intergenic regions of these 15 genes with the regulatory sequence alignment tools (http://rsat.ulb.ac.be/rsat) did not reveal sequences similar to the RpaR target sequence described above. To test whether these pC-HSL-controlled genes are directly regulated by RpaR, we tested DNA probes containing the entire upstream intergenic DNA sequence for each gene (see Materials and Methods for details), as targets for RpaR binding in EMSA experiments. Only the rpaI promoter fragment and none of the other probes bound RpaR (data not shown).
Additional RpaR-pC-HSL-regulated elements identified by NSR RNAseq analysis.
We were surprised to find RpaR bound to only one of 16 potential target DNA fragments. We wanted to demonstrate that pC-HSL regulation of the promoters in these fragments was dependent on RpaR. It seemed possible that the genes identified in the previous study (34) were pC-HSL dependent but not RpaR dependent or that an alternative transcriptomics technique would not confirm regulation of these genes. Because we constructed an RpaR mutant, we were able to compare the genes controlled by RpaR to those previously identified as pC-HSL controlled, and we used RNAseq rather than microarray technology.
A limitation of RNAseq is that rRNAs constitute the bulk of the reads, and only a small fraction of the cDNA pool is synthesized from mRNA. This limits RNAseq sensitivity. To circumvent this issue, protocols utilizing polyadenylated priming (for eukaryotic samples) or ribosome depletion have been used (49). Recently, we developed a new method to create cDNA pools from human RNA samples that are enriched for nonribosomal sequences by computational identification of hexamers predicted to bind rRNA sequences and omission of these hexamers from the final primer pool (4). These “not-so-random” (NSR) hexamers contained a universal 5′ tail for amplification annealing and were synthesized in both antisense (first strand cDNA synthesis) and sense (second strand DNA synthesis) orientations, allowing for the preservation of strand orientation (4). We created a similar NSR primer set for R. palustris (see Materials and Methods and see Table S2 in the supplemental material), and we successfully enriched our cDNA libraries for non-rRNA transcripts with ca. 25 to 30% of the mapped reads corresponding to unique DNA sequences rather than rRNA sequences (see Table S3 in the supplemental material).
By using NSR RNAseq, we compared the transcriptomes of the RpaR mutant grown with or without pC-HSL and found that no elements were differentially expressed (≥2.5-fold, Padj ≤ 0.05; see Table S4 in the supplemental material). This indicates that, as anticipated, genes regulated by pC-HSL are also regulated by RpaR. We also compared wild-type R. palustris grown on succinate with or without pC-HSL, an experiment analogous to the previously published microarray experiments (34). We identified 48 differentially expressed genes (≥2.5-fold difference in expression) of which 39 were induced and 9 repressed by pC-HSL (Table 2 and see Table S4 in the supplemental material). Of the 17 pC-HSL regulated genes identified in the previous microarray analysis, 10 were included in this list (5 of the remaining seven genes were differentially regulated but did not meet our fold cutoff or Padj criteria). Thus, there is reasonably good concordance between the microarray and RNAseq analyses.
Table 2.
RpaR-pC-HSL regulon as defined by NSR RNAseq analysis
| Elementa | Expression ratiob |
Annotationc | |
|---|---|---|---|
| WT (+) vs (-) pC-HSL | WT vs rpaR mutant with pC-HSL | ||
| Sense | |||
| RPA0318 | 11.6 | 67.1 | Putative Mg2+ chelatase family protein |
| RPA0320* | 35.8 | 27.3 | RpaI acyl-homoserine lactone synthase |
| RPA0543 | 3.1 | 4.1 | Unknown protein |
| RPA0745* | 8.2 | 8.8 | Possible outer membrane protein precursor |
| RPA1096* | 6.7 | 5.3 | Methyl-accepting chemotaxis sensory transducer |
| RPA1098* | 13.3 | 17.2 | Hypothetical protein |
| RPA2815 | 7.0 | 6.8 | Possible outer membrane protein |
| RPA3332 | 3.1 | 2.9 | Hypothetical protein |
| RPA3333 | 3.3 | 3.9 | Putative curli production assembly/transport component csgG precursor |
| RPA3599* | 16.4 | 36.2 | Hypothetical protein |
| RPA3751 | 2.5 | 3.1 | Unknown protein |
| RPA4296* | 3.7 | 4.4 | Unknown protein |
| RPA4639 | 2.9 | 8.8 | Methyl-accepting chemotaxis receptor/sensory transducer |
| RPA4691 | 3.4 | 4.3 | Methyl-accepting chemotaxis receptor/sensory transducer |
| RPA1845* | –4.0 | –3.7 | Putative TonB-dependent receptor protein |
| RPA1846 | –4.9 | –5.9 | Unknown protein |
| RPA1875 | –11.7 | –11.1 | Possible uncharacterized iron-regulated membrane protein |
| RPA1876* | –11.2 | –10.0 | Putative TonB-dependent iron siderophore receptor |
| RPA2124 | –2.7 | –3.3 | TonB-dependent iron siderophore receptor |
| RPA2843 | –5.9 | –6.6 | Pseudo gene of Fe3+ siderophore transport receptor |
| Antisense | |||
| RPA0321* | 23.6 | –d | RpaR, LuxR two-component transcriptional regulator |
| RPA0493 | 4.7 | 8.1 | 50S ribosomal protein L28 |
| Intergenic | |||
| RPA0494 | 7.1 | 27.5 | Positive strand |
| RPA1526 | 3.3 | 2.8 | Negative strand |
| RPA3332† | 2.6 | 3.1 | Negative strand |
| RPA3589† | 6.1 | 9.9 | Negative strand |
| RPA3877 | 5.4 | 5.4 | Negative strand |
| RPA3892† | 6.3 | 7.7 | Negative strand |
| RPA4296† | 4.8 | 7.1 | Positive strand |
| RPA1095† | –13.4 | –23.3 | Negative strand |
| RPA1846† | –19.2 | –20.9 | Negative strand |
| RPA2125† | –2.7 | –3.0 | Negative strand |
| RPA2130 | –6.4 | –8.3 | Negative strand |
| RPA3285 | –6.1 | –12.0 | Positive strand |
| RPA3821 | –2.8 | –3.7 | Positive strand |
Genes regulated by pC-HSL in Affymetrix chip experiments (34) are indicated in boldface. Elements with promoters that were examined by EMSA experiments are indicated by an asterisk. Intergenic sequences with adjacent genes exhibiting pC-HSL-RpaR regulation are indicated by a dagger (†).
The expression of elements listed was regulated by ≥2.5-fold and had an adjusted P (Padj)value of ≤0.05 in both comparisons. WT, wild type.
As defined by the Joint Genome Institute site (www.jgi.doe.gov).
–, An antisense transcript was detected in the WT but not in the RpaR mutant because this gene is deleted in the mutant strain.
We also compared the wild type and the RpaR mutant when both were grown in the presence of pC-HSL and identified 22 differentially expressed genes (≥2.5-fold difference in transcript levels) (see Table S4 in the supplemental material). When these results were compared to those from the comparison of wild type grown with or without pC-HSL, we found an overlap of 14 activated and 6 repressed genes (Table 2). Genes regulated by either pC-HSL in the wild type or by RpaR included rpaI and its downstream genes, rpa0319 (did not make the Padj cutoff) and rpa0318, and might be considered to define the R. palustris quorum-controlled regulon. As observed previously, genes annotated as chemotaxis functions are over-represented, and we also found iron metabolism genes (Table 2).
We next sought to determine whether some of the newly identified quorum-controlled genes were direct targets of RpaR by performing EMSA (rpa0745 and rpa1876). As with the previously identified pC-HSL-dependent genes (34), these genes did not appear to be direct targets of RpaR (data not shown).
Discovery of an rpaR antisense transcript.
One advantage of our NSR RNAseq protocol is the ability to track the strand specificity of transcripts (4). As expected, the majority of elements identified in the RpaR-pC-HSL regulon mapped to the sense transcripts of annotated ORFs (see Table S4 in the supplemental material). However, we identified two differentially expressed antisense transcripts (Table 2). The rpa0493 antisense transcript exhibited a 4- to 8-fold pC-HSL-RpaR-dependent increase. This gene codes for the L28 ribosomal protein of the 50S ribosomal subunit. The genes adjacent to rpa0493 did not show RpaR-pC-HSL regulation. Of particular interest was our observation that antisense reads from rpaR (rpa0321) were increased >20-fold by pC-HSL addition to R. palustris wild-type cells (Table 2 and Fig. 5). Based on the location of the RNAseq reads (Fig. 5), it appears that the antisense RNA originates in the 3′ region of the ORF (assuming a single antisense transcript). This discovery of rpaR antisense RNA is intriguing and opens up an interesting line of future pursuit.
Identification of potential regulatory RNAs.
In addition to tracking strand specificity, our RNAseq method, like many others, can detect transcripts, including small, untranslated RNAs, regardless of whether they are in annotated reading frames. We identified 13 intergenic (IG) regions that showed differential expression (≥2.5-fold, Padj ≤ 0.05) in both the comparison of the wild type with or without signal and the comparison of the RpaR mutant to the wild type. Seven of the thirteen are within about 100 bp of the predicted start or stop codons of quorum-controlled ORFs (rpa1846, rpa2125, rpa3332, and rpa4296, as well as rpa1095, rpa3589, and rpa3892, which have adjacent ORFs that were differentially expressed but did not meet our criteria for inclusion in Table 2) and could very well be part of the transcripts for these ORFs.
Two of the thirteen differentially expressed IG regions were larger than 1 kb, which suggested uncalled ORFs may be present. The first IG region is 1,053 bp in size, is located adjacent to rpa2130, and exhibits strong DNA identity to the analogous genome region in the R. palustris TIE-1 chromosome (>98% identity) and more limited similarity to a region in the R. palustris DX-1 chromosome (>50% identity). In these analogous DNA regions there are annotated ORFs in both TIE-1 (rpal_2421) and DX-1 (rpdx1DRAFT_1011 and rpdx1DRAFT_1012), which appear to be present but are not annotated in CGA009. Presumably, these ORFs were called in the more recently sequenced genomes (TIE-1 and DX-1) as a result of improved annotation software. The second IG region, 1,551 bp between fumA and rpa3877, showed no identity to similar chromosomal regions in other R. palustris strains. However, DNA sequence analysis of this region using the program ORFfinder (www.ncbi.nlm.nih.gov/gorf/gorf.html) identified a potential 636-bp ORF whose product shares 43% amino acid identity with the hypothetical gene rpdx1DRAFT_3098 from R. palustris DX-1. Thus, we believe that these two IG transcripts code for previously unrecognized polypeptides and their expression is influenced by RpaR-pC-HSL.
Of the remaining four IG regions, the region upstream of pufA (rpa1526) is only 13 bp. It seems unlikely that this is of biological significance. Of the remaining differentially expressed IG regions one, rpa0494, was activated by RpaR, and pC-HSL and two, rpa3285 and rpa3821, were repressed. An mfold analysis (52) of the IG regions for all of these three cases indicated putative transcripts that have predicted stable secondary structure (free energy values ranging from −55 to −109 kcal/mol). This analysis suggests that there might be quorum-sensing-controlled, untranslated transcripts in R. palustris with potential regulatory functions. Of course, further work is required to address this question.
DISCUSSION
Although previous experiments with recombinant bacteria suggested that RpaR was an rpaI repressor and that pC-HSL was required for derepression, we provide strong evidence here that RpaR functions as an rpaI activator. We constructed an RpaR-null mutant, which showed reduced rpaI transcript levels and reduced levels of the RpaI enzymatic product, pC-HSL (Fig. 1). This result is consistent with activator rather than repressor activity. We showed that RpaR-pC-HSL binds to rpaI promoter DNA, specifically (Fig. 3). Because pC-HSL is required for the induction of rpaI, this result is consistent with an activator function for RpaR. Finally, we mapped the RpaR binding site in the rpaI promoter region to an inverted repeat centered at −48.5 from the start of rpaI transcription (Fig. 3 and 4). This location of the RpaR binding site with respect to the start of transcription is common for ambidexterous transcriptional activators, including LuxR (11, 15, 30, 41).
Although pC-HSL has an aromatic rather than a fatty acyl side group, its receptor RpaR shows solubility and DNA-binding characteristics akin to many LuxR-type fatty-acyl-HSL receptors (Fig. 2 to 4). It is likely that pC-HSL binds a pocket similar to those described for fatty acyl-HSL compounds (21, 50). Consistent with this idea, an RpaR mutant protein with a substitution in a residue conserved among LuxR family members, which interacts with the imino group in the signal, D82N, forms inactive insoluble aggregates. It is likely that these aggregates form because the mutant protein cannot bind the stabilizing ligand pC-HSL. However, we assume that there are fundamental differences in the pC-HSL binding pocket of RpaR and the fatty acyl-HSL binding pockets of other known LuxR homologs. Our purification of RpaR sets the stage for future structural analyses aimed at identifying residues that distinguish pC-HSL recognition from fatty acyl-HSL recognition.
We previously identified 17 pC-HSL-dependent R. palustris genes (including rpaI and the gene cotranscribed with rpaI) by microarray analysis (34). To further define the pC-HSL-RpaR-controlled regulon, we used RNAseq not only to study the influence of pC-HSL on the transcriptome but also to compare the RpaR mutant we constructed for the present study to the parent strain. The RNAseq method was based on a protocol that we developed previously for mammalian transcript analysis (4). This method does not require a rRNA depletion step. By using the procedure we found that 25 to 30% of the mapped reads were non-rRNA (see Table S3 in the supplemental material). The use of RNAseq also detects RNAs that do not map to ORFs, allowing for the discovery of small regulatory RNAs.
We identified 20 genes that were regulated by pC-HSL and RpaR, 14 were activated by these factors and 6 were repressed. We tested the ability of the promoter regions of 18 of these quorum-controlled genes (including those identified previously in the microarray analysis) to serve as an RpaR binding target and, strikingly, RpaR bound only the promoter of the rpaI operon (rpa0320-0319). We cannot exclude the possibility that there may be additional cellular factors or physiological conditions required for RpaR binding to the elements that did not serve as targets in our EMSA (Table 2). It could also be that pC-HSL-RpaR-regulated elements (Table 2) are controlled via a feed-forward mechanism, perhaps through regulatory RNA or an RpaR-controlled transcription factor that that did not meet our cutoff criteria. Of note, we detected five particularly interesting apparently noncoding RNAs that were induced by pC-HSL and RpaR, including an rpaR antisense transcript. These RNAs deserve further study, particularly because small regulatory RNAs (sRNAs) have been implicated in the mediation of quorum-controlled gene activation in other bacteria (36, 42). We note a recent report of antisense psyR (a luxR homolog) transcripts in Pseudomonas syringae (10). This antisense RNA is thought to originate from readthrough of the convergently transcribed luxI homolog, psyI. However, in the case of R. palustris, which has a divergent orientation of rpaI and rpaR, readthrough of rpaI into rpaR is not a possibility (Fig. 3).
There are now available genome sequences for seven strains of Rhodopseudomonas (24). Whether or not these represent more than one species is a point of debate. Of these seven genomes rpaR-rpaI is conserved in five. Although we have identified a set of genes that is regulated by pC-HSL and RpaR in strain CGA009, there is no obvious phenotype, at least under the conditions we use to grow this bacterium. Perhaps another strain will show a dramatic phenotype, or perhaps other growth conditions are required to detect a phenotype. It will be of interest to study aryl-HSL signaling in other strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jean Huang for sharing unpublished plasmids.
This study was funded by grants from the U.S. Army Research Office (grant W911NF-09-1-0350) and the Office of Science (BER), U.S. Department of Energy (DE-FG02-08ER64688). H.H. received funding from the Japan Society for the Promotion of Science, the Uehara Memorial Foundation, and the Cell Science Research Foundation.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Anderson R. M., Zimprich C. A., Rust L. 1999. A second operator is involved in Pseudomonas aeruginosa elastase (lasB) activation. J. Bacteriol. 181:6264–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antunes L. C., Ferreira R. B., Lostroh C. P., Greenberg E. P. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J. Bacteriol. 190:4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antunes L. C., et al. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J. Bacteriol. 189:8387–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Armour C. D., et al. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods 6:647–649 [DOI] [PubMed] [Google Scholar]
- 5. Choi S. H., Greenberg E. P. 1991. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. U. S. A. 88:11115–11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi S. H., Greenberg E. P. 1992. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J. Bacteriol. 174:4064–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Devine J. H., Shadel G. S., Baldwin T. O. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC 7744. Proc. Natl. Acad. Sci. U. S. A. 86:5688–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duerkop B. A., Ulrich R. L., Greenberg E. P. 2007. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J. Bacteriol. 189:5034–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Egland K. A., Greenberg E. P. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol. Microbiol. 31:1197–1204 [DOI] [PubMed] [Google Scholar]
- 10. Filiatrault M. J., et al. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J. Bacteriol. 192:2359–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finney A. H., Blick R. J., Murakami K., Ishihama A., Stevens A. M. 2002. Role of the C-terminal domain of the alpha subunit of RNA polymerase in LuxR-dependent transcriptional activation of the lux operon during quorum sensing. J. Bacteriol. 184:4520–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuqua C., Greenberg E. P. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183–189 [DOI] [PubMed] [Google Scholar]
- 13. Hanzelka B. L., Greenberg E. P. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:815–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 15. Johnson D. C., Ishihama A., Stevens A. M. 2003. Involvement of region 4 of the sigma70 subunit of RNA polymerase in transcriptional activation of the lux operon during quorum sensing. FEMS Microbiol. Lett. 228:193–201 [DOI] [PubMed] [Google Scholar]
- 16. Kim M.-K., Harwood C. S. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 83:199–204 [Google Scholar]
- 17. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 18. Larimer F. W., et al. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55–61 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. H., Lequette Y., Greenberg E. P. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59:602–609 [DOI] [PubMed] [Google Scholar]
- 20. Li H., Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo Z. Q., Smyth A. J., Gao P., Qin Y., Farrand S. K. 2003. Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J. Biol. Chem. 278:13173–13182 [DOI] [PubMed] [Google Scholar]
- 22. More M. I., et al. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272:1655–1658 [DOI] [PubMed] [Google Scholar]
- 23. Moreno-Hagelsieb G., Collado-Vides J. 2002. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics 18(Suppl. 1):S329–S336 [DOI] [PubMed] [Google Scholar]
- 24. Oda Y., et al. 2008. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc. Natl. Acad. Sci. U. S. A. 105:18543–18548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan C., et al. 2008. Characterization of anaerobic catabolism of p-coumarate in Rhodopseudomonas palustris by integrating transcriptomics and quantitative proteomics. Mol. Cell Proteomics 7:938–948 [DOI] [PubMed] [Google Scholar]
- 26. Parsek M. R., Val D. L., Hanzelka B. L., Cronan J. E., Jr., Greenberg E. P. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 96:4360–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quandt J., Hynes M. F. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 28. Ramirez-Romero M. A., Masulis I., Cevallos M. A., Gonzalez V., Davila G. 2006. The Rhizobium etli sigma70 (SigA) factor recognizes a lax consensus promoter. Nucleic Acids Res. 34:1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rey F. E., Heiniger E. K., Harwood C. S. 2007. Redirection of metabolism for biological hydrogen production. Appl. Environ. Microbiol. 73:1665–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rhodius V. A., Busby S. J. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152–159 [DOI] [PubMed] [Google Scholar]
- 31. Richard C. L., Tandon A., Kranz R. G. 2004. Rhodobacter capsulatus nifA1 promoter: high-GC −10 regions in high-GC bacteria and the basis for their transcription. J. Bacteriol. 186:740–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rust L., Pesci E. C., Iglewski B. H. 1996. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J. Bacteriol. 178:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 34. Schaefer A. L., et al. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599 [DOI] [PubMed] [Google Scholar]
- 35. Schaefer A. L., Val D. L., Hanzelka B. L., Cronan J. E., Jr., Greenberg E. P. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. U. S. A. 93:9505–9509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schu D. J., Carlier A. L., Jamison K. P., von Bodman S., Stevens A. M. 2009. Structure/function analysis of the Pantoea stewartii quorum-sensing regulator EsaR as an activator of transcription. J. Bacteriol. 191:7402–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuster M., Urbanowski M. L., Greenberg E. P. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A. 101:15833–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seed P. C., Passador L., Iglewski B. H. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simon R., Priefer U., Puhler A. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. BioTechnology 1:37–45 [Google Scholar]
- 41. Stevens A. M., Fujita N., Ishihama A., Greenberg E. P. 1999. Involvement of the RNA polymerase alpha-subunit C-terminal domain in LuxR-dependent activation of the Vibrio fischeri luminescence genes. J. Bacteriol. 181:4704–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tsai C. S., Winans S. C. 2010. LuxR-type quorum sensing regulators that are detached from common scents. Mol. Microbiol. 77:1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urbanowski M. L., Lostroh C. P., Greenberg E. P. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagner V. E., Bushnell D., Passador L., Brooks A. I., Iglewski B. H. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waters C. M., Bassler B. L. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
- 46. Welch M., et al. 2000. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Whiteley M., Greenberg E. P. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson D. O., Johnson P., McCord B. R. 2001. Nonradiochemical DNase I footprinting by capillary electrophoresis. Electrophoresis 22:1979–1986 [DOI] [PubMed] [Google Scholar]
- 49. Yoder-Himes D. R., et al. 2009. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 106:3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang R. G., et al. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971–974 [DOI] [PubMed] [Google Scholar]
- 51. Zhu J., Winans S. C. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. U. S. A. 98:1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.